Abstract
Background
ATP-dependent chromatin remodelers are evolutionarily conserved complexes that alter nucleosome positioning to influence many DNA-templated processes, such as replication, repair, and transcription. In particular, chromatin remodeling can dynamically regulate gene expression by altering accessibility of chromatin to transcription factors.
Scope of review
This review provides an overview of the importance of chromatin remodelers in the regulation of metabolic gene expression. Particular emphasis is placed on the INO80 and SWI/SNF (BAF/PBAF) chromatin remodelers in both yeast and mammals. This review details discoveries from the initial identification of chromatin remodelers in Saccharomyces cerevisiae to recent discoveries in the metabolic requirements of developing embryonic tissues in mammals.
Major conclusions
INO80 and SWI/SNF (BAF/PBAF) chromatin remodelers regulate the expression of energy metabolism pathways in S. cerevisiae and mammals in response to diverse nutrient environments. In particular, the INO80 complex organizes the temporal expression of gene expression in the metabolically synchronized S. cerevisiae system. INO80-mediated chromatin remodeling is also needed to constrain cell division during metabolically favorable conditions. Conversely, the BAF/PBAF remodeler regulates tissue-specific glycolytic metabolism and is disrupted in cancers that are dependent on glycolysis for proliferation. The role of chromatin remodeling in metabolic gene expression is downstream of the metabolic signaling pathways, such as the TOR pathway, a critical regulator of metabolic homeostasis. Furthermore, the INO80 and BAF/PBAF chromatin remodelers have both been shown to regulate heart development, the tissues of which have unique requirements for energy metabolism during development. Collectively, these results demonstrate that chromatin remodelers communicate metabolic status to chromatin and are a central component of homeostasis pathways that optimize cell fitness, organismal development, and prevent disease.
Keywords: Metabolism, Chromatin-remodeling, INO80, SWI/SNF, BAF/PBAF, TOR
Highlights
-
•
Chromatin modifications facilitate responsive, rapid, and reversible gene expression.
-
•
Chromatin remodelers regulate metabolic gene expression and cell division in response to changing nutrient environments.
-
•
Metabolic signaling pathways, such as the TOR pathway, cooperate with chromatin-remodeling.
-
•
Chromatin remodelers control gene expression programs in developing tissues with diverse metabolic requirements.
1. Chromatin modifications elicit adaptive environmental responses
The coordination of cellular function with the environment is essential for adaptation and survival. For example, cells have a remarkable ability to sense diverse (that is, nutrient-rich or -limiting) environments and reprogram their energy metabolism and proliferative capacity accordingly. Dynamic nutrient environments are ubiquitous throughout nature and include competitive growth environments of proliferating microorganisms and tissue niches in multicellular organisms. Failure to adapt can lead to cell death, developmental defects, and disease. Indeed, energy metabolism alterations are a major contributing factor for many pathologies, including cancer, cardiovascular disease, and diabetes.
Adaptive cellular responses are often achieved by inducible changes in gene expression programs [1]. An ideal mechanism to achieve dynamic gene expression is through modification of chromatin because they are rapid, responsive, and reversible. Chromatin is a complex structure that is dynamically reorganized to facilitate DNA-templated processes such as transcription, chromosome segregation, DNA replication, and DNA repair. Enzymes that restructure the chromatin environment are critical components of epigenetic maintenance and can contribute to disease when disrupted. Included among chromatin modifiers are enzymes that post-translationally modify histones and ATP-dependent chromatin remodelers that alter the position and histone composition of nucleosomes. Chromatin modifiers are evolutionarily conserved and regulate diverse processes required for normal cell function and organismal development [2,3].
Moreover, chromatin modifications are intimately tied to metabolic processes, as many intermediary metabolites are required co-factors for histone post-translational modification [4]. For example, not only is acetyl-CoA, which is produced in the TCA cycle, required for biosynthesis of fatty acids, sterols, and amino acids, but it is also a required cofactor for histone acetylation. Histone acetyltransferases use nuclear acetyl-CoA during high glucose conditions to acetylate histones, creating a permissive state for transcription [5]. Histone acetylation also promotes the expression of genes involved in the TCA cycle [6], thereby creating a feedback mechanism that supports the acetyl-CoA production pathway. Furthermore, histone acetylation regulates the expression of genes involved in cell growth and proliferation, namely ribosome biogenesis, translation, and amino acid metabolism, further demonstrating the link between high nutrient environments and the promotion of cell proliferation.
