Abstract
SARS‐CoV‐2 pandemic and recurrent dengue epidemics in tropical countries have turned into a global health threat. While both virus‐caused infections may only reveal light symptoms, they can also cause severe diseases. Here, we review the possible antibody‐dependent enhancement (ADE) occurrence, known for dengue infections, when there is a second infection with a different virus strain. Consequently, preexisting antibodies do not neutralize infection, but enhance it, possibly by triggering Fcγ receptor‐mediated virus uptake. No clinical data exist indicating such mechanism for SARS‐CoV‐2, but previous coronavirus infections or infection of SARS‐CoV‐2 convalescent with different SARS‐CoV‐2 strains could promote ADE, as experimentally shown for antibodies against the MERS‐CoV or SARS‐CoV spike S protein. © 2020 International Society for Advancement of Cytometry
Keywords: coronavirus, 2019‐nCoV, SARS‐CoV‐2, COVID‐19, antibody‐dependent enhancement
SARS‐CoV‐2 pandemic and recurrent dengue epidemics in tropical countries are a global health threat. Both virus‐caused infections may only reveal light symptoms, but also cause severe diseases. As known for Dengue infections, Antibody‐Dependent Enhancement (ADE) can occur during a second infection with a different virus strain: Preexisting antibodies do not neutralize but enhance infection, possibly by triggering FcgR‐mediated virus uptake. Thus, previous coronavirus infections or infections with different SARS‐CoV‐2 strains could promote ADE and aggravate COVID‐19.
The new SARS‐CoV‐2 coronavirus pandemic has actually over 6.3 million confirmed cases and an uncertain but presumably much higher dark figure resulting from symptom free course or lack of analysis due to limited resources or reagents. Depending on the estimations so far, there may be globally clearly over 12 million infected persons, still climbing up. It is impossible to say at this point what percentage of the human population will finally have encountered a virus, but it will be a substantial proportion. All these individuals will have then developed immunity against one or more strains of the virus. The mortality rate of SARS‐CoV‐2 pandemic is so far clearly below that of the outbreaks of the coronaviruses SARS‐CoV or MERS‐CoV, and the vast majority of infected have already recovered from the disease. However, SARS‐CoV‐2 is transmitted more easily, infecting by far higher number of people than the two other coronavirus (CoV) (1).
In the last decade, dengue fever outbreaks had a comparable number of infected, several tens of millions. Most of them recovered and presented an adaptive immunity against the virus. Here, we will briefly discuss effects when individuals with an anti‐dengue antibody (Ab) are re‐infected by the dengue virus and relate the findings with the present SARS‐CoV‐2 pandemic. Our incentive is to discuss features in antibody‐dependent enhancement (ADE) already known for dengue and discuss them in regards of perspectives of SARS‐CoV‐2 infections.
Dengue and Ace
Dengue viruses (DENV) are members of the Flaviviridae family and present four serotypes (DENV‐1, DENV‐2, DENV‐3, and DENV‐4) (2). They are capable of causing classic dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS) and are the leading cause of child death in some countries (3). Commonly, DF presents only mildly symptoms, including fever, headache, rash, abdominal pain, and nausea. Basically, after inoculation by the mosquito, local dendritic cells and macrophages are infected, followed by virus entry into the bloodstream and infection of other blood cells. This context results in leukopenia and thrombocytopenia in laboratory tests of patients (4, 5). After some days, humoral and cellular immune responses are efficiently mounted against the virus, eliminating infection. This humoral response produces protective serotype‐specific antibodies. These antibodies cross‐react, but do not neutralize other virus subtypes, failing to offer protective immunity against them (6).
A second infection, with other DENV serotypes, can be more severe and lethal than the first one. Commonly, DHF and DSS occur in this context, presenting more severe forms of symptoms, such as fever, thrombocytopenia, hemorrhagic manifestations, and hypovolemia (7). Studies have demonstrated that the presence of cross‐reactive antibodies against different DENV serotypes predisposes the enhanced illness (8) and contributes to the development of DHF and DSS (9). These non‐neutralizing preexisting antibodies can be obtained from previous infection, maternal passive immunity, or vaccination.
