To the Editor,
Real‐time reverse transcription polymerase chain reaction (RT‐PCR) is the mainstay of coronavirus disease 2019 (Covid‐19) diagnosis. 1 Up to 30% of the patients clinically suspected of Covid‐19 may have initial or repeat RT‐PCR negative results before positive test conversion, most notably when upper respiratory tract (URT) specimens are processed. 2 , 3 , 4 , 5 , 6 , 7 False‐negative RT‐PCR results may hamper the clinical management of patients and hinder the adoption of epidemiological measures to control the pandemic. A number of pre‐analytical and analytical factors may impact on the diagnostic efficiency of RT‐PCR, including the type of and time to specimen processing, conservation before testing, quality of samples, the timing of sample collection after symptoms onset, or the intrinsic performance of the assay (ie, limit of detection [LOD]). 2 , 8 A large number of commercially available severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RT‐PCR assays targeting one or more SARS‐CoV‐2 genes has been launched and have reached global widespread use. 9 Most of these assays include a spike‐in control for RT‐PCR amplification, such as MS2 phage RNA genome, but provide no information on specimen cellularity, as no primers targeting human housekeeping genes (ie, RNase P) are usually included in the reaction. The current study was aimed at assessing whether amplification of β‐glucuronidase (GUSB) gene would help estimate the accuracy of SARS‐CoV‐2 RT‐PCR negative results in URT samples.
As shown in Figure 1, out of 401 patients admitted to our center until 14th April with clinical suspicion of Covid‐19, 202 were eventually diagnosed by microbiological means, either by RT‐PCR (n = 191) or serological methods (n = 11). The median age of these patients was 65 years (range, 3‐98 years); 115 were males and 87 females. A total of 199 patients received a final diagnosis of Covid‐19 on clinical, laboratory, and imaging grounds and without microbiological documentation.
Figure 1.
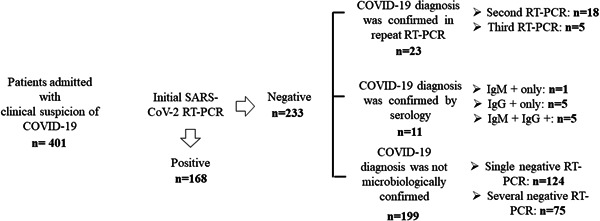
SARS‐CoV‐2 RT‐PCR and serology testing results in patients with clinical suspicion of Covid‐19 admitted to Hospital Clínico Universitario of Valencia during the study period. Nasopharyngeal or oropharyngeal swabs were obtained with flocked swabs in universal transport medium (Beckton Dickinson, Sparks, MD or Copan Diagnostics, Murrieta, CA) and conserved at 4°C until processed (within 6 hours). Nucleic acid extraction was performed using the Qiagen EZ1 Viral extraction kit or the DSP virus Pathogen Minikit on the EZ1 or QiaSymphony Robot instruments (Qiagen, Valencia, CA), respectively. Commercially available PCR assays used for SARS‐CoV‐19 testing included the LightMix Modular SARS‐CoV‐2 (COVID‐19) E‐gene/LightMix Modular SARS‐CoV‐2 (COVID‐19) RdRP gene from TIB MOLBIOL GmHD, distributed by Roche Diagnostics (Pleasanton, CA) on the Light Cycler 2.0 instrument, the SARS‐CoV‐2 Real‐time PCR Kit from Vircell Diagnostics (Granada, Spain), or the Realquality RQ‐2019‐nCoV from AB Analitica (Padua, Italy), both on the Applied Biosystems 7500 instrument and the SARS‐CoV‐2 (S gene)–BD Max System (Viasure Real‐Time PCR Detection Kits; CerTest, Zaragoza, Spain). Results were interpreted according to the respective manufacturer's instructions. Sera from these patients were drawn at a median of 12 days (range, 10‐21 days) after admission. The presence of either SARS‐CoV‐2 IgM (determined by the MAGLUMI 2019‐nCoV SARS‐CoV‐2‐ IgM assay on the fully automated Maglumi analyzers‐Snibe—Shenzhen New Industries Biomedical Engineering Co, Ltd, Shenzhen, China), IgG (Euroimmun anti‐SARS‐CoV‐2 IgG assay; Euroimmun, Luebeck, Germany), or both confirmed diagnosis of Covid‐19. Covid‐19, coronavirus disease; IgG, immunoglobulin G; IgM, immunoglobulin M; RT‐PCR, real‐time reverse transcription polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Of the 202 patients, 34 (16.8%) tested negative by RT‐PCR on first URT specimens, collected at a median of 5 days (range, 1‐14 days) after the onset of symptoms. In these patients, as per protocol, URT swabs were collected every 24 to 72 hours until RT‐PCR positive conversion. Twenty‐three patients tested positive by RT‐PCR in the second (n = 18) or third (n = 5) URT sample. Diagnosis of Covid‐19 was achieved by serological methods in the remaining 11 patients.
A total of 47 URT specimens from 21 patients testing positive by RT‐PCR in the second or third specimen were subjected to GUSB gene RT‐PCR analysis, which was performed in a parallel to viral RT‐PCR testing. Twenty‐six and 21 out of the 47 specimens yielded negative or positive SARS‐CoV‐2 RT‐PCR results, respectively. To this end, we used the HEQC one‐step kit (Seqplexing, Valencia, Spain), a one‐step real‐time RT‐PCR. RNA was extracted from clinical samples using the DSP virus Pathogen Minikit on the QiaSymphony Robot instruments (Qiagen, Valencia, CA), reverse‐transcribed to complementary DNA and subsequently amplified in the LightCycler 480 Real‐Time PCR System Version II (Roche Diagnostics, Pleasanton). Cy5 fluorescent signal (618‐660 nm) revealed amplification of the target gene.
