Abstract
Background
More than 180 single nucleotide polymorphisms (SNPs) associated with breast cancer susceptibility have been identified; these SNPs can be combined into polygenic risk scores (PRS) to predict breast cancer risk. Because most SNPs were identified in predominantly European populations, little is known about the performance of PRS in non-Europeans. We tested the performance of a 180-SNP PRS in Latinas, a large ethnic group with variable levels of Indigenous American, European, and African ancestry.
Methods
We conducted a pooled case-control analysis of US Latinas and Latin American women (4658 cases and 7622 controls). We constructed a 180-SNP PRS consisting of SNPs associated with breast cancer risk (P < 5 × 10–8). We evaluated the association between the PRS and breast cancer risk using multivariable logistic regression, and assessed discrimination using an area under the receiver operating characteristic curve. We also assessed PRS performance across quartiles of Indigenous American genetic ancestry. All statistical tests were two-sided.
Results
Of 180 SNPs tested, 142 showed directionally consistent associations compared with European populations, and 39 were nominally statistically significant (P < .05). The PRS was associated with breast cancer risk, with an odds ratio per SD increment of 1.58 (95% confidence interval [CI = 1.52 to 1.64) and an area under the receiver operating characteristic curve of 0.63 (95% CI = 0.62 to 0.64). The discrimination of the PRS was similar between the top and bottom quartiles of Indigenous American ancestry.
Conclusions
The 180-SNP PRS predicts breast cancer risk in Latinas, with similar performance as reported for Europeans. The performance of the PRS did not vary substantially according to Indigenous American ancestry.
More than 180 single nucleotide polymorphisms (SNPs) associated with breast cancer susceptibility have been discovered in genome-wide association studies (GWAS) (1–4). Though each SNP has a modest effect, multiple SNPs can be combined into a polygenic risk score (PRS) (5). PRS has emerged as a promising tool for breast cancer risk stratification. The risk associated with having a PRS in the upper 20–25th percentile is similar to that of strong clinical risk factors such as having extremely dense breasts (6), and adding PRS to risk models improves discrimination and reclassification (6–8). Ongoing clinical trials are studying the use of PRS to personalize breast cancer screening and prevention (9). Some commercial genetic testing laboratories are already returning PRS results to those who tested negative for pathogenic moderate- or high-penetrance mutations (10,11).
A major barrier to the widespread use of PRS is the paucity of knowledge regarding its performance in non-European populations. To date, SNP discovery has overwhelmingly occurred in European populations (12). However, the effect sizes, allele frequencies, and linkage disequilibrium patterns of SNPs vary by ancestry (12,13). Though relatively few studies have examined PRS performance in non-Europeans, they suggest that PRS constructed using European SNP summary statistics (effect size and allele frequency) perform worse in Latinas (14) and women of African ancestry (14,15). Currently, commercial testing laboratories report breast cancer PRS results only to women of European ancestry (10,11).
Disparities in the use and performance of PRS could especially affect Latinas. Latinos and Latinas comprise the largest minority group in the United States, representing 17.8% of the population in 2016 (16). This group includes genetically admixed individuals who have varying degrees of Indigenous American, European, African, and Asian ancestry (17–19). We previously identified SNPs in the 6q25 locus associated with breast cancer risk exclusively in Latinas (20). Most SNPs discovered in European populations display directional consistency in Latinas, with some being nominally statistically significant (20,21). One previous study assessed the performance of a breast cancer PRS in Latinas, finding that a 71-SNP PRS had worse prediction in Latinas than in Europeans (5,14). However, it included only 147 cases and did not account for genetic ancestry (14).
We sought to test the performance of PRS in US Latinas and Latin American women (collectively referred to hereafter as Latinas). To that end, we conducted a pooled case-control analysis of eight studies comprising 13 624 Latinas. We examined the predictive performance of a 71-SNP and a 180-SNP PRS, and whether PRS performance varies by genetic ancestry.
Methods
Participants
Our analysis included 13 624 self-identified Latinas, of whom 5697 women with invasive breast cancer were considered cases and 7927 without breast cancer were controls. Participants came from eight studies (Table 1 and Supplementary Table 1 [available online]). Recruitment details and patient characteristics have been previously reported for each study except for Peru Genetics and Genomics of Breast Cancer Study (PEGEN-BC). All studies obtained local institutional review board approval and written informed consent from participants.
Table 1.
