Abstract
Background
Many metabolites serve as important signalling molecules to adjust cellular activities and functions based on nutrient availability. Links between acetyl-CoA metabolism, histone lysine acetylation, and gene expression have been documented and studied over the past decade. In recent years, several additional acyl modifications to histone lysine residues have been identified, which depend on acyl-coenzyme A thioesters (acyl-CoAs) as acyl donors. Acyl-CoAs are intermediates of multiple distinct metabolic pathways, and substantial evidence has emerged that histone acylation is metabolically sensitive. Nevertheless, the metabolic sources of acyl-CoAs used for chromatin modification in most cases remain poorly understood. Elucidating how these diverse chemical modifications are coupled to and regulated by cellular metabolism is important in deciphering their functional significance.
Scope of review
In this article, we review the metabolic pathways that produce acyl-CoAs, as well as emerging evidence for functional roles of diverse acyl-CoAs in chromatin regulation. Because acetyl-CoA has been extensively reviewed elsewhere, we will focus on four other acyl-CoA metabolites integral to major metabolic pathways that are also known to modify histones: succinyl-CoA, propionyl-CoA, crotonoyl-CoA, and butyryl-CoA. We also briefly mention several other acyl-CoA species, which present opportunities for further research; malonyl-CoA, glutaryl-CoA, 3-hydroxybutyryl-CoA, 2-hydroxyisobutyryl-CoA, and lactyl-CoA. Each acyl-CoA species has distinct roles in metabolism, indicating the potential to report shifts in the metabolic status of the cell. For each metabolite, we consider the metabolic pathways in which it participates and the nutrient sources from which it is derived, the compartmentalisation of its metabolism, and the factors reported to influence its abundance and potential nuclear availability. We also highlight reported biological functions of these metabolically-linked acylation marks. Finally, we aim to illuminate key questions in acyl-CoA metabolism as they relate to the control of chromatin modification.
Major conclusions
A majority of acyl-CoA species are annotated to mitochondrial metabolic processes. Since acyl-CoAs are not known to be directly transported across mitochondrial membranes, they must be synthesized outside of mitochondria and potentially within the nucleus to participate in chromatin regulation. Thus, subcellular metabolic compartmentalisation likely plays a key role in the regulation of histone acylation. Metabolite tracing in combination with targeting of relevant enzymes and transporters will help to map the metabolic pathways that connect acyl-CoA metabolism to chromatin modification. The specific function of each acyl-CoA may be determined in part by biochemical properties that affect its propensity for enzymatic versus non-enzymatic protein modification, as well as the various enzymes that can add, remove and bind each modification. Further, competitive and inhibitory effects of different acyl-CoA species on these enzymes make determining the relative abundance of acyl-CoA species in specific contexts important to understand the regulation of chromatin acylation. An improved and more nuanced understanding of metabolic regulation of chromatin and its roles in physiological and disease-related processes will emerge as these questions are answered.
Keywords: Compartmentalisation, Acyl-CoA, Histone, Acylation, Metabolism
Highlights
-
•
Acyl-CoAs are intermediates in diverse metabolic pathways.
-
•
Acyl-CoAs participate in chromatin regulation by serving as substrates for histone lysine acylation.
-
•
Distinct biochemical properties of acyl-CoAs and their corresponding lysine acylation marks may determine function.
-
•
The spatial regulation of acyl-CoA metabolism is critical for linking metabolism and chromatin regulation.
-
•
Acyl-CoA metabolic pathways are largely annotated to mitochondria and nuclear production mechanisms are poorly understood.
1. Introduction
Adaptation to the nutritional environment, on both the cellular and organismal level, is essential for health and metabolic homeostasis. Accordingly, metabolites can exert powerful signalling functions to adjust energy usage and cellular behaviour in response to nutrient availability [1,2]. How metabolic signals influence physiology has important implications in diverse contexts in which metabolism is regulated, including development, cancer, metabolic diseases, immune function, stem cell biology, and ageing [3,4].
Acyl-CoAs are a family of metabolic intermediates comprised of an acyl group linked by a thioester bond to coenzyme A. These molecules are critical for acyl transfer in a wide range of metabolic processes, with the energy release upon hydrolysis of the thioester bond facilitating acyl transfer in thermodynamically unfavourable reactions [5]. Acyl-CoA metabolites are conserved throughout biological kingdoms and across evolution [6], and thioesters are proposed as a biochemical basis for primordial metabolism [7,8]. Although the acyl-groups in acyl-CoAs are commonly derived from short-chain mono- or di-carboxylic acids, they can also include longer chain hydrophilic acyl-chains and xenobiotic compounds [9]. Since acyl groups are transferred not only within metabolic pathways but are also used to modify amino acid side chains, such as cysteine and lysine, acyl-CoAs have the potential to exert signalling functions through post-translational protein modifications.
A direct relationship between the availability of the central metabolite acetyl-CoA and histone lysine acetylation, originally reported in yeast and mammalian cells over a decade ago [10,11] is now well-established. Acetyl-CoA metabolism and its roles in chromatin regulation have been extensively reviewed elsewhere [3,[12], [13], [14]]. The relatively recent discovery of at least 9 alternative histone lysine acylations [15,16] and the identification of 5 additional lysine modifications that may affect histones [17,18] offers the possibility of numerous previously unrecognised nodes of communication between cellular metabolism and chromatin modification. Acyl-CoAs are generated in diverse metabolic processes, including lipid metabolism, ketone body metabolism, amino acid catabolism, and metabolism of short chain fatty acids derived from intestinal microbiota, potentially fine-tuning metabolic control of chromatin in a manner responsive to these processes. Yet, the metabolic sources of acyl-CoAs as they relate to chromatin modification are often unknown, as are the levels and mechanisms of production of these acyl-CoAs within the nucleus and their relative abundance.
Cellular concentrations of different acyl-CoA metabolites span orders of magnitude and are positively correlated with the relative abundances of their respective histone acylations observed in cells, supporting the notion that histone acylations are closely linked to the metabolic state of the cell [19]. However, the relationship between acyl-CoA metabolism and histone acylation must be influenced by compartmentalisation, because many acyl-CoAs are generated within mitochondria and cannot directly cross mitochondrial membranes but are nonetheless required to be in the nucleus to participate in chromatin modification. In the case of acetyl-CoA, the mechanism allowing transport out of mitochondria is well established, occurring through citrate export via the mitochondrial citrate transporter and subsequent cleavage by the enzyme ATP-citrate lyase (ACLY) [[20], [21], [22], [23]]. For most histone acylations, however, the mechanisms of acyl-CoA generation in the nuclear-cytoplasmic compartment remain very poorly understood, a point we will elaborate within this article. Acyl-CoA metabolites may transition from the cytosol to the nucleus through nuclear pores, which are permeable to small molecules [24]. This is experimentally supported by the demonstration that the addition of acyl-CoAs to isolated nuclei impacts corresponding chromatin acylation [19,25]. Moreover, both metabolites and metabolic enzymes may gain access to chromatin when the nuclear envelope breaks down during mitosis. The extent to which nuclear availability of acyl-CoAs is derived from diffusion from the cytosol versus local production remains unclear. In the case of acetyl-CoA, growing evidence highlights the likely importance of onsite generation in the nucleus for processes, such as gene regulation and DNA repair [13].
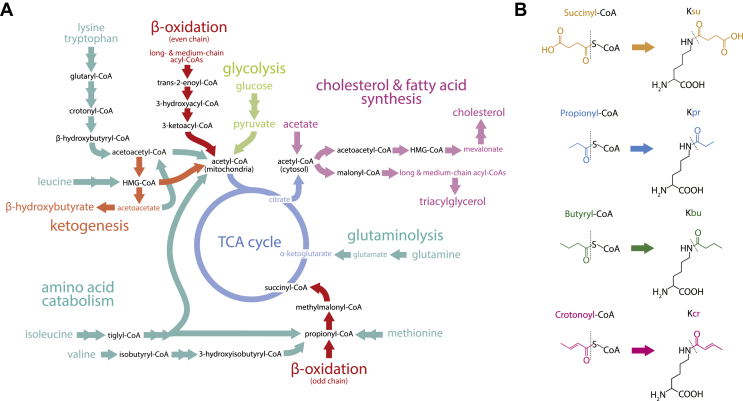
In this article, we review the metabolic pathways through which acyl-CoAs are produced (Figure 1A) and emerging evidence of the functional roles of diverse acyl-CoAs in chromatin regulation. We focus on 4 acyl-CoA metabolites integral to major metabolic pathways that are known to modify histones and have been linked to biological functions: succinyl-CoA, propionyl-CoA, crotonoyl-CoA and butyryl-CoA (Figure 1B). We also briefly mention several other acyl-CoA species, which present promising new opportunities for further research: malonyl-CoA, glutaryl-CoA, 3-hydroxybutyryl-CoA (also called β-hydroxybutyryl-CoA), 2-hydroxyisobutyryl-CoA and lactyl-CoA. Each acyl-CoA species has a distinct role in metabolism (connoting the potential to report shifts in metabolic environment), as well as distinct chemical properties that impact its biological regulation and function (facilitating response to metabolic environment). For each metabolite we will consider the metabolic pathways in which it participates and the nutrient sources from which it is derived, the compartmentalisation of its metabolism, the factors influencing its abundance and potential nuclear availability, its roles in the regulation of acylation and the biological functions of these metabolically-linked acylation marks. We aim to illuminate key questions in acyl-CoA metabolism as they relate to the control of chromatin modification.
Figure 1.
CoA World. A) Overview of metabolic pathways containing abundant acyl-CoA species. B) Chemical structures of four acyl-CoA molecules highlighted in this review and the corresponding lysine acylation marks.
2. Succinyl-CoA
2.1. Succinyl-CoA generation: metabolic pathways and compartmentalisation
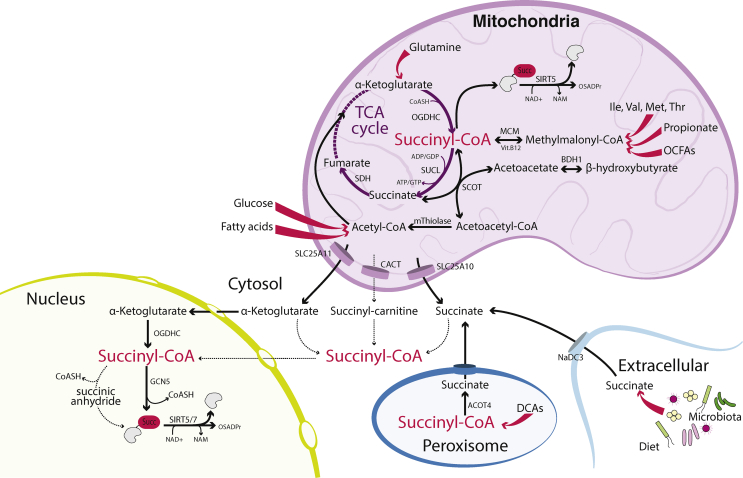
Succinyl-CoA is known for its role as an intermediate in the TCA cycle in the mitochondria, where it is generated from α-ketoglutarate (also known as oxoglutarate) and coenzyme A through the action of the oxoglutarate dehydrogenase complex (OGDH) and degraded to succinate through the action of the succinyl-CoA synthase complex (SUCL), which is subsequently converted to fumarate through succinate dehydrogenase (SDH) (Figure 2). Additionally, mitochondrial succinyl-CoA is produced from propionyl-CoA, which can be generated from isoleucine, valine, methionine and threonine catabolism, odd-chain fatty acid and cholesterol side chain oxidation and from propionate (discussed in greater detail in Section 3.1). Mitochondrial succinyl-CoA also participates in ketolysis in non-hepatic tissues by supplying CoA for the formation of acetoacetyl-CoA from the ketone body acetoacetate, a reversible reaction catalysed by succinyl-CoA:3-ketoacid CoA transferase (SCOT) (Figure 2) [26]. A mitochondrial thiolase converts acetoacetyl-CoA to 2 molecules of acetyl-CoA, which can then enter the TCA cycle [26]. Succinyl-CoA is also required for heme biosynthesis, which is initiated by the reaction, succinyl-CoA + glycine → δ-aminolevulinic acid, catalysed by the mitochondrial enzyme δ-aminolevulinic acid synthetase (ALAS) [27]. Thus, succinyl-CoA is generated and used in multiple mitochondrial metabolic processes.
Figure 2.
