Abstract
Background
The SARS-CoV-2 is the etiological agent causing COVID-19 which has infected more than 2 million people with more than 200000 deaths since its emergence in December 2019. In the majority of cases patients are either asymptomatic or show mild to moderate symptoms and signs of a common cold. A subset of patients, however, develop a severe atypical pneumonia, with the characteristic ground-glass appearance on chest x-ray and computerized tomography, which evolves into an acute respiratory distress syndrome, that requires mechanical ventilation and eventually results in multiple organ failure and death. The Molecular pathogenesis of COVID-19 is still unknown.
Aim of the study
In the present work we performed a stringent metanalysis from the publicly available RNAseq data from bronchoalveolar cells and peripheral blood mononuclear cells to elucidate molecular alterations and cellular deconvolution to identify immune cell profiles.
Results
Alterations in genes involved in hyaluronan, glycosaminoglycan and mucopolysaccharides metabolism were over-represented in bronchoalveolar cells infected by SARS-CoV-2, as well as potential lung infiltration with neutrophils, T CD4+ cell and macrophages. The blood mononuclear cells presented a proliferative state. Dramatic reduction of NK and T lymphocytes, whereas an exacerbated increase in monocytes.
Conclusions
In summary our results revealed molecular pathogenesis of the SARS-CoV-2 infection to bronchoalveolar cells inducing the hyaluronan and glycosaminoglycan metabolism that could shape partially the components of the ground-glass opacities observed in CT. And the potential immune response profile in COVID-19.
Key Words: Hyaluronan, Bronchoalveolar transcriptome, COVID-19, SARS-CoV-2
Introduction
Since its emerging in Wuhan, China in December 2019 the coronavirus disease (COVID-19) epidemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has progressed rapid to pandemic (1). As for April 27, 2020 more than 2 million people worldwide have been diagnosed with COVID-19 (2). It is associated with significant mortality due to the development of acute respiratory distress syndrome (ARDS) and eventually in multiple organ failure, particularly in certain high-risk populations such as the elderly, as well as patients with obesity, diabetes and hypertension. This complicated course is thought to be due to a so-called “cytokine storm” characterized by over production of TNF, IL6 and IL1ß, which together lead to vascular hyperpermeability, multiorgan failure and eventually death (3).
The SARS-CoV-2 viral entry depends upon the binding of viral spike (S) protein to the host cell surface protein angiotensin-converting enzyme 2 (ACE2). Once internalized, the S protein undergoes proteolytic processing by transmembrane serine protease 2 (TMPRSS2), as well as by cathepsin L and B (CTSL and CTSB, respectively), resulting in cleavage at the S1/S2 site, and the ensuing S2-dependent fusion of viral and cellular membranes (4). Coronaviruses such as SARS-CoV and MERS-CoV have evolved strategies to dampen or delay IFN production, which usually trigger exacerbated inflammatory host response leading to severe lung pathology (5). Among these mechanisms is the disruption of the pathogen associated molecular patterns recognition by the viral nonstructural protein 15 (nsp15) (6). Despite the global treats of COVID-19, the host immune response against virus infection remains poorly understood (5). Therefore, in the present work we carried out comprehensive and stringent transcriptomic metanalysis of SARS-CoV-2 infected bronchoalveolar cells and peripheral blood mononuclear cells to unveil the molecular alterations caused by viral infection as well as deconvolution analysis to identify the immune cell profiles in COVID-19 patients.
Materials and Methods
Bronchoalveolar Lavage Fluid and Peripheral Blood Mononuclear Cells RNAseq Files Analysis
Paired end fastq files were downloaded from BIGD-GSA and SRI from NCBI. Accession numbers are CRA002390 and SRR10571724, SRR10571730, and SRR10571732. Data files consisted in 3 COVID-19 patients bronchoalveolar fluid lavage (BALF) and 3 bronchoalveolar fluid lavage controls, and also 3 COVID-19 peripheral blood mononuclear cells (PBMC) and 3 control peripheral blood mononuclear cells. Fastq files quality control of was performed using FastQC v. 0.11.9. Samples adapter were trimmed with the Cutadapt v tool. 2.10.