Participant characteristics by study and case-control status
| Characteristic | SFBCS/NC-BCFR |
Kaiser RPGEH |
MEC |
CAMA |
COLUMBUS (Colombia) |
COLUMBUS (Mexico) |
PEGEN-BC/Peru |
COH/CCGCRN |
All |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | |
| Number of individuals | 589 | 942 | 3563 | 222 | 1469 | 532 | 702 | 709 | 761 | 954 | 453 | 481 | 85 | 818 | 305 | 1039 | 7927 | 5697 |
| Age at diagnosis (cases) or interview (controls) in years, mean (SD) | 53 (11) | 50 (11) | 55 (13) | 57 (10) | 67 (8) | 66 (8) | 52 (9) | 52 (10) | 64 (10) | 52 (10) | 35 (12) | 57 (13) | ND | 50 (11) | 52 (11) | 43 (9) | 57 (13) | 52 (12) |
| Positive family history of breast cancer, No. (%) | 55 (9)* | 190 (20)* | 211 (6)‡ | 38 (17)‡ | 141 (10)* | 73 (14)* | 27 (4) | 50 (7) | ND | 49 (5)† | 34 (8)‡ | 23 (5)‡ | ND | 54 (7) | 26 (9)† | 348 (33)† | 494 (6) | 825 (14) |
| Genetic ancestry, mean % (SD) | ||||||||||||||||||
| Indigenous American | 39.1 (16.9) | 35.9 (17.3) | 29.1 (18.1) | 26.9 (17.0) | 38.6 (14.2) | 36.4 (13.5) | 63.9 (18.1) | 59.0 (18.2) | 43.2 (10.8) | 43.3 (10.0) | 57.0 (14.8) | 57.7 (17.7) | 76.3 (15.1) | 76.3 (16.3) | 40.8 (18.8) | 43.2 (19.2) | 38.6 (20.2) | 48.7 (21.6) |
| European | 52.6 (17.4) | 56.0 (17.8) | 62.4 (19.8) | 64.5 (19.4) | 54.3 (15.2) | 56.2 (14.1) | 30.9 (16.2) | 35.5 (17.1) | 50.0 (10.6) | 49.5 (9.9) | 36.2 (14.0) | 37.3 (17.0) | 17.8 (11.2) | 17.0 (11.4) | 48.0 (18.0) | 45.4 (18.1) | 53.8 (20.4) | 44.3 (20.7) |
| African | 6.3 (7.5) | 6.2 (6.8) | 5.9 (8.3) | 5.0 (4.4) | 4.9 (2.8) | 5.2 (3.4) | 3.7 (3.2) | 3.8 (3.0) | 6.2 (5.5) | 6.7 (5.1) | 65.8 (2.5) | 4.2 (3.2) | 3.6 (5.9) | 4.4 (7.8) | 4.4 (3.8) | 4.9 (5.8) | 5.5 (6.5) | 5.2 (5.6) |
| Asian | 2.0 (3.7) | 1.9 (3.2) | 2.6 (6.7) | 3.6 (9.1) | 2.3 (3.2) | 2.1 (4.4) | 1.5 (1.3) | 1.7 (2.0) | 0.6 (0.8) | 0.6 (0.7) | 1.3 (2.5) | 0.8 (0.8) | 2.3 (4.5) | 2.3 (4.7) | 2.4 (2.1) | 2.9 (3.8) | 2.1 (4.9) | 1.9 (3.7) |
| Estrogen receptor status, No. (%) | ||||||||||||||||||
| Positive§ | NA | 593 (72.1)‖ | NA | 161 (84.7) | NA | 303 (73.7) | NA | 116 (69.0) | NA | 354 (66.7) | NA | 140 (77.3) | NA | 548 (69.0) | NA | 585 (71.5) | NA | 2800 (71.5) |
| Negative§ | NA | 230 (27.9) | NA | 29 (15.3) | NA | 108 (26.3) | NA | 52 (31.0) | NA | 177 (33.3) | NA | 41 (22.7) | NA | 246 (31.0) | NA | 233 (28.5) | NA | 1116 (28.5) |
| Unknown | NA | 119 | NA | 32 | NA | 121 | NA | 541 | NA | 423 | NA | 300 | NA | 24 | NA | 221 | NA | 1781 |
| 71-SNP PRS, mean (SD) | 0.99 (0.49) | 1.19 (0.57) | 1.01 (0.50) | 1.21 (0.55) | 1.01 (0.47) | 1.18 (0.52) | 0.93 (0.48) | 1.07 (0.49) | 0.99 (0.45) | 1.20 (0.59) | 0.93 (0.45) | 1.08 (0.55) | 0.93 (0.46) | 1.16 (0.54) | 0.99 (0.48) | 1.16 (0.55) | 0.99 (0.48) | 1.16 (0.55) |
| 180-SNP PRS, mean (SD)# | 1.00 (0.68) | 1.26 (0.79) | 0.98 (0.61) | 1.27 (0.64) | 1.03 (0.64) | 1.29 (0.72) | 0.99 (0.71) | 1.23 (0.77) | 1.02 (0.61) | 1.32 (0.80) | 1.00 (0.61) | 1.24 (0.79) | 1.08 (0.73) | 1.44 (0.95) | NA | NA | 1.00 (0.64) | 1.30 (0.81) |
Positive family history of breast cancer in a first-degree relative only. CAMA = Cancer de Mama; COH/CCGCRN = City of Hope/Clinical Cancer Genetics Community Research Network; COLUMBUS = Colombian Study of Environmental and Heritable Causes of Breast Cancer; MEC = Multiethnic Cohort; NC-BCFR ¼ Northern California Breast Cancer Family Registry; ND = not determined; PEGEN-BC = Peru Genetics and Genomics of Breast Cancer Study; PRS = polygenic risk score; RPGEH = Research Project on Genes, Environment, and Health; SFBCS = San Francisco Bay Area Breast Cancer Study; SNP = single nucleotide polymorphism.
Positive family history of breast cancer in a first- or second-degree relative.
Positive family history of breast cancer in any relative.
Percentage within cases with known ER status.
Includes two cases with borderline ER status.
Calculated for all datasets.
Calculated for all datasets except COH/CCGCRN.