IgG antibodies against the specific DENV serotype could cross the placenta and enter into the blood to fetuses, resulting in a detrimental immune response against other serotypes after the birth. In fact, children with passive immunity from immunized mothers tend to present DHF during their first DENV infection (10, 11). Similar to natural infection and passive‐acquired immunity, vaccines against one specific serotype produce cross‐reactive non‐neutralizing antibodies against other serotypes, predisposing the enhanced illness in secondary heterotypic infection (12). In order to overcome this harmful effect, the tetravalent live‐attenuated vaccine was produced from chimeric structures (13). This vaccine produces protective neutralizing antibodies (NAbs) against the four serotypes and has been administered in endemic areas of 20 countries (14). However, the vaccine presented adverse effects in certain groups, enhancing illness. For this reason, vaccinating is now recommended for seropositive subjects aging between 9 and 45 years (14).
The phenomenon, in which preexisting non‐neutralizing antibodies lead to enhanced infection, is termed ADE. Beyond studies with patients suggesting this phenomenon in DENV infection, highlighting those described above with newborn infants and children (10, 11, 15), studies in vitro and with animal model also suggest ADE upon secondary DENV infections. Growth curves of DENV in vitro with peripheral blood leukocytes from non‐immunized and immunized animals indicated that preexisting antibodies play a role in ADE (16). Similarly, rhesus monkeys,which had received DENV‐immune cord blood sera, presented higher viremias compared to animals that had received non‐immune sera. Such data also suggest ADE in secondary DENV infections (17).
In DENV‐ADE, non‐NAbs bind DENV, and this complex is recognized and internalized by Fcγ receptor (FcγR)‐bearing cells, resulting in increased virus load and possibly enhanced illness. In fact, FcγRIIA‐expressing BHK cells cultured with sera from patients after secondary infection presented 10‐fold higher virus titers compared to cells without this receptor (18). Moreover, in ADE‐DENV mediated by FcγRs, there was a decrease of antiviral type‐I interferon (IFN) and IFN‐stimulated gene, such as interferon regulatory factor 1 (IRF‐1), NOS2, RIG‐1, and MDA‐5, whereas IL‐6 and IL‐10 levels increased (19, 20). These alterations in levels of cytokines and molecules of antiviral response play a role in DENV‐enhanced illness triggered by ADE.
Potential Consequences Regarding the New Coronavirus Pandemic SARS‐CoV‐2 and COVID‐19
The critical question is, whether ADE is relevant in SARS‐CoV‐2 infection and COVID‐19. Would morbidity and mortality increase in individuals with immunity against one SARS‐CoV‐2 strain when becoming infected a second time with the same or another virus strain? Could individuals vaccinated with an active vaccine, who had been previously “naïve” to SARS‐CoV‐2 develop ADE when infected? There are epidemiologic similarities between dengue fever and SARS‐CoV‐2 infection. For both, fatalities substantially increase in adults >65 years of age (at least during the primary infection), and for most infected the disease course is asymptomatic or displays only mild symptoms so that the number of infected is generally underestimated.
Coronaviruses belong to the Coronavirida family (Orthocornavirinae subfamily) and are RNA viruses like the dengue virus. Specific for Coronaviridae is their “corona” of spikes (S) that has the function of docking to specific receptors on the host cell and inducing the entry and thereafter the replication of the virus. Coronaviruses can be grouped into at least seven strains (21) with SARS‐CoV‐2 as the latest member of this family. Most of these strains induce common cold, but three (SARS‐CoV, MERS‐CoV, SARS‐CoV‐2) may induce more severe disease courses.
Spike (S) proteins of SARS‐CoV and SARS‐CoV‐2 are binding with the receptor‐binding domain (RBD) to the enzymatic domain of angiotensin‐converting enzyme 2 (ACE2) on the host cell surface. The S protein is then hydrolyzed by transmembrane protease, serine 2 (TMPRSS2) and can enter the host cell. Data from the two SARS‐CoV outbreaks in 2003 and 2004 indicate that infectivity and disease severity are positively correlated with RBD/ACE2 binding affinity (22, 23, 24). Presently, the RBD binding affinity of SARS‐CoV‐2S compared to SARS‐CoV S is not unambiguous, ranging from10‐fold to 20‐fold higher for SARS‐CoV‐2 S (25) to similar to SARS‐CoV S (26). CD147 has also been identified as second S‐binding receptor for SARS‐COV‐2 cell entry (reviewed in ref. (27)).