Compartmentalisation of metabolic pathways involving succinyl-CoA and protein succinylation indicates distinct regulation of succinyl-CoA metabolism in the mitochondria, cytosol, peroxisomes and nucleus. While multiple substrates are known contributors to mitochondrial succinyl-CoA and peroxisomes can generate succinyl-CoA from oxidation of di-carboxylic acids (DCAs), the metabolic origin of precursors of succinyl-CoA in the nucleus and cytosol are not defined. Enzymes are indicated in bold type. Tapered arrows denote multiple reactions. Solid black arrows denote known reactions. Dashed arrows denote proposed or potential transformations. Abbreviations: ACOT4 (acyl-CoA thioesterase 4), BDH1 (3-hydroxybutyrate dehydrogenase 1), β-hydroxybutyrate (also known as 3-hydroxybutyrate), CACT (carnitine/Acyl-carnitine translocase), DCAs (dicarboxylic acids), GCN5 (general control of amino acid synthesis protein 5-like 2, or KAT2A), Ile (isoleucine), MCM (methylmalonyl-CoA mutase), Met (methionine), mThiolase (mitochondrial thiolase - there are several enzymes capable of performing this reaction, including ACAT1: acetyl-CoA acetyltransferase, mitochondrial, or acetoacetyl-CoA thiolase), NaDC-3 (sodium dicarboxylate transporter-3), NAD+ (nicotinamide adenine dinucleotide, oxidized form), NADH (nicotinamide adenine dinucleotide, reduced form), NAM (nicotinamide), OCFAs (odd chain fatty acids), OGDHC (oxoglutarate dehydrogenase complex), OSADPr (O-succinyl-ADP-ribose), SCOT (succinyl-CoA: 3-ketoacid CoA transferase), SDH (succinate dehydrogenase), SIRT5 (sirtuin 5), SIRT7 (sirtuin 7), SLC25A1 (solute carrier 25A1), SLC25A10 (solute carrier 25A10), SUCL (succinyl-CoA ligase), Thr (threonine), Val (valine).
Despite the clear mitochondrial localisation of relevant enzymes and pathways in succinyl-CoA metabolism, mounting evidence from ‘succinylome’ studies have established that protein succinylation occurs abundantly on resident cytosolic proteins, as well as on histones, indicating the presence of succinyl-CoA in the cytosol and nucleus [[28], [29], [30]]. Indeed, effective export of succinyl-CoA precursors from the mitochondria is supported by studies in SDH-deficient cells, which exhibit elevated cellular succinate and succinyl-CoA levels, as well as increased lysine succinylation in multiple compartments, including the cytosol and nucleus [31]. Through what mechanisms is succinyl-CoA generated outside of mitochondria? A direct transport mechanism for acyl-CoAs across the mitochondrial membranes is not known to exist, indicating that succinyl-CoA is likely generated extra-mitochondrially. A large number of the 26-member family of acyl-CoA synthetase enzymes, whose substrates are not fully defined, are present in the cytosol [32,33], suggesting that succinyl-CoA can likely be generated from succinate within the cytosol (discussed in following paragraph). Succinyl-CoA production has also been reported in the nucleus upon nuclear localization of the OGDH complex, allowing α-ketoglutarate conversion to succinyl-CoA, which can then be used by the acetyltransferase GCN5 to succinylate histones [34]. α-ketoglutarate is transported out of mitochondria through the mitochondrial oxoglutarate carrier SLC25A11 [20,35]. Another mechanism of succinyl group export from the mitochondria is via the carnitine shuttle, as evidenced by elevated succinyl-carnitine levels in the blood of patients carrying mutations in the genes encoding subunits of SUCL [36]. However, a mechanism for the conversion of succinyl-carnitine back to succinyl-CoA within the nucleo-cytosolic compartment is not known, and succinyl-CoA is not a substrate of carnitine acetyltransferase (CrAT) for interconversion to succinyl-carnitine [37]. Thus, possible precursors to succinyl-CoA in the cytosol and nucleus include α-ketoglutarate, succinyl-carnitine and succinate (Figure 2).
Succinate is produced in or transported to the cytosol via several mechanisms. It can be exported from mitochondria via the dicarboxylate carrier SLC25A10 [21,38]. Peroxisomes may be another source of succinate in the nuclear/cytosolic compartment. The peroxisomal thioesterase ACOT4, which is highly expressed in the kidney, liver and intestine, hydrolyses succinyl-CoA to succinate with high specificity compared to a panel of other acyl-CoA substrates [39,40]. Succinyl-CoA production in peroxisomes is likely via the β-oxidation of dicarboxylic acids [39,41]. Succinate may transition from peroxisomes to the cytosol via non-selective peroxisomal membrane channels, such as PXMP2 [42], which permit passage of small solutes, but not larger CoA-conjugated molecules. An additional source of succinate in the cytosol is via its uptake from the environment. Circulating succinate is derived, at least in part, from microbiota [43]. In humans, succinate concentration is 1–3 mM in the contents of large intestines and faeces, which corresponds to approximately 2–4% of the total concentration of organic ions [44]. Succinate has recently been found to mediate the beneficial metabolic effects of a high fibre diet in mice and improve glucose homeostasis through its use as a substrate for intestinal gluconeogenesis [45]. In an analysis of metabolite production and consumption across tissues in pigs; the pancreas, colon and portal circulation were found to be significant producers of succinate, consistent with its production from gastrointestinal microbiota, while the liver and kidney exhibited significant succinate uptake [46]. Brown adipose tissue also has a remarkable capacity to capture circulating succinate to promote thermogenesis [47]. Plasma succinate has been reported in a range from 2 to 55 μM in rodents and humans [[48], [49], [50]]. Succinate, taken up from the extracellular environment can be transported into the cell via sodium dicarboxylate transporter-3 (NaDC-3 or Slc13a3), which has a Km value for succinate in the range of 20–102 μM and is highly expressed in the brain, liver, kidney, placenta and pancreas [[51], [52], [53]]. Thus, multiple sources of succinate exist but remain to be defined in terms of their roles in provisioning succinyl-CoA for histone and other nuclear-cytosolic protein succinylation (Figure 2). Genetic and pharmacological targeting of candidate enzymes and transporters in combination with isotope tracing approaches are needed to resolve the origin and regulation of nuclear and cytosolic succinyl-CoA.
2.2. Lysine succinylation of histones: roles and regulation
Succinylation occurs prominently on mitochondrial proteins, and both succinyl-CoA and lysine succinylation are highest in tissues with a high mitochondrial content, including the heart, brown adipose tissue and liver [28,[54], [55], [56]]. However, succinylation is also observed on histone and non-histone proteins outside of mitochondria [[28], [29], [30],34,57,58]. Succinyl-CoA can be of equal concentration or even higher than acetyl-CoA in some conditions and cell types [54]. Succinylation is highly responsive to metabolic conditions and has been shown to be altered by dietary factors, including fat and ethanol consumption [[59], [60], [61]], hypoxia [62], tricarboxylic acid cycle (TCA) cycle disruption [31] and activation of thermogenesis in brown adipose tissue [55]. Salient biochemical features of succinyl-CoA distinguish it in terms of its roles in post-translational modification. First, the reactivity of succinyl-CoA makes succinylation likely to occur by non-enzymatic addition [18,63,64], although it is enzymatically removed by sirtuin 5 (SIRT5) [65] and SIRT7 [66]. Secondly, succinylation is chemically distinct from acetylation because it is bulkier (C4O3H5 vs C2OH3) and imparts a negative charge under physiological pH, while acetylation is neutral [67].
2.2.1. Non-enzymatic succinylation
Substantial evidence indicates that succinyl-CoA allows for the non-enzymatic succinylation of proteins. At least two distinct mechanisms for non-enzymatic acylation have been described. The first mechanism involves a deprotonated lysine ε-amino group performing a nucleophilic attack on acyl-CoA carbonyl carbon [63,68]. Since the pKa of the lysine ε-amino group is 10.5, this process is highly pH dependent and more prevalent in the relatively alkaline (pH 7.9–8.0) environment of the mitochondria. Notably, the pKa may depend on the lysine residue's context within a protein, with reactive lysines found in close proximity to basic residues [69]. Peptide array analysis revealed that basic residues preceding a lysine residue, typically in the −7 to −3 position, increase the likelihood of non-enzymatic lysine acetylation, potentially via reducing the pKa of the lysine ε-amino group [70]. The high concentration of succinyl-CoA in the mitochondria in combination with the alkaline pH is thought to drive the substantial lysine succinylation observed in this compartment. Peroxisomes also have an elevated pH of (pH 8.2) compared to the cytosol (pH 7.2) [71] and a high capacity for acyl-CoA generation as intermediates through lipid oxidation. Peroxisomes are therefore likely to be affected by non-enzymatic acylation by the same mechanism, although acylation in peroxisomes has not been extensively studied. A second mechanism proposed to drive non-enzymatic acylation, which is particularly important for succinyl-CoA, is autocatalysis to a reactive intermediate. Succinyl-CoA is highly unstable because it is subject to self-hydrolysis to form succinic anhydride, a reactive cyclic intermediate that subsequently acylates lysine residues on proteins over a wide pH range [18]. Other dicarboxylate species, including HMG-CoA, glutaryl-CoA and 3-methylglutaryl-CoA [18], as well as malonyl-CoA [64], are also reported to be highly reactive and prone to engage in non-enzymatic acylation. The relative importance of succinic anhydride formation in chromatin modification in the nucleus is not known, although the propensity for self-hydrolysis to occur even at neutral pH suggests its potential to contribute to lysine succinylation in the nuclear and cytosolic compartments.
2.2.2. Enzymatic regulation of succinylation by succinyl transferases
Enzyme-catalysed lysine succinylation has also been reported. Wang et al. made the intriguing discovery that all 3 components of the succinyl-CoA generating OGDH complex, classically known to localise to the mitochondria, are also present in the nucleus; this complex was also shown to interact with the histone acyl transferase (HAT) GCN5 (also known as KAT2A) [34]. This allows nuclear generation of succinyl-CoA, which is proposed to directly supply substrate to GCN5 for histone succinylation. Given the reactivity of succinyl-CoA (discussed above), the provisioning of succinyl-CoA directly to the transferase enzyme could enable the targeting of succinylation to specific loci. GCN5-catalysed histone succinylation was implicated in supporting tumour cell proliferation. ChIP-seq analysis revealed that H3K79 succinylation (H3K79Suc) as well as GCN5 and OGDH were significantly enriched at gene promoter regions. Furthermore, RNA sequencing (RNA-seq) analysis revealed suppression of specific pathways involved in cell signalling and cell cycle upon depletion of H3K79Suc through expression of mutant GCN5 inactive for succinylation (but not acetylation) capacity or mutant OGDH with the inactivated nuclear localisation sequence of the dihydrolipoyl succinyltransferase (DLST) subunit. This indicates that H3K79Suc may act as an activating mark for gene transcription [34]. In independent support of enzymatic succinylation, nuclear protein extracts from HepG2 cells separated by strong cation exchange chromatography showed that a specific fraction (not those containing the HAT p300/CBP) of purified proteins could catalyse succinylation of purified histone proteins in a manner that was sensitive to heat inactivation and was decreased by competition with acetyl-CoA, supporting the idea of protein dependent catalysis of histone succinylation [72]. Lysine succinyl–transferase activity has also been proposed for OGDH [73] in the mitochondria and carnitine palmitoyltransferase 1 (CPT1) [30] on the outer mitochondrial outer membrane, although the physiological relevance of these activities is not clear.
The balance between enzymatic and non-enzymatic succinylation in chromatin regulation will require further clarification. Simithy et al. reported histone succinylation is closely correlated with increasing total cellular succinyl-CoA. However, seemingly in contrast with the Wang study, in vitro assays testing a panel of acyl-CoAs with acyl-transferases, including GCN5 (GCN5, CBP, p300, PCAF, NatA, Tip60 and MOF), against purified histones found no increase in succinylation with the addition of enzymes compared to control, in contrast to acetylation, butyrylation and propionylation, which were dramatically increased by enzyme addition [19]. These different findings may be influenced by site specificity, assay conditions (including different substrates, co-factors, reducing agents, and purification assays) or the quantification methods used. Wang et al. focused on GCN5-mediated succinylation of H3K79 and used both succinyl-lysine and H3K79Suc antibodies for Western blot analyses, as well as qualitative identification by mass spectrometry. Simithy et al. reported an average of acyl modifications of 14 Lysine sites on histone 3, including H3K79 and used quantitative mass spectrometry to detect histone acylation. Considering these studies together, one possibility is that much of succinylation is non-enzymatic, but that enzymatic succinylation may occur at specific loci or under particular biological conditions.
2.2.3. De-succinylation
Because lysine succinylation may be largely non-enzymatic, regulation of the modification can be achieved through its removal [65,74,75]. The identification of lysine de-succinylase activity in the bacterial enzyme CobB indicates that regulation of protein succinylation is a conserved function [76]. Two members of the mammalian sirtuin NAD+ dependent de-acetylase family, SIRT5 and SIRT7, have de-succinylase activity. Studies in murine SIRT5 knockout models show dramatic increases in lysine succinylation, particularly in mitochondria, which is the primary localisation site of SIRT5, suggesting the importance of this enzyme in regulating bulk succinylation levels [77]. Histone succinylation levels are also increased in SIRT5 knockout models, implying a role for SIRT5 in regulating de-succinylation in the nucleus [28]. SIRT7 was characterised as a histone desuccinylase that functions in the DNA damage response [66]. Li et al. demonstrated that SIRT7 is recruited to double-strand break (DSB) sites in a poly [ADP-ribose] polymerase 1 (PARP1)-dependent manner and catalyses desuccinylation of H3K122 at DSB sites, thereby promoting chromatin condensation and efficient DSB repair. SIRT7 depletion also led to increased succinylation of specific histone sites (H2BK46, H2BK108, H4K31 and H4K77), impaired chromatin compaction and DNA repair and sensitised cells to genotoxic stresses, suggesting a more widespread role in regulation of histone succinylation and chromatin regulation.