In particular, the chromatin of ribosomal DNA (rDNA) loci needs to be highly modulated in coordination metabolic pathways and stress responses, as ribosome biogenesis is one of the most energy demanding processes in eukaryotic organisms [7,8]. It has been proposed that approximately 60% of total Saccharomyces cerevisiae transcription is devoted to rRNA production and 50% of Pol II transcription is involved in ribosomal protein expression in nutrient-rich environments [9]. While acetyl-CoA production and histone acetylation of rDNA loci increases in high glucose environments, in glucose-limiting environments, NAD + begins to accumulate as the TCA cycle slows. High NAD + levels activate SIRT1 histone deacetylase (HDAC) to deacetylate histones at the rDNA loci [10], thereby slowing growth in coordination with limiting nutrients.
2. Chromatin-remodeling complexes regulate energy metabolism
Not only are histone modifications directly linked to energy metabolism, but chromatin remodelers are as well. Chromatin remodelers are part of superfamily 2 (SF2) helicases that contain DEAD-box ATPase subunits [11]. These complexes utilize the energy of ATP to alter the contacts between histones and DNA to reposition or edit nucleosome composition [12]. Current research demonstrates that chromatin remodelers have diverse roles in many DNA-templated processes, such as transcription, DNA repair, and replication [2,13]. The first characterized ATP-dependent remodeling complex was the S. cerevisiae SWI/SNF complex [14,15]. However, subunits of the SWI/SNF (switch/sucrose non-fermenting) complex were originally identified as transcriptional regulators of genes involved in growth in the presence of alternative fermentable carbon sources, such as sucrose [16,17].
The SWI/SNF complex is highly conserved and regulates energy metabolism in both yeast and mammals. Mammalian SWI/SNF complexes are a family of BRG-/BRM-associated factor (BAF) and polybromo-associated BAF (PBAF) complexes. The link between BAF/PBAF and mammalian disease has been repeatedly demonstrated, as loss of function contributes to developmental abnormalities and many subunits are mutated in cancer [3,18,19].
In mammalian skeletal muscle, the subunit Baf60c regulates glycolytic metabolism [20]. Skeletal muscle contains both slow-twitch and fast-twitch myofibers that generate ATP through diverse mechanisms. Slow-twitch myofibers are mitochondria-rich and utilize oxidative phosphorylation, while fast-twitch fibers utilize glycolysis for ATP production [21]. In mice, muscle-specific transgenic expression of BAF60C results in increased expression in fast-twitch fibers compared to slow-twitch fibers [20]. Transgenic BAF60C mice also displayed elevated levels of glycolytic capacity and reduced mitochondrial mass. Interestingly, transgenic mice were less susceptible to diet-induced insulin resistance and glucose intolerance, demonstrating the disease relevance of Baf60c's role in glucose homeostasis and diabetes.
Glycolytic metabolism is not only important for muscle metabolism, but has also been observed in cancer cells where aerobic glycolysis is utilized to feed growth pathways, such as lipid and protein biogenesis, to increase proliferative capacity and make new cells [22]. In other studies, cancer cells have been found to exhibit plasticity in energy metabolism, as they can switch their metabolism between fermentation and respiration depending on nutrient and oxygen availability [23,24].
There are intriguing similarities between the metabolism of cancer cells and that of S. cerevisiae in that both are optimized for rapid proliferation in diverse environments. S. cerevisiae have also evolved metabolic diversity in carbon catabolism pathways. Specifically, in glucose-rich environments, budding yeast preferentially utilize glycolysis followed by fermentation. When glucose is limiting, cells undergo a diauxic shift to respiration [25]. Growth in high glucose results in “glucose repression,” which is characterized by transcriptional repression of genes involved in alternate carbon source metabolism, including those in sucrose metabolism and respiration [26]. The state of “glucose repression” is not unlike that of the “Warburg effect,” where cancer cells utilize aerobic glycolysis to feed growth pathways, such as lipid and protein biogenesis, over energy production via respiration [22].
One type of cancer that is dependent on the Warburg effect is clear cell renal cell carcinoma (ccRCC), named after its cellular histological appearance caused by elevated glycogen and lipid storage caused increased glycolysis. Approximately 46% of ccRCCs have mutations in polybromo-1 (PBRM1) [27], another subunit of the PBAF complex. It is the second most mutated gene after the Von Hippel-Lindau (VHL) tumor suppressor gene, which is mutated in 48% of ccRCCs. A mouse model of ccRCC caused by inactivation of both PBRM1 and VHL recapitulated the histological features of patient-derived ccRCCs [28]. Importantly, increased glycogen storage and decreased expression of genes in the oxidative phosphorylation pathway are dependent on loss of PBRM1 and VHL. In addition, reintroduction of wild type PBRM1 into a ccRCC cell line decreased glucose uptake and proliferation [29], further demonstrating that PBRM1 plays a critical role in the regulation of cancer cell metabolism.