Wrapp and co‐workers (25) conclude from results using biolayer interferometry that vaccines or neutralizing antibodies (NAbs) developed against SARS‐CoV may not be appropriate for SARS‐CoV‐2. Walls and co‐workers (26) in contrast demonstrated in a cell culture model that NAbs derived from SARS‐CoV immunized mice prevented SARS‐CoV‐2 pseudovirus entry. Even though, mutations at critical sites of the RBD domain could lead to an increasing binding affinity to ACE2 (22) and/or CD147, and already existing NAbs in seropositive individuals could bind with reduced affinity to the RBD domain. However, at this stage, the relevance of ADE in SARS‐CoV‐2 infection is speculative (28, 29), but we can learn from SARS‐CoV and MERS‐CoV research of the last 20 years.
A possible mechanism for viral entry based on the ADE mechanism has been postulated (30). NAbs recognizing the RBD of the S protein of the MERS coronavirus bind to the Fc receptor and allow virus entry. The NAb‐Fc receptor complex would mimic the cell surface virus receptor and promote virus entry pathways into IgG Fc receptor‐expressing cells (30). If this was confirmed for SARS‐CoV‐2, the aim to block virus entry by neutralizing antibodies against S protein domains could have the opposite of the desired effect. Instead of blocking virus entry, cell infection would be facilitated, and the spectrum of infected cell types possibly extended. Such a mechanism needs to be considered for therapeutic antibody and vaccine design (30).
In theory, SARS‐CoV or MERS‐CoV survivors, who still carry CoV specific antibodies, could have been infected for a second time by an altered CoV. These antibodies may recognize regions of the S protein and induce a mechanism of virus entry, such as laid out above.
In line with such mechanism, cell entry of SARS‐CoV spike‐pseudo‐typed lentiviral particles was facilitated in cell lines expressing FcγRII, but not ACE2, in the presence of anti‐spike antisera from mice immunized with inactivated SARS‐CoV or S‐protein (31). The human HL‐CZ promyelotic cell line, co‐expressing ACE2 and FcγRII, was effectively infected by SARS‐CoV. Dilution of human or murine anti‐spike antisera or anti‐spike Abs increased viral replication in HL‐CZ significantly (32) indicating ADE. In infected Chinese Rhesus Macaques, anti‐spike IgG Nabs, but not control IgG induced fatal lung disease (33). All animals actively vaccinated with Ankara virus encoding full‐length SARS‐CoV S protein developed anti‐spike Nabs. Both, active vaccination or passive vaccination with an anti‐spike antibody, but not mock immunization, induced severe lung injury after SARS‐CoV infection. The authors also show elevated titer of IgG NAbs in deceased SARS‐CoV patients compared to recovered patients.
It is well known that IgG subclasses (IgG1, IgG2, IgG3, and IgG4) present varied affinities to different FcγR, resulting in different immune responses. In this context, experimental results with SARS‐CoV suggest that FcγRIIa and FcγRIIb, but not FcγRI and FcγRIII, promote ADE (31, 34). Moreover, polymorphisms in FcγRIIa correlated with SARS‐CoV severity (35). The allelic polymorphism, in a single G or A nucleotide, results in an arginine (R) or histidine (H) at residue 131 of the Ig binding domain of FcγRIIa. This polymorphism modifies receptor affinity and specificity to the binding of different IgG subclasses. It has been demonstrated that this polymorphism affects susceptibility or severity of several infections, including Streptococcus pneumoniae infections, dengue fever, HIV infection, and SARS infection (35, 36, 37, 38). In fact, FcγRIIa‐R131 polymorphism seems to have clinical implication in SARS‐CoV pathology and possibly ADE. Yuan and co‐workers availed 180 infected patients and 200 controls and observed a correlation between the FcγRIIa‐R/R131 (homozygotes) and severe SARS‐CoV (patients requiring treatment in an intensive care unit) (35). Thus, individual composition of cellular expression of polymorphic variants of FcγRII, which can vary with geographic region and ethnicity (39), could define susceptibility to SARS‐CoV, COVID‐19 severity, and ADE (34, 35).