2.2.4. Lysine-succinyl readers
Current evidence for direct lysine-succinyl reader activity is limited. One proposed reader is the YEATS domain of glioma-amplified sequence-41 (GAS41), which is reported to bind succinylated histones in a pH-dependent manner [78]. Bromodomains on the other hand, appear unlikely to recognise succinyl-lysine, since a comprehensive survey of 49 bromodomains failed to identify any with affinity for succinyl-lysine [79].
2.3. Perspectives and future directions
Succinyl-CoA is a highly abundant metabolite, and although it is mainly localised to mitochondria, lysine succinylation of histones may play a significant role in gene regulation. A key question is whether histone succinylation occurs primarily non-specifically as a metabolic stress response or if it is used as a specific regulatory mechanism. Moreover, if it is used as a specific regulatory mechanism, what advantages are conferred by succinylation over other modifications? Does histone succinylation convey information about specific metabolic conditions, triggering appropriate responses? Investigation of carbon sources that contribute to nuclear lysine succinylation and the mechanisms of nuclear succinyl-CoA production will help to address these questions. Given the propensity for succinylation to occur non-enzymatically, highly localised production of succinyl-CoA in the nucleus may be required for it to serve in a specific regulatory capacity, making the localisation of relevant succinyl-CoA producing enzymes of particular interest. Thus, compartmentalisation is likely key to the metabolic signals transduced by histone succinylation.
3. Propionyl-CoA
3.1. Propionyl-CoA generation: metabolic pathways and compartmentalisation
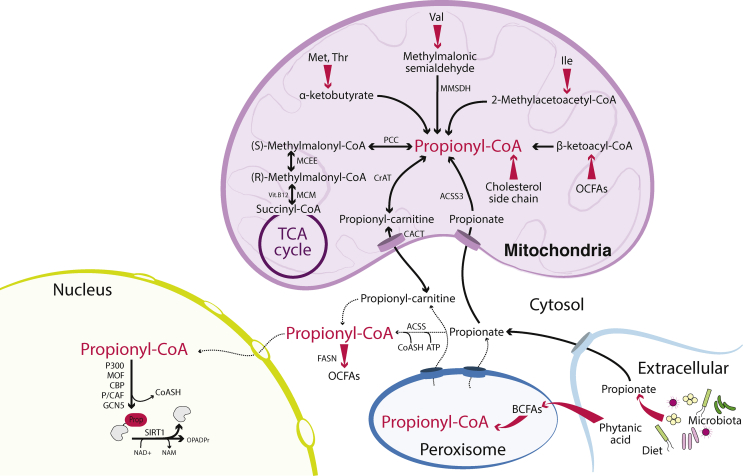
Propionyl-CoA has a 3-carbon acyl group (Figure 1B) and is a catabolic product of several mitochondrial pathways. Catabolism of diverse substrates converge on propionyl-CoA, including certain amino acids – methionine, threonine and the branched chain amino acids (BCAAs) isoleucine and valine [80] – as well as oxidation of odd chain fatty acids (OCFAs) and oxidation of the side chain of cholesterol [81]. The primary metabolic fate of propionyl-CoA in mitochondria is anaplerosis via a multi-step reaction that favours moving excess propionyl-CoA into the TCA cycle [82]. This occurs via the carboxylation of propionyl-CoA to form (S)-methylmalonyl-CoA catalysed by propionyl-CoA carboxylase (PCC), conversion of (S)- to (R)-methylmalonyl-CoA by methylmalonyl-CoA epimerase and subsequent conversion of (R)-methylmalonyl-CoA to succinyl-CoA catalysed by the vitamin B12 dependent enzyme methylmalonyl-CoA mutase (MCM) [83,84]. Propionyl-CoA may also be generated in peroxisomes through oxidation of branched chain fatty acids, notably from dietary phytanic acid, which is enriched in dairy products from ruminants [85,86]. Peroxisomal propionyl-CoA is likely converted to propionyl-carnitine by peroxisomal CrAT or to propionate through peroxisomal thioesterases [87] for export through the PXMP2 porin, although the precise enzymes and transporters involved are not well defined [85] (Figure 3).
Figure 3.
Metabolic regulation of propionyl-CoA is annotated to the mitochondria despite histone propionylation occurring in the nucleus. Propionyl-CoA can be generated in the mitochondria through a variety of metabolic sources. It is also generated in peroxisomes from branched-chain fatty acid oxidation. Nuclear and cytosolic sources of propionyl-CoA are not defined. Enzymes are indicated in bold type. Tapered pink arrows denote multiple reactions. Solid black arrows denote known reactions. Dashed arrows denote proposed or potential transformations. Abbreviations: ACSS (acyl-CoA synthase short-chain family member, ACSS2 is localised to the cytosol, ACSS1 and 3 are localised to mitochondria), BCFAs (branched-chain fatty acids), CBP (CREB binding protein), CrAT (carnitine acyl-transferase), FASN (fatty acid synthase), GCN5(general control of amino acid synthesis protein 5-like 2, or KAT2A), Ile (isoleucine), MMSDH (methylmalonic semialdehyde dehydrogenase), MCM (methylmalonyl-CoA mutase), MCEE (methylmalonyl-CoA epimerase), Met (methionine), MOF (males absent on the first; also known as MYST1 or KAT8), NAM (nicotinamide), NAD+ (nicotinamide adenine dinucleotide, oxidised form), OCFAs (odd-chain fatty acids), OPADPr (O-propionyl-ADP-ribose), P/CAF (P300/CBP-associated factor, or KAT2B), PCC (propionyl-CoA carboxylase), SIRT1 (sirtuin 1), Thr (threonine), Val (valine), Vit. B12 (vitamin B12). (For interpretation of the references to color/colour in this figure legend, the reader is referred to the Web version of this article.)
Propionyl-CoA can also be produced from the short chain fatty acid propionate via one or more mammalian acyl-CoA synthetases [32,88,89]; the mitochondrial acyl-CoA synthetase short chain family member 3 (ACSS3) from rats has been shown to preferentially use propionate as a substrate over acetate and butyrate [90], and yeast acetyl-CoA synthetase can catalyse the production of propionyl CoA from propionate, although its Km for propionate is higher than that for acetate [91,92]. Propionate is produced by gut microbiota and is one of the 3 most abundant short chain fatty acids (SCFAs) in the gastrointestinal tract and in circulation (along with acetate and butyrate), although it is largely metabolised in the liver, with levels of 17–194 μM in portal circulation compared to peripheral levels of 1–13 μM in humans [93]. Propionate generation can be modulated by dietary fibre [94] and can also be directly ingested in the diet, since it is a common and unrestricted food additive used as an antimicrobial and flavouring agent [95,96].
Propionyl-CoA cannot directly cross the inner mitochondrial membrane, but it may be transported to the cytosol via the carnitine shuttle after conversion to propionyl-carnitine by mitochondrial CrAT, which displays efficient interconversion of propionyl-CoA and propionylcarnitine [37]. Propionyl-carnitine levels are 2000-fold higher in the context of inborn errors of metabolism affecting the mitochondrial enzymes PCC or MCM [97]. Propionyl-carnitine can, presumably, be reconverted back to propionyl-CoA in the cytosol, since propionyl-CoA has been shown to be used by cytosolic fatty acid synthase (FASN) to build odd chain fatty acids in a manner dependent on CrAT [80,98,99]. A CrAT-dependent mechanism could therefore also potentially supply propionyl-CoA for nuclear histone propionylation, although this remains to be tested. Interestingly, CrAT nuclear localization has been reported, and is proposed to generate nuclear acetyl-CoA from acetylcarnitine [100]. In sum, metabolic pathways and enzymes producing propionyl-CoA are annotated to the mitochondrion and peroxisomes [[101], [102], [103]], and although metabolic sources of propionyl-CoA used for histone modification remain to be defined, CrAT has been implicated in its transport from mitochondria to cytosol.
3.2. Lysine propionylation of histones: roles and regulation
Lysine propionylation (Kpr) was first identified as a protein modification on histones H3 and H4 [104,105], and cellular propionyl-CoA abundance correlates with histone propionylation in a variety of contexts [19,106]. Unlike lysine succinylation, lysine propionylation appears to be primarily enzymatically driven [19] and the similar biochemical properties of lysine-propionylation and acetylation (i.e., both are short-chain hydrophobic modifications that neutralise the lysine positive charge [15]) suggest that these two modifications may serve similar or overlapping functions.
3.2.1. Histone-propionylation function
Histone lysine-propionylation, like lysine acetylation, has been described as a mark of active gene transcription. ChIP-sequencing of histone 3 lysine 14 propionylation (H3K14pr) revealed 31% of H3K14Pr peaks were located at promoter-transcriptional start site (TSS) regions. Comparison of ChIP-seq and RNA-seq data demonstrated a positive correlation between abundance of H3K14Pr marks and gene expression, and showed that H3K14pr, H3K9ac and H3K14 butyrylation (bu) were commonly present at the same loci. Moreover, in a cell-free in vitro transcription system, propionyl-CoA could act as a substitute for acetyl-CoA to stimulate transcription [107]. In terms of its roles in biological regulation, histone propionylation has been found to be regulated during cell differentiation. Histone propionylation levels decrease during myogenic differentiation, coincident with a decrease in levels of propionyl-CoA [19]. Analogously, U937 leukaemia cells exhibited propionylation at 7% of histone H3K23 residues and lost propionylation during monocytic differentiation [106]. These data are correlative, and a specific functional role for propionylation in maintaining cell identity or regulating differentiation remains to be clarified.
3.2.2. Enzymatic regulation of propionylation and propionyl-histone readers
Propionylation can be added to and removed from histones by many of the same enzymes that control acetylation, a function conserved in bacterial GCN5-related N-acetyltransferase enzymes and the deacetylase sirtuin CobB [108], as well as eukaryotic acetyltransferases p300 [109], CREB-binding protein (CBP) [110], P/CAF [111], GCN5 [107,112] and MOF [113] and the deacetylases SIRT1 and SIRT2 [108,110]. Peptide pulldown experiments performed to determine proteins that bind to H3K14pr compared with H3K14ac revealed a very similar set of bromodomain-containing proteins, including components of the (P)BAF chromatin remodelling complex [114]. Thus, histone propionylation is linked to transcriptional activation, mediated enzymatically by acyltransferases, and appears to be bound by a similar set of bromodomain-containing proteins as acetylated histones, pointing to similar or overlapping biological functions of propionylation and acetylation.
3.2.3. Propionyl-CoA abundance affects lysine-propionylation
Histone and non-histone protein propionylation appear to be closely linked to the abundance of propionyl-CoA. Direct evidence for this comes from experiments conducted in isolated nuclei, which demonstrated that propionyl-CoA supplementation increased histone propionylation in a dose-dependent manner [19]. In this system, propionyl-CoA supplementation resulted in a simultaneous increase in H3K14pr and a decrease in H3K14ac, suggesting reciprocal interplay between acetylation and propionylation at this site [19], possibly by competing for the same acyltransferases. Of note, the stoichiometry of H3K14 acetylation and propionylation are comparable in both HeLa cells and myogenic cells, an interesting observation considering that acetyl-CoA is 10–15-fold more abundant than propionyl-CoA in whole cell measurements [19,25].
Links between propionyl-CoA abundance and propionylation are evident in the context of propionic acidaemia, an inherited metabolic disorder characterized by a deficiency in PCC, resulting in elevated propionyl-CoA levels and systemic complications impacting multiple body systems [115]. Fibroblasts from patients with propionic acidaemia show increased protein propionylation [116]. Livers of mice deficient for the PCC alpha subunit exhibit elevated H3K14pr levels [107]. Propionyl-CoA is reported be most abundant in the liver, followed by kidney, heart, muscle and brain [54]. While the relationship between whole cell propionyl-CoA abundance (which may reflect predominantly mitochondrial pools) and levels in the nucleus remains unclear, these data nevertheless indicate that propionylation is dynamically regulated and metabolically sensitive.
3.2.4. Interactions between diet propionyl-CoA and histone propionylation
Propionate metabolism occurs predominantly in the liver and gastrointestinal tract, and propionate administration profoundly impacts liver metabolism [117]. Propionate consumption has been linked to metabolic changes in mice and humans with effects on lipid metabolism, insulin sensitivity, appetite and athletic performance [95,[118], [119], [120]], raising the intriguing possibility that some effects of propionate supplementation may, at least in part, be mediated through changes in histone propionylation, although such a role remains to be demonstrated. High fat feeding has been shown to influence both propionyl-CoA and histone propionylation levels. Four weeks of high-fat diet (HFD) feeding in mice suppressed propionyl-CoA levels in the liver and perigonadal white fat [121], consistent with an independent analysis of histone marks altered in HFD (8 week regimen) compared to chow fed mice, which found that H3K23pr was suppressed in the livers of the HFD group [122]. Conversely, another study reported increased propionyl-CoA levels in the liver of mice fed HFD for 12 weeks [123], possibly reflecting either differential effects of the duration of HFD feeding or microbiome differences, as SCFA production by the gut microbiota is heavily influenced by diet [124]. Further work is needed to elucidate the mechanisms through which diet may influence propionyl-CoA metabolism and chromatin modification.