BAF/PBAF are not the only chromatin remodelers known to regulate energy metabolism. The INO80 complex, originally identified for its role in inositol metabolism [30,31], also regulates the glycolytic and respiration pathways in budding yeast. Mutant strains of the INO80 complex exhibit deregulated expression of nearly 15% of the transcriptome with significant enrichment in biological processes that pertain to cellular metabolism [32]. Similar to the SWI/SNF complex, the INO80 complex also regulates “glucose repression” in yeast cells. In nutrient-rich environments, mutants of the INO80 complex display increased expression of nearly every gene involved in the electron transport chain, while genes in glycolysis are decreased [32]. Accordingly, mitochondrial potential and oxygen consumption are increased in INO80 mutants. These alterations of metabolic activity in the mutants are observed in both glucose-containing media when oxidative phosphorylation is normally repressed and ethanol-containing media when oxidative phosphorylation is normally active. Thus, INO80 mutants have constitutively increased rates of respiration, regardless of carbon source availability.
INO80's role in cellular metabolism was again confirmed in a genetic screen that elucidated the functional composition of different subunits in the INO80 complex [33]. Known functions of INO80 subunits were captured in the genetic screen, such as chromatin modification, transcriptional regulation, and chromatin assembly. However, genetic interactions of specific INO80 subunits were significantly enriched in metabolic processes, such as amino acid biosynthesis and mitochondrial inheritance. Indeed, defects in mitochondrial inheritance were found in INO80 mutant cells compared to wild type [33]. Thus, INO80 regulates glucose repression [32], which is largely dependent on glycolysis, and it also influences mitochondrial inheritance, which is necessary for respiration [33]. Taken together, INO80 appears to be critical for the maintenance of diverse energy metabolism pathways to improve survival and proliferation in fluctuating nutrient environments.
3. Chromatin remodeling links metabolic homeostasis with periodic gene expression
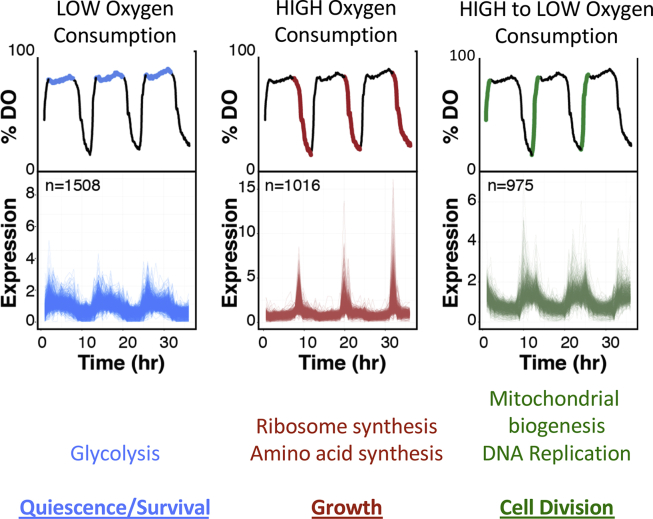
The connection between metabolic status and gene expression can be systematically examined using a metabolically synchronized continuous growth system in S. cerevisiae [34]. In this system, cells are grown in a chemostat, or bioreactor, and are metabolically synchronized through nutrient deprivation, followed by the constant perfusion of glucose-containing media. Following reintroduction of glucose to the culture, cells collectively undergo continuous respiration oscillations that characterize the yeast metabolic cycle (YMC) [35,36]. The YMC is defined by temporal separation of glycolysis during low oxygen consumption, oxidative phosphorylation during high oxygen consumption, followed by DNA replication and cell division during the subsequent transition from low oxygen to high oxygen consumption [36] (Figure 1). Remarkably, these diverse metabolic states are largely driven by periodic fluctuations in metabolic gene expression that are exquisitely in tune with environmental conditions (Figure 1). Indeed, over 50% of all genes are periodically expressed at different phases in the YMC.
Figure 1.
Respiration and gene oscillations in the yeast metabolic cycle (YMC). Metabolically, synchronization is facilitated by growing cells to saturation followed by continuous batch feeding of the culture with low glucose media in a closed bioreactor environment. The YMC is divided into three phases depending on respiration as monitored by dissolved oxygen (DO) in the culture. mRNA expression plots are shown as determined from reference [36]. n = number of genes previously determined to undergo periodic gene expression [36].
An analogous system that coordinates periodic gene expression with metabolic status and cell division is the circadian cycle that is driven by day/night cycles [[37], [38], [39], [40]]. Indeed, periodic gene expression is evolutionarily conserved and has been observed in individual yeast cells, plants, flies, and mammalian tissues [41]. This temporal organization has distinct energy conservation advantages, in that gene expression can be activated just prior to the time at which the encoded protein is needed in a specific metabolic or cell cycle pathway [[42], [43], [44], [45]]. An example of this is seen during the regulated expression of genes involved in protein synthesis (for example, ribosome biogenesis), which are energetically demanding as they are highly expressed and involve several rDNA genomic loci [9,46]. During the YMC, the expression of ribosome biogenesis genes occurs at the same time when ATP levels are highest, to provide energy for synthesis, and just prior to cell division, to constrain biomass production until commitment to new cell production [36,47].