BALF and PBMC samples duplicates were spotted using Picard v. 2.22.4, the RNAseq reads alignments were done using STAR tools v. 2.7.3a, with the parameters previously described (Xiong Y, et al, 2020). The RNAseq reading count was carried out using the Rsubread package with the annotations of Homo sapiens GRCh.38.99 downloaded from Ensembl. Subsequent analyzes were performed using the stable Gene ID identifier with BioMart annotations: Ensembl Genes 100, Human genes (GRCh38.p13), the identifier was translated to HGNC symbol to facilitate biological interpretation. The BALF and PBMC counts were subjected to quality control using the NOISeq package, biases associated with length, GC content, RNA composition and batch effect, were identified and removed. BALF and PBMC samples were processed using full-quantile normalization for GC content and length, finally, the ARSyN function was used to eliminate batch effect. The normalization process was carried out using the NOISeq and EDASeq packages in R. Differentially expressed genes were calculated on the normalized counts using the noiseqbio function of the NOISeq package, using the predefined parameters and filtered by a probability greater than 0.99.
Deconvolution and Gene Ontology Analysis
The deconvolution analysis was performed using the online tool CIBERSORT (Cell Type Identification By Estimating Relative Samples Of RNA Transcripts), which solves the equation m = f × B, where m is a mixture of mRNA, f is a vector that denotes the Cell fractions that make up the mixture and B is an array of gene expression profiles characteristic of each cell subtype. CIBERSORT is based on the application of v-SVR (nu-support vector regression) where limits of a hyperplane are determined that fits most of the possible points at a constant distance and where a regression is performed. Support vectors are genes selected from B. CIBERSORT was used for the present work with the default parameters, using the LM22 expression profile matrix (22 types of immune cells) and 100 permutations to calculate the p-value associated by Monte Carlo Sampling. The mRNA mixture used was a matrix constructed with the expression values of each sample obtained by bronchoalveolar lavage fluid and peripheral blood mononuclear cells RNAseq files analysis.
WebGestalt (http://www.webgestalt.org) was used for understanding the biological meaning behind the resulting list of genes, to obtain gene ontology and pathway information for significantly de-regulated genes in COVID-19 patients.
Results
SARS-CoV-2-infected Bronchoalveolar Molecular Alterations
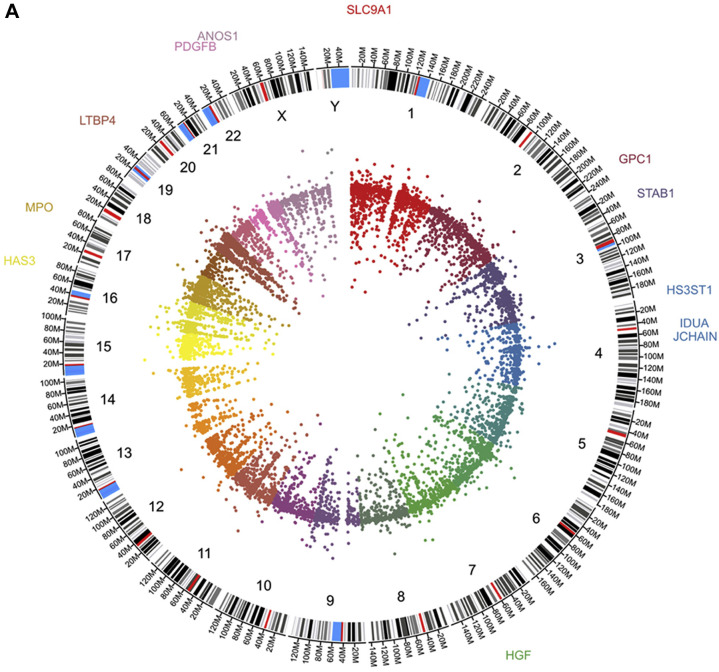
In order to identify potential cellular and molecular alterations in lung tissue we analyzed top 100 genes up- and down regulated. The gene ontology (GO) at biological process level of the 100 most up-regulated genes showed participation in hyaluronan metabolic process (HGF, PDGFB and SLC9A1), glycosaminoglycan metabolic process (HS3ST1, IDUA and GPC1), aminoglycan metabolic process (HS3ST1, PDGFB, and GPC1) and mucopolysaccharide metabolic process (HGF, IDUA and SLC9A1) among others. The GO at molecular function showed again participation in glycosaminoglycan binding (MPO, JCHAIN and ANOS1) and chemoattractant activity (HGF and PDGFB). Very interestingly, not only the up-regulated genes showed participation in COVID-19 relevant events. The gene ontology (GO) at biological process level of the 100 most down-regulated genes showed participation in primarily in chemokine events such as chemokine-mediated signaling pathway, response to chemokine, (CCL17, CXCL9 and SLIT3), and antimicrobial humoral response (AZU1, PPBP and ITLN1). The GO at molecular function level showed involvement in heparin binding (PTCH1, RSPO3 and SAA1) and G protein-coupled receptor binding (CRH, CXCL9 and GNAS14) (Figure 1 ).