3.3. Perspectives and future directions
More research is needed to clearly delineate the connections between the metabolism of food components such as propionate and branched chain amino acids, the compartmentalized production of propionyl-CoA, and the modification of proteins including histones and subsequent changes in gene expression. Key questions include whether histone propionylation plays distinct roles from acetylation in modulating cellular behaviour under specific environmental conditions and whether it may play compensatory roles under conditions of acetyl-CoA limitation. A critical evaluation of the distinct roles of these modifications under physiological conditions that alter the balance of acetyl-CoA and propionyl-CoA will help to shed light on these issues.
4. Crotonoyl-CoA
4.1. Crotonoyl-CoA generation: metabolic pathways and compartmentalisation
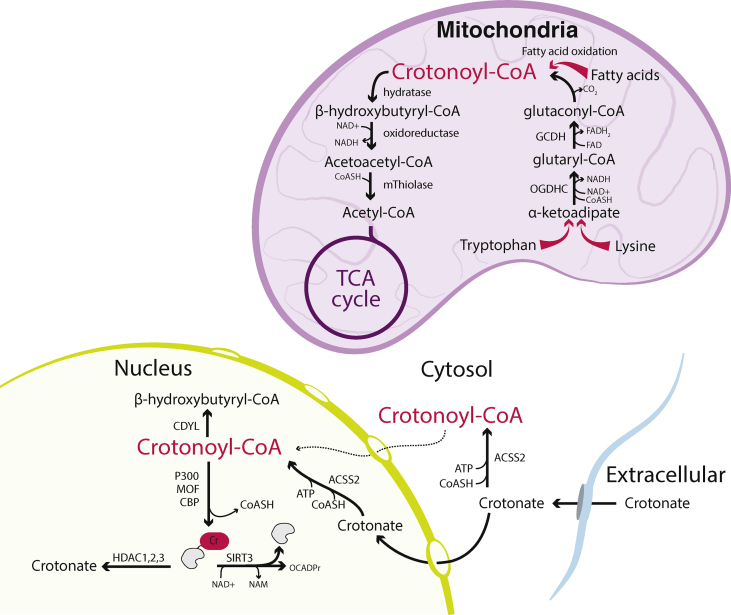
Crotonoyl-CoA is of low abundance compared to other acyl-CoA species, comprising <5% of the total short chain acyl-CoA pool in a range of cells types (HeLa cells, myoblasts and myotubes) [19]. It is generated in mitochondria from several substrates, including tryptophan, lysine, and fatty acids (Figure 4). Catabolism of tryptophan and lysine converges at α-ketoadipate, which is converted to glutaryl-CoA by the OGDH. The FAD-dependent enzyme glutaryl-CoA dehydrogenase (GCDH) converts glutaryl-CoA to the enzyme-bound intermediate glutaconyl-CoA, generating FADH2, which can be used in the electron transport chain [125]. Glutaconyl-CoA is decarboxylated to crotonoyl-CoA. Crotonoyl-CoA is subsequently converted to L-β-hydroxybutyryl-CoA (also called L-3-hydroxybutyryl-CoA), then to acetylacetyl-CoA, and finally to acetyl-CoA, which can then enter the TCA cycle. Fatty acid oxidation in the mitochondria also produces 2,3-enoyl-CoA intermediates, including crotonoyl-CoA. Although these immediate steps in the pathway for crotonoyl-CoA generation and degradation occur in the mitochondria, the nuclear localised enzyme chromodomain Y-like transcription corepressor (CDYL) was found to act as a crotonyl-CoA hydratase. CDYL can convert crotonyl-CoA to β-hydroxybutyryl-CoA in the nucleus and negatively regulates histone lysine crotonylation [126]. Mechanisms of crotonoyl-CoA generation from endogenous precursors in the nucleus or cytosol are not defined (Figure 4).
Figure 4.
Endogenous crotonoyl-CoA is generated in the mitochondria from amino acid and fatty acid catabolism, while histone crotonylation is associated with exogenous crotonate. Crotonoyl-CoA is an intermediate in tryptophan and lysine catabolism as well as fatty acid oxidation, which produces 2,3-enoyl-CoA intermediates, including crotonoyl-CoA in the mitochondria. Physiological exogenous sources of crotonate are not well defined, but addition of extracellular crotonate results in increased intracellular crotonoyl-CoA and histone crotonylation. Crotonoyl-CoA can be converted to L-β-hydroxybutyryl-CoA (also called L-3-hydroxybutyryl-CoA) by CDYL in the nucleus. Enzymes are indicated in bold type. Tapered arrows denote multiple reactions. Solid black arrows denote verified transformations. Dashed arrows denote proposed transformations. Abbreviations: CDYL (chromodomain Y-like transcription corepressor), HDAC (histone deacetylase), GCDH (glutaryl-CoA dehydrogenase), mThiolase (mitochondrial thiolase – there are several enzymes capable of performing this reaction, including ACAT1: acetyl-CoA acetyltransferase, mitochondrial or acetoacetyl-CoA thiolase), NAM (nicotinamide), NAD+ (nicotinamide adenine dinucleotide, oxidised form), NADH (nicotinamide adenine dinucleotide, reduced form), OGDHC (oxoglutarate dehydrogenase complex), OCADPr (O-crotonyl-ADP-ribose), SIRT1, 2, 3 (sirtuin 1, 2, 3).
The short-chain fatty acid (SCFA) crotonate is a direct precursor to crotonoyl-CoA. Addition of exogenous crotonate to mammalian cells results in crotonoyl-CoA generation, at least in part by the cytoplasmic/nuclear localised enzyme acyl-CoA synthetase 2 (ACSS2), corresponding with increased histone crotonylation [127]. ACSS2 expression is under the control of SREBP [128], and conditions that suppress ACSS2, such as HFD, in liver and white adipose tissue [121], or that alter the availability of crotonate potentially impact the production of crotonoyl-CoA in the nuclear-cytosolic compartment. Crotonyl-CoA levels were found to be suppressed in white adipose tissue upon high-fat feeding [121]. However, physiological sources of crotonate to mammalian cells are unclear.
4.2. Lysine crotonylation of histones: roles and regulation
Histone lysine crotonylation (Kcr) has been shown to be a positive regulator of gene transcription and to be associated with active regions of chromatin [127,129,130], although the modification generally occurs at very low stoichiometry in comparison to acetylation or propionylation, consistent with the low abundance of crotonyl-CoA [19]. Histone Kcr is also reported to be regulated over the cell cycle [129] and to be enriched in embryonic stem cells, as well as on sex chromosomes in post-meiotic male germ cells [129,131]. Crotonoyl-CoA has a 4-carbon acyl chain containing an α,β-unsaturated carbonyl [15]. Thus, crotonylation is chemically distinct from other 4-carbon acylation marks in that it lacks the flexibility of butyryl-CoA and the carboxylic acid and charge of succinyl-CoA (Figure 1B). Lysine crotonylation displays the lowest non-enzymatic modification amongst a panel of short-chain acyl-CoAs [19], consistent with a low chemical reactivity toward lysine nucleophiles attributed to its resonance properties [132].
4.2.1. Enzymes involved in crotonylation, de-crotonylation and crotonyl-lysine readers
P300 was shown to act as a crotonyl-transferase [127]. Consistently, structural analyses of P300 show that the active site can accommodate the crotonoyl acyl chain and that it can catalyse crotonylation in vitro, although Kcr activity is much less efficient (by 64-fold) than Kac activity [109]. The acetyl-transferases MOF and CBP have also been reported to display crotonoyl–transferase activity [130]. Interestingly, P300 dependent crotonylation activates gene transcription more potently than acetyl-CoA in cell-free assays. Moreover, treatment of macrophages with crotonate to increase intracellular crotonyl-CoA levels and histone crotonylation enhanced transcriptional response to inflammatory gene activation [127]. Thus, despite being a low-abundance modification, crotonylation may have potent capacity to support transcription.
Response to crotonylation can be mediated through YEATS domain proteins. YEATS domains of a number of transcriptional regulators have been shown to bind preferentially to Kcr marks versus a panel of alternative acylation marks including Kac, Kpr, Kbu and Khib (lysine hydroxyisobutyrylation) [[133], [134], [135]]. The PHD finger domains of human MOZ (also known as KAT6A) and DPF2 (also known as BAF45d) also display a preference for Kcr [136]. Bromodomains also bind Kcr marks but with lower affinity compared to Kac marks, as shown for TAF1 [79].
Removal of Kcr has been proposed for a range of enzymes. Sirt1, Sirt2 and Sirt3 can catalyse the hydrolysis of lysine crotonylated histone peptides and proteins in vitro, and the decrotonylase function of Sirt3 has been demonstrated in living cells [137]. Histone decrotonylase activity has also been shown in Class I histone deacetylases: HDAC1, HDAC2 and HDAC3 [129,131]. Inhibition of HDAC function by the SCFA butyrate derived from microbiota inhibits de-crotonylation in gut epithelial cells, illustrating an interesting interaction of Kcr marks with SCFA metabolism [129].
4.3. Perspectives and future directions
Given the low abundance of crotonoyl-CoA in many cell types, its poor ability to compete with acetyl-CoA for use by KATs and the resultant low stoichiometry of crotonylation, it is important to better define whether there are contexts in which crotonylation plays distinct roles. The preferential recognition of Kcr by a number of YEATS and PHD finger domains indicates the possibility that Kcr marks may be able to compete with Kac marks to modulate specific functions despite being of much lower abundance. One challenge in testing this is how to specifically modulate crotonylation independent of acetylation or other acylations, considering that many of the same writer and eraser enzymes are employed. Defining the endogenous metabolic sources and mechanisms of generation of crotonyl-CoA within the nucleus may provide one avenue to investigate this question. Moreover, if crotonyl-CoA is specifically important in certain settings, this raises the question of what unique adaptations in metabolism result in the generation of crotonoyl-CoA in the nuclear-cytoplasmic compartment in these contexts.
5. Butyryl-CoA
5.1. Butyryl-CoA generation: metabolic pathways and compartmentalisation
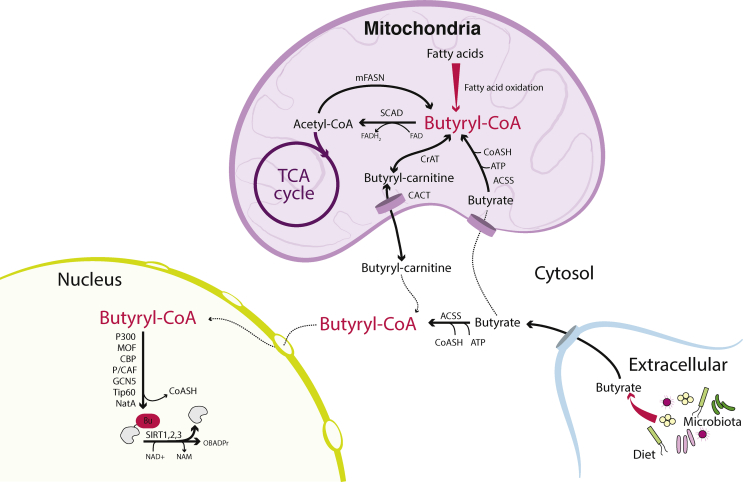
Endogenous butyryl-CoA is generated in the mitochondria through β-oxidation of fatty acids, as well as in the mitochondrial fatty acid synthesis pathway (Figure 5). Butyryl-CoA can accumulate under conditions of defective fatty acid oxidation, such as in deficiency of short-chain acyl-CoA dehydrogenase (SCAD) [116]. In livers of Scad−/− mice, butyryl-CoA levels can accumulate to levels as high or higher than acetyl-CoA [138]. Mitochondrial CrAT displays high activity toward butyryl-CoA and likely facilitates the interconversion of butyryl-carnitine and butyryl-CoA for mitochondrial export [37]. Consistent with this, SCAD-deficient mouse models accumulate butyryl-carnitine in tissue [139], and elevated plasma butyryl-carnitine is a diagnostic feature in human patients with SCAD deficiency [140]. Global butyryl-lysine is increased in the livers of Scad−/− mice and in fibroblasts from human patients with SCAD deficiency, although the specific effect on histone butyrylation has not been explicitly determined [116].
Figure 5.
Butyryl-CoA can be generated from the microbial fermentation product butyrate and as an intermediate in fatty acid synthesis and oxidation in the mitochondria. Histone butyrylation can be catalysed by a variety of acetyl-transferases and may be removed by sirtuin 1, 2 or 3. Enzymes are indicated in bold type. Tapered arrows denote multiple reactions. Solid black arrows denote verified transformations. Dashed arrows denote proposed transformations. Abbreviations: ACSS (acyl-CoA synthetase short-chain family member- ACSS2 is localised to the cytosol, ACSS1 and 3 are localised to mitochondria), CACT (carnitine/acyl-carnitine translocase), CBP (CREB-binding protein), CrAT (carnitine acyl-transferase), GCN5 (general control of amino acid synthesis protein 5-like 2, or KAT2A), mFASN (mitochondrial fatty acid synthase), MOF (males absent on the first; also known as MYST1 or KAT8), NAD+ (nicotinamide adenine dinucleotide, oxidised form), NADH (nicotinamide adenine dinucleotide, reduced form), NAM (nicotinamide), OBADPr (O-butyryl-ADP-ribose), NatA (N-terminal acetyl-transferase A), P/CAF (P300/CBP-associated factor, or KAT2B), PCC (propionyl-CoA carboxylase), SCAD (short-chain acyl-CoA dehydrogenase), SIRT1, 2, 3 (sirtuin 1, 2, 3), Tip60 (60 kDa Tat-interactive protein, or KAT5).