In the YMC, genes encoding proteins with similar metabolic function are activated and repressed at approximately the same time, suggesting that they are under tight control by shared transcriptional mechanisms. One demonstrated transcriptional mechanism that regulates YMC periodic gene expression is chromatin remodeling. Nucleosome repositioning by chromatin remodelers is a critical feature of dynamic gene expression in the YMC [48]. Specifically, chromatin remodelers, such as SWI/SNF, reposition nucleosomes to regulate the accessibility of promoters to transcription factors. The extent of nucleosome repositioning closely reflects the amplitude of dynamic gene expression. Moreover, nucleosomes at genes involved in growth pathways, such as those at the rDNA locus, undergo acute nucleosome repositioning just before DNA replication and cell division [48].
The INO80 complex also plays critical roles regulating the YMC. Cells lacking subunits of the INO80 complex exhibit disrupted YMC respiration cycles and gene expression oscillations [49]. More specifically, while the periodic timing of many gene oscillations remains intact, the amplitude is substantially dampened. The YMC gene expression defects observed in INO80 mutant cells are linked to alterations in chromatin accessibility [49]. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) was used to determine chromatin accessibility throughout the YMC in both wild type and INO80 mutants. ATAC-seq uses a hyperactive Tn5 transposase that preferentially inserts adaptors in regions with accessible chromatin [50]. Global defects in the chromatin structure were observed in INO80 mutants, in that wild type cells exhibited periodic oscillations in accessibility that could be observed along entire chromosomes [49]. However, accessibility in INO80 mutants is much more static, with large chromosomal domains exhibiting persistent increased or decreased accessibility throughout the YMC. Similar results were observed when investigating transcription factor motif accessibility. Wild type cells displayed oscillations in accessibility that corresponded to gene activation, while in INO80 mutants, these regions did not oscillate and periodic expression was dampened. Taken together, these results demonstrate the importance of chromatin remodeling for the regulation of pathways that control adaptive gene expression and metabolic homeostasis.
4. Chromatin remodelers function in metabolic signaling pathways
Interestingly, the accessibility of several transcription factor motifs regulated by the INO80 complex are downstream effectors of the metabolic signaling TORC1 pathway. The target of the rapamycin (TOR) signaling pathway responds to environmental cues and coordinates stress and growth responses in both yeast and mammals. The mammalian TOR (mTOR) kinase is deregulated in numerous metabolic disorders [51]. In S. cerevisiae, TOR complex 1 (TORC1) and TOR complex 2 (TORC2) connect nutrient availability to cell growth and division [51]. TOR signaling also regulates adaptive transcription in response to nutrient availability. For example, during nutrient deprivation, TOR alters carbon metabolism gene expression [52] and influences chromatin structure and the expression of ribosomal genes [53].
INO80 regulates the accessibility of Msn2 and 4 transcription factor motifs [49] that are phosphorylated by TORC1 to regulate their nuclear localization [52]. Msn2 and 4 activate the expression of genes involved in carbon metabolism and stress responses [[54], [55], [56], [57], [58]]. In addition, INO80 controls the accessibility of Dot6 and Tod6 binding, which are phosphorylated in a TORC1-dependent manner to increase the expression of ribosome biogenesis genes [59]. Interestingly, the global transcriptional profiles of cells lacking INO80 subunits are similar to those of cells treated with rapamycin, a potent inhibitor of TORC1, and include dramatic alterations of genes in Msn2/4-response genes, ribosome biogenesis, and nitrogen metabolism [33,49].
The genetic screens that confirmed the roles of INO80 in cell metabolism also identified novel connections between INO80 and TOR signaling [33]. Specifically, subunits of both TORC1 and TORC2 and the Sch9 downstream signaling kinase have positively correlated genetic interaction profiles with INO80 subunits. Moreover, INO80 subunits are located within highly connected genetic nodes with TORC1 signaling components, supporting a central role for INO80 in TORC1 signaling to chromatin [33]. Treatment with rapamycin markedly reduced the strength of these genetic interactions, confirming that they are specific to nutrient-rich conditions and are significantly reduced when TORC1 signaling is inhibited.