Studies are briefly described here and in more detail in the Supplementary Methods (available online): They include the San Francisco Bay Area Breast Cancer Study (SFBCS) and the Northern California Breast Cancer Family Registry (NC-BCFR), a population-based case-control study recruiting from the San Francisco Bay Area (22,23); the Kaiser Permanente Research Project on Genes, Environment, and Health (RPGEH), a biobank recruiting from Northern California and the Pacific Northwest (24); the Multiethnic Cohort (MEC) study, a prospective cohort study recruiting from Southern California and Hawaii (25); the Cancer de Mama (CAMA) study, a population-based case-control study in Mexico (26); the Post–Columbian Study of Environmental and Heritable Causes of Breast Cancer (COLUMBUS-Colombia), a population-based case-control study in southern Colombia (20); the Post–Columbian Study of Environmental and Heritable Causes of Breast Cancer (COLUMBUS-Mexico), a population-based case-control study in Mexico (20); the, a case series from a Peruvian cancer center with unrelated Peruvian individuals from 1000 Genomes (27) included as controls; and the City of Hope Clinical Cancer Genetics Community Research Network (COH/CCGCRN), the Southern California site of a multisite cancer center and community-based registry for familial breast cancer (28). Of note, the COLUMBUS substudies (Colombia and Mexico) were analyzed as separate datasets, given differences in study populations and genotyping methods.
Genotyping and Genetic Ancestry
For all studies except COH/CCGCRN, genotyping was performed using high-density arrays (Supplementary Table 1 [available online]). Genotyping of COH/CCGCRN was performed using next-generation sequencing with a targeted capture kit that included all 89 breast cancer susceptibility SNPs identified as of 2016, before publication of the OncoArray GWAS results (3). Further information about genotyping is provided in the Supplementary Methods (available online).
We estimated genetic ancestry from genome-wide markers using the program ADMIXTURE (29) in unsupervised mode with a model containing four ancestral populations: European, Indigenous American (IA), African, and East Asian. We used genotype data from 90 European Americans (CEU) and 90 Nigerian Yorubans (YRI) from HapMap (30) to represent European and African populations, respectively. We also included a subset of 504 East Asian individuals from 1000 Genomes (27) and 71 IAs previously genotyped on the Affymetrix Axiom LAT1 array (31,32). Women with greater than 75% East Asian ancestry were excluded.
PRS
We used a 180-SNP PRS for our primary analysis (Supplementary Table 2 [available online]). We considered for inclusion 184 SNPs associated with invasive breast cancer with genome-wide statistical significance (P < 5 × 10–8) in previous studies (1–4). These included 172 SNPs from the discovery (n = 65) and replication (n = 107) phases of the Breast Cancer Association Consortium OncoArray study (3), which took place in European (discovery and replication) and Asian (replication only) populations. These SNPs also included nine nonoverlapping SNPs from GWAS of estrogen receptor–negative breast cancer (3) and three SNPs from 6q25 discovered in GWAS (rs140068312) (20) and fine-mapping studies (rs3778609, rs851984) (21) in Latinas. Of these 184 SNPs, one pair (rs35054928 and rs2981578) was in linkage disequilibrium (LD) using an r2 cutoff of 0.3, and we excluded the latter based on a lower β coefficient with breast cancer. We also excluded rs17879961, given that it was not polymorphic in our study, and rs2016394 and rs554219 because of a missing call rate of less than 5%. We included all SNPs regardless of imputation quality, given there were no substantive differences in the associations with breast cancer between the 180-SNP PRS and PRSs constructed with imputation r2 thresholds of 0.5 and 0.8, respectively (Supplementary Table 3 [available online]).
Since targeted genotyping was performed within COH/CCGCRN, genotypes were available for 89 SNPs. We dropped one SNP because of missing data. Of the remaining 88 SNPs, 63 overlapped, and eight had LD proxies (r2 > 0.7), with the 180 SNPs comprising the main PRS. We used these 71 SNPs to construct a PRS within COH/CCGCRN. We then constructed a 71-SNP PRS in the seven remaining datasets using the 63 shared SNPs and eight respective LD proxies, and we pooled all eight datasets to evaluate the performance of the 71-SNP PRS.
We constructed the PRS as previously described (7,33). Briefly, the PRS represents the product of the likelihood ratios across multiple SNPs, assuming each SNP has an independent effect. The likelihood ratio for each SNP was calculated based on the number of risk alleles present and the allele frequency and odds ratio (OR) of the risk allele. We used risk-allele frequencies derived from the Latin American (AMR) population in 1000 Genomes (27) and published odds ratios for overall breast cancer (3). The latter predominantly reflects the effect of the SNP within a European population, except for those discovered in Latina studies (Supplementary Table 2 [available online]) (20,21).
Statistical Analysis
First, we tested the associations between individual SNPs and breast cancer risk using multivariable logistic regression models adjusted for genetic ancestry and study. Using METAL (34), we performed an inverse variance-based meta-analysis of 180 SNPs across three studies: COLUMBUS-Colombia, COLUMBUS-Mexico, and pooled SFBCS/NC-BCFR, Kaiser RPGEH, MEC, CAMA, and PEGEN-BC studies. The Cochran Q test for heterogeneity was used within METAL to test for differences in associations between the three studies (34).