Exogenous butyrate is another metabolic source of intracellular butyryl-CoA. Butyrate is an abundant SCFA produced by the microbiota in the gastrointestinal tract, with levels reaching 24 mmol/kg of contents in the human large intestine [93], and its generation can be modulated by dietary fibre [94]. The majority of butyrate metabolism occurs in the intestinal epithelia and liver. Transport into the cell is mediated by a number of transporters, including monocarboxylate transporter 1 (MCT1), monocarboxylate transporter 4 (MCT4), the sodium-coupled monocarboxylate transporter 1 (SMCT1) and breast cancer resistance protein (BCRP), also known as ABCG2. These transporters are specifically localised to the apical surface in gut epithelial cells, and their expression can be regulated by signalling through cell surface receptors sensing butyrate [141]. Butyrate is actively metabolised by gut epithelial cells in a manner that maintains a spatial concentration gradient, limiting butyrate exposure of stem cells at the base of colonic crypts [142]. Circulating butyrate levels are generally low due to uptake, and metabolism in the liver. Butyrate occurs at levels ranging 14–64 μM in portal circulation compared to peripheral levels of 1–12 μM in humans [93].
Generation of butyryl-CoA from exogenous butyrate can occur through the action of acyl-CoA synthetase short-chain enzymes [89]. Entry of butyrate into the mitochondria, where it is oxidized to produce acetyl-CoA, is believed to be mediated both via carnitine-dependent and -independent transport [143]. In intestinal cells, butyrate oxidation increases histone acetylation in a manner dependent on mitochondrial citrate export and acetyl-CoA generation in the nuclear/cytosolic compartment via ACLY [144]. Notably, butyrate is also an effective Class I HDAC inhibitor, and in cells that are not actively oxidizing butyrate, it increases histone acetylation through its HDAC activity [144,145]. Thus, butyrate can potentially contribute to chromatin modification via HDAC inhibition, its oxidation to produce acetyl-CoA for histone acetylation, or its conversion to butyryl-CoA to facilitate butyrylation.
5.2. Butyryl-CoA: roles in histone modification and gene regulation
Histone lysine butyrylation (Kbu), like acetylation and propionylation, is predominantly enzymatic [19] and is generally an activating mark for transcription [107]. Histone butyrylation is reported to be catalysed to varying extents by KATs, including PCAF, GCN5, CBP, p300, NatA, Tip60 and MOF [19,104]. The efficiency of enzymatic histone butyrylation is generally lower than for acetylation and propionylation – decreasing with increasing acyl chain length – although efficiency for different acyl groups varies site-specifically [19]. Butyryl-CoA has also been reported to inhibit GCN5, with structural analyses showing binding of the butyryl acyl chain to the active site in a manner that occludes its enzymatic activity [112]. Butyryl-CoA also inhibits PCAF-dependent histone acetylation in in vitro assays, although it is unclear whether this is through competition as a substrate or through inhibition of its enzymatic activity [121]. Interestingly, a positive correlation between the acetyl-CoA: (iso)butyryl-CoA ratio and several histone Kac marks was noted in mouse tissue [121]. In terms of removal, SIRT1, 2 and 3 have been shown to exert weak debutyrylase activity in vitro [146].
Histone butyrylation can be recognized by bromodomain-containing proteins. The bromodomains of BRD9 and CECR2 and the second bromodomain of TAF1 have been identified as Kbu binders in vitro [79]. A distinct functional role for histone butyrylation was identified in the context of spermatogenesis. Kbu at H4K5 and H4K8 was shown to compete with Kac at these sites. Kbu, as opposed to Kac, delays histone turnover and prevents binding of the transcriptional regulator BRDT, which in turn controls gene expression and chromatin reorganisation during spermatogenesis [147]. Thus, histone butyrylation appears to serve a similar role to acetylation and propionylation in terms of mediation by similar enzymes and association with transcriptional activation, although distinct binding properties to chromatin readers may confer specific functions to the modification.
5.3. Perspectives and future directions
Butyryl-CoA is notable for its impact on histone acylation through multiple mechanisms: as a direct butyryl donor for histone butyrylation, as a precursor for acetyl-CoA production for histone acetylation and through inhibition of acetyltransferases. Further, its SCFA precursor butyrate acts as a class I HDAC inhibitor, making the metabolic interplay between butyrate and butyryl-CoA a regulator of both acylation and deacetylation. The direct metabolic link between the abundant microbiota-derived butyrate and butyryl-CoA means that butyryl-CoA directly responds to diet and changes in microbiota, at least in tissues proximal to the gastrointestinal tract (GIT), including gut epithelial cells and the liver. Notably, inflammatory bowel disease has been associated with altered butyrate levels and metabolism in the GIT [148], suggesting that histone butyrylation and its metabolic regulation could plausibly participate in the pathogenesis of such diseases. Specific regulation of butyryl-CoA metabolism in the nucleus has not yet been defined, and elucidating its nuclear abundance and regulation compared to acetyl-CoA and other acyl-CoAs will aid in understanding the role of butyryl-CoA in communicating nutritional cues to chromatin.
6. Other acyl-CoAs that modify histones
6.1. Malonyl-CoA
Malonyl-CoA is a three-carbon product of acetyl-CoA carboxylation by acetyl-CoA carboxylase (ACC) in the cytosol and a major substrate for fatty acid synthesis [149]. Malonyl-CoA is also an inhibitor of carnitine palmitoyltransferase 1 (CPT1). CPT1 is located on the outer mitochondrial membrane and catalyses the initial step in the mitochondrial import of long-chain fatty acids for β-oxidation. Thus, malonyl-CoA plays a critical role in preventing fatty acid synthesis and oxidation from occurring simultaneously in a futile cycle [150]. In the mitochondria, malonyl-CoA is an intermediate in the less well-recognised process of mitochondrial fatty acid synthesis [151], and mitochondrial malonyl-CoA can be generated from malonate by Acyl-CoA synthetase family member 3 (ACSF3) [152]. Decarboxylation of malonyl-CoA to acetyl-CoA by malonyl CoA decarboxylase (MCD) may also occur in the mitochondrial and cytosolic compartments [153].
Lysine malonylation occurs predominantly non-enzymatically and is removed by SIRT5 [64,67,75,154,155]. Functional studies highlight malonyl-CoA as a reactive thioester metabolite that can modify and inhibit glycolytic enzyme activity [64], although examination of hypermalonylation in isolated mitochondria from SIRT5 and MCD deletion models showed no impact on mitochondrial respiratory function [156]. Malonylation has also been proposed to play a role in nutrient signalling via mammalian target of rapamycin (mTOR) regulation [157]. The functional role and regulation of histone malonylation remains to be elucidated.
6.2. Glutaryl-CoA
Glutaryl-CoA is a product of tryptophan and lysine catabolism preceding crotonoyl-CoA production in the mitochondria [125] (see Figure 4 and Section 4.1). Increased total protein glutarylation was observed in mammalian cells deficient in GCDH, the FAD-dependent enzyme that converts glutaryl-CoA to crotonoyl-CoA, as well as in Drosophila fed increasing amounts of tryptophan [158]. Changes in the distribution of glutarylated proteins were also observed in mice under fed or fasted conditions, indicating that this modification is sensitive to dietary changes [158]. Glutaryl-CoA is proposed to form a reactive anhydride intermediate [18], and lysine glutarylation can occur non-enzymatically, with bulk protein glutarylation regulated through its removal by SIRT5 [158]. A recent study also showed that the specific mark H4K91glu is added by KAT2A and removed by Sirt7 and that this mark is regulated over the cell cycle and may control chromatin condensation required for cell cycle progression [159].
6.3. 3-Hydroxybutyryl-CoA
3-Hydroxybutyryl-CoA (also known as β-hydroxybutyryl-CoA, BHB-CoA) may occur as D and L enantiomers. D-BHB-CoA may be produced during ketone body metabolism, presumably through the action of short-chain acyl-CoA synthetases on the ketone body D-β-hydroxybutyrate (BHB). L-BHB-CoA is a product of fatty acid oxidation, and lysine and tryptophan catabolism generated through crotonoyl-CoA hydatase activity (Figure 4). Histone hydroxybutyrylation (Kbhb) marks are dramatically induced in response to elevated ketogenesis in livers of mice subjected to prolonged fasting or streptozotocin-induced diabetic ketoacidosis [160]. ChIP-seq and RNA-seq analysis revealed that histone Kbhb is enriched in active gene promoters. Additionally, the increased H3K9bhb levels during starvation are associated with genes upregulated in starvation-responsive metabolic pathways [160], indicating specificity of Kbhb in regulation to achieve an appropriate response. The specific reader enzymes that bind histone Kbhb marks to mediate this response, however, remain to be determined. Kbhb has been shown to be removed by HDAC enzymes and by SIRT3 in a manner that is preferential for specific sites [161]. Recent interest in the effects of intermittent fasting and ketogenic diets inspires speculation on the potential involvement of BHB-CoA and Kbhb. Much remains to be uncovered about the mechanisms regulating BHB-CoA in the nucleus, and the physiological relevance of histone Kbhb, especially in non-hepatic tissues.
6.4. 2-Hydroxyisobutyryl-CoA
2-Hydroxyisobutyryl-CoA is not a known intermediate in mammalian metabolic pathways but may be generated by acyl-CoA synthetases acting upon 2-hydroxyisobutyrate (2-HIB) (not to be confused with 3-HIB, an intermediate in valine catabolism). 2-HIB derived from the microbial degradation of dietary proteins that escape digestion in the upper gastrointestinal tract has been associated with the presence of specific microbial species in the colon [162]. 2-HIB levels in urine were increased in obesity in humans [[163], [164], [165]], decreased in sleep deprivation in humans [166], and varied according to gender in obese mouse models [167]. 2-HIB is a major metabolic product detected in urine and plasma following exposure to the organic compound tert-butyl methyl ether (MTBE) [168,169], which is used as a gasoline additive. Addition of deuterated 2-HIB to cells was used to demonstrate that 2-HIB is a precursor for histone 2-hydroxyisobutyrylation (Khib) marks [170], presumably via a 2-HIB-CoA intermediate, since Khib is catalysed by the acyl-CoA transferase p300 [171]. Khib was originally identified as a mark associated with genes with high transcriptional activity in male germ cells [170]. Histone Khib is believed to be a high stoichiometry mark with specific sites reaching 5.33% (H4K91) and 7.79% (H1.2K62) in HeLa cells [170]. P300-dependent Khib also targets glycolytic enzymes, regulating their activity in response to nutrient availability [171]. The xenobiotic origin of HIB potentially connects 2-HIB-CoA and histone Khib marks to environmental exposures, including gut microbiota and environmental contaminants, although much remains to be uncovered about the metabolic regulation of Khib and its physiological roles in these contexts.
6.5. Lactyl-CoA
Lactyl-CoA (also known as lactoyl- or (R/S)-2-hydroxypropanoyl-CoA) has been shown in in vitro assays to to be used for a newly identified histone modification derived from lactate, lysine lactylation [16]. Histone lysine lactylation increases under conditions in which lactate production rises, including hypoxia and M1 macrophage polarisation [16], and the modification is associated with active chromatin regions [16]. Lactyl-CoA itself has not been detected endogenously in cells to date, however, and lactoylglutathione has been demonstrated to facilitate the non-enzymatic lactylation of glycolytic enzymes [172]. Regardless, the modulation of lactate production through glycolysis under a number of important physiological conditions, including cancer [173] and immune cell activation [174,175], make lactylation a potentially exciting signalling link between metabolism and cell signalling, with further research into its biochemistry and physiological roles needed [176].
6.6. More species
Increasing sensitivity and specificity of mass spectrometry-based methods has resulted in the identification of multiple novel histone acyl modifications [177]. The recent identification of lysine hydroxymethylglutarylation [18], acetoacetylation, isovalerylation, 2-methylbutyrylation, tiglylation [17] and lactylation [16,172] brings the number of identified short-chain (less than 6 carbon) lysine acyl modifications to 15 species, although not all of these have not been specifically reported as histone modifications. Considering the diversity of potential acyl groups in the “CoA-ome” there are almost certainly more acylations derived from endogenous and even exogenous and xenobiotic substrates. Regulation of histone lysine acylation marks other than acetylation has been implicated in a variety of important disease settings, physiological processes and responses to environmental modifications (summarised in Table 1). Data so far suggests that many of these modifications particularly affect cell differentiation; however, a clear understanding of their biological significance has yet to be achieved. Critically, many unanswered questions remain regarding the mechanisms of nuclear regulation of the many species that comprise the acyl-CoA world (Figure 1) and their impact on histone modification and gene regulation.