As mentioned, INO80 chromatin remodeling is needed to regulate chromatin accessibility of transcription factors in the TOR signaling pathway. Another way in which INO80 can facilitate TOR-dependent gene expression is by regulating histone acetylation status, thus transcriptional potential. As previously described, histone acetylation requires metabolic intermediate acetyl-CoA and high levels of histone acetylation are present on genes that regulate the energy metabolome [60,61]. Thus, a feedback loop likely exists, whereby the expression of metabolome regulators promotes acetyl-CoA production, which then subsequently increases histone acetylation and gene expression of metabolome regulators. INO80 genetically interacts with both acetyltransferases and deacetylases [33]. In particular, both Rpd3L deacetylase and acetylated H3K56, the product of Rtt109 acetyltransferase activity, are in the TORC1 signaling pathway [59,62,63] and genetically interact with the INO80 complex [33]. In addition, the genome occupancy of INO80 and acetylated H3K56 correlate, as do many histone acetyl marks, and loss of INO80 reduces histone acetylation at metabolic loci. Thus, INO80 may also function to promote histone acetylation on growth genes downstream of TORC1 signaling [33]. A summary of INO80's function in the regulation of metabolic gene expression is shown in Figure 2.
Figure 2.
INO80 complex is a component of the TOR signaling pathway that regulates metabolic gene expression. The model illustrates the INO80 complex at promoters targeted by Msn2/4, Tod6, and Dot6 that are phosphorylated in the TORC1 signaling pathway and regulate ribosome biogenesis, nitrogen metabolism, and stress responses. The INO80 complex is a model of the electron microscopy-derived structure bound to a nucleosome (PDB and 6FML) [84]. DNA is shown in green and the INO80-bound nucleosome in red. The INO80 complex regulates promoter accessibility and histone acetylation at target genes. TORC1 signaling regulates Msn2/4 subcellular localization and Tod6/Dot6 chromatin recruitment.
Metabolic signaling is also needed for tissue-specific regulation of BAF/PBAF function. Specifically, the previously described role of Baf60c in muscle glycolytic metabolism induces Akt signaling [20], a key metabolic signaling kinase that enhances glycolytic metabolism in muscle [64,65]. In addition, in ccRCC, where PBRM1 is commonly mutated, mTOR activation was found as a critical contributor to malignant transformation [28]. Collectively, these results demonstrate how chromatin remodelers, such as INO80 and BAF/PBAF, cooperate with signaling pathways to reprogram metabolic gene expression for proper tissue function and disease prevention. Chromatin remodelers have been previously found to be integral components of signaling pathways in DNA damage responses [66], thus these complexes are ideally positioned to enact rapid changes in the chromatin environment in response to different cellular stimuli and environments.
5. INO80 chromatin remodeling connects cell division with metabolic homeostasis
The coordination of energy metabolism with cell division is essential to regulate proliferation in competitive nutrient environments. The interdependence of these processes has long been recognized and several yeast mutants originally identified as having defects in cell cycle progression were later also found to have metabolic deficiencies [[67], [68], [69]]. Indeed, many cell division cycle (CDC) genes are involved in energy metabolism and the biosynthetic pathways [6]. For example, Cdc53 is a both a component of the SCF complex involved in the G1-S transition and also a regulator of methionine biosynthesis genes that produce S-adenosylmethionine (SAM), a required cofactor for histone methylation [70].
Furthermore, diverse biosynthetic pathways are needed to successfully accomplish cell division. For example, the TCA cycle provides essential intermediary metabolites for nitrogen utilization in building amino acids and nitrogenous bases in nucleic acids. In addition, ATP is needed for the energy-demanding process of ribosome biogenesis [9,46] as previously mentioned. In the YMC, glycolysis, TCA cycle activation and oxidative phosphorylation are all temporally compartmentalized. The timing of DNA replication and cell division in the YMC is restricted to a specific temporal window following biomass production [36]. DNA replication that occurs outside of this window results in genome instabilities and reduced fitness [71].
INO80 mutants not only exhibit altered metabolic gene expression, but also defects in the timing of cell division in the YMC [49]. In mutant cells, genes involved in the cell division pathways exhibited peak expression in a different phase of the YMC compared to wild type cells [49]. As a result, cell division occurred throughout the YMC and was no longer coupled to metabolic status, resulting in dramatically impaired cell fitness. The result is not unlike that of cancer cells that prioritize proliferation over metabolic and genome stability.
6. Insights into chromatin remodeling, metabolism, and development
The INO80 complex coordinates the timing of cell division not only in the YMC, but also during mammalian development. While INO80-deficient mice are embryonically lethal [72], tissue-specific knockouts can be utilized to investigate its role in development. INO80 was individually deleted from three different cell types in the heart [73]. Similar to many organs, the development of the heart is extremely complex as it transforms from a single tube into a mature structure consisting of endothelial, epicardial, and myocardial tissues.
In tissue-specific INO80-deficient mice, the most apparent phenotype occurred in embryos with endothelial-specific deletions. These mutant mice displayed a dramatic cardiac phenotype resembling ventricular non-compaction (LVNC), the third most common human cardiomyopathy, the etiology of which is not entirely known [74]. LVNC patients have increased risk of heart failure due to compromised heart function. LVNC is believed to arise from incomplete developmental maturation of the heart muscle, which normally transitions from a loose meshwork of fibers early in embryogenesis to dense and compact layers of cells that are capable of pumping blood late in development.
