Abstract
The allosteric coupling constant in K-type allosteric systems is defined as a ratio of the binding of substrate in the absence of effector to the binding of the substrate in the presence of a saturating concentration of effector. As a result, the coupling constant is itself an equilibrium value comprised of a ΔH and a TΔS component. In the scenario in which TΔS completely compensates ΔH, no allosteric influence of effector binding on substrate affinity is observed. However, in this “silent coupling” scenario, the presence of effector causes a change in the ΔH associated with substrate binding. A suggestion has now been made that “silent modulators” are ideal drug leads because they can be modified to act as either allosteric activators or inhibitors. Any attempt to rationally design the effector to be an allosteric activator or inhibitor is likely to be benefitted by knowledge of the mechanism that gives rise to coupling. Hydrogen/deuterium exchange with mass spectrometry detection has now been used to identify regions of proteins that experience conformational and/or dynamic changes in the allosteric regulation. Here, we demonstrate the expected temperature dependence of the allosteric regulation of rabbit muscle pyruvate kinase by Ala to demonstrate that this effector reduces substrate (phosphoenolpyruvate) affinity at 35°C and at 10°C but is silent at intermediate temperatures. We then explore the use of hydrogen/deuterium exchange with mass spectrometry to evaluate the areas of the protein that are modified in the mechanism that gives rise to the silent coupling between Ala and phosphoenolpyruvate. Many of the peptide regions of the protein identified as changing in this silent system (Ala as the effector) were included in changes previously identified for allosteric inhibition by Phe.
Significance
One outcome from high throughput drug screens is the identification of lead molecules that bind to the protein but do not alter the targeted protein function in initial assays. Ligand binding without a functional consequence is consistent with entropy-enthalpy compensation in the allosteric coupling constant (i.e., silent coupling) that relates protein function (substrate binding, catalysis) and binding of the lead molecule. A proposal has been made that silent lead molecules can be chemically modified to develop either allosteric activators or allosteric inhibitors. Understanding the coupling mechanism in the silent system is expected to guide those drug designs. We report a hydrogen/deuterium exchange with mass spectrometry characterization of the regions of pyruvate kinase that contribute to a silent coupling mechanism.
Introduction
K-type allosteric regulation is most often identified when the binding of one ligand to a protein results in altered binding affinity of the protein for a second ligand. This classic method of defining allostery relies on the detection of altered binding (and therefore altered free energy for ligand binding) as a monitor of energetic coupling between two binding events on the protein. However, several studies have indicated energetic coupling between binding events at distinct sites, even though binding affinities of ligands are not modified (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13).
In a linked-function evaluation of allostery, the allosteric coupling constant is defined as a ratio of the binding of substrate in the absence of effector to the binding of the substrate in the presence of a saturating concentration of effector (14, 15, 16, 17, 18, 19, 20). This allosteric coupling constant is itself an equilibrium constant that can be converted into free energy (ΔGax). When considered as free energy with both enthalpy (ΔH) and entropy (TΔS) contributions, we can imagine a scenario in which the ΔH and TΔS have equal magnitudes such that they completely compensate each other to result in a ΔGax equal to zero. Therefore, the historic study of enthalpy-entropy compensation (21,22) applies to allosteric coupling and serves as a basis for how two ligand-binding events can be energetically coupled even though binding affinities are not altered as an allosteric effect.
Energetic coupling between two binding events that does not result in altered binding (i.e., complete enthalpy-entropy compensation) has been named by a range of descriptors. In a homotropic cooperative example, the Fisher laboratory used “isoergonic cooperativity” based on calorimetry measurements of the enthalpy associated with binding (1,2). The Reinhart laboratory, who evaluated heterotropic allosteric coupling as a function of temperature, published “temperature-induced inversion” and “obfuscation of allostery” to describe this compensation phenomenon (3,4). “Silent agonists” has been used in descriptions of receptors (5, 6, 7, 8, 9). Three laboratories have used “hidden allostery” to describe a related phenomenon (although not always exactly consistent with our favored definition of allostery because the coupling was not between two ligand-binding events) (10,11,13). In a review of the thermodynamic nature of the compensation phenomenon, Fisher adopted “silent coupling,” crediting that nomenclature to Reinhart (12). Here, we will follow this final example and use “silent coupling” to refer to complete compensation with the acknowledgment that partial compensations are more broadly recognized (e.g., (23,24)). In choosing to use the “silent coupling” nomenclature, we also acknowledge that the allosteric coupling in any enthalpy-entropy compensated system is only “silent” at certain temperatures and that those temperatures will be unique to each system. Interestingly, although the observation of this complete compensation phenomenon has been sufficiently rare to avoid standardized nomenclature, it was foreseen by Gregorio Weber in the 1970s: “Therefore, it is impossible for the binding of two ligands to the same protein to be really independent” (17). (Perhaps, in Dr. Weber’s quote, “impossible” should be replaced with “improbable.”)
It has now been proposed that the identification of “silent modulators” is an ideal outcome from a small molecule library screen because those molecules can be modified to be either activating or inhibiting allosteric drugs (25). Consistent with this idea, we previously demonstrated that the magnitude of an allosteric effect could be modulated with subtle chemical changes in the effector (26). It follows that knowledge of the molecular mechanism that gives rise to the silent coupling might provide insights to aid rational modification of the silent modulator to derive allosteric effectors and/or allosteric activators. In this study, we explore the use of hydrogen/deuterium exchange with mass spectrometry (H/DX-MS), a methodology that has now been used to characterize a range of allosteric systems (27, 28, 29, 30, 31), to identify regions of a protein that experience structural or dynamic changes as a result of the silent coupling mechanism.
The protein system with silent coupling used herein was rabbit muscle pyruvate kinase (M1PYK). The identification of silent coupling between alanine (Ala) binding in the effector site and phosphoenolpyruvate (PEP) binding in the active site was serendipitous. To identify what allosteric changes occur in the protein that are in addition to effector binding (in the absence of allostery), we previously compared structural changes that result when M1PYK binds the allosteric effector, phenylalanine (Phe), to those changes that occur when the enzyme binds the analog, Ala (19,26,27,32). The design of those earlier studies was to consider Ala as a nonallosteric analog. (Use of the nomenclature “nonallosteric” must now be reconsidered given the outcome of this study demonstrating that Ala is an allosteric effector at temperatures below and above those used in the previous studies.) Using that previous design to compare one ligand that elicits an allosteric response with a second similar ligand that does not cause allostery in combination with small-angle x-ray scattering (SAXS), we demonstrate that the large conformational changes observed upon Phe binding are also elicited with Ala binding. Hence, the large structural changes observed upon effector binding (causd by both Ala and Phe binding) are not sufficient to cause the observed Phe-dependent inhibition of PEP affinity (32). In that same study, we collected SAXS profiles for the four enzyme complexes that constitute the allosteric energy cycle when Ala is treated as an effector. Although Ala causes negligible change in the protein’s affinity for PEP (at the 25–30°C range; see figures below), the ΔP(r) function derived from comparing the P(r) data for the free enzyme versus PEP-bound enzyme is very different from the ΔP(r) function derived from comparing the P(r) data for the Ala-bound complex with the ternary PEP-M1PYK-Ala complex. This difference implied that Ala alters the PEP-enzyme interactions, despite the lack of an effect of Ala on the observed PEP affinity. It was this “silent coupling” in M1PYK with Ala as the effector that was used as a test case in this study to explore what insights might be learned by an H/DX-MS evaluation. Similar to the previous SAXS study, the design used here was to collect H/DX-MS data for all four enzyme complexes that constitute the allosteric energy cycle: free enzyme, PEP-M1PYK, M1PYK-Ala, and PEP-M1PYK-Ala.