Table 1.
Regulation of histone lysine acylation marks.
| Histone acylation affected | Disease/physiological process/environmental modification | Reference |
|---|---|---|
| Succinylation | TCA cycle disruption | [31] |
| Brown adipose thermogenic activation | [55] | |
| Tumour proliferation | [34] | |
| Chromatin compaction, DNA repair, genotoxic stress | [66] | |
| Myogenic differentiation | [19] | |
| Propionylation | Myogenic differentiation | [19] |
| Monocytic differentiation | [106] | |
| Propionic acidaemia/propionyl-CoA carboxylase (PCC) deficiency | [107,116] | |
| High-fat diet | [122] | |
| Crotonylation | Spermatogenesis | [178] |
| Cell cycle | [129] | |
| Gut epithelia response to microbiota metabolic products (butyrate)/antibiotic treatment | [129] | |
| Embryonic stem cell self-renewal | [131] | |
| Macrophage activation | [127] | |
| Myogenic differentiation | [19] | |
| Acute kidney injury | [179] | |
| Butyrylation | Spermatogenesis | [147] |
| Myogenic differentiation | [19] | |
| Glutarylation | Myogenic differentiation | [19] |
| Dietary tryptophan (Drosophila) | [158] | |
| 3-hydroxybutyrylation | Prolonged fasting, diabetic ketoacidosis | [160] |
| Lactylation | Macrophage activation | [16] |
| Hypoxia | [16] |
7. Conclusion
Compartmentalisation of acyl-CoA metabolism is key to connecting histone acylation in the nucleus to metabolic pathways. The metabolic role of many acyl-CoA species has been described only in mitochondria. Mechanisms for transport of precursors and enzymes required for synthesis of the relevant acyl-CoAs into the cytosolic and nuclear compartments likely play an important role in the regulation of acyl modification, as well as the availability of SCFAs in the bloodstream or immediate microenvironment. Emerging evidence for nuclear localisation of metabolic enzymes canonically found in the mitochondria highlights one possible mechanism for site-specific production of acyl-CoAs within the nucleus. We propose that metabolite tracing in combination with targeting of key enzymes and transporters will help to illuminate the metabolic pathways that connect histone modifications to acyl-CoA metabolism. Dissecting the biological function of specific acyl-CoA species in chromatin acylation will require the consideration of their specific biochemical properties. In particular, it will be critical to consider their propensity for enzymatic versus non-enzymatic protein modification, and to identify the enzymes mediating addition, removal and binding of each acyl modification. Determination of the relative abundance of acyl-CoA species in specific contexts is also important since different acyl-CoA species can, to varying degrees, compete as substrates of acyltransferase enzymes. Additionally, unique signals may be conveyed through combinatorial effects of different modifications. The discovery of multiple histone acylations over the past decade has opened numerous questions about the roles of these modifications in physiological and disease-related processes. An improved understanding of the metabolic sources of acyl-CoAs used for chromatin modification is needed to elucidate the extent to which acyl-CoAs serve as metabolic signals linking nutrient utilisation to transcriptional responses.
Author contributions
KEW and ST conceptualised the scope. ST and CL wrote the manuscript. ST prepared the figures. KEW, NWS, ST and CL revised and corrected the manuscript.
Acknowledgements
KEW is supported by NIH grants R01CA174761, R01CA228339 and R01DK116005. NWS is supported by the NIH grant R01GM132261. ST is supported by the American Diabetes Association through post-doctoral fellowship 1-18-PDF-144. CDL is supported by the NIH grant T32 GM07170. We thank Eliana von Krusenstiern for assistance with figures.
Contributor Information
Nathaniel W. Snyder, Email: NateWSnyder@temple.edu.
Kathryn E. Wellen, Email: wellenk@upenn.edu.
Conflict of interest
None declared.
References
- 1.Gut P., Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502(7472):489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 2.Campbell S.L., Wellen K.E. Metabolic signaling to the nucleus in cancer. Molecular Cell. 2018;71(3):398–408. doi: 10.1016/j.molcel.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nature Reviews Molecular Cell Biology. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 4.Ryan D.G., Murphy M.P., Frezza C., Prag H.A., Chouchani E.T., O'Neill L.A. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nature Metabolism. 2019;1(1):16–33. doi: 10.1038/s42255-018-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C.T., Tu B.P., Tang Y. Eight kinetically stable but thermodynamically activated molecules that power. Cell Metabolism. 2018;118(4):1460–1494. doi: 10.1021/acs.chemrev.7b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imachi H., Nobu M.K., Nakahara N., Morono Y., Ogawara M., Takaki Y. Isolation of an archaeon at the prokaryote–eukaryote interface. BioRxiv. 2019:726976. doi: 10.1101/726976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracher P.J., Snyder P.W., Bohall B.R., Whitesides G.M. The relative rates of thiol–thioester exchange and hydrolysis for alkyl and aryl thioalkanoates in water. Origins of Life and Evolution of Biospheres. 2011;41(5):399–412. doi: 10.1007/s11084-011-9243-4. [DOI] [PubMed] [Google Scholar]
- 8.Chevallot-Beroux., Elodie G., Jan Moran J. Energy conservation via thioesters in a non-enzymatic metabolism-like reaction network. CemRxiv. 2019 [Google Scholar]
- 9.Aires C.C.P., Ruiter J.P.N., Luís P.B.M., ten Brink H.J., IJlst L., de Almeida I.T. Studies on the extra-mitochondrial CoA-ester formation of valproic and Δ4-valproic acids. Biochimica et Biophysica Acta (BBA) – Molecular and Cell Biology of Lipids. 2007;1771(4):533–543. doi: 10.1016/j.bbalip.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi H., McCaffery J.M., Irizarry R.A., Boeke J.D. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Molecular Cell. 2006;23(2):207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 11.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Current Opinion in Cell Biology. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivanand S., Viney I., Wellen K.E. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends in Biochemical Sciences. 2018;43(1):61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G., Abozguia K. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metabolism. 2015;21(6):805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Sabari B.R., Zhang D., Allis C.D., Zhao Y. Metabolic regulation of gene expression through histone acylations. Nature Reviews Molecular Cell Biology. 2017;18(2):90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldensperger T., Sanzo S. Di., Ori A., Glomb M.A. Quantitation of reactive acyl-CoA species mediated protein acylation by HPLC–MS/MS. Analytical Chemistry. 2019 doi: 10.1021/acs.analchem.9b02656. [DOI] [PubMed] [Google Scholar]
- 18.Wagner G.R., Bhatt D.P., O'Connell T.M., Thompson J.W., Dubois L.G., Backos D.S. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metabolism. 2017;25(4):823–837. doi: 10.1016/j.cmet.2017.03.006. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simithy J., Sidoli S., Yuan Z.-F., Coradin M., Bhanu N.V., Marchione D.M. Characterization of histone acylations links chromatin modifications with metabolism. Nature Communications. 2017;8(1):1141. doi: 10.1038/s41467-017-01384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Archiv: European Journal of Physiology. 2004;447(5):689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 21.Palmieri F., Stipani I., Quagliariello E., Klingenberg M. Kinetic study of the tricarboxylate carrier in rat liver mitochondria. European Journal of Biochemistry. 1972;26(4):587–594. doi: 10.1111/j.1432-1033.1972.tb01801.x. [DOI] [PubMed] [Google Scholar]
- 22.SRERE P.A. The citrate cleavage enzyme. I. Distribution and purification. Journal of Biological Chemistry. 1959;234:2544–2547. [PubMed] [Google Scholar]
- 23.Watson J.A., Lowenstein J.M. Citrate and the conversion of carbohydrate into fat. Fatty acid synthesis by a combination of cytoplasm and mitochondria. Journal of Biological Chemistry. 1970;245(22):5993–6002. [PubMed] [Google Scholar]
- 24.Paine P.L., Moore L.C., Horowitz S.B. Nuclear envelope permeability. Nature. 1975;254(5496):109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.V., Carrer A., Shah S., Snyder N.W., Wei S., Venneti S. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metabolism. 2014;20(2):306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metabolism. 2017;25(2):262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kafina M.D., Paw B.H. Intracellular iron and heme trafficking and metabolism in developing erythroblasts. Metallomics: Integrated Biometal Science. 2017;9(9):1193–1203. doi: 10.1039/c7mt00103g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Chen Y., Tishkoff D.X.X., Peng C., Tan M., Dai L. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molecular Cell. 2013;50(6):919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H., Lou Y., Fu Y., Zhang Y., Liu N., Liu Z. Involvement of the vitamin D receptor in energy metabolism revealed by profiling of lysine succinylome of white adipose tissue. Scientific Reports. 2017;7(1):14132. doi: 10.1038/s41598-017-14477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurmi K., Hitosugi S., Wiese E.K., Boakye-Agyeman F., Gonsalves W.I., Lou Z. Carnitine palmitoyltransferase 1A has a lysine succinyltransferase activity. Cell Reports. 2018;22(6):1365–1373. doi: 10.1016/j.celrep.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smestad J., Erber L., Chen Y., Maher L.J. Chromatin succinylation correlates with active gene expression and is perturbed by defective TCA cycle metabolism. iScience. 2018;2:63–75. doi: 10.1016/j.isci.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins P.A., Maiguel D., Jia Z., Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. Journal of Lipid Research. 2007;48(12):2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Grevengoed T.J., Klett E.L., Coleman R.A. Acyl-CoA metabolism and partitioning. Annual Review of Nutrition. 2014;34(1):1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Guo Y.R., Liu K., Yin Z., Liu R., Xia Y. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017 doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monné M., Miniero D.V., Bisaccia F., Fiermonte G., Fiermonte G. The mitochondrial oxoglutarate carrier: from identification to mechanism. Journal of Bioenergetics and Biomembranes. 2013;45(1–2):1–13. doi: 10.1007/s10863-012-9475-7. [DOI] [PubMed] [Google Scholar]
- 36.Rizzo C., Boenzi S., Inglese R., la Marca G., Muraca M., Martinez T.B. Measurement of succinyl-carnitine and methylmalonyl-carnitine on dried blood spot by liquid chromatography–tandem mass spectrometry. Clinica Chimica Acta. 2014;429:30–33. doi: 10.1016/j.cca.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Violante S., IJlst L., Ruiter J., Koster J., van Lenthe H., Duran M. Substrate specificity of human carnitine acetyltransferase: implications for fatty acid and branched-chain amino acid metabolism. Biochimica et Biophysica Acta – Molecular Basis of Disease. 2013;1832(6):773–779. doi: 10.1016/j.bbadis.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Fiermonte G., Palmieri L., Dolce V., Lasorsa F.M., Palmieri F., Runswick M.J. The sequence, bacterial expression, and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. Journal of Biological Chemistry. 1998;273(38):24754–24759. doi: 10.1074/jbc.273.38.24754. [DOI] [PubMed] [Google Scholar]
- 39.Westin M.A.K., Hunt M.C., Alexson S.E.H. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. Journal of Biological Chemistry. 2005 doi: 10.1074/jbc.M508479200. [DOI] [PubMed] [Google Scholar]