While the probability of SARS‐CoV or MERS‐CoV recovered for a second infection was extremely low because of low prevalence of SARS‐CoV with roughly 8,000 cases worldwide (1, 40) and MERS‐CoV with ~2,000 cases, such a possibility is real in the case of SARS‐CoV‐2. Concurrently, our search did not yield any reliable epidemiological and clinical studies on CoV‐related ADE in humans; however, in case of secondary infection with a mutated virus, ADE cases might be observed (28). The authors of this article suggest that the concentration of severe cases of COVID‐19 in certain geographic areas results from ADE responses induced by antibodies developed against similar epitopes of local viruses. This interpretation conflicts with experimental results from Ou et al.(41), who found that antisera from recovered SARS or COVID‐19 patients had only limited cross neutralization effects of 293/hACE2 cells' infection with SARS‐CoV S or SARS‐CoV‐2 S pseudovirons.
Several studies with SARS‐CoV anti‐spike S‐protein antisera tested in cell lines or in laboratory animals indicate ADE (31, 32, 33). Furthermore, the immune response and production of anti‐spike NAbs are accelerated in patients with fatal outcome (42, 43). In addition, enhancement of viral uptake dependent on RBD specific NAbs as well as on FcγRII is reported in vitro in MERS‐CoV (24, 30).
In summary, in both SARS‐CoV and MERS‐CoV infected anti‐spike S‐protein NAbs are present. They can induce experimentally in vitro and in vivo ADE mediated by FcγRII receptor virus entry. Previous immunization by vaccination or infection might aggravate symptoms of a follow‐up infection as described above for dengue fever, such complications were also observed for common influence and other viral infections (24).
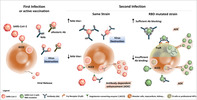
Are there indications that ADE might be relevant in the SARS‐CoV‐2 pandemic? By nature, at this time point, trustworthy proofs for ADE playing clinically a role are not available and might be expected earliest in fall of 2020. Even so, the potential threat of ADE has to be investigated more closely, particularly in view of the vast number of recovered individuals who have already developed immunity. Thus, ADE is of relevance for the strategies for vaccine development and therapeutic agents. For example, ADE can play a role in individuals with immunity and NAbs against strain A if the next infection is by RBD mutated strain B. Antistrain A NAbs may not neutralize strain B, or antibody titers are too low for sufficient neutralization, or NAbs have efficient neutralizing capabilities, but cell entry occurs via FcγRII into APCs or B cells (43) (see Fig. 1).
Figure 1.
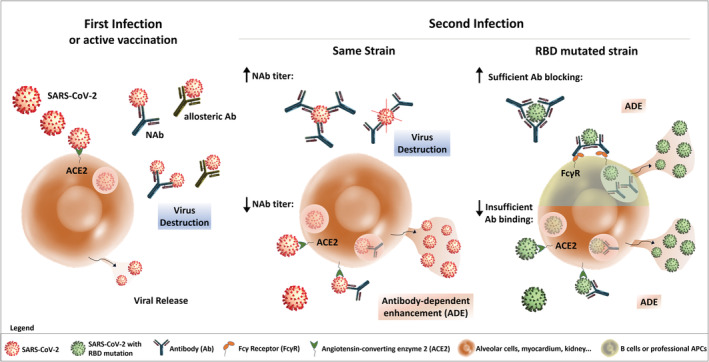
Potential pathways of aggravation of SARS‐CoV‐2 infection by ADE. Initially, the spike protein of SARS‐CoV‐2 binds through the RBD to angiotensin‐converting enzyme 2 (ACE2) on the host cell surface for virus invasion. After some days, humoral responses develop against the virus, eliminating infection through allosteric and neutralizing antibodies (NAbs). After a second infection with the same SARS‐CoV‐2 strain virus destruction may occur, if the NAb titer is high enough. However, in case of a low NAb titer, ADE may be observed following binding of the SARS‐CoV‐2/antibody complex to ACE2, internalization of the complex and IgG induced stimulation. In addition, immunized patients may be re‐infected with a different SARS‐CoV‐2 strain, such as a RBD‐mutated strain. In this context, already existing Abs could bind with reduced affinity to mutated RBD, inducing low levels of SARS‐CoV‐2/antibody complexes, following by internalization through the ACE2 receptor and ADE . On the other hand, immunized patients re‐infected by an RBD‐mutated strain may present sufficient Ab blockade of the heterotypic SARS‐CoV‐2 strain. In this case, SARS‐CoV‐2 covered by Abs may connect to Fcγ receptors II (FcγRII) on the surface of B cells or other professional antigen‐presenting cells (APCs). This receptor mediates SARS‐CoV‐2 invasion into immune cells, further spreading viral infection into all organs and ADE. [Color figure can be viewed at wileyonlinelibrary.com]
Yet unconfirmed studies (44, 45) performed early this year reported isolation of various SARS‐CoV‐2 strains from patients with mutations in the RBD. Some of these mutations apparently have increased RBD‐binding affinities. Furthermore, mutated strains seem to be globally unevenly distributed with focuses in distinct global regions. New studies show that all recovered SARS‐CoV‐2 patients (n = 29) presented IgG/IgM response to spike protein S1 and the titer of S1 specific IgG increases with age (46, 47). Some authors also speculated on the possibility that ADE was induced by SARS‐CoV‐2 in SARS‐CoV convalescent individuals (1, 28), although this view is not unanimously accepted (29), and antisera as well as monoclonal NAbs isolated from eight SARS‐CoV‐2 patients did not bind to SARS‐CoV or MERS‐CoV RBD (47).