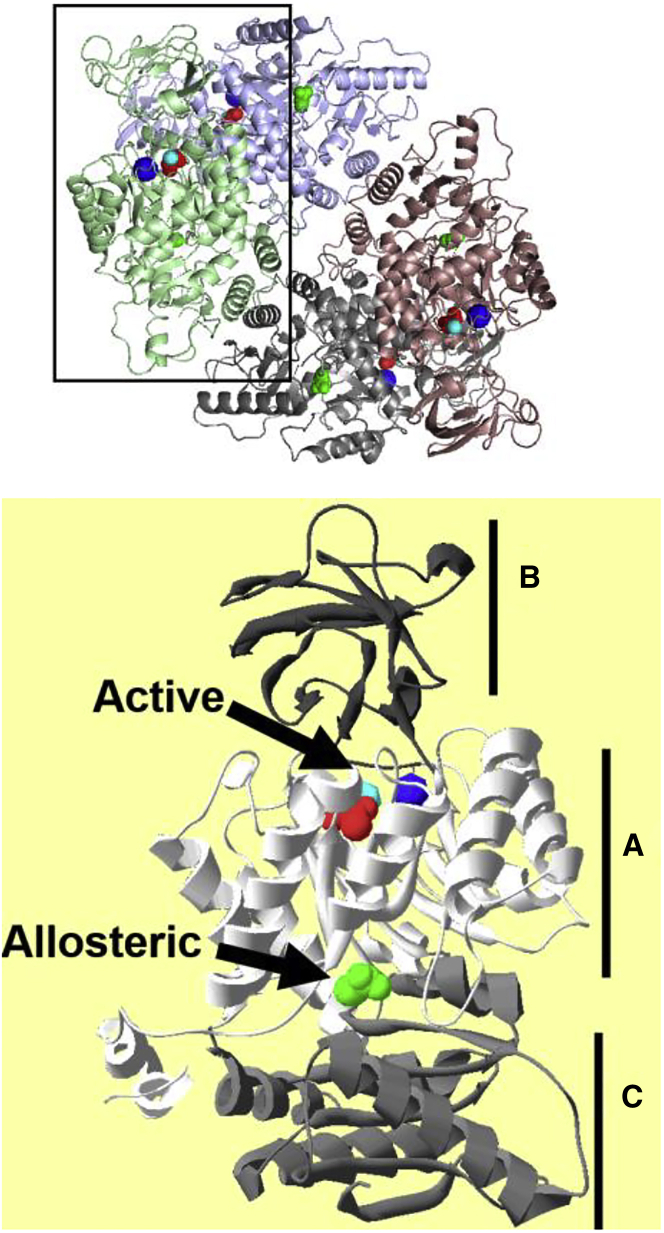
To facilitate the description of H/DX-MS data, consider that M1PYK is a homotetramer. Each subunit includes three domains (Fig. 1; recreated to match (27)). The active site lies between the A and B domains. The amino-acid effector binding site is between the A and C domains. Subunits assemble first as a dimer across the A-A interface and then as a dimer of dimers across the C-C interface.
Figure 1.
The homotetrameric (top) and subunit (bottom) structures of rM1-PYK as determined by x-ray crystallography (PDB: 2G50). Subunits within the homotetramer are colored red, light blue, green, and gray. Each subunit contains three domains labeled as A, B, and C. The active site lies between the A and B domains, whereas the amino acid allosteric site is located at the A-C domain interface. The presentation of the subunit (bottom) distinguishes the three domains with various shades of gray. In both homotetrameric and subunit views, the active site is occupied by spacefill views of potassium (blue), Mn2+ (cyan), and pyruvate (red). The allosteric amino-acid binding site is occupied by Ala (green spacefill). This figure and legend are reproduced from (27). To see this figure in color, go online.
In this study, we demonstrated that Ala binding influences the enthalpy of PEP binding using isothermal titration calorimetry (ITC) and the temperature dependence of the allosteric coupling constant between PEP and Ala binding to M1PYK. We found that this PEP-M1PYK-Ala system is “silently coupled” (i.e., completely entropy-enthalpy compensated) at both 15 and 30°C. We also determined a peptide resolution map of which regions of the protein contribute to this silent coupling mechanism. Finally, we initiate a conversation of what the observation of silent coupling might imply within the evolution of a protein family, with the hope that future studies will further consider these evolutionary questions.
Conceptual framework for silent coupling
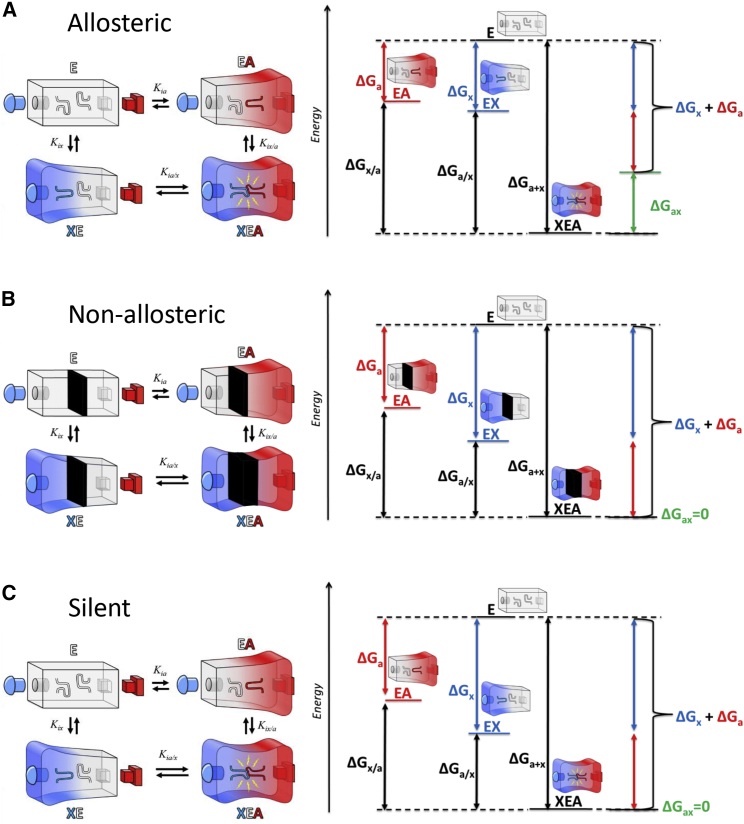
Because of the rarity of reports of silent coupling, there have been few efforts to conceptualize what molecular mechanisms might give rise to this regulation. As an example, given that the ΔG associated with allosteric coupling is zero, what is driving the process of silent coupling? The answer to that question can be explained using energy diagrams (18,33, 34, 35): primarily, individual binding events continue to have negative free energies (Fig. 2). Energy diagrams can also be used to provide a conceptual framework to relate allosteric, nonallosteric, and silent coupling examples.
Figure 2.
A free-energy diagram presentation for an (A) allosteric system, (B) nonallosteric system, and (C) silent coupling system. ΔGa and ΔGx are the free energies associated with substrate (A) and effector (X) binding in the absence of other ligands. ΔGx/a is the free energy for the binding of X when A is present. ΔGa/x is the free energy for the binding of A when X is present. ΔGax is the free energy associated with allosteric coupling. This value is zero in both a nonallosteric system and a silent coupling system. Structural representations have been added in each of the three panels. As diagrammed, in both the allosteric and the silent coupling systems, binding of one ligand alters the second ligand-binding site. To see this figure in color, go online.
The free enzyme (E) can bind substrate (A) first to derive the EA complex, and that binding event has an associated ΔGa. As a second “first” option, the free enzyme (E) can bind effector (X) first to derive the EX complex and that binding event has an associated ΔGx. EA can then bind X with an associated ΔGx/a or EX can then bind A with an associated ΔGa/x. On an energy diagram, the free energy associated with allostery (Fig. 2 A) is ΔGax, which can be expressed as the difference between (ΔGa + ΔGx) and (ΔGa + ΔGx/a) or the difference between (ΔGa + ΔGx) and (ΔGx + ΔGa/x). Again, this is a free-energy presentation for classical allosteric systems. We can emphasize the allosteric nature by annotating the energy diagram with images of proteins to indicate that each ligand-binding site is modified when the second ligand is present. Note the binding of X does not necessarily cause change directly in the A binding site and the binding of A does not necessarily cause change directly in the X binding site (19). In the cartoon used in Fig. 2, it is the modification of two internal loops by the respective ligand-binding events that are relevant to allostery, such that allostery is only realized in the ternary complex.
For nonallosteric systems that bind two ligands, one might assume that the two binding events are completely isolated from each other (Fig. 2 B). Using the same annotated energy diagram, this scenario results in a ΔGax equal to zero. To imply a lack of interaction between the two binding sites, no ligand-induced changes in the protein schematic propagate beyond the center of the protein (black division wall in the protein).
However, as a challenge to the assumption that the two binding events are completely isolated, a description of silent coupling (Fig. 2 C) on the energy diagram requires a fusion of ideas from both the allostery example and the nonallosteric scenario. In this case, ΔGax is equal to zero, similar to the nonallosteric example. However, the protein schematics must be modified to reflect that one ligand-binding event alters the second ligand-binding event. Overall, this energy diagram indicates that the energy difference between the E and EA complex is equal to the difference between the XE and XEA complexes, even if the contributing interactions to the binding event between E and A are modified when X is present.