Next, we tested the crude and adjusted associations between the PRS with breast cancer. Given that genetic ancestry and the study were possible confounders of this association (Table 2 and Supplementary Table 4 [available online]), we adjusted for both in our main analysis. To do so, we performed linear regression of the study and ancestry on the PRS (dependent variable). We then used the residual as the main predictor in univariate logistic regression with breast cancer as the outcome. We analyzed the residual as a continuous variable normalized to the mean and SD in controls. We tested the discrimination of the adjusted PRS by estimating the area under the receiver operating characteristic curve (AUROC). We tested calibration using the Hosmer-Lemeshow test across deciles of the adjusted PRS, with the 40–50th and 50–60th deciles combined and used as the reference group.
Table 2.
Association between 180-SNP and 71-SNP PRS and breast cancer risk
| PRS category | 180-SNP PRS* |
71-SNP PRS† |
||||||
|---|---|---|---|---|---|---|---|---|
| Controls | Cases | odds ratio (95% CI)‡ | P § | Controls | Cases | odds ratio (95% CI)‡ | P § | |
| Continuous PRS (per SD) | 7622 | 4658 | 1.58 (1.52 to 1.64) | 7927 | 5697 | 1.51 (1.46 to 1.56) | ||
| Percentiles of PRS | <.001 | <.001 | ||||||
| <10 | 762 | 196 | 0.46 (0.39 to 0.55) | 793 | 278 | 0.54 (0.47 to 0.64) | ||
| 10–20 | 763 | 223 | 0.52 (0.44 to 0.62) | 792 | 345 | 0.68 (0.58 to 0.79) | ||
| 20–30 | 762 | 340 | 0.80 (0.69 to 0.93) | 793 | 379 | 0.74 (0.64 to 0.86) | ||
| 30–40 | 761 | 335 | 0.79 (0.68 to 0.92) | 792 | 430 | 0.84 (0.73 to 0.97) | ||
| 40–60 | 1525 | 850 | 1 (Referent) | 1587 | 1021 | 1 (Referent) | ||
| 60–70 | 763 | 498 | 1.17 (1.02 to 1.35) | 791 | 656 | 1.29 (1.13 to 1.47) | ||
| 70–80 | 762 | 593 | 1.40 (1.22 to 1.60) | 793 | 694 | 1.36 (1.20 to 1.55) | ||
| 80–90 | 761 | 728 | 1.72 (1.50 to 1.96) | 793 | 832 | 1.63 (1.44 to 1.85) | ||
| >90 | 763 | 895 | 2.10 (1.85 to 2.39) | 793 | 1062 | 2.08 (1.84 to 2.35) | ||
Calculated in case-control analysis of seven datasets, excluding City of Hope/Clinical Cancer Genetics Community Research Network (COH/CCGCRN) (n = 12 280). CI = confidence interval; OR = odds ratio; PRS = polygenic risk score; SNP = single nucleotide polymorphism.
Calculated in case-control analysis of all datasets (n = 13 624).
odds ratio from multivariable logistic regression of PRS adjusted for study and genetic ancestry.
Two-sided P value for the test of the linear trend between per-decile estimates.
To examine the ancestry-specific performance of the PRS, we divided the pooled dataset into quartiles of IA ancestry. We performed logistic regression within each quartile of IA ancestry and compared the resulting coefficients using a Wald test of linear hypothesis. To compare AUROC estimates, we performed a test of equality of AUROC as described by DeLong et al. (35). Given differences in the population structures between US Latina and Latin American studies, we also examined ancestry-specific performance of the PRS by geographic origin of study, specifically the United States (SFBCS/NC-BCFR, RPGEH, and MEC) vs Latin America (CAMA, COLUMBUS, and PEGEN-BC).
All tests for statistical significance (eg, Wald, DeLong, Cochran) used a two-sided alpha of 0.05. When testing the associations between individual SNPs and breast cancer risk, we also used an alpha of P < 2.8 × 10–4, reflecting Bonferroni correction for multiple testing of 180 SNPs. We developed the script to calculate the PRS using R (R Foundation). We performed all statistical analyses using Stata 14.2 (StataCorp, College Station, TX).
Results
Study Characteristics
Our pooled data included 13 624 women from eight studies, for a total of 5697 cases and 7927 controls (Table 1). Across all studies, ancestry was predominantly European and IA. There was substantial variation in ancestry within and across studies (Supplementary Figure 2 [available online]). For instance, PEGEN-BC in Peru had the highest average IA ancestry (76.3% in cases and controls), whereas RPGEH in Northern California had the lowest (26.9% in cases and 29.1% in controls). Within each study, cases tended to have similar or lower IA ancestry than did controls, as previously reported (36,37). In the pooled analysis, cases had higher IA ancestry because nearly half the controls came from RPGEH, the study with the lowest IA ancestry.
Association of PRS with Breast Cancer Risk
We first examined the associations between individual SNPs and breast cancer risk. Of 180 SNPs, 142 had associations that were directionally consistent with those reported in European populations (Supplementary Table 2 [available online]) (3). Forty-four SNPs were nominally statistically significant (P < .05), with 39 also directionally consistent. Six SNPs remained statistically significant to P less than 2.8 × 10–4 after Bonferroni correction for multiple testing. Nineteen SNPs displayed heterogeneous associations across studies (Phet < .05). For both PRS, the mean unadjusted PRS was higher in cases than in controls (Table 1, Supplementary Figure 1 [available online]).