Figure 1.
Panel A depict circos plot from the bronchoalveolar lavage fluid cells transcriptome from COVID-19 patients. Inner ring is the scatter plot representing each transcript identified followed by chromosome number and size, the outer ring present genes identified participating in hyaluronan, glycosaminoglycan and mucopolysaccharide metabolisms. B and C panels show Gene Ontology at biological process and molecular function terms, respectively, from top 100 up-regulated genes. D and E panels show Gene Ontology at biological process and molecular function terms, respectively, from top 100 down-regulated genes.
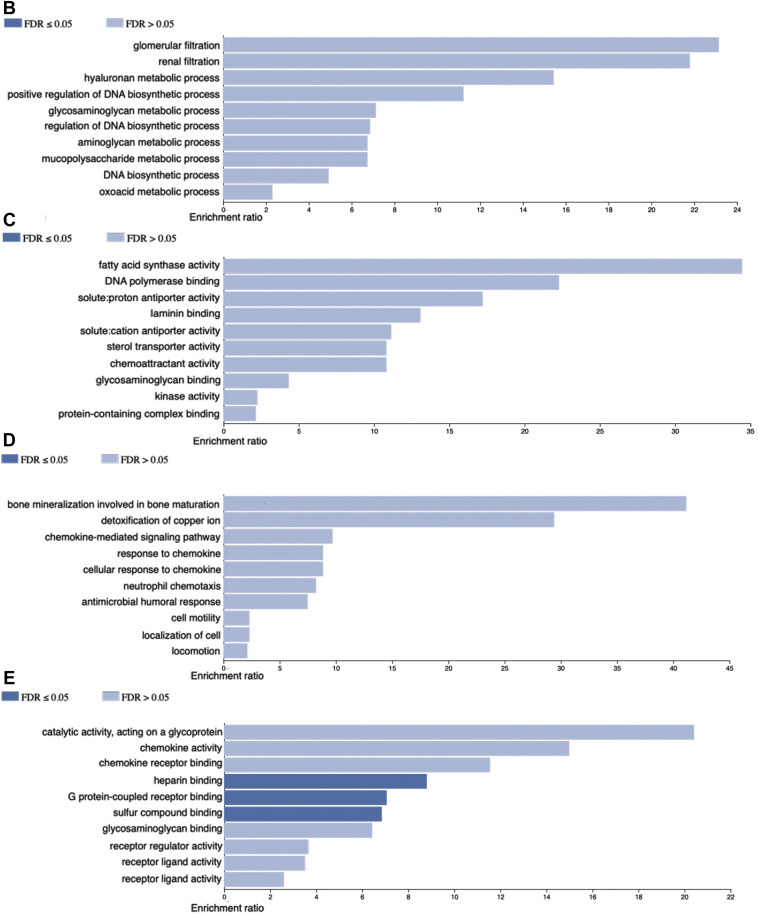
Further characterization of the molecular alterations was followed. Once we identify the transcriptome profile of the SARS-CoV-2 infected bronchoalveolar cells we carried out cellular deconvolution to identify potential infiltrating immune cells. We observed more presence of Neutrophils, T CD4+ lymphocytes and macrophages in COVID-19 patients BALF compared to control (Figure 2 ).
Figure 2.