Materials and Methods
ITC
ITC has been used extensively by the Lee laboratory to characterize allosteric regulation of M1PYK, and the methods presented here have been detailed elsewhere (36, 37, 38). A brief summary of those designs is presented here. All the calorimetric experiments were performed in the buffer 50 mM Tris-HCl (pH 9.0), 500 mM KCl, 10 mM MgCl2, and 0.1 mM EDTA at 25°C, unless otherwise specified, to minimize aggregation of the protein-ligand complex. Titrations were carried out on the VP-ITC MicroCalorimeter (MicroCal, Northampton, MA), which was calibrated with electrically generated heat pulses as recommended by the manufacturer. The initial protein concentration range was between 35 and 55 μM. Initial concentration of PEP in the syringe was 10 mM. When present, Ala was at 10 mM. All solutions were thoroughly degassed by stirring under vacuum for 5 min before use. Right after degassing, the concentration of protein was measured to eliminate the potential change in concentration during the degassing process. Ligands (PEP, Ala) were prepared in the dialysate of the protein solution to minimize artifacts due to minor differences in buffer composition. Even though the buffer was 50 mM Tris, its buffering capacity was not sufficient to maintain the pH at 9.0 in preparing PEP or Ala solutions greater than 3 mM. Therefore, adjustment of pH was needed. The pH of another aliquot of the buffer dialysate was adjusted to pH 10.0 with 10 M KOH. This solution was then used to adjust the pH of the ligand solution to 9.0. In most cases, the pH differences between buffer and ligand were less than 0.05 pH units. The reaction cell contained 1.4372 mL of protein in buffer, and the reference cell contained water only. The injection syringe was filled with PEP solution and was rotating at 459 rpm—previously determined as optimal (36, 37, 38)—during temperature equilibration and experiment. A titration experiment consisted of 25 injections. The first injection was 1 μL, and the subsequent injections were 1–5 (1 μL), 6–10 (2 μL), 11–15 (4 μL), 16–20 (8 μL), and 21–25 (10 μL). Injection speed was 0.5 μL/s with a 4 min interval between injections. A separate titration of the ligand solution into the buffer was performed to determine the heat of dilution of the ligand, which was then subtracted from the heat obtained during the titration of the ligand solution into the protein solution. The first titration point was discarded because of dilution with buffer. The Leverber-Marquardt algorithm performed by Microcal Origin scientific plotting software was used to fit the incremental heat of the ith titration (ΔQ(i)) of the total heat, Qt, to Eq. 1.
| (1) |
where V0 is the volume of the sample solution. For a sequential binding model as represented in Eq. 2,
| (2) |
where [P]t is the total M1PYK tetramer concentration in the sample vessel; [L] is represented in Eq. 3,
| (3) |
[L]t is the total ligand concentration; ΔHi-values are the binding enthalpies; and Ki-values are the macroscopic binding constants for the high-affinity binding sites. Standard deviations for ΔHi and Ki from multiple titration runs were calculated while fitting to four PEP binding sites, consistent with the tetrameric structure and the stoichiometry previously determined (36, 37, 38).
Temperature dependence of the coupling constant
Because the ligands for the M1PYK system (PEP, ATP, Mg2+, K+, and Ala) all can undergo changes in ionization, we chose to use an open system design to evaluate the temperature dependence of the allosteric coupling constant (39). The open system design maintains constant pH of 9.0 at each temperature (i.e., both temperature and total proton concentration change) and results in ΔH and ΔCp estimates consistent with those expected in a nonionizable buffer system but will differ from ΔH and ΔCp determined by ITC (39).
Allosteric coupling was determined as previously described (26). Briefly, enzyme velocity was determined over a concentration range of PEP to determine a Kapp-PEP value. The assay used a lactate dehydrogenase coupled system that included 50 mM Tris-HCl or TAPS (pH 9.0), 10 mM MgCl2, 0.1 mM EDTA, 0.18 mM NADH, 19.6 U/mL lactate dehydrogenase, and 5 mM ADP and was completed in 96-well plates. Changes in the A340 associated with NADH concentration were used to determine reaction rates. Fits of the change in initial velocity (v) at various concentrations of PEP to Eq. 4 were used to determine Kapp-PEP.
| (4) |
where Vmax is the maximum velocity associated with the high-affinity PEP phase, Kapp-PEP is the concentration of substrate that yields a rate equal to one-half the Vmax, and nH is the Hill coefficient associated with the high-affinity PEP phase. In many cases, c was evaluated at high effector concentration and held constant across all other concentrations of effector.
Kapp-PEP was then determined over a concentration range of the effector to determine the allosteric coupling constant and fitted to Eq. 5 (16):
| (5) |
where Ka = Kapp-PEP when [Effector] = 0, Kix = the dissociation constant for effector (X) binding to the protein in the absence of substrate (A), and Qax is the allosteric coupling constant (16). Effectors evaluated here include Ala, Phe, and 2-amino butyric acid (2AB). Error estimates for Kapp-PEP-values determined from data fitting to Eq. 4 were propagated and used to weight data fits to Eq. 5.
At temperatures below 30°C, the response of Kapp-PEP to Phe included a steeper transition than could be accommodated by Eq. 5. That type of steeper transition indicates a change in the cooperativity of the effector. However, because of the detection of allosteric coupling based on monitoring Kapp-PEP, we did not directly assess the cooperativity of effector binding. To accommodate this steep transition, the response of Kapp-PEP to Phe at temperatures below 30°C were fitted to Eq. 6:
| (6) |
Importantly, unlike Eq. 5, Eq. 6 is not based on a rigorous derivation and is used here as an empirical fit to gain an estimate of Qax. The response of Kapp-PEP to Phe concentration at 30°C was fit to Eq. 6.
Qax was determined at a range of temperatures from 10 to 35°C. Effector concentration was varied from 0 to 100 mM. For each effector type and in both Tris and TAPS buffers, the coupling constant at each temperature was determined either in duplicate or triplicate. A precipitant was visible in the Tris buffer system at 35°C (likely Mg(OH)2), and therefore, 30°C was the highest temperature used for that system. The temperature dependence of Qax for Ala and 2AB were fitted to Eq. 7 (40,41).
| (7) |
where Q is the measured Qax value; R is the gas constant (8.314 × 10−3 kJ/mol K); T is the temperature at which Qax is evaluated (the independent variable); Tref is the reference temperature (298 K used here; equal to 25°C); Qref is the fit value for Qax at the reference temperature of 298 K (equal to 25°C); ΔHref is ΔH associated with Qax at the reference temperature; and ΔCp is the heat capacity associated with Qax. Error estimates for Qax-values determined from data fitting to Eq. 5 or Eq. 6 were propagated and used to weight data fits to Eq. 7.
The allosteric coupling can artificially be influenced if temperature modulate causes a sufficient change in the binding affinity for ADP, Mg2+, or K+ sufficiently to cause reduced occupancy (i.e., the concentration used is no longer saturating) at a given temperature and if any one of those ligands also influences PEP binding. To account for these possibilities, K+ is kept high at 500 mM so that any changes in concentration would represent a minimal percent change in concentration. Titrations of activity with MgCl2 in the TAPS buffered assay indicate that the 10 mM concentration used here was saturating at 10, 25, and 35°C (data not shown). Titrations with ADP resulted in negative cooperativity, making it more challenging to gain high confidence that saturation is maintained. Nonetheless, there was minimal change in the presence versus absence of effectors at the three temperatures evaluated (data not shown). One exception was that at 35°C, enzyme activity was not observed when titrating ADP concentration and in the absence of effectors. Presumably, this indicates that the enzyme is not stable at this temperature in the absence of effector and ADP. This observation is not likely to influence our study, given that any time the enzyme was exposed to this highest temperature, ADP was present to provide the necessary stability. However, the instability in the absence of ADP did prevent a confirmation of ADP saturation at the 35°C condition.