Paradoxically, in mice with endothelial-specific deletion of INO80, the most pronounced defects were observed in the neighboring myocardial tissue [73]. Some endocardial-derived paracrine factors have been identified that support myocardial development [75,76], although these were not found to be obviously defective in INO80-deficient hearts [73]. Instead, in INO80-deleted endothelial cells, coronary vascularization was defective with incomplete sprouting and migration of vessels. Mechanistically, INO80 deletion increased the expression of E2F-activated genes and S-phase occupancy of endothelial cells, demonstrating that cell cycle regulation is critical to the execution of developmental transitions in the heart.
Further research is needed to determine if metabolic dysfunction accompanies the cell cycle alterations observed in INO80-deficient endothelial cells. Indeed, energy metabolism can regulate the development of tissues within complex organs and contribute to developmental abnormalities when disrupted. This is particularly true of tissues in the developing heart. For example, early in development, myocardial muscle cells rely on glycolysis to fuel the production of new cell building blocks during proliferation. As the myocardium tissue develops, oxidative phosphorylation and fatty acid oxidation are utilized to fuel energy requirements for active muscle function [77]. In addition, the endothelial cells that induce vessel sprouting during cardiac vasculature development rely heavily on glycolysis despite their proximity to oxygenated blood [78]. These endothelial cells exhibit profound metabolic plasticity in that they can remain quiescent for prolonged periods followed by quick transition to glycolysis to promote proliferation and migration. Notably, glucose consumption of cardiac endothelial cells in vitro is comparable to that of the “Warburg effect” in cancer cells that exhibit glucose addiction to feed glycolytic pathways [79].
Similar to the ability of chromatin remodelers to enact rapid gene expression programs in changing nutrient environments, remodelers can also facilitate developmental transitions coordinated with tissue-specific metabolic demands. Mutations in chromatin modifiers were found in human patients with congenital heart defects (CHD) [80]. Moreover, defects in BAF/PBAF chromatin remodeling have been shown to cause defective mouse cardiac development [3,81]. For example, the previously introduced Baf60c regulates glycolytic metabolism in muscle [20] as well as embryonic cardiac development. Specifically, knockdown of Baf60c during development impairs heart expansion and cardiac muscle differentiation [82].
Although metabolic specification is critical for developmental transitions and organ function, very little is known about epigenetic programming of metabolism in different tissues. However, it is known that the consequences of deregulated metabolic signaling often result in disease. Indeed, energy metabolism alterations are a major contributing factor for many pathologies, including cancer, cardiovascular disease, and diabetes, which together account for half of all deaths in industrialized nations [83]. Future research will help illuminate the intersection of epigenetic-regulated metabolic homeostasis and development and disease prevention.
Acknowledgments
Funding was provided by NIH grant GM119580 to AJM. Figure 1 illustration courtesy of Devin A. King (Stanford University). Figure 2 was created using BioRender.com.
Conflict of interest
None declared.
References
- 1.López-Maury L., Marguerat S., Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nature Reviews Genetics. 2008;9(8):583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 2.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annual Review of Biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 3.Alfert A., Moreno N., Kerl K. The BAF complex in development and disease. Epigenetics & Chromatin. 2019;12(1):1–15. doi: 10.1186/s13072-019-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gut P., Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502(7472):489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 5.Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Current Opinion in Cell Biology. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai L., Tu B.P. Driving the cell cycle through metabolism. Annual Review of Cell and Developmental Biology. 2012;28:59–87. doi: 10.1146/annurev-cellbio-092910-154010. [DOI] [PubMed] [Google Scholar]
- 7.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes & Development. 2003;17(14):1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 8.Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cellular and Molecular Life Sciences. 2007;64(1):29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 10.Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133(4):627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Hopfner K.-P., Gerhold C.-B., Lakomek K., Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. - PubMed - NCBI. Current Opinion in Structural Biology. 2012;22(2):225–233. doi: 10.1016/j.sbi.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C.Y., Johnson S.L., Gamarra N.I., Narlikar G.J. Mechanisms of ATP-dependent chromatin remodeling motors. Annual Review of Biophysics. 2016;45:153–181. doi: 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison A.J., Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nature Reviews Molecular Cell Biology. 2009;10(6):373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns B.R., Kim Y.J., Sayre M.H., Laurent B.C., Kornberg R.D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proceedings of the National Academy of Sciences. 1994;91(5):1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson C.L., Dingwall A., Scott M.P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proceedings of the National Academy of Sciences. 1994;91(8):2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stern M., Jensen R., Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. - PubMed - NCBI. Journal of Molecular Biology. 