The existence of ADE in SARS‐CoV‐2 might aggravate the COVID‐19 pandemic with higher morbidity and mortality, when convalescent patients will encounter infections in the future by possibly mutated strains (44, 45). However, the evidence for distinct types or strains in the present stage of evolution of SARS‐CoV‐2, which could account for differences in morbidity and RBD binding affinity, is conflicting (48).
ADE development should also be considered as possible adverse effects of hyperimmune globulin therapy and vaccine development. Duan and co‐workers (49) successfully used convalescent plasma collected from cured patients with a titer of 1:640 SARS‐CoV‐2‐NAbs for transfusion into COVID‐19 patients. While this study was successful in terms of abolishing viral load in 7 of 10 patients together with augmenting saturation by oxyhemoglobin, declining immune response, minimally increased lymphocyte count and partially reduced lung lesions (49), the efficacy is unclear as no control treatment was included.
On the other hand, hyperimmune globulin and vaccine development may carry the risk of ADE, as discussed in detail by de Alwis and co‐workers (50). Obviously, adaptative immune responses depend on the vaccine antigen load. NAbs of convalescent plasma from recovered COVID‐19 patients are supposedly different from those produced as consequence of vaccine presentation. NAbs are produced under proinflammatory conditions, which does not necessarily occur under vaccination conditions.
In summary, while innate immune responses could be caused by vaccination, uncontrolled immune responses and ADE are much more probable upon transfusion of hyperimmune globulin. The study of Duan and co‐workers (49) needs to be extended by robust clinical data from double‐blinded studies with control treatments for confirming or not the safety of transfer of convalescent plasma obtained from recovered patients.
Although it is too early to accept or reject the hypothesis and clinical relevance of ADE in SARS‐CoV‐2, actual research in clinical diagnosis and vaccine and therapy development needs to consider such possibility. Even after nearly 20 years of vaccine research for SARS‐CoV and 10 years for MERS‐CoV, there are still not any approved vaccines and antiviral treatments available (51), particularly due to severe complications encountered during animal testing (24).
What Cytometry Can and Needs to Do
What can be learned from dengue fever and ADE is that carefully designed tests are at need to perform informative clinical diagnosis and reliable quantitative and reproducible studies for basic science as well as vaccine and drug development and diagnostic assays to fight the pandemic. Cytometry has been an important technology to facilitate investigation in SARS‐CoV and MERS‐CoV as well as in dengue research and diagnosis.
Flow and image cytometry are an important part of this endeavor. Quantitative flow cytometry assays for primary cells and cell culture experiments for testing mechanisms of antisera, NAbs and virus attachment and entry have been already applied for different coronaviruses (32, 41, 47). Measuring cell and cell systems' responses after viral challenge including apoptosis induction and cytokine production (52) as well as multiplexed polychromatic single‐cell analysis are important for characterizing cell phenotypes, immune responses and cytokine production and their relation to disease course and severity (52, 53, 54). Such studies have already started and are urgently needed for a deeper understanding of the underlying immune responses and early identification of patients with severe disease course, following up therapeutic interventions. They are important for decision‐making, which part of the population would profit from active vaccination and which would be at risk to develop ADE. Sorting of RB‐specific memory B cells for the production of therapeutic monoclonal antibodies (47) is an important contribution to vaccine development.