H/DX-MS
The control to confirm that a change in solvent from H2O to D2O does not alter allosteric coupling between Ala and PEP binding events has previously been reported (42). Other methods follow those previously described (27). Data for the free enzyme and PEP-bound enzyme were taken directly from that previous study (27). Briefly, the protein was desalted into a Tris-based buffer at pH 9.0 with the appropriate ligands. When added, final concentrations for both PEP and Ala were 10 mM; that concentration was maintained with the protein both before and after transfer to D2O. The protein was diluted in an equivalent buffer made in 99% D2O, and the sample was incubated for the designated time at 24°C. Exchange was quenched by removing aliquots and diluting each with cold ammonium phosphate buffer to result in a final pH of 2.4 at 0°C. The protein was digested with pepsin, and peptide fragments were detected using online HPLC coupled to an LTQ-FT mass spectrometry. Peptides were analyzed with HDXFinder (43) and HDExaminer (Sierra Analytics) and manually using Qual Browser (Thermo-Finnigan) and Mag-Tran (44). Outputs from all approaches were combined. The number of deuteriums incorporated (Dt) at a given time (t) was calculated using Eq. 8:
| (8) |
where Mt is the total average mass observed for a peptide at a given time, M0 is the average mass obtained for peptide in the unlabeled sample, [ΔD] is the mass difference of deuterium relative to hydrogen, and z is the charge of the peptide.
Deuterium content at various times was fitted to Eq. 9:
| (9) |
where D is the deuterium content at time t, N1 and N2 are the number of fast and slow exchanging amide atoms, and k1 and k2 are the respective rate constants. Only the rates for the slow phase were reported in this study (Data S1). The total amount of deuterium was calculated as a sum of N1 and N2 from Eq. 9.
Errors were propagated when H/DX data were combined for multiple enzyme complexes. When propagated error values of differences were greater than the magnitude of the difference, those peptides were excluded from further analysis. Nonetheless, all fit parameters for individual peptides are included in the Supporting Material.
Results
To our knowledge, our identification of silent coupling in M1PYK by comparing a lack of energetic coupling between two ligand-binding events with a SAXS signature that indicates coupling was the first example of detection using a technique other than the determination of enthalpy (i.e., ITC or van’t Hoff analysis) (1, 2, 3, 4,12). In contrast, previous detections of energetic coupling have most often been via directly monitoring enthalpy or determining the allosteric coupling constant over a temperature range.
ITC titration
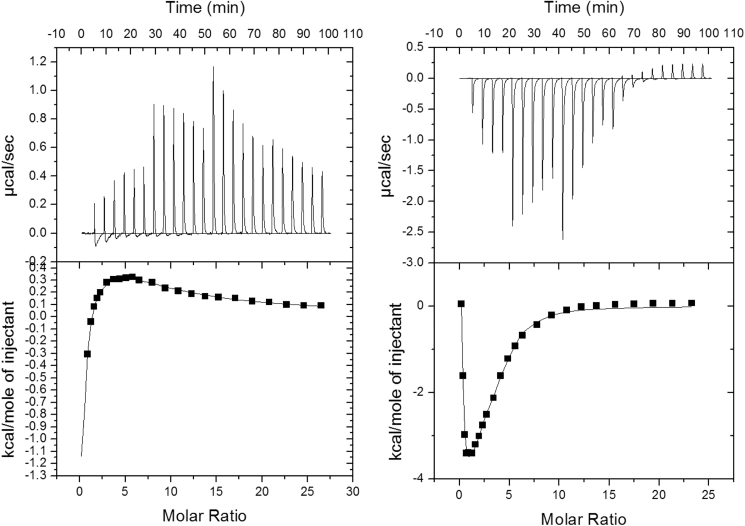
Because of the novelty of the identification method, we first used ITC titrations to evaluate whether the presence of Ala altered the enthalpy signature of PEP binding to M1PYK. The enthalpy of PEP binding to M1PYK was evaluated in the absence and presence of Ala (Fig. 3). The presence of Ala causes a very different binding signature in PEP titrations. The fact that Ala changes the PEP response is consistent with silent coupling. The binding of PEP is very complex, with both exothermic and endothermic reactions present in one titration. Rather than focusing on the fit parameters from the complex responses due to four different PEP binding events in the tetramer (see Table S1), the very different shape caused by the presence of Ala is presented as qualitative confirmation that the presence of Ala influences the enthalpy associated with PEP binding.
Figure 3.
ITC titrations of PEP binding to M1PYK. (Left) Titration in the absence of Ala is shown. The initial concentration of M1PYK was 52 μM, and the stock concentration of PEP in the syringe was 10 mM. Titration was performed in 50 mM Tris-HCl buffer (pH 9.0), 500 mM KCl, 10 mM MgCl2, and 0.1 mM EDTA at 25°C. The first titration point was discarded because of dilution with buffer. (Right) Titration in the presence of Ala is shown. Initial concentrations of M1PYK and Ala were 39 μM and 10 mM, respectively. The stock concentration of PEP in the syringe was 10 mM.
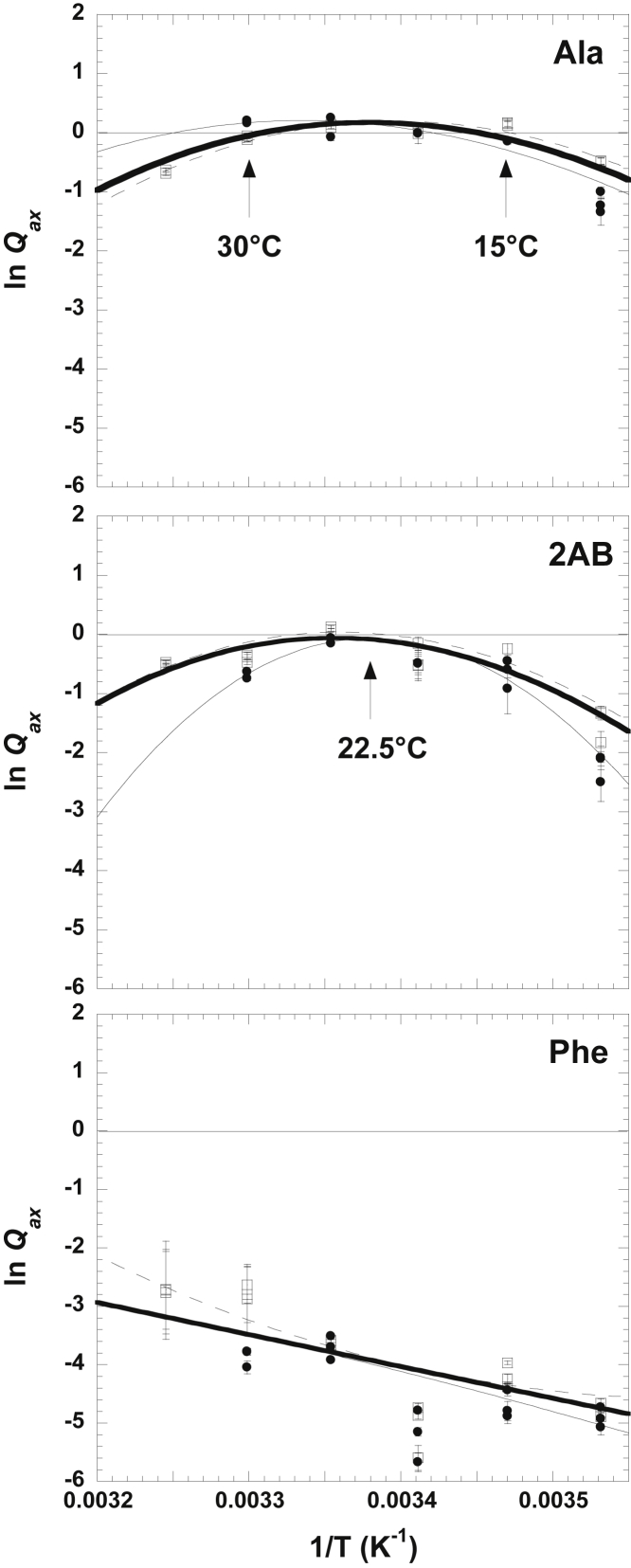
The temperature dependence of the allosteric coupling constant
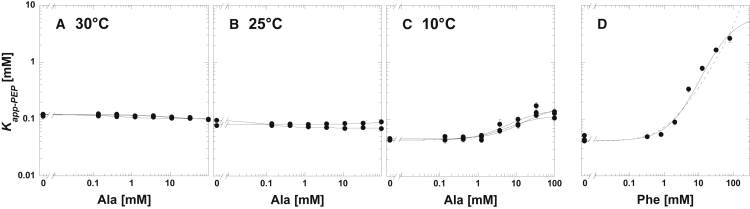
Given the ITC evidence to support the silent coupling of this system, we next determined the temperature dependence of the allosteric regulation by Ala. To accomplish this, the Ala/PEP allosteric coupling constant was determined across a range of temperatures from 10 to 35°C. As noted earlier, inhibition by Ala was negligible at 30°C (Fig. 4). However, inhibition was apparent at both below 15°C and above 30°C. In the response of Kapp-PEP over a concentration range of effector, that inhibition is seen as the difference between a horizontal plateau at low effector concentration and a horizontal plateau at high concentrations of effector (Fig. 4). When the resulting coupling constants are graphed as a van’t Hoff plot, that inhibition is seen as values below zero (Fig. 5). Curvature in the temperature dependence indicates that ΔH is itself temperature dependent (i.e., ΔCp is not zero) (Fig. 5). In the case of the allosteric coupling between Ala and PEP, it is this curvature that results in Ala acting as an allosteric inhibitor at both temperatures above 30°C and below 15°C. At these two temperatures, no allosteric influence of Ala on PEP binding is apparent (i.e., silent coupling). Between these two temperatures, the fit of the temperature dependence of the allosteric coupling constant to Eq. 7 indicates that the presence of Ala causes a very subtle increase in PEP affinity (i.e., allosteric activation). However, the minimal activation predicted at the intermediate temperatures, the error estimates associated with the experimentally determined Qax-values, and the scatter represented by the replicate Qax-values make it challenging to have full confidence that activation is realized (Fig. 4 B).