Deconvolution results by CIBERSORT algorithm show probability percentage of presence of Neutrophils, NK cells, T CD4+ cell and macrophages in COVID-19 patients BALF. SIB036, 030 and 028 correspond to BALF control and CRR119894, −119895 and −119897 correspond to COVID-19 BALF.
Transcriptome Profiles from COVID-19 Patients’ Peripheral Blood Mononuclear Cells
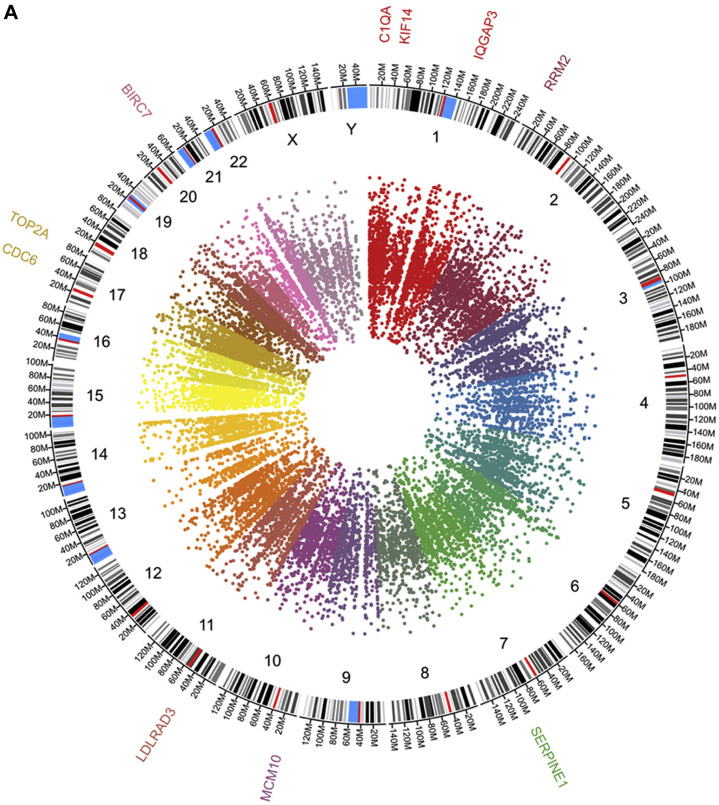
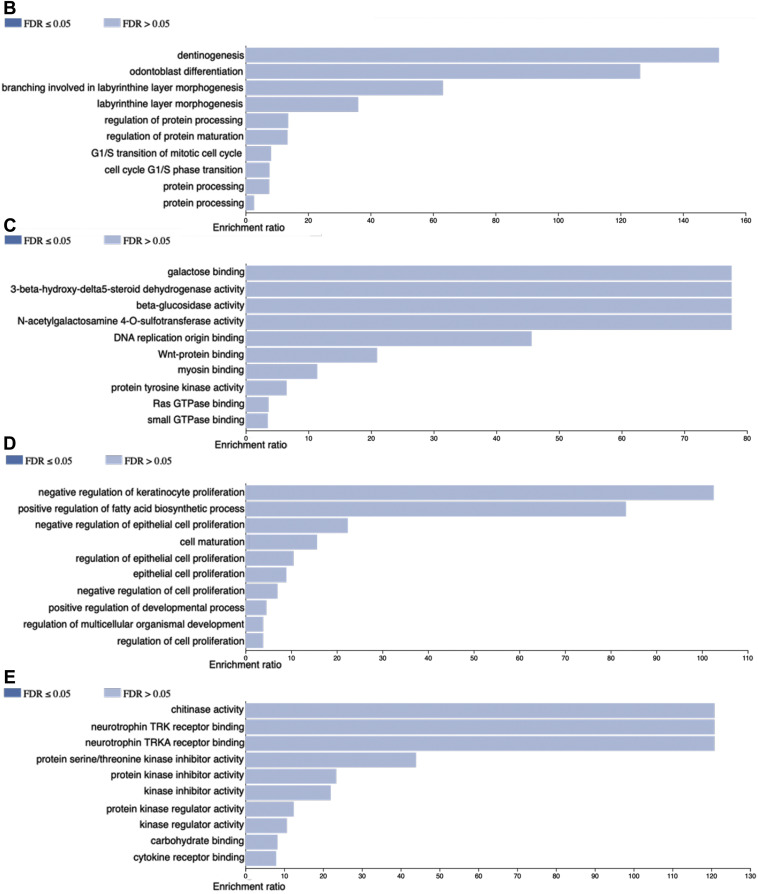
The PBMC transcriptomic profiles from COVID-19 differs from the control group (Figure 3 ). The top 100 up-regulated genes GO at biological process level showed involvement in G1/S transition of mitotic cell cycle (CDC6, MCM10 and RRM2) and regulation of protein processing (BIRC7, SERPINE1 and C1QA) among others. WNT-protein binding (FZD5 and MERTK), RAS GTPase binding (NRG1, SRGAP1 and IQGAP3), was observed at the molecular function terms of the GO. Consistently with the observed in the up-regulated genes, the down-regulated genes showed involvement in negative regulation of cell proliferation (CDKN1C, SFN and RUNX3) and cell maturation (SOX8, RHEX and GATA2) at GO biological process. In the GO molecular function terms cytokine receptor binding (CXCL1, EFNA5 and RHEX) among others (Figure 3).
Figure 3.
Panel A depict circos plot from the peripheral blood mononuclear cells transcriptome from COVID-19 patients. Inner ring is the scatter plot representing each transcript identified followed by chromosome number and size, the outer ring present genes identified participating in G1/S transition of mitotic cell cycle and protein processing. B and C panels show Gene Ontology at biological process and molecular function terms, respectively, from top 100 up-regulated genes. D and E panels show Gene Ontology at biological process and molecular function terms, respectively, from top 100 down-regulated genes.
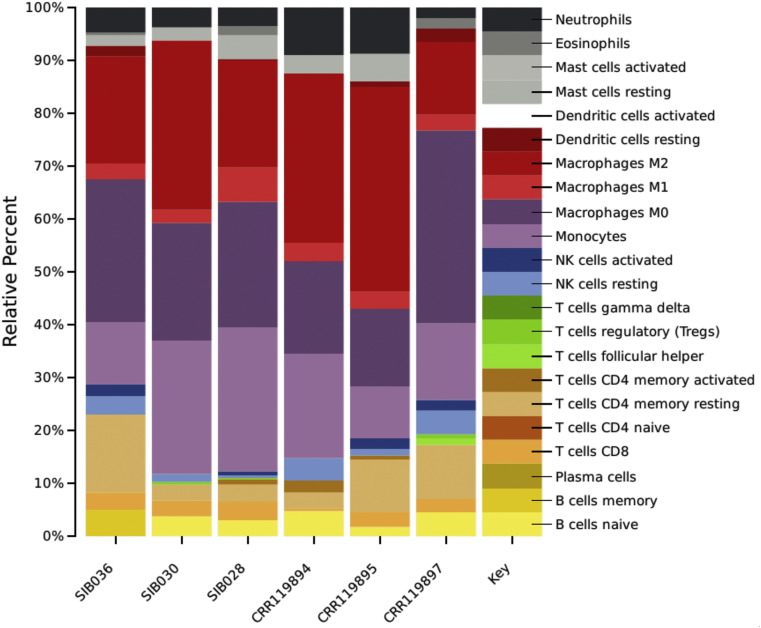
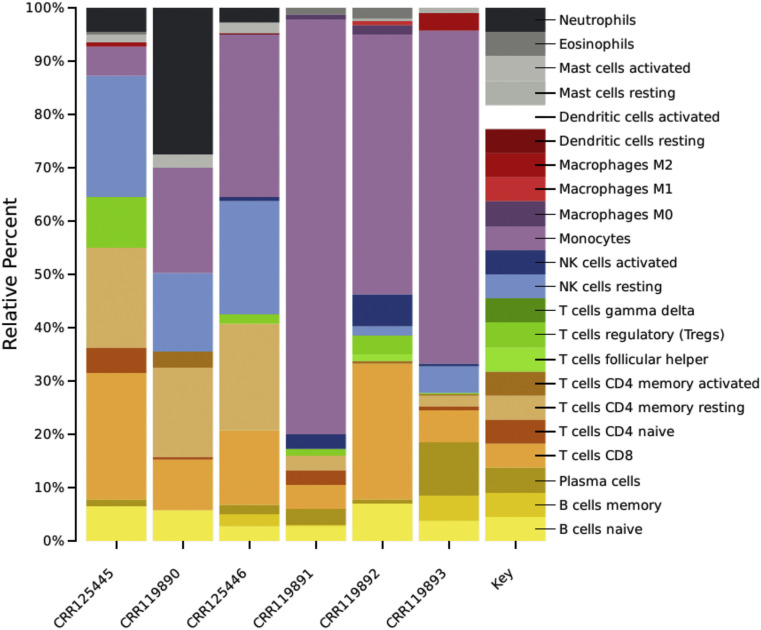
Very interestingly the cellular deconvolution analysis showed dramatic reduction of NK and T cells, whereas an exacerbated increase in monocytes (Figure 4 ).
Figure 4.
Deconvolution results by CIBERSORT algorithm show probability percentage of reduction of neutrophils, NK and T cells, whereas an exacerbated increase in monocytes in COVID-19 patients PBMC. CRR125445, 119890 and 125446 correspond to control PBMC whereas CRR119891, 119892 and 119893 correspond to COVID-19 PBMC.
Discussion
The present study constitutes a comprehensive meta-analysis of the available RNA sequencing data from pulmonary and blood mononuclear cells from patients with COVID-19 infection. In order to be able to design effective therapeutic strategies against SARS-CoV-2 infection, it is crucial to understand the molecular basis of its interaction with host cells. The infection of SARS-CoV-2 can cause severe pulmonary disease and complications with severe mortalities, currently there is no effective drug (5).
The acute respiratory distress syndrome that complicates a substantial number of patients with COVID-19 infection is characterized by a generalized endothelial dysfunction with a substantial fluid leak into the interstitial space thereby reducing lung compliance and thus, effective ventilation (7,8). This is the basis for the so-called ground-glass appearance on chest CT (9) and correlates with recent autopsy findings showing filling of the lungs with a clear fluid or gel-like material, much resembling what occur in wet drowning (10). Although the fluid filling components of the lungs are not yet defined, hyaluronan accumulation has been documented in patients with ARDS, and the potential role of polysaccharides for the interstitial edema of the lung have not received much attention (11). Hyaluronan is glycosaminoglycan component of the extracellular matrix found in lung, and can absorb water 1000 times its molecular weight, and the alteration in hyaluronan synthesis and accumulation in ARDS is established (12). Our molecular findings in bronchoalveolar cells regarding the upregulation of genes encoding proteins involved in hyaluronan, glycosaminoglycan, aminoglycan and mucopolysaccharide metabolism support the notion that these complex polysaccharides play indeed a role in the genesis of this severe form of interstitial pulmonary edema. Thus, inhibition of hyaluronan synthesis could represent a useful measure in the management of these patients (13).
Several inflammatory cytokines produced in COVID-19 could be strong inducers of hyaluronan metabolism (10) therefore the relevance to identify immune cells infiltrating lung tissue. Using molecular deconvolution analysis, we identified the presence of neutrophils, T CD4+ lymphocytes and macrophages infiltrating the lungs of COVID-19 patients, consistently our findings with the previously reported (14). Increased infiltrating macrophages are identified in autopsies and are thought to be responsible for fueling and perpetuating the inflammatory process (14). Neutrophils recruited to the site of infection can induce a prominent proinflammatory response directly by recognizing and clearing pathogens or by recruiting other immune cells (15). In summary, potentially the ARDS could be prompted by SARS-CoV-2 and fueled by host immune response.
Our in-silico findings revealed that peripheral mononuclear blood cells are in a proliferative state, perhaps, these proliferation events could represent clonal expansion of the remaining immune cells specific to viral infection. Interestingly, the molecular deconvolution analysis showed an evident shift of the immune profiles between COVID-19 and controls showing dramatic reduction of NK and T lymphocytes, whereas an exacerbated increase in monocytes. Changes in myeloid, NK and B cells in the COVID-19 patients has been reported (16). Important lymphopenia is also observed, indicating an impairment of the immune system during the SARS-CoV-2 infection (17). During viral infections it is generally accepted that host immune responses determine both protection against viral infections and the pathogenesis of respiratory injury (16). Coordinated immune response, innate and adaptative, may lead to rapid control of virus, whereas failed immune response may lead to viral spreading, cytokine storm and eventually death (18).