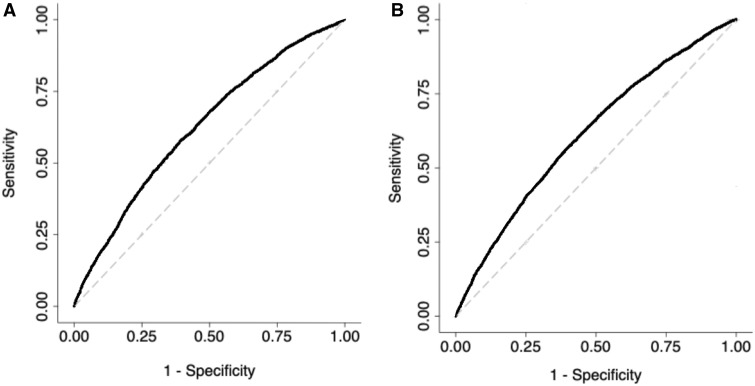
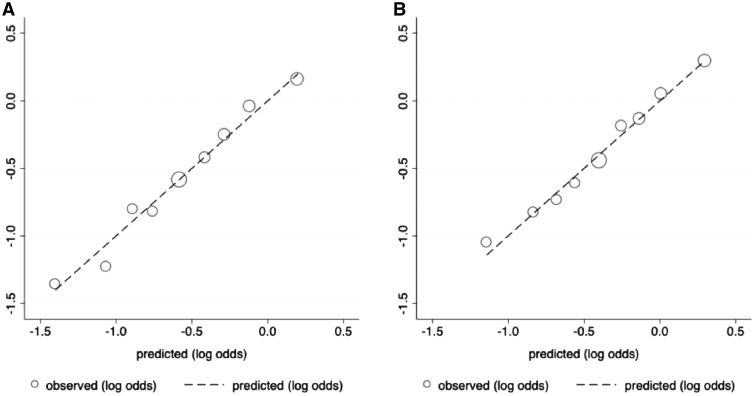
Our main analysis evaluated the performance of a 180-SNP PRS in 12 280 women (4658 cases and 7622 controls) from seven studies, excluding COH/CCGCRN, given that only 89 SNPs were genotyped in that study. The unadjusted 180-SNP PRS was strongly associated with breast cancer risk, odds ratio per SD increment = 1.70 (95% CI = 1.63 to 1.78). Adjusting for genetic ancestry and study slightly attenuated the association, odds ratio = 1.58 (95% CI = 1.52 to 1.64) (Table 2). The associations with breast cancer risk were especially pronounced among extremes of the PRS. Compared with women with a PRS in the 40–60th percentile, women with a PRS in the bottom decile had an odds ratio of 0.46 (95% CI = 0.39 to 0.55), whereas those with a PRS in the top decile had an odds ratio of 2.10 (95% 1.85 to 2.39). The AUROC for the 180-SNP PRS was 0.63 (95% CI = 0.62 to 0.64) (Figure 1A). The Hosmer-Lemeshow test suggested good fit, χ2 = 10.45 (P = .32), (Figure 2A).
Figure 1.
Receiver operating characteristic curves for two polygenic risk scores (PRS) containing 180 and 71 single nucleotide polymorphisms (SNPs), respectively. The 180-SNP PRS (A) had an area under the receiver operating characteristic curve (AUROC) of 0.63 (95% CI = 0.62 to 0.64) in seven datasets, excluding City of Hope/ Clinical Cancer Genetics Community Research Network, n = 12 280. The 71-SNP PRS (B) had an AUROC of 0.61 (95% CI = 0.61 to 0.62) in all datasets, n = 13 624.
Our secondary analysis evaluated the performance of a 71-SNP PRS in 13 624 women (5697 cases and 7927 controls) from eight studies, including COH/CCGCRN. Compared with the 180-SNP PRS, the unadjusted 71-SNP PRS had a similar association with breast cancer risk (odds ratio = 1.70, 95% CI = 1.62 to 1.79), although adjusting for study and genetic ancestry resulted in larger attenuation of its effect (Table 2, Figure 1B). The discrimination of the 71-SNP PRS was slightly lower, and the Hosmer-Lemeshow test was again suggestive of good fit, χ2 = 6.59 (P = .68) (Figure 2B). To assess whether inclusion of COH/CCGCRN participants affected these associations, we tested the 71-SNP PRS with COH/CCGCRN excluded and found similar results (Supplementary Table 5 [available online]).
Figure 2.
Calibration of two polygenic risk scores (PRS) containing 180 and 71 single nucleotide polymorphisms (SNPs), respectively. Calibration plots for (A) the 180-SNP PRS in seven datasets, excluding City of Hope/ Clinical Cancer Genetics Community Research Network, n = 12 280, and (B) the 71-SNP PRS (B) in all datasets n = 13 624. The graph depicts the predicted vs observed proportions of cases within each decile of the log-normalized PRS. Each circle corresponds to a decile of the PRS, with the middle (largest) circle representing the 40–60th percentile. Two-sided Hosmer-Lemeshow P value = .32 for 180-SNP PRS and .68 for 71-SNP PRS.