1984;178(4):853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 17.Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodges C., Kirkland J.G., Crabtree G.R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harbor Perspectives in Medicine. 2016;6(8):a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadoch C., Crabtree G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Science Advances. 2015;1(5):e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Z.-X., Li S., Wang L., Ko H.J., Lee Y., Jung D.Y. Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through Deptor-mediated Akt activation. Nature Medicine. 2013;19(5):640–645. doi: 10.1038/nm.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiological Reviews. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 22.Diaz-Ruiz R., Rigoulet M., Devin A. The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochimica Et Biophysica Acta (BBA) - Bioenergetics. 2011;1807(6):568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Enríquez S., Gallardo-Pérez J.C., Avilés-Salas A., Marín-Hernández A., Carreño-Fuentes L., Maldonado-Lagunas V. Energy metabolism transition in multi-cellular human tumor spheroids. Journal of Cellular Physiology. 2008;216(1):189–197. doi: 10.1002/jcp.21392. [DOI] [PubMed] [Google Scholar]
- 24.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., Capaldi R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Research. 2004;64(3):985–993. doi: 10.1158/0008-5472.CAN-03-1101. [DOI] [PubMed] [Google Scholar]
- 25.Fiechter A., Fuhrmann G.F., Käppeli O. Regulation of glucose metabolism in growing yeast cells. Advances in Microbial Physiology. 1981;22:123–183. doi: 10.1016/s0065-2911(08)60327-6. [DOI] [PubMed] [Google Scholar]
- 26.Gancedo J.M. Yeast carbon catabolite repression. Microbiology and Molecular Biology Reviews : Microbiology and Molecular Biology Reviews. 1998;62(2):334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Drill E., Shen R. Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nargund A.M., Pham C.G., Dong Y., Wang P.I., Osmangeyoglu H.U., Xie Y. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Reports. 2017;18(12):2893–2906. doi: 10.1016/j.celrep.2017.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhury B., Porter E.G., Stewart J.C., Ferreira C.R., Schipma M.J., Dykhuizen E.C. PBRM1 regulates the expression of genes involved in metabolism and cell adhesion in renal clear cell carcinoma. PloS One. 2016;11(4):e0153718. doi: 10.1371/journal.pone.0153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebbert R., Birkmann A., Schüller H.J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Molecular Microbiology. 1999;32(4):741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 31.Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 32.Yao W., King D.A., Beckwith S.L., Gowans G.J., Yen K., Zhou C. The INO80 complex requires the arp5-ies6 subcomplex for chromatin remodeling and metabolic regulation. Molecular and Cellular Biology. 2016;36(6):979–991. doi: 10.1128/MCB.00801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckwith S.L., Schwartz E.K., Garcia-Nieto P.E., King D.A., Gowans G.J., Wong K.M. The INO80 chromatin remodeler sustains metabolic stability by promoting TOR signaling and regulating histone acetylation. PLoS Genetics. 2018;14(2) doi: 10.1371/journal.pgen.1007216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick A., Szilard L. Description of the chemostat. Science. 1950;112(2920):715–716. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- 35.Klevecz R.R., Bolen J., Forrest G., Murray D.B. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(5):1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu B.P., Kudlicki A., Rowicka M., McKnight S.L. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310(5751):1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 37.Tu B.P., McKnight S.L. Metabolic cycles as an underlying basis of biological oscillations. 2006;7(9):696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 38.Panda S. Circadian physiology of metabolism. Science. 2016;354(6315):1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 40.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Doherty C.J., Kay S.A. Circadian control of global gene expression patterns. Annual Review of Genetics. 2010;44(1):419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang Z., Cai L., Zhang X., Ji H., Tu B.P., Boeke J.D. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nature Structural & Molecular Biology. 2014;21(10):854–863. doi: 10.1038/nsmb.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAdams H.H., Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301(5641):1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 44.Wang G.-Z., Hickey S.L., Shi L., Huang H.-C., Nakashe P., Koike N. Cycling transcriptional networks optimize energy utilization on a genome scale. Cell Reports. 2015;13(9):1868–1880. doi: 10.1016/j.celrep.2015.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaslaver A., Mayo A.E., Rosenberg R., Bashkin P., Sberro H., Tsalyuk M. Just-in-time transcription program in metabolic pathways. Nature Genetics. 2004;36(5):486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 46.Wagner A. Energy constraints on the evolution of gene expression. Molecular Biology and Evolution. 2005;22(6):1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- 47.Machné R., Murray D.B. The yin and yang of yeast transcription: elements of a global feedback system between metabolism and chromatin. PloS One. 2012;7(6):e37906. doi: 10.1371/journal.pone.0037906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nocetti N., Whitehouse I. Nucleosome repositioning underlies dynamic gene expression. Genes & Development. 2016;30(6):660–672. doi: 10.1101/gad.274910.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gowans G.J., Schep A.N., Wong K.M., King D.A., Greenleaf W.J., Morrison A.J. INO80 chromatin remodeling coordinates metabolic homeostasis with cell division. Cell Reports. 