Figure 4.
Kapp-PEP values as functions of effector concentrations and at various temperatures. All representative data are in Tris buffer. (A) Triplicate data sets for the response of Kapp-PEP to Ala at 30°C are given. (B) Duplicate data sets for the response of Kapp-PEP to Ala at 25°C are shown. Although the fit of Qax as a function of temperature in Fig. 5 indicates a very slight allosteric activation between Ala and PEP in the M1PYK system, data in (B) are not completely convincing that activation is ever realized. (A) Triplicate data sets for the response of Kapp-PEP to Ala at 10°C are shown. In this presentation, Qax is indicated by the difference between the horizontal plateau at low Ala concentration and the horizontal plateau that is approached at high Ala concentration. At 10°C, Ala clearly acts as an allosteric inhibitor. For (A)–(C), lines represent the best fits to Eq. 5. (D) The response of Kapp-PEP to Phe at 10°C is shown. Only one of the replicates is included. These data demonstrate the rapid transition in the response to Phe that cannot be accommodated by the fit to Eq. 5 (dashed line) but can be accommodated by a fit to Eq. 6 (solid line). This rapid transition in the response to Phe occurred at all temperatures below 30°C. This example is at 10°C in Tris buffer. Error bars in all panels are error estimates made during the fitting of enzymatic rate as a function of PEP concentration to Eq. 4. When error bars are not apparent, they are smaller than the data points.
Figure 5.
A van’t Hoff plot of the dependencies of allosteric coupling constants on temperature. Temperature dependence was determined in an “open system” experimental design in which pH was held constant at pH 9.0. Data were collected both in Tris (solid circles and thin solid fit line) and TAPS (open squares and dashed line) buffers. Lines represent the best fits to Eq. 7. Error bars are error estimates made during the fitting of Kapp-PEP to Eq. 5. Because of the “open system” design, which maintains constant pH at all temperatures, buffer ionization should not influence the data trend, and therefore, data collected in both Tris and TAPS buffers can be treated equally. The thick solid line in each panel represents data fits for the combined Tris and TAPS data, and parameters obtained from the fitting of this combined data are included in Table S2.
We chose to extend the characterization of the temperature dependence of Qax to the study of regulation by both 2AB and Phe (Fig. 5). The response to 2AB was very similar to that for Ala, only 2AB never became an activator but had silent coupling between the 20 and 25°C assay conditions. Unlike regulation by Ala and 2AB, the response of allosteric regulation to Phe elicited a strong allosteric inhibition throughout the full temperature range evaluated.
H/DX
Next, H/DX-MS was evaluated for the four enzyme complexes in the Ala-PEP allosteric energy cycle (see Fig. 2 A: free enzyme, PEP-M1PYK, M1PYK-Ala, and PEP-M1PYK-Ala). After subtracting the initial methionine that is cleaved in vivo, M1PYK has 530 amino acids. Peptides that were observed in each of the four enzyme complexes encompass 361 positions. Thus, this study reports on 68% of the protein in the evaluation of the Ala-PEP silent coupling.
The total number of exchangeable protons evaluated at 48 h was one parameter used to evaluate ligand-induced changes (Fig. S1). As previously reported, most peptides indicated two exchange rates (27). The faster of the two rates is too rapid for accurate determination without a quench flow design. As a result, the rates for the slow phase were the second parameter compared among complexes. Using the same methodologies and instrumentation used here, we previously established a 2% error in the reproducibility of the number of protons that exchange and an 8% error in rate constants (27). All rates reported here exceed those error values.
Proton protection and deprotection due to ligand binding
PEP binding in the absence of Ala (Fig. S2, across top) protects a limited region of the protein located in the subunit interface near the PEP binding site. However, in the presence of Ala, PEP binding (Fig. S2, across the bottom) protects several regions that are broadly spread throughout the protein.
Ala binding in the absence of PEP (Fig. S2, left side) causes deprotection of two types of subunit interfaces and in the B-domain. However, when PEP is present (Fig. S2, right side), the only identified change upon Ala binding is minimal deprotection in a helix near the PEP binding site.
Altered rates of exchange due to ligand binding
PEP binding in the absence of Ala (Fig. S3, across top) results in only slightly increased rates of exchange in areas that are broadly centered around the active site. Much larger increases in exchange rates are seen when PEP binds in the presence of Ala (Fig. S3, across the bottom), and those increased rates are concentrated in the B-domain.
Ala binding in the absence of PEP (Fig. S3, left side) causes increased rates of exchange in a few peptides in the C-domain. When PEP is present, Ala binding (Fig. S3, right side) causes only modest increases in the rates of exchange in peptides isolated in the B-domain.
Surprisingly, we did not see peptides with reduced rates of exchange (blue) as the liganded state of the protein became more complex.
Proton protection and deprotection and altered rates of exchange due to silent coupling
In Figs. S2 and S3, we identified changes caused by PEP binding both in the absence (across top) or in the presence (across bottom) of bound Ala. Allosteric coupling is defined as a comparison of these two differences (substrate binding in the absence versus presence of effector). Therefore, by comparing these two groups of changes, we can identify changes that are associated with the allosteric coupling or, in this case, the silent coupling. Changes that are only related to ligand binding should be subtracted out in this evaluation, leaving only coupling-relevant changes. Fig. 6, A and B present changes in protection and deprotection of proton exchange and changes in rates of exchange. Clearly, there are example areas in M1PYK that experience protection against proton exchange, deprotection against proton exchange, slower rates of exchange, and faster rates of exchange in response to silent coupling. Two helixes near the PEP binding site and peptides in the B-domain were also previously highlighted by our H/DX study comparing the M1PYK-Phe complex with the M1PYK-Ala complex (27). However, the identification methods for each of those peptides (e.g., protection and deprotection identified a peptide in the study with Phe, but that same peptide was identified with a change in the rate of exchange in this study) were opposite in this study compared with that previous work. Furthermore, the presence of Phe caused additional changes in the A-A interface.
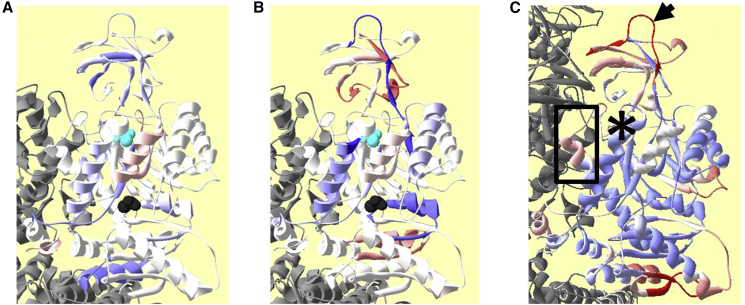
Figure 6.
H/DX-MS data and COREX/BEST calculations mapped onto the M1PYK structure. (A and B) Changes associated with silent coupling as determined by comparing the changes between E and EA (comparison across the top of Figs. S2 and S3) with the changes between XE and XEA (comparison across the bottom of Figs. S2 and S3) are shown. In both (A) and (B), neighboring subunits in the homotetramer are in gray. The active site includes pyruvate in cyan spacefill. The amino acid binding site includes Ala in black spacefill. (A) includes differences in total numbers of exchanged protons. (B) includes differences in rates of exchange. (C) lnKif-values calculated for each position in one subunit of the PDB: 2G50 (26) structure of M1PYK using COREX/BEST (45) are shown. The average lnKif-value was color-coded white. A white to red gradient was used to indicate positions predicted to have a range of lower than average stability (deeper red is less stable/more dynamic), and a white to blue gradient was used to indicate positions predicted to have a range of higher than average stability (deeper blue is more stable/less dynamic). There is no designed coordination between color codes used in this figure with color codes used in other structural figures. To aid discussion, a highly stable helix and a highly flexible loop are marked with an asterisk and an arrow, respectively. Additionally, a helix-loop that was identified in the previous Phe-binding versus Ala-binding study, but not in the current silent coupling by Ala, is shown. To see this figure in color, go online.