Performance of PRS by IA Ancestry
The 180-SNP PRS displayed similar performance regardless of IA ancestry, with comparable ORs and AUROCs across quartiles of IA ancestry (Table 3). In contrast, the 71-SNP PRS performed worse in the top compared to the bottom quartile (odds ratio 1.46 [95% CI = 1.36 to 1.56] vs odds ratio 1.68 [95% CI = 1.55 to 1.83], P = .01). This corresponded to top versus bottom quartile AUROCs of 0.61 (95% CI = 0.59 to 0.63) and 0.64 (95% CI = 0.62 to 0.66), respectively (P = .02). Given differences in ancestry structure between US Latinas and Latin American women, we stratified the analysis by geographic origin of study. Among 7317 women from the US studies, the 180-SNP PRS performed best in the bottom quartile of IA ancestry (Supplementary Table 6 [available online]). However, among the 4963 women from the Latin American studies, the 180-SNP PRS performed similarly across quartiles of IA ancestry (Supplementary Table 6 [available online]).
Table 3.
AUROCs and ORs per SD of the 71-SNP PRS and 180-SNP PRS in Hispanics, by quartiles of IA ancestry
| IA ancestry category | 180-SNP PRS* |
71-SNP PRS† |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | AUROC (95% CI)‡ | P § | odds ratio (95% CI)‖ | P ¶ | Controls | Cases | AUROC (95% CI)‡ | P § | odds ratio (95% CI)‖ | P¶ | |
| All | 7622 | 4658 | 0.63 (0.62 to 0.64) | 1.58 (1.52 to 1.64) | 7927 | 5697 | 0.61 (0.61 to 0.62) | 1.51 (1.46 to 1.56) | ||||
| Quartiles of IA ancestry | .56 | .28 | .02 | .01 | ||||||||
| Q1, <0.29 | 2349 | 721 | 0.63 (0.61 to 0.66) | 1.67 (1.52 to 1.83) | 2455 | 951 | 0.64 (0.62 to 0.66) | 1.68 (1.55 to 1.83) | ||||
| Q2, 0.29–0.42 | 2049 | 1021 | 0.61 (0.59 to 0.63) | 1.51 (1.39 to 1.64) | 2117 | 1289 | 0.60 (0.58 to 0.62) | 1.44 (1.34 to 1.55) | ||||
| Q3, 0.42–0.55 | 1820 | 1250 | 0.63 (0.61 to 0.65) | 1.57 (1.45 to 1.69) | 1869 | 1537 | 0.62 (0.60 to 0.63) | 1.52 (1.41 to 1.63) | ||||
| Q4, >0.55 | 1404 | 1666 | 0.63 (0.61 to 0.65) | 1.56 (1.45 to 1.68) | 1486 | 1920 | 0.61 (0.59 to 0.63) | 1.46 (1.36 to 1.56) | ||||
Calculated in case-control analysis of seven datasets, excluding City of Hope/Clinical Cancer Genetics Community Research Network (n = 12 280). AUROC = area under the receiver operating characteristic curve; CI = confidence interval; IA = Indigenous American; OR = odds ratio; PRS = polygenic risk score; SNP single nucleotide polymorphism.
Calculated in case-control analysis of all datasets (n = 13 624).
AUROC from multivariable logistic regression of PRS adjusted for study and genetic ancestry.
Two-sided P value from DeLong’s test of equality of AUROCs between Q1 and Q4 of IA ancestry.
Odds ratio of breast cancer per SD increment of PRS relative to the mean in controls. Calculated from multivariable logistic regression adjusted for study and genetic ancestry.
Two-sided P value for comparison of odds ratio of PRS between Q1 and Q4 of IA ancestry using the Wald test of linear hypothesis.
Discussion
We found that PRSs primarily consisting of SNPs identified in European populations were predictive of breast cancer risk in Latinas. Our 180-SNP PRS had an adjusted odds ratio per SD increment of 1.58 (95% CI = 1.52 to 1.64) and an AUROC of 0.63 (95% CI = 0.62 to 0.64). These results are comparable to those of European studies, which tested PRSs including 77 to 3820 SNPs, and reported odds ratios per SD between 1.46 and 1.66 and AUROCs between 0.60 and 0.64 (5,38). Our 71-SNP PRS performed worse than the 180-SNP PRS did, though the difference was modest.
Ours is the largest study to date on breast cancer PRS in Latinas, and it extends the literature by refining estimates of PRS performance in this population. Allman et al. (14) reported that a 71-SNP PRS had an odds ratio per SD increment of 1.39 (95% CI = 1.18 to 1.64) and AUROC of 0.59 (95% CI = 0.54 to 0.64) among US Latinas, but this study included only 147 cases and did not account for ancestry.
We could not definitively determine whether PRS performance varies by ancestry. Differential PRS performance by genetic ancestry might be expected given differences in LD structures between European and non-European populations, which can attenuate the associations between GWAS hits discovered in Europeans and causal SNPs only in LD. Additionally, causal alleles may be present only in certain populations. However, the 180-SNP PRS performed similarly across quartiles of IA ancestry. In contrast, the 71-SNP PRS performed better in the bottom quartile of IA ancestry, corresponding to higher European ancestry. This analysis included 1039 additional cases from COH/CCGCRN, and may have had greater statistical power to detect differences in performance by IA ancestry.