New protocols need to be developed and experiments performed applying best practices for performing carefully reproducible flow cytometry under safety conditions (52, 55). Because of risk of infection, many research groups are not capable of performing necessary flow cytometric analysis and sorting of the virus (33, 56) and are often restricted to the counting of fixed virus particles (57). In the past, virus size and limitations of staining were a limiting problem for most flow cytometers. In view of that, an alternative method was the coupling of fluorescence emitting reporter coding to virus particles for experiments on virus infection in cell culture (58).
Interestingly, a novel application of flow cytometry, flow virometry, benefits from improving staining methods and technical advances of flow cytometers, which are capable of analyzing very small virus particles, including maturating dengue virions (59, reviewed in ref. 60).
Protocols for biosafety guidelines for flow laboratories and equipment protecting the personal and environment have to be applied and existing guidelines possibly adapted to the particular situation (53, 55, 56). Flow and image cytometry is of importance for the analysis of cells and tissues of SARS‐CoV‐2 infected humans and laboratory animals. So far many of the studies qualitatively analyzed microscopic images (32), but also semi‐automated and automated image analysis was performed for virus characterization (31, 33). As ACE2 is expressed in many organs of the human body, including intestinal tract, kidney, heart and brain (61), infection of these organs is likely. Quantitative image cytometry is important for supporting histopathology research and better understanding and treatment of this new pandemic.
Conflict of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgments
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; Project No. 2018/08426–0).
All authors contributed equally to the manuscript.
Literature Cited
- 1. Raoult D, Zumla A, Locatelli F, Ippolito G, Kroemer G. Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses. Cell Stress 2020;4:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Diamond MS, Pierson TC. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 2015;162(3):488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . Dengue Haemorrhagic Fever: Diagnosis, Treatment Prevention and Control. Geneva: WHO, 1997. https://www.who.int/csr/resources/publications/dengue/Denguepublication/en/. [Google Scholar]
- 4. Kalayanarooj S, Vaughn DW, Nimmannitya S, Green S, Suntayakorn S, Kunentrasai N, Viramitrachai W, Ratanachu‐eke S, Kiatpolpoj S, Innis BL, et al. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis 1997;176(2):313–321. [DOI] [PubMed] [Google Scholar]
- 5. Kuo CH, Tai DI, Chang‐Chien CS, Lan CK, Chiou SS, Liaw YF. Liver biochemical tests and dengue fever. Am J Trop Med Hyg 1992;47(3):265–270. [DOI] [PubMed] [Google Scholar]
- 6. Halstead SB. Dengue antibody‐dependent enhancement: Knowns and unknowns. Microbiol Spectr 2014;2(6):1–18. [DOI] [PubMed] [Google Scholar]
- 7. Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 1998;11(3):480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morens DM, Venkateshan CN, Halstead SB. Dengue 4 virus monoclonal antibodies identify epitopes that mediate immune infection enhancement of dengue 2 viruses. J Gen Virol 1987;68(Pt1):91–98. [DOI] [PubMed] [Google Scholar]
- 9. Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody‐dependent enhancement of infection. Arch Virol 2013;158(7):1445–1459. [DOI] [PubMed] [Google Scholar]
- 10. Halstead SB, Lan NT, Myint TT, Shwe TN, Nisalak A, Kalyanarooj S, Nimmannitya S, Soegijanto S, Vaughn DW, Endy TP. Dengue hemorrhagic fever in infants: Research opportunities ignored. Emerg Infect Dis 2002;8(12):1474–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simmons CP, Chau TN, Thuy TT, Tuan NM, Hoang DM, Thien NT, Lien Le B, Quy NT, Hieu NT, Hien TT, et al. Maternal antibody and viral factors in the pathogenesis of dengue virus in infants. J Infect Dis 2007;196(3):416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taylor A, Foo SS, Bruzzone R, Dinh LV, King NJ, Mahalingam S. Fc receptors in antibody‐dependent enhancement of viral infections. Immunol Rev 2015;268(1):340–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halstead SB. Safety issues from a phase 3 clinical trial of a live‐attenuated chimeric yellow fever tetravalent dengue vaccine. Hum Vaccin Immunother 2018;14(9):2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. WHO . Immunization, Vaccines and Biological: Questions and Answers on Dengue Vaccines 2018. http://www.who.int/immunization/research/development/dengue_q_and_a/en/