Next, we questioned whether the regions identified by H/DX (changes related to both ΔH and ΔS) as associated with coupling (Fig. 6, A and B) were isolated from or primarily associated with more dynamic regions of the M1PYK protein. To evaluate this possibility, Fig. 6 C includes energetic/stability calculations from COREX/BEST (45) (a calculation of ΔG) for each position in one subunit of the Protein Data Bank, PDB: 2G50 structure of M1PYK (26). Please note that calculations of the energetics of the protein were initiated with a single structure of the M1PYK protein. We acknowledge that a better approach might be to use calculations of energetics and stability that start with protein structure complexes that are specific for each of the four corners of the energy cycles represented in each panel of Fig. 2 (46). Unfortunately, not all of those structures are available for M1PYK. As an alternative, we can imagine that silent coupling or allostery causes changes in the energetics and stability of highly energetic regions of the protein, but in each of the four corners of the energy cycle (Fig. 2), those highly energetic regions continue to be overall more energetic than the most stable regions of the protein. Thus, simply identifying the most dynamic regions of the protein in any available structure might correlate with those areas of the protein that contribute to silent or allosteric mechanisms. Therefore, we include this simpler design of asking whether the H/DX outcomes correlate with less stable regions of the M1PYK protein represented in the PDB: 2G50 structure. Regions of the protein identified by H/DX-MS did not correlate with more or less stable regions of the protein. This can be appreciated by considering that both a highly stabilized helix (see ∗ in Fig. 6 C) and a highly dynamic loop (see arrow in Fig. 6 C) were both highlighted by H/DX-MS studies.
Discussion
On the study of the silent coupling mechanism
At first introduction to the concept of silent coupling, the most immediate question that arises is why study the mechanism of something that causes a “silent” outcome? The answer to that question has been provided by Schann et al. by the suggestion that “silent modulators” are ideal drug leads because they can be modified to be either allosteric activators or inhibitors (25). The next question is how silent coupling effectors are most likely to be identified. In the design that uses a binding assay to screen a small molecule library for leads that bind to a target protein, followed by assays to evaluate modified protein function, it is now common to identify leads that bind but fail to alter function. Therefore, identification of silent couplers is now common in many small molecule library screens, again, exemplified by the Schann et al. study (25).
If indeed, silent coupling leads are an ideal starting point for drug design, it follows that understanding the molecular mechanisms that give rise to silent coupling is expected to facilitate rational drug design of both allosteric activators and inhibitors from a common lead molecule. Our laboratory has a long history of evaluating allosteric mechanisms using a linked-function analysis (14, 15, 16, 17,19,20). In functional studies of the structure and function questions of allosteric mechanisms, we have routinely monitored binding of one ligand over a concentration range of the second ligand (26,47, 48, 49). However, with no change in binding affinities, this design will clearly not be informative in the study of a silent coupling mechanism. Therefore, it is worth noting that silent coupling mechanisms (considered at the temperature where silent coupling is observed) will best be studied using structural techniques (e.g., H/DX-MS) and techniques that evaluate ΔH (i.e., ITC and van’t Hoff analysis).
Given this reasoning for why studies of silent coupling are useful, it also stands to reason that studies of silent coupling mechanisms and the mechanisms that give rise to allosteric regulation may complement each other. To further appreciate this idea, consider the examples that show that the magnitude of the allosteric coupling constant is dependent on effector chemistry (26,48,50), cofactors (e.g., metal type in the active site (47)), and changes in solution conditions (e.g., temperature (3,4)). In particular, the influence of effector chemistry on the coupling constant is highlighted in our M1PYK system by Phe eliciting an allosteric response in the affinity of the enzyme for PEP (when assayed at 30°C). That contrasts with the lack of a change in PEP binding when Ala is present (again, at 30°C) (26,32). Therefore, it is reasonable that in the study of a single protein, an allosteric mechanism can be compared to a silent coupling system to distinguish subtle changes in the protein that control the magnitude of the allosteric coupling. That design was a primary motivation for the initiation of this study.
In the study of M1PYK, we previously asked what changes are elicited by Phe (allosteric effector) that are in addition to changes caused by Ala (a silent modulator at the 30°C condition). We previously reported an H/DX-MS study that compared the binding of Phe to M1PYK and the binding of Ala to M1PYK (27). In contrast, the comparison included in Fig. 6, A and B identifies what regions of M1PYK that are modified by Ala binding are differentially altered when PEP is already bound as Ala binds (i.e., associated with silent coupling). Unfortunately, the accessible solubility range of Phe limits evaluation of the PEP-M1PYK-effector complex (26,32), and therefore, we cannot make the latter comparisons when Phe is the effector ligand. With the acknowledgment that we are comparing the outcomes of two different types of comparisons (i.e., an apples-to-oranges comparison), we find it surprising that the two different methods of comparing H/DX-MS data for the various enzyme complexes identify many (but not all) of the same regions of the protein. We can then ask 1) whether regions that are unique between the study of Phe and Ala are the regions that cause differences in the magnitude of the allosteric coupling (e.g., see helix/loop included in a box in Fig. 6 C), 2) whether regions that are commonly identified experience a different range of dynamic and structural changes when Phe is the effector versus when Ala is added (e.g., see loop marked by an arrow in Fig. 6 C), or 3) whether the inability to study the Phe/M1PYK/PEP complex prevents us from identifying the region of the protein that determines the magnitude of the allosteric response. Therefore, although questions remain, our study identifies specific regions of the M1PYK protein that are likely to contribute as determinants of the magnitude of the allosteric coupling.
Temperature as a variable in structure and function studies of allosteric mechanisms
The ITC results in Fig. 3 confirm that the presence of Ala causes a change in the enthalpy of PEP binding, and that observation is consistent with the proposed entropy-enthalpy compensation in the allosteric coupling constant. The possibility that Ala simply destabilizes the protein without binding specifically as an allosteric effector can be argued against by several observations in addition to the cocrystalization of Ala in the allosteric binding site (26). There is a long history of using Ala as a competitive binder when Phe is the allosteric inhibitor (26,51, 52, 53, 54). Ala binding as a function of PEP concentration has been determined in conjunction with H/DX and SAXS studies (32,42). In this study, the upper plateau observed in the response of the Kapp-PEP to Ala concentration at 10 and 35°C indicates that the influence of Ala on PEP affinity has a maximum, and that, in turn, is consistent with the saturated binding of an allosteric site, but not a nonspecific effect. In addition, Ala binding does not cause the general increase in numbers of exchangeable protons across all regions of the protein that would be expected for a general protein destabilization.
Our characterization of the temperature dependence of Qax for the Ala-PEP coupling in M1PYK indicates that there are two regions of silent coupling at 30 and 15°C (Fig. 5). Above 30°C and below 15°C, Ala acts as an allosteric inhibitor of PEP binding to M1PYK. At temperatures intermediate to these two silent coupling temperatures, a very subtle allosteric activation of Ala on PEP binding is possible. Interestingly, this trend is not completely consistent with previously reported linear temperature responses of allosteric coupling constants in other systems (1,2) because the ΔH for the allosteric coupling constant in those other systems seems to have little, if any, dependence on temperature (i.e., ΔCp is zero).
Nonetheless, the temperature-dependent trend characterized here for M1PYK can be used to suggest future studies: primarily, structural studies (including H/DX-MS) can be completed at temperatures that support allosteric inhibition, and outcomes can be compared with equivalent studies completed at silent coupling temperatures. That same design can be used when 2AB is the effector, especially given that 2AB elicits a stronger allosteric inhibition (compared to Ala) at the 10°C condition. Silent coupling was not observed for the allosteric regulation by Phe at any temperature tested. The reduced inhibitory response needed to study the Phe-M1PYK-PEP complex is favored at higher temperatures. However, the potential of protein instability at 35°C indicates that the 30°C used in our previous studies (26,32) is likely the most ideal temperature, even though the solubility limits of Phe limit the percentage of the protein that is in the Phe-M1PYK-PEP complex at that most ideal temperature. Nonetheless, the experimental designs that can be suggested based on the temperature dependence of regulation by Ala and 2 AB are likely to be useful in identifying which regions of M1PYK experience temperature-dependent changes to contribute to the ΔG associated with the allosteric coupling constant.