URT specimens that tested negative by SARS‐CoV‐2 RT‐PCR displayed higher GUSB RT‐PCR cycle thresholds (C T) (P = .070; the Mann‐Whitney U test) than those testing positive (median, 30.7; range, 27.0‐40.0, and median 29.7; range 25.5‐36.8, respectively), thus reflecting poorer cellularity. Receiver operating characteristic (roc) curve analysis (not shown) indicated that a C T threshold of 31.2 discriminated best between positive and negative SARS‐CoV‐2 RT‐PCRs (area under the curve, 0.66; 95% CI, 0.50‐0.81; P = .08). This cut‐off yielded a true negative ratio of 89% and an accuracy of 70% (Table 1).
Table 1.
Accuracy of negative SARS‐CoV‐2 RT‐PCR results in upper respiratory tract specimens from patients with microbiological diagnosis of Covid‐19 upon β‐glucuronidase gene RT‐PCR C T value
C T, RT‐PCR cycle threshold.
The Realquality RQ‐2019‐nCoV was used in seven specimens. The LightMix Modular SARS‐CoV‐2 (COVID‐19) E‐gene/LightMix Modular SARS‐CoV‐2 (COVID‐19) RdRP gene was used in six specimens. The SARS‐CoV‐2 Real‐Time PCR Kit was used in four specimens. The SARS‐CoV‐2 (S gene)–BD Max System (Viasure Real‐Time PCR Detection Kits) was used in four specimens.
The LightMix Modular SARS‐CoV‐2 (COVID‐19) was used in 17 specimens. The Realquality RQ‐2019‐nCoV was used in six specimens. The SARS‐COV‐2 Real‐Time PCR Kit was used in three specimens.
The current study has several limitations that should be acknowledged. First, a relatively scarce number of specimens were subjected to GUSB gene analysis. Second, consecutive specimens from a given patient could have been tested by different SARS‐CoV‐2 RT‐PCR assays displaying distinct LOD. Third, regardless of the specimen cellularity, SARS‐CoV‐2 load may have been intrinsically lower in patients testing negative than in those testing positive, although this possibility seems unlikely given the dynamics of viral load in URT specimens during the course of Covid‐19, which peaks within the first week after onset of symptoms. 10 , 11 In this sense, as stated, first and second SARS‐CoV‐2 RT‐PCR negative specimens were usually collected within this time frame. In summary, our data suggested that amplification of the GUSB gene by RT‐PCR may help to appraise the accuracy of negative SARS‐CoV‐2 RT‐PCR results on nasopharyngeal or oropharyngeal swabs from patients in whom Covid‐19 is eventually diagnosed. Further studies are warranted to validate this assumption.
CONFLICT OF INTERESTS
The authors declare no conflicts of interests.
AUTHOR CONTRIBUTIONS
EA, BF, IT, AS, and CS performed the GUSB gene RT‐PCR analyses and designed the study. MJA performed the serological tests. JB, JC, FB, DH, BO, and AV performed SARS‐CoV‐2 RT‐PCR tests. DN designed the study, analyzed the data, and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
ACKNOWLEDGMENTS
No public or private funds were used for the current study. We are grateful to all personnel working at Microbiology Service of Clinic University Hospital for their unwavering commitment in the fight against Covid‐19. We are indebted to all colleagues who attended the patients, in particular to Josep Redón, María Luisa Blasco, Jaime Signes‐Costa, María José Galindo, and María José Forner.
REFERENCES
- 1. World Health Organization (2020). Laboratory testing strategy recommendations for COVID‐19: interim guidance, 21 March 2020. World Health Organization. WHO/2019‐nCoV/lab_testing/2020.1.
- 2. Arevalo‐Rodriguez I, Buitrago‐Garcia D, Simancas‐Racines D, et al. False‐negative results of initial RT‐PCR assays for Covid‐19: a systematic review. MedRxiv Preprint. 2020. 10.1101/2020.04.16.20066787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing 448 in coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;Feb 26:200642. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernheim A, Mei X, Huang M, et al. Chest CT findings in 451 coronavirus disease‐19 (COVID‐19): relationship to duration of infection. Radiology. 2020;295:200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID‐19: comparison to RT‐PCR. Radiology. 2020;Feb 19:200432. 10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Y, Yao L, Li J, et al. Stability issues of RT‐PCR testing of SARS‐CoV‐2 for hospitalized patients clinically diagnosed with COVID‐19. J Med Virol. 2020;Mar 26:jmv.25786. 10.1002/jmv.25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winichakoon P, Chaiwarith R, Liwsrisakun C, et al. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID‐19. J Clin Microbiol. 2020;58(5):e00297‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lowe CF, Matic N, Ritchie G, et al. Detection of low levels of SARS‐CoV‐2 RNA from nasopharyngeal swabs using three commercial molecular assays. J Clin Virol. 2020;128:104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Find Evaluation Update : SARS‐COV‐2 Molecular Diagnostics. https://www.finddx.org/covid-19/sarscov2-eval-molecular/. Accessed April 24, 2020.
- 10. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect Dis. 2020;20:656‐657. pii: S1473‐3099(20)30232‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]