- 15. Kliks SC, Nimmanitya S, Nisalak A, Burke DS. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg 1988;38(2):411–419. [DOI] [PubMed] [Google Scholar]
- 16. Marchette NJ, Halstead SB, Chow JS. Replication of dengue viruses in cultures of peripheral blood leukocytes from dengue‐immune rhesus monkeys. J Infect Dis 1976;133(3):274–282. [DOI] [PubMed] [Google Scholar]
- 17. Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis 1979;140(4):527–533. [DOI] [PubMed] [Google Scholar]
- 18. Moi ML, Lim CK, Takasaki T, Kurane I. Involvement of the fc gamma receptor IIA cytoplasmic domain in antibody‐dependent enhancement of dengue virus infection. J Gen Virol 2010;91(Pt1):103–111. [DOI] [PubMed] [Google Scholar]
- 19. Khandia R, Munjal A, Dhama K, Karthik K, Tiwari R, Malik YS, Singh RK, Chaicumpa W. Modulation of dengue/Zika virus pathogenicity by antibody‐dependent enhancement and strategies to protect against enhancement in Zika virus infection. Front Immunol 2018;9:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolph MS, Zaid A, Rulli NE, Mahalingam S. Downregulation of interferon‐β in antibody‐dependent enhancement of dengue viral infections of human macrophages is dependent on interleukin‐6. J Infect Dis 2011;204(3):489–491. [DOI] [PubMed] [Google Scholar]
- 21. Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses 2010;2(8):1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS coronavirus. J Virol. 2020;94(7):e0017–20. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, et al. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J 2005;24(8):1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ricke DO, Malone RW. Medical countermeasures analysis of 2019‐nCoV and vaccine risks for antibody dependent enhancement (ADE). Preprints (www.preprints.org) 2020. doi:10.20944/preprints 202,003.0138.v1.
- 25. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 2020;181(2):281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulrich H, Pillat MM. CD147 as a target for COVID‐19 treatment: Suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep 2020;20:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tetro JA. Is COVID‐19 receiving ADE from other coronaviruses? Microbes Infect 2020;22(2):72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma A. It is too soon to attribute ADE to COVID‐19. Microbes Infect. 2020;22(2):72–73.32092539 [Google Scholar]
- 30. Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular mechanism for antibody‐dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015–19. 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jaume M, Yip MS, Cheung CY, Leung HL, Li PH, Kien F, Dutry I, Callendret B, Escriou N, Altmeyer R, et al. Anti‐severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH‐ and cysteine protease‐independent FcγR pathway. J Virol 2011;85(20):10582–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YM, et al. Antibody‐dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun 2014;451(2):208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. Anti‐spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS‐CoV infection. JCI Insight. 2019;4(4):e123158. 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID‐19. Nate Rev Immunol 2020;20:339–341. 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuan FF, Tanner J, Chan PKS, Biffin S, Dyer WB, Geczy AF, Tang JW, Hui DSC, Sung JJY, Sullivan JS, et al. Influence of FcγRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens 2005;66:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan FF, Wong M, Pererva N, Keating J, Davis AD, Bryant JA, Sullivan JS. FcgRIIA polymorphisms in Streptococcus pneumoniae infection. Immunol Cell Biol 2003;81:192–195. [DOI] [PubMed] [Google Scholar]
- 37. Loke H, Bethell D, Phuong CX, Day N, White N, Farrar J, Hill A. Susceptibility to dengue hemorrhagic fever in Vietnam: Evidence of an association with variation in the vitamin D receptor and fc receptor IIA genes. Am J Trop Med Hyg 2002;67:102–106. [DOI] [PubMed] [Google Scholar]
- 38. Brouwer KC, Lal RB, Mirel LB, Yang C, Eijk AM, Ayisi J, Otieno J, Nahlen BL, Steketee R, Lal AA, et al. Polymorphism of fc receptor Iia for IgG in infants is associated with susceptibility to perinatal HIV‐1 infection. AIDS 2004;18:1187–1194. [DOI] [PubMed] [Google Scholar]