The influence of silent coupling on the evolution of an allosteric mechanism in a protein family
Beyond the specific study of silent coupling in M1PYK, the recognition that silent coupling exists causes us to ponder how silent coupling influences thoughts on the evolution of allostery within a protein family and what functional or evolutionary pressures might influence the existence of silent coupling. First, consider that several studies of allosteric systems compare structure and function outcomes with an isozyme of the same protein that lacks energetic coupling between the two binding events (i.e., likely labeled as nonallosteric). However, the knowledge that silent coupling is a potential mechanism that can exist makes it more challenging to use a simple effector influence on substrate affinity to identify if one isozyme in a protein family truly lacks energetic coupling between binding events. Next, we can propose that silent coupling may be a mechanism for obtaining specificity in the allosteric effector. In other words, rather than an effector binding or not binding to obtain the necessary ligand specificity, the regulatory ligand can cause an allosteric effect by which the silent modulator binds but results in a compensated (i.e., silent) response. Hence, only the regulatory ligand causes modified function. The competitive binding between one ligand that elicits an allosteric response and one that does not may provide an even more nuanced control mechanism. As a second consideration, we find it possible that although a silent modulator may not influence substrate affinity in isolation, that same ligand might influence the regulation of substrate affinity by a third ligand (i.e., a three-ligand-coupled event).
A final consideration about the evolution of silent coupling comes from the idea that a binding site may be more challenging to derive in a protein family than an allosteric mechanism. To support this idea, first consider that many pyruvate kinase isozymes are regulated by phosphorylated sugars (55), although the identity of those effectors varies considerably among isozymes (56). Our previous study used extensive numbers of effector analogs, point mutations, and crystallographically determined structures to characterize which regions of the fructose-1,6-bisphosphate (Fru-1,6-BP) allosteric site of human liver PYK (LPYK) contribute to allostery (50,57). One of the two PYK isozymes from Escherichia coli (EcPYK) is also regulated by Fru-1,6-BP (58). However, at both the sequence and structural levels, the region identified to be most relevant to regulation in LPYK is not conserved in EcPYK. At the same time, the loop that interacts with a phosphate on most known sugar-phosphate effectors is highly conserved. Therefore, in the PYK family of proteins, it appears that the binding site for the sugar-phosphate is highly conserved, but the allosteric mechanism is less conserved. Returning to the influence of silent coupling on considerations of how allosteric mechanisms evolve over time, it remains possible that silent coupling offers a mechanism by which a binding site can be conserved while the resulting regulation can be modified to best benefit organism survival. Although we use observations from the PYK family that includes changes in the allosteric binding site, the more general idea that allostery can evolve more easily than a binding site is also consistent with a large fraction of a protein that might evolve to modulate allostery as opposed to a much smaller percentage of a protein that constitutes an allosteric binding site.
Conclusions
In this study, we confirm that at 30°C, the PEP-M1PYK-Ala system represents a silent coupled system (i.e., complete entropy-enthalpy compensation in the allosteric coupling constant). H/DX-MS indicates that many of the regions identified to be important for allosteric inhibition by Phe also contribute to the silent coupling mechanism by Ala. The temperature dependence of the allosteric coupling constant PEP-M1PYK-Ala allows experimental designs that may be useful to further identify regions of the M1PYK protein that contribute to determining allosteric function. Furthermore, appreciating that silent coupling systems exist influences thoughts on the evolution of allosteric mechanisms within a protein family.
Acknowledgments
We appreciate consultation from Dr. James R. Horn (Northern Illinois University) for data fitting the temperature-dependent data. Max Fairlamb generated the cartoon representations of protein conformational changes, and we are grateful for his efforts to make such illustrative figures.
This research was supported by National Institutes of Health grant GM115340 to A.W.F. H/DX-MS data were collected in the University of Kansas Medical Center Mass Spectrometry/Proteomics Core Laboratory. The ITC experiments completed at the University of Texas Medical Branch were supported by The Robert A. Welch Foundation H-0013 to JCL.
Editor: Doug Barrick.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.05.012.
Author Contributions
C.B.P. and A.W.F. designed and performed H/DX studies in collaboration with A.A. Primary analyses of that data were performed by C.B.P. An independent analysis was performed by M.T.V., and all analyses were further confirmed by A.A. A.G. and J.C.L. performed and evaluated the calorimetry data and determined COREX values. T.A.M. generated the temperature-dependent data. C.B.P. and A.W.F. wrote the initial manuscript draft and all authors commented on and contributed to revisions. J.C.L., A.A., and A.W.F. supervised the project.
Supporting Material
H/D Exchange Data
References
- 1.Fisher H.F., Tally J. Isoergonic cooperativity in glutamate dehydrogenase complexes: a new form of allostery. Biochemistry. 1997;36:10807–10810. doi: 10.1021/bi9708388. [DOI] [PubMed] [Google Scholar]
- 2.Fisher H.F., Tally J. Isoergonic cooperativity: a novel form of allostery. Methods Enzymol. 1998;295:331–349. doi: 10.1016/s0076-6879(98)95047-9. [DOI] [PubMed] [Google Scholar]
- 3.Braxton B.L., Tlapak-Simmons V.L., Reinhart G.D. Temperature-induced inversion of allosteric phenomena. J. Biol. Chem. 1994;269:47–50. [PubMed] [Google Scholar]
- 4.Tlapak-Simmons V.L., Reinhart G.D. Obfuscation of allosteric structure-function relationships by enthalpy-entropy compensation. Biophys. J. 1998;75:1010–1015. doi: 10.1016/S0006-3495(98)77589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blunt C.E.W., Dougherty D.A. Investigating the binding interactions of NS6740, a silent agonist of the nicotinic receptor. Mol Pharmacol. 2019;97 doi: 10.1124/mol.119.116244. mol.119.116244. [DOI] [PubMed] [Google Scholar]
- 6.Papke R.L., Peng C., Stokes C. NS6740, an α7 nicotinic acetylcholine receptor silent agonist, disrupts hippocampal synaptic plasticity. Neurosci. Lett. 2018;677:6–13. doi: 10.1016/j.neulet.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quadri M., Papke R.L., Horenstein N.A. Dissection of N,N-diethyl-N'-phenylpiperazines as α7 nicotinic receptor silent agonists. Bioorg. Med. Chem. 2016;24:286–293. doi: 10.1016/j.bmc.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papke R.L., Bagdas D., Damaj M.I. The analgesic-like properties of the alpha7 nAChR silent agonist NS6740 is associated with non-conducting conformations of the receptor. Neuropharmacology. 2015;91:34–42. doi: 10.1016/j.neuropharm.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chojnacka K., Papke R.L., Horenstein N.A. Synthesis and evaluation of a conditionally-silent agonist for the α7 nicotinic acetylcholine receptor. Bioorg. Med. Chem. Lett. 2013;23:4145–4149. doi: 10.1016/j.bmcl.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit C.M., Zhang J., Lee A.L. Hidden dynamic allostery in a PDZ domain. Proc. Natl. Acad. Sci. USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hegazy U.M., Musdal Y., Mannervik B. Hidden allostery in human glutathione transferase p1-1 unveiled by unnatural amino acid substitutions and inhibition studies. J. Mol. Biol. 2013;425:1509–1514. doi: 10.1016/j.jmb.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Fisher H.F. Detecting “silent” allosteric coupling. Methods Mol. Biol. 2012;796:71–96. doi: 10.1007/978-1-61779-334-9_5. [DOI] [PubMed] [Google Scholar]
- 13.Roche J., Girard E., Madern D. The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization. J. Struct. Biol. 2019;208:7–17. doi: 10.1016/j.jsb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart G.D. The determination of thermodynamic allosteric parameters of an enzyme undergoing steady-state turnover. Arch. Biochem. Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 15.Reinhart G.D. Linked-function origins of cooperativity in a symmetrical dimer. Biophys. Chem. 1988;30:159–172. doi: 10.1016/0301-4622(88)85013-0. [DOI] [PubMed] [Google Scholar]
- 16.Reinhart G.D. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 17.Weber G. Energetics of ligand binding to proteins. Adv. Protein Chem. 1975;29:1–83. doi: 10.1016/s0065-3233(08)60410-6. [DOI] [PubMed] [Google Scholar]
- 18.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 19.Fenton A.W. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem. Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson G.M., Fenton A.W. What mutagenesis can and cannot reveal about allostery. Biophys. J. 2016;110:1912–1923. doi: 10.1016/j.bpj.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 22.Horn J.R., Russell D., Murphy K.P. Van’t Hoff and calorimetric enthalpies from isothermal titration calorimetry: are there significant discrepancies? Biochemistry. 2001;40:1774–1778. doi: 10.1021/bi002408e. [DOI] [PubMed] [Google Scholar]
- 23.Jin L., Yang J., Carey J. Thermodynamics of ligand binding to trp repressor. Biochemistry. 1993;32:7302–7309. doi: 10.1021/bi00079a029. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y., Ackers G.K. Enthalpic and entropic components of cooperativity for the partially ligated intermediates of hemoglobin support a “symmetry rule” mechanism. Biochemistry. 1995;34:6316–6327. doi: 10.1021/bi00019a009. [DOI] [PubMed] [Google Scholar]
- 25.Schann S., Mayer S., Neuville P. Chemical switch of a metabotropic glutamate receptor 2 silent allosteric modulator into dual metabotropic glutamate receptor 2/3 negative/positive allosteric modulators. J. Med. Chem. 2010;53:8775–8779. doi: 10.1021/jm101069m. [DOI] [PubMed] [Google Scholar]
- 26.Williams R., Holyoak T., Fenton A.W. Differentiating a ligand’s chemical requirements for allosteric interactions from those for protein binding. Phenylalanine inhibition of pyruvate kinase. Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 27.Prasannan C.B., Villar M.T., Fenton A.W. Identification of regions of rabbit muscle pyruvate kinase important for allosteric regulation by phenylalanine, detected by H/D exchange mass spectrometry. Biochemistry. 2013;52:1998–2006. doi: 10.1021/bi400117q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckett D. Hydrogen-deuterium exchange study of an allosteric energy cycle. Methods Mol. Biol. 2012;796:261–278. doi: 10.1007/978-1-61779-334-9_14. [DOI] [PubMed] [Google Scholar]
- 29.Frantom P.A., Zhang H.M., Blanchard J.S. Mapping of the allosteric network in the regulation of alpha-isopropylmalate synthase from Mycobacterium tuberculosis by the feedback inhibitor L-leucine: solution-phase H/D exchange monitored by FT-ICR mass spectrometry. Biochemistry. 2009;48:7457–7464. doi: 10.1021/bi900851q. [DOI] [PubMed] [Google Scholar]
- 30.Tischer A., Brown M.J., Auton M. Arabinose alters both local and distal H-D exchange rates in the Escherichia coli AraC transcriptional regulator. Biochemistry. 2019;58:2875–2882. doi: 10.1021/acs.biochem.9b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donovan K.A., Zhu S., Dobson R.C. Conformational dynamics and allostery in pyruvate kinase. J. Biol. Chem. 2016;291:9244–9256. doi: 10.1074/jbc.M115.676270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenton A.W., Williams R., Trewhella J. Changes in small-angle X-ray scattering parameters observed upon binding of ligand to rabbit muscle pyruvate kinase are not correlated with allosteric transitions. Biochemistry. 2010;49:7202–7209. doi: 10.1021/bi100147w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilser V.J. Biochemistry. An ensemble view of allostery. Science. 2010;327:653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilser V.J. Modeling the native state ensemble. Methods Mol. Biol. 2001;168:93–116. doi: 10.1385/1-59259-193-0:093. [DOI] [PubMed] [Google Scholar]
- 35.Motlagh H.N., Wrabl J.O., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herman P., Lee J.C. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 2. Fluorescence study. Biochemistry. 2009;48:9456–9465. doi: 10.1021/bi900280u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herman P., Lee J.C. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 3. Mechanism. Biochemistry. 2009;48:9466–9470. doi: 10.1021/bi900281s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman P., Lee J.C. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 1. Calorimetric study. Biochemistry. 2009;48:9448–9455. doi: 10.1021/bi900279x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horn J.R., Brandts J.F., Murphy K.P. van’t Hoff and calorimetric enthalpies II: effects of linked equilibria. Biochemistry. 2002;41:7501–7507. doi: 10.1021/bi025626b. [DOI] [PubMed] [Google Scholar]
- 40.Ha J.H., Spolar R.S., Record M.T., Jr. Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J. Mol. Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- 41.Tretyachenko-Ladokhina V., Ross J.B., Senear D.F. Thermodynamics of E. coli cytidine repressor interactions with DNA: distinct modes of binding to different operators suggests a role in differential gene regulation. J. Mol. Biol. 2002;316:531–546. doi: 10.1006/jmbi.2001.5302. [DOI] [PubMed] [Google Scholar]
- 42.Prasannan C.B., Artigues A., Fenton A.W. Monitoring allostery in D2O: a necessary control in studies using hydrogen/deuterium exchange to characterize allosteric regulation. Anal. Bioanal. Chem. 2011;401:1083–1086. doi: 10.1007/s00216-011-5133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller D.E., Prasannan C.B., Artigues A. HDXFinder: automated analysis and data reporting of deuterium/hydrogen exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 2012;23:425–429. doi: 10.1007/s13361-011-0234-5. Published online November 15, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Marshall A.G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 45.Vertrees J., Barritt P., Hilser V.J. COREX/BEST server: a web browser-based program that calculates regional stability variations within protein structures. Bioinformatics. 2005;21:3318–3319. doi: 10.1093/bioinformatics/bti520. [DOI] [PubMed] [Google Scholar]
- 46.Johnson L.E., Ginovska B., Raugei S. Chokepoints in mechanical coupling associated with allosteric proteins: the pyruvate kinase example. Biophys. J. 2019;116:1598–1608. doi: 10.1016/j.bpj.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenton A.W., editor. Allostery: Methods and Protocols. Humana Press: Springer Science; New York: 2012. [Google Scholar]
- 48.Urness J.M., Clapp K.M., Fenton A.W. Distinguishing the chemical moiety of phosphoenolpyruvate that contributes to allostery in muscle pyruvate kinase. Biochemistry. 2013;52:1–3. doi: 10.1021/bi301628k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alontaga A.Y., Fenton A.W. Effector analogues detect varied allosteric roles for conserved protein-effector interactions in pyruvate kinase isozymes. Biochemistry. 2011;50:1934–1939. doi: 10.1021/bi200052e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishwar A., Tang Q., Fenton A.W. Distinguishing the interactions in the fructose 1,6-bisphosphate binding site of human liver pyruvate kinase that contribute to allostery. Biochemistry. 2015;54:1516–1524. doi: 10.1021/bi501426w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kayne F.J., Price N.C. Amino acid effector binding to rabbit muscle pyruvate kinase. Arch. Biochem. Biophys. 1973;159:292–296. doi: 10.1016/0003-9861(73)90455-4. [DOI] [PubMed] [Google Scholar]
- 52.Carminatti H., Jiménez de Asúa L., Rozengurt E. Allosteric properties of skeletal muscle pyruvate kinase. J. Biol. Chem. 1971;246:7284–7288. [PubMed] [Google Scholar]
- 53.Ibsen K.H., Trippet P. Effects of amino acids on the kinetic properties of three noninterconvertible rat pyruvate kinases. Arch. Biochem. Biophys. 1974;163:570–580. doi: 10.1016/0003-9861(74)90516-5. [DOI] [PubMed] [Google Scholar]
- 54.Tsao M.U. Kinetic properties of pyruvate kinase of rabbit brain. Mol. Cell. Biochem. 1979;24:75–81. doi: 10.1007/BF00314888. [DOI] [PubMed] [Google Scholar]
- 55.Muñoz M.E., Ponce E. Pyruvate kinase: current status of regulatory and functional properties. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003;135:197–218. doi: 10.1016/s1096-4959(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 56.Schormann N., Hayden K.L., Chattopadhyay D. An overview of structure, function, and regulation of pyruvate kinases. Protein Sci. 2019;28:1771–1784. doi: 10.1002/pro.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McFarlane J.S., Ronnebaum T.A., Lamb A.L. Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-π bond. Acta Crystallogr. F Struct. Biol. Commun. 2019;75:461–469. doi: 10.1107/S2053230X19007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattevi A., Valentini G., Coda A. Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition. Structure. 1995;3:729–741. doi: 10.1016/s0969-2126(01)00207-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
H/D Exchange Data