A major strength of our study was the size and diversity of our study population. Additionally, we accounted for genetic ancestry, which can bias associations in genetic studies (39). Given that ancestry was a confounder and an independent predictor of breast cancer risk, we used a novel approach to calculate an “ancestry-adjusted” PRS. We also examined PRS performance by IA ancestry, which has not been previously done. Another strength was the inclusion of several large, diverse breast cancer studies representing populations from several geographic areas (Western United States and Central and South America) and including women with varying degrees of IA vs European ancestry.
Our results should be interpreted in light of three limitations. First, the generalizability of our findings is limited to Latina populations with similar distributions of genetic ancestry. Although the ancestry composition of our study resembled that of other large studies of Latinas from the western United States and Central and South America (19,40), our results may not be generalizable to Caribbean Latinas, whose population structures have higher proportions of African ancestry (17–19). We did not test the performance of PRS according to African ancestry, given that our study population was predominantly Latina with limited African ancestry. Second, our analysis included women from community-based and familial breast cancer clinics and may include moderate or high-penetrance mutation carriers. Although PRS is associated with breast cancer risk in mutation carriers and women with elevated familial risk, the magnitudes of these associations vary slightly from those in the average-risk population (41). Finally, we tested a PRS containing 180 SNPs representing known GWAS hits at the time of analysis. However, others have constructed PRSs comprising 313 and 3820 SNPs by including SNPs that did not have genome-wide statistically significant associations with breast cancer (38). Though these expanded PRS performed better than a 77-SNP PRS did, there was little difference in performance between the 313-SNP and 3820-SNP PRS (38). We included only SNPs with genome-wide statistically significant associations in our PRS because these signals may be more robust across ancestry. The AUROC for our 180-SNP PRS (0.63) was similar to that of the 313-SNP PRS (38).
Our results suggest that the PRS has predictive value in Latinas, a large and rapidly growing population in the United States. Although studies on the ability of the PRS to inform decisions around screening and prevention are underway (9), several commercial genetic testing laboratories already return PRS results to women of European descent who tested negative for deleterious mutations (10,11). If this practice were extended to Latinas, one could expect the PRS to perform comparably well. Even if the performance of the PRS were slightly attenuated in Latinas of higher IA ancestry, this should not preclude its use in this population. Instead, results could account for this attenuation and model the joint effects of PRS and ancestry.
Though our findings suggest that the PRS can predict breast cancer risk in Latinas, they do not nullify the prospect of disparities in genetic discovery research (42,43). Whereas we studied mostly common variants, rare variants display more geographic clustering (44). As genetic association studies identify more rare variants, those discovered in European populations will be less generalizable to other populations. Thus, high-quality genetic studies in non-European populations remain a priority. Fine mapping in large datasets may enhance the identification of causal SNPs associated with breast cancer risk. Likewise, GWAS should be intentional about including Latinas, particularly those with higher IA and/or African ancestry. In addition, future studies should prospectively assess prediction and examine the contribution of PRS to clinical risk models. Though one such trial is currently using the PRS to tailor decision making around breast cancer screening and prevention (9), similar clinical effectiveness studies also should aim to recruit diverse women.
Funding
This work was funded in part by grants from the National Cancer Institute (K24CA169004, R01CA120120 to EZ and R01CA184545 to EZ and SLN). YS was supported by the National Center for Advancing Translational Sciences under award KL2TR001870 and the National Cancer Institute under award K08CA237829.
The Northern California Breast Cancer Family Registry was supported by grant UM1 CA164920 from the National Cancer Institute. The SFBCS was funded by grants R01CA063446 and R01CA077305 from the National Cancer Institute, grant DAMD17-96–1-6071 from the US Department of Defense, and grant 7PB-0068 from the California Breast Cancer Research Program.
The Kaiser Permanente Research Program on Genes, Environment and Health was supported by Kaiser Permanente national and regional Community Benefit programs, and grants from the Ellison Medical Foundation, the Wayne and Gladys Valley Foundation, and the Robert Wood Johnson Foundation. Genotyping in the GERA cohort was supported by grant RC2AG036607 from the National Institutes of Health.
The Multiethnic Cohort Study was supported by the National Institutes of Health grants R01CA63464 and R37CA54281, R01CA132839, and 5UM1CA164973.
The CAMA Study was funded by Consejo Nacional de Ciencia y Tecnología, México (SALUD-2002-C01–7462).
The PEGEN-BC study was supported by the National Cancer Institute (R01CA204797 [LF]) and the Instituto Nacional de Enfermedades Neoplásicas (Lima, Peru).
The COLUMBUS Consortium was supported by grants from School of Medicine (Dean’s Fellowship in Precision Health Equity to LGC-C) and support from the Office of the Provost for LGC-C’s Latinos United for Cancer Health Advancement (LUCHA) Initiative); The V Foundation for Cancer Research (V Foundation Scholarship to LGC-C); GSK Oncology (Ethnic Research Initiative to LGC-C and ME); The US National Institutes of Health (Cancer Center Support Grant P30CA093372 from the National Cancer Institute). LGC-C, MEB, and ME are also grateful for support from Colciencias (Graduate Studentship to Jennyfer Benavides, member of COLUMBUS, from Convocatoria para la Formación de Capital Humano de Alto Nivel para el Departamento de Tolima— COLCIENCIAS — 755/2016), Universidad del Tolima, Colombia (Grants to MEB and ME, project 10112), and Sistema Nacional de Regalías, Gobernación del Tolima (grants to MEB and ME, project 520115). JT was supported by Coordinación Nacional de Investigación en Salud, IMSS, México, grant FIS/IMSS/PROT/PRIO/13/027, and by the Consejo Nacional de Ciencia y Tecnología (Fronteras de la Ciencia grant 773), México.