- 39. de Taeye SW, Rispens T, Vidarsson G. The ligands for human IgG and their effector functions. Antibodies (Basel) 2019;8(2):E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. https://www.who.int/csr/sars/country/table2004_04_21/en/
- 41. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang L, Zhang F, Yu W, He T, Yu J, Yi CE, Ba L, Li W, Farzan M, Chen Z, et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol 2006;78(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fu Y, Cheng Y, Wu Y. Understanding SARS‐CoV‐2‐mediated inflammatory responses: From mechanisms to potential therapeutic tools. Virol Sin. 2020;3. 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ou J, Zhou Z, Zhang J, Lan W, Zhao S, Wu J, Seto D, Zhang G, Zhang Q. RBD mutations from circulating SARS‐CoV‐2 strains enhance the structure stability and 1 infectivity of the spike protein. bioRxiv preprint, 2020. 10.1101/2020.03.15.991844. [DOI]
- 45. Jin X, Xu Kangli, J Penglei, L Jiangshan, H Shaorui, J Hongyu, Y Hangping, Z Yimin, Z Ruoheng, C Dong, Y Jinmei et al. Virus strain of a mild COVID‐19 patient in Hangzhou represents a new trend in SARS‐CoV‐2 evolution related to Furin cleavage site. medRxiv preprint, 2020. 10.1101/2020.03.10.20033944. [DOI] [PMC free article] [PubMed]
- 46. Jiang H‐W, Li Y, Zhang H‐N, Wang W, Men D, Yang X, Qi H, Zhou J, Tao J‐C. Global profiling of SARS‐CoV‐2 specific IgG/ IgM responses of convalescents using a proteome microarray. medRxiv preprint (not peer‐reviewed). 2020. 10.1101/2020.03.20.20039495. [DOI] [PMC free article] [PubMed]
- 47. Ju B, Zhang Q, Ge X, Wang R, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, et al. Potent human neutralizing antibodies elicited 1 by SARS‐CoV‐2 infection. 2020. BioRxiv preprint (not peer‐reviewed). 10.1101/2020.03.21.990770. [DOI] [PubMed]
- 48. MacLean OA, Orton RJ, Singer JB, Robertson DL. No evidence for distinct types in the evolution of SARS‐CoV‐2. Virus Evolution 2020;6(1):veaa034. 10.1093/ve/veaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, et al. Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proc Natl Acad Sci U S A 2020;117(17):9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Alwis R, Chen S, Gan ES, Ooi EE. Impact of immune enhancement on Covid‐19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine 2020;55:102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh PK, Chaicumpa W. COVID‐19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother 2020;1–7. 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C. Modena Covid‐19 Working Group (MoCo19)#. SARS‐CoV‐2, the virus that causes COVID‐19: Cytometry and the new challenge for global health. Cytometry A 2020;97A(4):340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cossarizza A, Gibellini L, De Biasi S, Lo Tartaro D, Mattioli M, Paolini A, Fidanza L, Bellinazzi C, Borella R, Castaniere I, et al. Handling and processing of blood specimens from patients with COVID‐19 for safe studies on cell phenotype and cytokine storm. Cytometry A 2020. 10.1002/cyto.a.24009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng H, Zhang M, Yang C, Zhang N, Wang X, Yang X, Dong X, Zheng Y. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID‐19 patients. Cell Mol Immunol 2020;17:541–543. 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez PA, Hulspas R. Special issue on enhancement of reproducibility and rigor. Cytometry A 2020;97A(2):105–106. [DOI] [PubMed] [Google Scholar]
- 56. Perfetto S, Reifel K, Swan B, Jellison E, Ambrozak D, Baijer J, Nguyen R, Monard S, Lyon G, Fontes B. Procedures for flow cytometry‐based sorting of unfixed SARS‐CoV‐2 infected cells and other infectious agents. Cytometry A 2020; (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brussaard CP, Marie D, Bratbak G. Flow cytometric detection of viruses. J Virol Methods 2000;85(1–2):175–182. [DOI] [PubMed] [Google Scholar]
- 58. McSharry JJ. Analysis of virus‐infected cells by flow cytometry. Methods 2000;21(3):249–257. [DOI] [PubMed] [Google Scholar]
- 59. Zicari S, Arakelyan A, Fitzgerald W, Zaitseva E, Chernomordik LV, Margolis L, Grivel JC. Evaluation of the maturation of individual dengue virions with flow virometry. Virology 2016;488:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lippé R. Flow Virometry: A powerful tool to functionally characterize viruses. J Virol 2018;92(3):e01765–e01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The human Protein Atlas. Available online at: https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue


