Abstract
SARS-CoV-2 was first detected in the city of Wuhan, Hubei Province, China. In this report, we describe the complete genome sequence of the first imported SARS-CoV-2, detected in a Mexican patient who had traveled to Bergamo, Italy. Phylogenetic analysis showed that this isolate belongs to subclade A2a (lineage G) and is closely related to isolates from Finland, Germany and Brazil, all of which were from patients with a history of travel to Italy. This is the first report of the complete genome sequence of this virus in Mexico.
Virus name: hCoV/Mexico/CDMX/InDRE_01/2020
GISAID accession ID: EPI_ISL_412972
A novel coronavirus, recently named severe acute respiratory syndrome-related coronavirus type 2 (SARS-CoV-2), was identified in the city of Wuhan, Hubei Province, China, in December of 2019 It is betacoronavirus that affects the lower respiratory tract and manifests as pneumonia in humans [1, 2]. According to the Johns Hopkins University Coronavirus Resource Center and the Daily Technical Statement on New Coronavirus Cases Worldwide (COVID-19) issued by the Secretary of Health of Mexico, as of March 20, 2020, 234,072 confirmed cases have been reported worldwide with 9840 deaths. In Mexico, 203 cases have been confirmed, with two deaths reported [3, 4]. At the time of this writing, 768 complete genomic sequences of SARS Cov-2 have been obtained and deposited in the GISAID database, and 10 clades have been defined from phylogenetic studies carried out with these sequences (A1, A1a, A2, A2a, A3, A5, B, B1, B2 and B4) [5].
Here, we report the complete genome sequence of SARS-CoV-2 from the first imported case of COVID-19 detected in Mexico. The sample hCoV/Mexico/CDMX/InDRE_01/2020 was obtained directly from a nasopharyngeal swab from a 35-year-old male patient (InDRE 1198) residing in Mexico City who had a confirmed diagnosis of SARS-CoV-2 on February 27, 2020 and had traveled to Bergamo, Italy, between February 16 and 22, 2020.
A diagnostic test for COVID-19 was first conducted at the Instituto Nacional de Enfermedades Respiratorias, Mexico City, on February 27, 2020. Following the national guidelines of the Mexican Ministry of Health [6], a sample was sent to the National Influenza Center at the Instituto de Diagnostico y Referencia Epidemiologicos (InDRE); the confirmatory diagnosis was conducted using a real-time RT-PCR protocol to detect SARS-CoV-2 [7], and of a 221-bp fragment of the region coding for the orf1ab polyprotein was sequenced by the Sanger method and will be published elsewhere. The sample from the patient had a cycle threshold value of 19.5 for the E gene and 20.4 for the RdRP gene.
The complete virus genome from the positive case was amplified using the novel coronavirus sequencing multiplex primers [8] (https://artic.network/ncov-2019). cDNA synthesis was conducted using Super Script IV Reverse Transcriptase (Invitrogen) and the concentration of PCR the product was measured using a Qubit dsDNA High Sensitivity Kit on a Qubit 3.0 fluorometer (Thermo Fisher) and an Agilent 2100 Bioanalyzer (Agilent Technologies).
A library without barcodes was generated and sequenced using an Ion Torrent system, using an Ion Xpress™ Plus Fragment Library Kit for sequencing on an Ion 318™ v2 chip. A total of 1,445,751 reads were obtained for hCov/Mexico/CDMX/InDRE_01/2020. The complete genome sequence was assembled using CLC Genomics Workbench 20 (QIAGEN) and SPAdes [9] and mapped against the reference sequence Wuhan-Hu-1 (GenBank accession number NC_045512.2).
The consensus sequence was assembled into a single contig with a total length of 29,849 bp, corresponding to 99.8% of genome, and a deep average coverage of 3,983X. The sequence was deposited in the GISAID database [5] under accession ID EPI_ISL_412972.
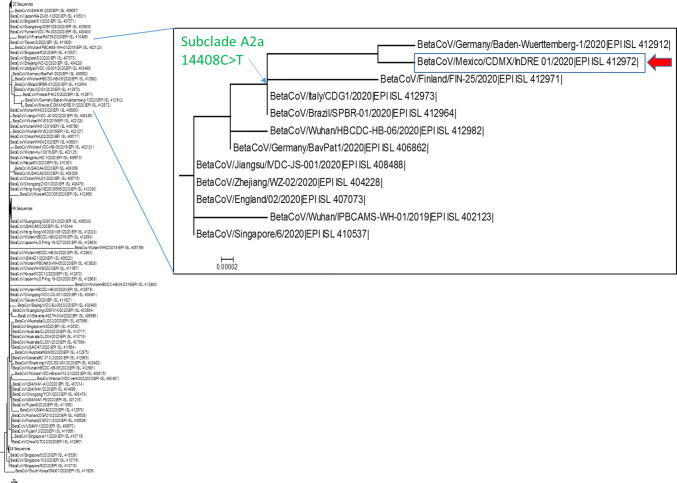
A database was constructed including 178 genomic sequences available on the GISAID platform, obtained from December 31, 2019 to March 05, 2020, reported in 22 countries. A phylogenetic tree was constructed in MEGA 6 software using the neighbor-joining method and the Kimura 2-parameter evolution model with 100 bootstrap replicates [10]. The strain hCoV/Mexico/CDMX/InDRE_01/2020 was grouped in the clade A2 subclade A2a (also called lineage G) and is closely related to isolates from Finland, Italy, and Germany as well as an isolate from the first imported SARS-CoV-2 case in Brazil [11] (Fig. 1).
Fig. 1.
Neighbor-joining tree constructed in MEGA using the Kimura 2-parameter evolution model with 100 bootstrap replicates. hCoV/Mexico/CDMX/InDRE_01/2020 (indicated by an arrow) is grouped in clade A2a with sequences previously reported from Brazil, Germany, Italy, and Finland. The 14408C>T substitution is characteristic of subclade A2a
Genetic analysis showed that the hCoV/Mexico/CDMX/InDRE_01/2020 genome differs by seven nucleotide substitutions compared to the Wuhan-Hu-1 reference strain: 241C>T, 3037C>T, 14408C>T, 23403A>G, 28881G>A, 28882G>A, and 28883G>C. We confirmed the presence of the 14408C>T substitution, which is characteristic of the sequences clustered in subclade A2a.
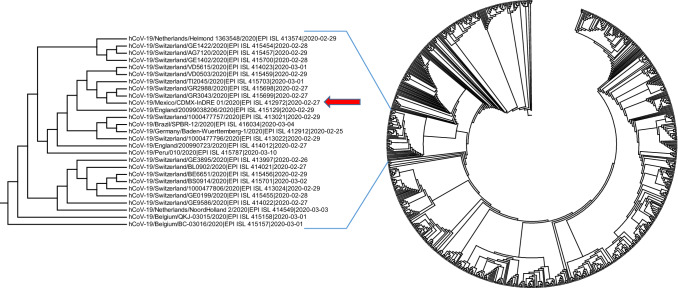
A second database of 768 sequences deposited until March 20, 2020, was used to construct a phylogenetic tree. This analysis showed that hCoV/Mexico/CDMX/InDRE_01/2020 clusters with European lineage G sequences, mainly from Belgium, Switzerland, and England (Fig. 2). This finding correlates with the transmission events reported by GISAID [5], which suggest that this strain was first introduced into Belgium, then England, and finally, Italy, where the Mexican patient had been traveling.
Fig. 2.
Neighbor-joining tree constructed with 768 sequences available in the GISAID database on March 20, 2020. The analysis shows that the Mexican strain (indicated by an arrow) clusters with European sequences, corroborating the transmission events reported by GISAID (Belgium-Germany-Italy-Mexico) for this first imported case in our country
This is the first report of the complete genome of this virus in our country, and these data will contribute to the molecular epidemiology of the SARS-CoV-2.
Acknowledgements
We thank all the staff of the Technological Development and Molecular Research Unit (Susana Serrano, Lidia García, Estela Corona, América Mandujano, Araceli Rodríguez, Isaura Martinez, Elizabeth Andrade, Juan Luis Tellez Mario Torres, Claudia Gómez, Guadalupe Montiel, Aída García), Virology Department, and Sample Control and Services Department at InDRE for technical assistance; we also thank Blanca Taboada for bioinformatical assistance, and Nuno Faria for sharing multiplex primers. No external funding was received for this work.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official opinion of the Ministry of Health of Mexico.
Funding
None.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gisela Barrera-Badillo, Email: gisela.barrera20@yahoo.com.mx.
José Ernesto Ramírez-González, Email: ernesto.ramirez@salud.gob.mx.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Secretaria de Salud (2020) Comunicado Técnico Diario Nuevo Coronavirus en el Mundo (COVID-19). 2020, March 20. https://www.gob.mx/cms/uploads/attachment/file/542599/Comunicado_Tecnico_Diario_COVID-19_2020.03.20.pdf. Accessed 20 Mar 2020
- 4.Coronavirus Resource Center Johns Hopkins University. 2020. https://coronavirus.jhu.edu/. Accessed 20 Mar 2020
- 5.Global Initiative on Sharing All Influenza Data. 2020. https://www.gisaid.org/epiflu-applications/next-sars-cov2-app/. Accessed 20 Mar 2020
- 6.Secretaría de Salud. Lineamiento Estandarizado para la Vigilancia Epidemiológica y por Laboratorio de Enfermedad por COVID-2019. https://www.gob.mx/cms/uploads/attachment/file/537944/Lineamiento_COVID-19_2020.02.27.pdf. Accessed 29 Feb 2020
- 7.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders D, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 doi: 10.2807/1560-7917.es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artic Network n-CoV 2019. https://artic.network/ncov-2019. Accessed 22 Jan 2020
- 9.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.First Report of Covid-19 in south America. http://virological.org/t/first-report-of-covid-19-in-south-america/409. Accessed 29 Feb 2020