2018;22(3):611–623. doi: 10.1016/j.celrep.2017.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer. Diabetes and Ageing. 2010;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck T., Hall M.N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402(6762):689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 53.Tsang C.K., Bertram P.G., Ai W., Drenan R., Zheng X.F.S. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. The EMBO Journal. 2003;22(22):6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elfving N., Chereji R.V., Bharatula V., Björklund S., Morozov A.V., Broach J.R. A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Research. 2014;42(9):5468–5482. doi: 10.1093/nar/gku176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuang Z., Pinglay S., Ji H., Boeke J.D. Msn2/4 regulate expression of glycolytic enzymes and control transition from quiescence to growth. eLife. 2017;6 doi: 10.7554/eLife.29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G. Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Pastor M.T., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) The EMBO Journal. 1996;15(9):2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt A.P., McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(12):5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. The EMBO Journal. 2011;30(15):3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mülleder M., Calvani E., Alam M.T., Wang R.K., Eckerstorfer F., Zelezniak A. Functional metabolomics describes the yeast biosynthetic regulome. Cell. 2016;167(2):553–565. doi: 10.1016/j.cell.2016.09.007. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai L., Sutter B.M., Li B., Tu B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular Cell. 2011;42(4):426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humphrey E.L., Shamji A.F., Bernstein B.E., Schreiber S.L. Rpd3p relocation mediates a transcriptional response to rapamycin in yeast. Chemistry & Biology. 2004;11(3):295–299. doi: 10.1016/j.chembiol.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Chen H., Fan M., Pfeffer L.M., Laribee R.N. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Research. 2012;40(14):6534–6546. doi: 10.1093/nar/gks345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izumiya Y., Hopkins T., Morris C., Sato K., Zeng L., Viereck J. Fast/glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metabolism. 2008;7(2):159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morrison A.J., Kim J.-A., Person M.D., Highland J., Xiao J., Wehr T.S. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130(3):499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Johnston G., Pringle J., Hartwell L. Coordination of growth with cell division in the yeast. Experimental Cell Research. 1977;105(1):79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen P., Tyers M. How cells coordinate growth and division. Current Biology. 2004;14(23):R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 69.Jorgensen P., Nishikawa J.L., Breitkreutz B.-J., Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297(5580):395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 70.Patton E.E., Willems A.R., Sa D., Kuras L., Thomas D., Craig K.L. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes & Development. 1998;12(5):692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Z., Odstrcil E.A., Tu B.P., McKnight S.L. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316(5833):1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 72.Qiu Z., Elsayed Z., Peterkin V., Alkatib S., Bennett D., Landry J.W. Ino80 is essential for proximal-distal axis asymmetry in part by regulating Bmp4 expression. BMC Biology. 2016;14(1):368. doi: 10.1186/s12915-016-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rhee S., Chung J.I., King D.A., D'Amato G., Paik D.T., Duan A. Endothelial deletion of Ino80 disrupts coronary angiogenesis and causes congenital heart disease. Nature Communications. 2018;9(1):368. doi: 10.1038/s41467-017-02796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Towbin J.A., Lorts A., Jefferies J.L. Left ventricular non-compaction cardiomyopathy. Lancet. 2015 doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 75.Zhang W., Chen H., Qu X., Chang C.-P., Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) American Journal of Medical Genetics Part C, Seminars in Medical Genetics. 2013;163C(3):144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grego-Bessa J., Luna-Zurita L., del Monte G., Bolós V., Melgar P., Arandilla A. Notch signaling is essential for ventricular chamber development. Developmental Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. Journal of Cardiovascular Pharmacology. 2010;56(2):130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 78.De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B.W., Cantelmo A.R. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 79.Eelen G., de Zeeuw P., Simons M., Carmeliet P. Endothelial cell metabolism in normal and diseased vasculature. Circulation Research. 2015;116(7):1231–1244. doi: 10.1161/CIRCRESAHA.116.302855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bevilacqua A., Willis M.S., Bultman S.J. SWI/SNF chromatin-remodeling complexes in cardiovascular development and disease. Cardiovascular Pathology : The Official Journal of the Society for Cardiovascular Pathology. 2014;23(2):85–91. doi: 10.1016/j.carpath.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lickert H., Takeuchi J.K., Bothvon I., Walls J.R., McAuliffe F. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 83.Centers for Disease Control and Prevention . 2015. Leading causes of death and numbers of deaths, by sex, race, and Hispanic origin: United States, 1980 and 2014. [Google Scholar]
- 84.Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556(7701):386–390. doi: 10.1038/s41586-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]