An additional interesting observation was the potential alteration in mast cells in both BALF and PBMC data. Mast cells could be found in submucosa of the respiratory tract and the nasal cavity. Mast cells secrete L-6, IL-1, IL-31, IL-33 and TNF, and chemokines CC5, CCL2, MCP-1 and CXCL8 which attract white blood cells to the inflammatory sites. Experiments with coronavirus in mice have shown that this pathogenic RNA microorganism can activate the cells of both innate and adaptive immune systems, including MCs (19,20).
In conclusion the present data shows several molecular events that are potentially involved in the pathogenesis of SARS-CoV-2 infection and its pulmonary and systemic complications. Specifically, our finding that genes encoding enzymes involved in hyaluronan and glycosaminoglycan metabolism are upregulated in pulmonary cells obtained by BAL of these patients could very well be the basis of the severe interstitial pneumonitis that characterizes this disease. Whether if our transcriptomic findings in PBMC indicating a proliferative state of immune cells reflect a clonal expansion resulting from recognition of specific viral antigens or simply a compensatory mechanism to replenish the number of immune cells, remains to be established.
Competing Interest
The authors declare no conflict of interest.
Acknowledgments
DMR is a recipient of the National Council for Science and Technology “Catedra CONACyT” program. This work was partially supported by grants 289499 from Fondos Sectoriales del Consejo Nacional de Ciencia y Tecnologia Mexico and R-2015-785-015 from Instituto Mexicano del Seguro Social (MM). The authors particularly thanks to the Instituto Mexicano del Seguro Social for the support to the formation of the Scientific Coalition for COVID-19 research.
(ARCMED_2020_627)
References
- 1.Bi Q., Wu Y., Mei Y. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020;20:30287–30295. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan N., Prevost B., Corvol H. Atypical presentation of COVID-19 in young infants. Lancet. 2020;10235:30980–30986. doi: 10.1016/S0140-6736(20)30980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jose R., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Y., Liu Y., Cao L. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackbarta M., Denga X., Bakera S. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee E., Ng M., Khong P. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 2020;4:384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Wang Y., Shao C. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hällgren R., Samuelsson T., Laurent T., Modig J. Accumulation of hyaluronan (hyaluronic acid) in the lung in adult respiratory distress syndrome. Am Rev Respir Dis. 1989;139:682–687. doi: 10.1164/ajrccm/139.3.682. [DOI] [PubMed] [Google Scholar]
- 12.Bell T., Brand O., Morgan D. Defective lung function following influenza virus is due to prolonged, reversible hyaluronan synthesis. Matrix Biol. 2019;80:14–28. doi: 10.1016/j.matbio.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collum S., Chen N., Hernandez A. Inhibition of hyaluronan synthesis attenuates pulmonary hypertension associated with lung fibrosis. Br J Pharmacol. 2017;174:3284–3301. doi: 10.1111/bph.13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao M., Liu Y., Yuan J. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. Preprint at medRxiv. 2020 doi: 10.1101/2020.02.23.20026690. [DOI] [Google Scholar]
- 15.Naumenko V., Turk M., Jenne C. Neutrophils in viral infection. Cell Tissue Res. 2018;371:505–516. doi: 10.1007/s00441-017-2763-0. [DOI] [PubMed] [Google Scholar]
- 16.Wen W., Su W., Tang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Preprint at medRxiv. 2020 doi: 10.1101/2020.03.23.20039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F., Nie J., Wang H. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z., Wan Y., Qiu C. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8⁺ T cells. Nat Commun. 2015;6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kritas S., Ronconi G., Caraffa A. Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. J Biol Regul Homeost Agents. 2020;34:1–14. doi: 10.23812/20-Editorial-Kritas. [DOI] [PubMed] [Google Scholar]
- 20.Theoharides T. COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. Biofactors. 2020;46:306–308. doi: 10.1002/biof.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]