- 40.Ellis J.J.M., Bowman C.E., Wolfgang M.M.J., Prentki M., Corkey B., Mitchell G. Metabolic and tissue-specific regulation of acyl-CoA metabolism. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0116587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt M.C., Tillander V., Alexson S.E.H.H. Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie. 2014;98:45–55. doi: 10.1016/J.BIOCHI.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 42.Antonenkov V.D., Hiltunen J.K. Transfer of metabolites across the peroxisomal membrane. Biochimica et Biophysica Acta – Molecular Basis of Disease. 2012;1822(9):1374–1386. doi: 10.1016/J.BBADIS.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 44.Meijer-Severs G.J., van Santen E. Short-chain fatty acids and succinate in feces of healthy human volunteers and their correlation with anaerobe cultural counts. Scandinavian Journal of Gastroenterology. 1987;22(6):672–676. doi: 10.3109/00365528709011141. [DOI] [PubMed] [Google Scholar]
- 45.De Vadder F., Kovatcheva-Datchary P., Zitoun C., Duchampt A., Bä F., Correspondence G.M. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metabolism. 2016;24:151–157. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 46.Jang C., Hui S., Zeng X., Cowan A.J., Wang L., Chen L. Metabolite exchange between mammalian organs quantified in pigs. Cell Metabolism. 2019 doi: 10.1016/j.cmet.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills E.L., Pierce K.A., Jedrychowski M.P., Garrity R., Winther S., Vidoni S. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature. 2018;560(7716) doi: 10.1038/s41586-018-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadagopan N., Li W., Roberds S.L., Major T., Preston G.M., Yu Y. Circulating succinate is elevated in rodent models of hypertension and metabolic disease. American Journal of Hypertension. 2007;20(11):1209–1215. doi: 10.1016/j.amjhyper.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kushnir M.M., Komaromy-Hiller G., Shushan B., Urry F.M., Roberts W.L. Analysis of dicarboxylic acids by tandem mass spectrometry. High-throughput quantitative measurement of methylmalonic acid in serum, plasma, and urine. Clinical Chemistry. 2001;47(11):1993–2002. [PubMed] [Google Scholar]
- 51.Burckhardt B.C., Lorenz J., Kobbe C., Burckhardt G. Substrate specificity of the human renal sodium dicarboxylate cotransporter, hNaDC-3, under voltage-clamp conditions. American Journal of Physiology – Renal Physiology. 2005;288(4):F792–F799. doi: 10.1152/ajprenal.00360.2004. [DOI] [PubMed] [Google Scholar]
- 52.Chen X., Tsukaguchi H., Chen X.Z., Berger U.V., Hediger M.A. Molecular and functional analysis of SDCT2, a novel rat sodium-dependent dicarboxylate transporter. Journal of Clinical Investigation. 1999;103(8):1159–1168. doi: 10.1172/JCI5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Fei Y.J., Kekuda R., Yang-Feng T.L., Devoe L.D., Leibach F.H. Structure, function, and genomic organization of human Na(+)-dependent high-affinity dicarboxylate transporter. American Journal of Physiology – Cell Physiology. 2000;278(5):C1019–C1030. doi: 10.1152/ajpcell.2000.278.5.C1019. [DOI] [PubMed] [Google Scholar]
- 54.Sadhukhan S., Liu X., Ryu D., Nelson O.D., Stupinski J.A., Li Z. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(16):4320–4325. doi: 10.1073/pnas.1519858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G., Meyer J.G., Cai W., Softic S., Li M.E., Verdin E. Regulation of UCP1 and mitochondrial metabolism in brown adipose tissue by reversible succinylation. Molecular Cell. 2019 doi: 10.1016/j.molcel.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rardin M.J., He W., Nishida Y., Newman J.C., Carrico C., Danielson S.R. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metabolism. 2013;18(6):920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y. Lysine succinylation and lysine malonylation in histones. Molecular & Cellular Proteomics. 2012;11(5):100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nature Chemical Biology. 2011;7(1):58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer J.G., Softic S., Basisty N., Rardin M.J., Verdin E., Gibson B.W. Temporal dynamics of liver mitochondrial protein acetylation and succinylation and metabolites due to high fat diet and/or excess glucose or fructose. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0208973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawai Y., Fujii H., Okada M., Tsuchie Y., Uchida K., Osawa T. Formation of Nε-(succinyl)lysine in vivo: a novel marker for docosahexaenoic acid-derived protein modification. Journal of Lipid Research. 2006;47(7):1386–1398. doi: 10.1194/jlr.M600091-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Ali H.R., Assiri M.A., Harris P.S., Michel C.R., Yun Y., Marentette J.O. Quantifying competition among mitochondrial protein acylation events induced by ethanol metabolism. Journal of Proteome Research: 2019 doi: 10.1021/acs.jproteome.8b00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H., Xu H., Potash S., Starkov A., Belousov V.V., Bilan D.S. Mild metabolic perturbations alter succinylation of mitochondrial proteins. Journal of Neuroscience Research. 2017;95(11):2244–2252. doi: 10.1002/jnr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner G.R., Payne R.M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. Journal of Biological Chemistry. 2013;288(40):29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulkarni R.A., Worth A.J., Zengeya T.T., Shrimp J.H., Garlick J.M., Roberts A.M. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chemical Biology. 2017;24(2):231–242. doi: 10.1016/j.chembiol.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science (New York, N.Y.) 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L., Shi L., Yang S., Yan R., Zhang D., Yang J. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nature Communications. 2016;7(1):12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirschey M.D., Zhao Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Molecular & Cellular Proteomics. 2015;14(9):2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paik W.K., Pearson D., Lee H.W., Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochimica et Biophysica Acta (BBA) – Nucleic Acids and Protein Synthesis. 1970;213(2):513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 69.Baeza J., Smallegan M.J., Denu J.M. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chemical Biology. 2015;10(1):122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olia A.S., Barker K., McCullough C.E., Tang H.-Y., Speicher D.W., Qiu J. Nonenzymatic protein acetylation detected by NAPPA protein arrays. ACS Chemical Biology. 2015;10(9):2034–2047. doi: 10.1021/acschembio.5b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dansen T.B., Wirtz K.W.A., Wanders R.J.A., Pap E.H.W. Peroxisomes in human fibroblasts have a basic pH. Nature Cell Biology. 2000;2(1):51–53. doi: 10.1038/71375. [DOI] [PubMed] [Google Scholar]
- 72.Yokoyama A., Katsura S., Sugawara A. Biochemical analysis of histone succinylation. Biochemistry Research International. 2017:1. doi: 10.1155/2017/8529404. –7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibson G.E., Xu H., Chen H.-L., Chen W., Denton T.T., Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. Journal of Neurochemistry. 2015;134(1):86–96. doi: 10.1111/jnc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osborne B., Bentley N.L., Montgomery M.K., Turner N. The role of mitochondrial sirtuins in health and disease. Free Radical Biology and Medicine. 2016;100:164–174. doi: 10.1016/j.freeradbiomed.2016.04.197. [DOI] [PubMed] [Google Scholar]
- 75.Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M. The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & Cellular Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colak G., Xie Z., Zhu A.Y., Dai L., Lu Z., Zhang Y. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Molecular & Cellular Proteomics. 2013;12(12):3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carrico C., Meyer J.G., He W., Gibson B.W., Verdin E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metabolism. 2018;27(3):497–512. doi: 10.1016/j.cmet.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Jin J., Chung M.W.H., Feng L., Sun H., Hao Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(10):2365–2370. doi: 10.1073/pnas.1717664115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flynn E.M., Huang O.W., Poy F., Oppikofer M., Bellon S.F., Tang Y. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015;23(10):1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Crown S.B., Marze N., Antoniewicz M.R. Catabolism of branched chain amino acids contributes significantly to synthesis of odd-chain and even-chain fatty acids in 3T3-L1 adipocytes. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halarnkar P.P., Blomquist G.J. Comparative aspects of propionate metabolism. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1989;92(2):227–231. doi: 10.1016/0305-0491(89)90270-8. [DOI] [PubMed] [Google Scholar]
- 82.Reszko A.E., Kasumov T., Pierce B.A., David F., Hoppel C.L., Stanley W.C. Assessing the reversibility of the anaplerotic reactions of the propionyl-CoA pathway in heart and liver. Journal of Biological Chemistry. 2003;278(37):34959–34965. doi: 10.1074/jbc.M302013200. [DOI] [PubMed] [Google Scholar]
- 83.Flavin M., Ochoa S. Metabolism of propionic acid in animal tissues. I. Enzymatic conversion of propionate to succinate. Journal of Biological Chemistry. 1957;229(2):965–979. [PubMed] [Google Scholar]
- 84.Jeffrey F.M., Storey C.J., Sherry A.D., Malloy C.R. 13C isotopomer model for estimation of anaplerotic substrate oxidation via acetyl-CoA. American Journal of Physiology-Endocrinology and Metabolism. 1996;271(4):E788–E799. doi: 10.1152/ajpendo.1996.271.4.E788. [DOI] [PubMed] [Google Scholar]
- 85.Wanders R.J.A., Waterham H.R., Ferdinandusse S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Frontiers in Cell and Developmental Biology. 2016;3:83. doi: 10.3389/fcell.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wanders R.J.A., Komen J., Ferdinandusse S. Phytanic acid metabolism in health and disease. Biochimica et Biophysica Acta (BBA) – Molecular and Cell Biology of Lipids. 2011;1811(9):498–507. doi: 10.1016/J.BBALIP.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Hunt M.C., Siponen M.I., Alexson S.E.H. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochimica et Biophysica Acta – Molecular Basis of Disease. 2012;1822(9):1397–1410. doi: 10.1016/j.bbadis.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 88.Reijnierse G.L.A., Veldstra H., Van den Berg C.J. Radioassay of acetyl-CoA synthetase, propionyl-CoA synthetase and butyryl-CoA synthetase in brain. Analytical Biochemistry. 1976;72(1–2):614–622. doi: 10.1016/0003-2697(76)90574-1. [DOI] [PubMed] [Google Scholar]
- 89.Scaife J.R., Tichivangana J.Z. Short chain acyl-CoA synthetases in ovine rumen epithelium. Biochimica et Biophysica Acta. 1980;619(2):445–450. doi: 10.1016/0005-2760(80)90097-1. [DOI] [PubMed] [Google Scholar]
- 90.Yoshimura Y., Araki A., Maruta H., Takahashi Y., Yamashita H. Molecular cloning of rat acss3 and characterization of mammalian propionyl-CoA synthetase in the liver mitochondrial matrix. Journal of Biochemistry. 2017;161(3):279–289. doi: 10.1093/jb/mvw067. [DOI] [PubMed] [Google Scholar]
- 91.Patel S.S., Walt D.R. Substrate specificity of acetyl coenzyme A synthetase. Journal of Biological Chemistry. 1987;262(15):7132–7134. [PubMed] [Google Scholar]
- 92.Frenkel E.P., Kitchens R.L. Purification and properties of acetyl coenzyme A synthetase from bakers' yeast. Journal of Biological Chemistry. 1977;252(2):504–507. [PubMed] [Google Scholar]
- 93.Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.-J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of Lipid Research. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tirosh A., Calay E.S., Tuncman G., Claiborn K.C., Inouye K.E., Eguchi K. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Science Translational Medicine. 2019;11(489) doi: 10.1126/SCITRANSLMED.AAV0120. [DOI] [PubMed] [Google Scholar]
- 96.Food and Drug Administration, U.S., 2019. 21CFR184.1081.
- 97.Wikoff W.R., Gangoiti J.A., Barshop B.A., Siuzdak G. Metabolomics identifies perturbations in human disorders of propionate metabolism. Clinical Chemistry. 2007;53(12):2169–2176. doi: 10.1373/clinchem.2007.089011. [DOI] [PubMed] [Google Scholar]
- 98.Wallace M., Green C.R., Roberts L.S., Lee Y.M., McCarville J.L., Sanchez-Gurmaches J. Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nature Chemical Biology. 2018;14(11):1021–1031. doi: 10.1038/s41589-018-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Green C.R., Wallace M., Divakaruni A.S., Phillips S.A., Murphy A.N., Ciaraldi T.P. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nature Chemical Biology. 2015;12(1):15–21. doi: 10.1038/nchembio.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Madiraju P., Pande S.V., Prentki M., Madiraju S.R.M. Mitochondrial acetylcarnitine provides acetyl groups for nuclear histone acetylation. Epigenetics. 2009;4(6):399–403. doi: 10.4161/epi.4.6.9767. [DOI] [PubMed] [Google Scholar]
- 101.Harper A.E., Miller R.H., Block K.P. Branched-chain amino acid metabolism. Annual Review of Nutrition. 1984;4(1):409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 102.Bartlett K., Eaton S. Mitochondrial beta-oxidation. European Journal of Biochemistry. 2004;271(3):462–469. doi: 10.1046/j.1432-1033.2003.03947.x. [DOI] [PubMed] [Google Scholar]
- 103.Houten S.M., Wanders R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. Journal of Inherited Metabolic Disease. 2010;33(5):469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & Cellular Proteomics. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang K., Chen Y., Zhang Z., Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. Journal of Proteome Research. 2009;8(2):900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu B., Lin Y., Darwanto A., Song X., Xu G., Zhang K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. Journal of Biological Chemistry. 2009;284(47):32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kebede A.F., Nieborak A., Shahidian L.Z., Le Gras S., Richter F., Gómez D.A. Histone propionylation is a mark of active chromatin. Nature Structural & Molecular Biology. 2017;24(12):1048–1056. doi: 10.1038/nsmb.3490. [DOI] [PubMed] [Google Scholar]
- 108.Garrity J., Gardner J.G., Hawse W., Wolberger C., Escalante-Semerena J.C. N -lysine propionylation controls the activity of propionyl-CoA synthetase. Journal of Biological Chemistry. 2007;282(41):30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 109.Kaczmarska Z., Ortega E., Goudarzi A., Huang H., Kim S., Márquez J.A. Structure of p300 in complex with acyl-CoA variants. Nature Chemical Biology. 2017;13(1):21–29. doi: 10.1038/nchembio.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S.S.C. Molecular characterization of propionyllysines in non-histone proteins. Molecular & Cellular Proteomics. 2009;8(1):45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leemhuis H., Packman L.C., Nightingale K.P., Hollfelder F. The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. ChemBioChem. 2008;9(4):499–503. doi: 10.1002/cbic.200700556. [DOI] [PubMed] [Google Scholar]
- 112.Ringel A.E., Wolberger C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallographica Section D Structural Biology. 2016;72(7):841–848. doi: 10.1107/S2059798316007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Han Z., Wu H., Kim S., Yang X., Li Q., Huang H. Revealing the protein propionylation activity of the histone acetyltransferase males absent on the first (MOF) Journal of Biological Chemistry. 2018 doi: 10.1074/jbc.RA117.000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vollmuth F., Geyer M. Interaction of propionylated and butyrylated histone H3 lysine marks with Brd4 bromodomains. Angewandte Chemie. 2010;49(38):6768–6772. doi: 10.1002/anie.201002724. [DOI] [PubMed] [Google Scholar]
- 115.Pena L., Franks J., Chapman K.A., Gropman A., Ah Mew N., Chakrapani A. Natural history of propionic acidemia. Molecular Genetics and Metabolism. 2012;105(1):5–9. doi: 10.1016/j.ymgme.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 116.Pougovkina O., te Brinke H., Wanders R.J.A., Houten S.M., de Boer V.C.J. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. Journal of Inherited Metabolic Disease. 2014;37(5):709–714. doi: 10.1007/s10545-014-9684-9. [DOI] [PubMed] [Google Scholar]
- 117.Perry R.J., Borders C.B., Cline G.W., Zhang X.-M., Alves T.C., Petersen K.F. Propionate increases hepatic pyruvate cycling and anaplerosis and alters mitochondrial metabolism. Journal of Biological Chemistry. 2016;291(23):12161–12170. doi: 10.1074/jbc.M116.720631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song B., Zhong Y.Z., Zheng C.B., Li F.N., Duan Y.H., Deng J.P. Propionate alleviates high-fat diet-induced lipid dysmetabolism by modulating gut microbiota in mice. Journal of Applied Microbiology. 2019;14389 doi: 10.1111/jam.14389. [DOI] [PubMed] [Google Scholar]
- 119.Chambers E.S., Viardot A., Psichas A., Morrison D.J., Murphy K.G., Zac-Varghese S.E.K. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scheiman J., Luber J.M., Chavkin T.A., MacDonald T., Tung A., Pham L.-D. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nature Medicine. 2019;1 doi: 10.1038/s41591-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carrer A., Parris J.L.D., Trefely S., Henry R.A., Montgomery D.C., Torres A. Impact of a high-fat diet on tissue acyl-CoA and histone acetylation levels. Journal of Biological Chemistry. 2017;292(8):3312–3322. doi: 10.1074/jbc.M116.750620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nie L., Shuai L., Zhu M., Liu P., Xie Z.-F., Jiang S. The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Molecular & Cellular Proteomics. 2017;16(7):1324–1334. doi: 10.1074/mcp.M117.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu X., Sadhukhan S., Sun S., Wagner G.R., Hirschey M.D., Qi L. High-resolution metabolomics with acyl-CoA profiling reveals widespread remodeling in response to diet. Molecular & Cellular Proteomics. 2015;14(6):1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dalby M.J., Ross A.W., Walker A.W., Morgan P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Reports. 2017;21(6):1521–1533. doi: 10.1016/j.celrep.2017.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rao K.S., Albro M., Dwyer T.M., Frerman F.E. Kinetic mechanism of glutaryl-CoA dehydrogenase. Biochemistry. 2006;45(51):15853–15861. doi: 10.1021/bi0609016. [DOI] [PubMed] [Google Scholar]
- 126.Liu S., Yu H., Liu Y., Liu X.X., Zhang Y., Bu C. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Molecular Cell. 2017;67(5):853–866. doi: 10.1016/j.molcel.2017.07.011. e5. [DOI] [PubMed] [Google Scholar]
- 127.Sabari B.R.R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., Kong H.E.E. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Molecular Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luong A., Hannah V.C., Brown M.S., Goldstein J.L. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. Journal of Biological Chemistry. 2000;275(34):26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- 129.Fellows R., Denizot J., Stellato C., Cuomo A., Jain P., Stoyanova E. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nature Communications. 2018;9(1):105. doi: 10.1038/s41467-017-02651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu X., Wei W., Liu Y., Yang X., Wu J., Zhang Y. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discovery. 2017;3(1):17016. doi: 10.1038/celldisc.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wei W., Liu X., Chen J., Gao S., Lu L., Zhang H. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Research. 2017;27(7):898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang W., Drueckhammer D.G. Understanding the relative acyl-transfer reactivity of oxoesters and thioesters: computational analysis of transition state delocalization effects. Journal of the American Chemical Society. 2001;123(44):11004–11009. doi: 10.1021/ja010726a. [DOI] [PubMed] [Google Scholar]
- 133.Zhao D., Guan H., Zhao S., Mi W., Wen H., Li Y. YEATS2 is a selective histone crotonylation reader. Cell Research. 2016;26(5):629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Y., Sabari B.R., Panchenko T., Wen H., Zhao D., Guan H. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Molecular Cell. 2016;62(2):181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andrews F.H., Shinsky S.A., Shanle E.K., Bridgers J.B., Gest A., Tsun I.K. The Taf14 YEATS domain is a reader of histone crotonylation. Nature Chemical Biology. 2016;12(6):396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xiong X., Panchenko T., Yang S., Zhao S., Yan P., Zhang W. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nature Chemical Biology. 2016;12(12):1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bao X., Wang Y., Li X., Li X.-M., Liu Z., Yang T. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. ELife. 2014;3 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Palladino A.A., Chen J., Kallish S., Stanley C.A., Bennett M.J. Measurement of tissue acyl-CoAs using flow-injection tandem mass spectrometry: acyl-CoA profiles in short-chain fatty acid oxidation defects. Molecular Genetics and Metabolism. 2012;107(4):679–683. doi: 10.1016/j.ymgme.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wood P.A., Amendt B.A., Rhead W.J., Millington D.S., Inoue F., Armstrong D. Short-chain acyl-coenzyme A dehydrogenase deficiency in mice. Pediatric Research. 1989;25(1):38–43. doi: 10.1203/00006450-198901000-00010. [DOI] [PubMed] [Google Scholar]
- 140.Nochi Z., Olsen R.K.J., Gregersen N. Short-chain acyl-CoA dehydrogenase deficiency: from gene to cell pathology and possible disease mechanisms. Journal of Inherited Metabolic Disease. 2017;40(5):641–655. doi: 10.1007/s10545-017-0047-1. [DOI] [PubMed] [Google Scholar]
- 141.Sivaprakasam S., Bhutia Y.D., Yang S., Ganapathy V. Short-chain fatty acid transporters: role in colonic homeostasis. Comprehensive Physiology. 2017;8(1):299–314. doi: 10.1002/cphy.c170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaiko G.E., Ryu S.H., Koues O.I., Collins P.L., Solnica-Krezel L., Pearce E.J. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165(7):1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Han A., Bennett N., MacDonald A., Johnstone M., Whelan J., Donohoe D.R. Cellular metabolism and dose reveal carnitine-dependent and -independent mechanisms of butyrate oxidation in colorectal cancer cells. Journal of Cellular Physiology. 2016;231(8):1804–1813. doi: 10.1002/jcp.25287. [DOI] [PubMed] [Google Scholar]
- 144.Donohoe D.R., Collins L.B., Wali A., Bigler R., Sun W., Bultman S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular Cell. 2012;48(4):612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Steliou K., Boosalis M.S., Perrine S.P., Sangerman J., Faller D.V. Butyrate histone deacetylase inhibitors. BioResearch Open Access. 2012;1(4):192–198. doi: 10.1089/biores.2012.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smith B.C., Denu J.M. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. Journal of Biological Chemistry. 2007;282(51):37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 147.Goudarzi A., Zhang D., Huang H., Barral S., Kwon O.K., Qi S. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Molecular Cell. 2016;62(2):169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Parada Venegas D., De la Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G. Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Frontiers in Immunology. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cellular and Molecular Life Sciences. 2005;62(16):1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Foster D.W. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. Journal of Clinical Investigation. 2012;122(6):1958–1959. doi: 10.1172/jci63967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Witkowski A., Joshi A.K., Smith S. Coupling of the de novo fatty acid biosynthesis and lipoylation pathways in mammalian mitochondria. Journal of Biological Chemistry. 2007;282(19):14178–14185. doi: 10.1074/jbc.M701486200. [DOI] [PubMed] [Google Scholar]
- 152.Bowman C.E., Rodriguez S., Selen Alpergin E.S., Acoba M.G., Zhao L., Hartung T. The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chemical Biology. 2017;24(6):673–684. doi: 10.1016/j.chembiol.2017.04.009. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kerner J., Hoppel C.L. Radiochemical malonyl-CoA decarboxylase assay: activity and subcellular distribution in heart and skeletal muscle. Analytical Biochemistry. 2002;306(2):283–289. doi: 10.1006/abio.2002.5696. [DOI] [PubMed] [Google Scholar]
- 154.Colak G., Pougovkina O., Dai L., Tan M., te Brinke H., Huang H. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Molecular & Cellular Proteomics. 2015;14(11):3056–3071. doi: 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nishida Y., Rardin M.J., Carrico C., He W., Sahu A.K., Gut P. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Molecular Cell. 2015;59(2):321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fisher-Wellman K.H., Draper J.A., Davidson M.T., Koves T.R., Grimsrud P.A., Muoio D.M. Respiratory phenomics across multiple models of protein hyperacylation in cardiac mitochondria reveals a marginal impact on bioenergetics. Cell Reports. 2019;26:1557–1572. doi: 10.1016/j.celrep.2019.01.057. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bruning U., Morales-Rodriguez F., Kalucka J., Goveia J., Taverna F., Queiroz K.C.S. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metabolism. 2018;28(6):866–880. doi: 10.1016/j.cmet.2018.07.019. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tan M., Peng C., Anderson K.A.A., Chhoy P., Xie Z., Dai L. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metabolism. 2014;19(4):605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bao X., Liu Z., Zhang W., Gladysz K., Fung Y.M.E., Tian G. Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Molecular Cell. 2019 doi: 10.1016/j.molcel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 160.Xie Z., Zhang D., Chung D., Tang Z., Huang H., Dai L. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Molecular Cell. 2016;62(2):194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang X., Cao R., Niu J., Yang S., Ma H., Zhao S. Molecular basis for hierarchical histone de-β-hydroxybutyrylation by SIRT3. Cell Discovery. 2019;5(1):35. doi: 10.1038/s41421-019-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li M., Wang B., Zhang M., Rantalainen M., Wang S., Zhou H. Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(6):2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Calvani R., Miccheli A., Capuani G., Tomassini Miccheli A., Puccetti C., Delfini M. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. International Journal of Obesity. 2010;34(6):1095–1098. doi: 10.1038/ijo.2010.44. [DOI] [PubMed] [Google Scholar]
- 164.Friedrich N., Budde K., Wolf T., Jungnickel A., Grotevendt A., Dreßler M. Short-term changes of the urine metabolome after bariatric surgery. OMICS: A Journal of Integrative Biology. 2012;16(11):612–620. doi: 10.1089/omi.2012.0066. [DOI] [PubMed] [Google Scholar]
- 165.Elliott P., Posma J.M., Chan Q., Garcia-Perez I., Wijeyesekera A., Bictash M. Urinary metabolic signatures of human adiposity. Science Translational Medicine. 2015;7(285) doi: 10.1126/scitranslmed.aaa5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Giskeødegård G.F., Davies S.K., Revell V.L., Keun H., Skene D.J. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Scientific Reports. 2015;5 doi: 10.1038/srep14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Won E.Y., Yoon M.K., Kim S.W., Jung Y., Bae H.W., Lee D. Gender-specific metabolomic profiling of obesity in leptin-deficient ob/ob mice by 1H NMR spectroscopy. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bernauer U., Amberg A., Scheutzow D., Dekant W. Biotransformation of 12C- and 2-13C-labeled methyl tert-butyl ether, ethyl tert-butyl ether, and tert-butyl alcohol in rats: identification of metabolites in urine by 13C nuclear magnetic resonance and gas chromatography/mass spectrometry. Chemical Research in Toxicology. 1998;11(6):651–658. doi: 10.1021/tx970215v. [DOI] [PubMed] [Google Scholar]
- 169.McGregor D. Tertiary-butanol: a toxicological review. Critical Reviews in Toxicology. 2010:697–727. doi: 10.3109/10408444.2010.494249. [DOI] [PubMed] [Google Scholar]
- 170.Dai L., Peng C., Montellier E., Lu Z., Chen Y., Ishii H. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature Chemical Biology. 2014;10(5):365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 171.Huang H., Tang S., Ji M., Tang Z., Shimada M., Liu X. p300-Mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Molecular Cell. 2018;70(4):663–678. doi: 10.1016/j.molcel.2018.04.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gaffney D.O., Jennings E.Q., Anderson C.C., Marentette J.O., Shi T., Schou Oxvig A.-M. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chemical Biology. 2019:1–8. doi: 10.1016/j.chembiol.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.O'Neill L.A.J., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nature Reviews Immunology. 2016:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jung J., Zeng H., Horng T. Metabolism as a guiding force for immunity. Nature Cell Biology. 2019:85–93. doi: 10.1038/s41556-018-0217-x. [DOI] [PubMed] [Google Scholar]
- 176.Izzo L.T., Wellen K.E. Histone lactylation links metabolism and gene regulation. Nature. 2019:492–493. doi: 10.1038/d41586-019-03122-1. [DOI] [PubMed] [Google Scholar]
- 177.Huang H., Lin S., Garcia B.A., Zhao Y. Quantitative proteomic analysis of histone modifications. Chemical Reviews. 2015;115(6):2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ruiz-Andres O., Sanchez-Niño M.D., Cannata-Ortiz P., Ruiz-Ortega M., Egido J., Ortiz A. Histone lysine crotonylation during acute kidney injury in mice. Disease Models and Mechanisms. 2016;9(6):633–645. doi: 10.1242/dmm.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]