Notes
The study funders and sponsors did not participate in the collection, analysis, or interpretation of data, or in the writing of the manuscript. The contents of this article are solely the responsibility of the authors and do not reflect the official views of the National Institutes of Health. The authors declare no competing interests.
We thank the participants of the SFBCS/NC-BCFR, Kaiser Permanente RPGEH, MEC, CAMA, COLUMBUS, PEGEN-BC, and COH-CCGCRN studies.
The contributors from the COLUMBUS Consortium (in alphabetical order) include Jennyfer Benavides (Universidad del Tolima, Ibagué, Colombia), Mabel E. Bohórquez (Universidad del Tolima, Ibagué, Colombia), Fernando Bolaños (Hospital Hernando Moncaleano Perdomo, Neiva, Colombia), Luis G Carvajal-Carmona (Universidad del Tolima, Ibagué, Colombia, University of California Comprehensive Cancer Center, Sacramento, USA, Fundación de Genética y Genómica, Medellín, Colombia, Genome Center and Department of Biochemistry and Molecular Medicine, University of California, Davis, Davis, CA, USA), Jenny Carmona (Dinámica IPS, Medellín, Colombia), Ángel Criollo (Universidad del Tolima, Ibagué, Colombia), Magdalena Echeverry (Universidad del Tolima, Ibagué, Colombia), Ana Estrada-Flórez (Universidad del Tolima, Ibagué, Colombia), Gilbert Mateus (Hospital Federico Lleras Acosta, Ibagué, Colombia), Raúl Murillo (Pontificia Universidad Javeriana, Bogotá, Colombia), Justo Ramírez (Hospital Hernando Moncaleano Perdomo, Neiva, Colombia), Yesid Sánchez (Universidad del Tolima, Ibagué, Colombia), Carolina Sanabria (Instituto Nacional de Cancerología, Bogotá, Colombia), Martha Lucia Serrano (Instituto Nacional de Cancerología, Bogotá, Colombia), John Jairo Suárez (Universidad del Tolima, Ibagué, Colombia), and Alejandro Vélez (Dinámica IPS, Medellín, Colombia, Hospital Pablo Tobón Uribe, Medellín, Colombia).
Supplementary Material
References
- 1. Michailidou K, Hall P, Gonzalez-Neira A. , et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361, 361.e1–e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Michailidou K, Lindstrom S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lilyquist J, Ruddy KJ, Vachon CM, et al. Common genetic variation and breast cancer risk—past, present, and future. Cancer Epidemiol Biomarkers Prev. 2018;27(4):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mavaddat N, Pharoah PDP, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5):812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vachon CM, Pankratz VS, Scott CG, et al. The contributions of breast density and common genetic variation to breast cancer risk. J Natl Cancer Inst. 2015;107(5):dju397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shieh Y, Hu D, Ma L, et al. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res Treat. 2016;159(3):513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuzick J, Brentnall AR, Segal C, et al. Impact of a panel of 88 single nucleotide polymorphisms on the risk of breast cancer in high-risk women: results from two randomized tamoxifen prevention trials. J Clin Oncol. 2017;35(7):743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shieh Y, Eklund M, Madlensky L, et al. Breast cancer screening in the precision medicine era: risk-based screening in a population-based trial. JNCI J Natl Cancer Inst. 2017;109(5):djw290.. [DOI] [PubMed] [Google Scholar]
- 10. Hughes E, Judkins T, Wagner S. , et al. Development and validation of a residual risk score to predict breast cancer risk in unaffected women negative for mutations on a multi-gene hereditary cancer panel. J Clin Oncol. 2017;35(15 suppl):1579. [Google Scholar]
- 11. Black MH, Li S, LaDuca H. , et al. Polygenic risk score for breast cancer in high-risk women. J Clin Oncol. 2018;36(15 suppl):1508. [Google Scholar]
- 12. Park SL, Cheng I, Haiman CA.. Genome-wide association studies of cancer in diverse populations. Cancer Epidemiol Biomarkers Prev. 2018;27(4):405.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fejerman L, Stern MC, Ziv E, et al. Genetic ancestry modifies the association between genetic risk variants and breast cancer risk among Hispanic and non-Hispanic white women. Carcinogenesis. 2013;34(8):1787–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allman R, Dite GS, Hopper JL, et al. SNPs and breast cancer risk prediction for African American and Hispanic women. Breast Cancer Res Treat. 2015;154(3):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S, Qian F, Zheng Y, et al. Genetic variants demonstrating flip-flop phenomenon and breast cancer risk prediction among women of African ancestry. Breast Cancer Res Treat. 2018;168(3):703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.United States Census Bureau. Facts for Features: Hispanic Heritage Month 2017 Maryland: United States Census Bureau. https://www.census.gov/newsroom/facts-for-features/2017/hispanic-heritage.html (2018). Accessed October 31, 2018.
- 17. Bertoni B, Budowle B, Sans M, et al. Admixture in Hispanics: distribution of ancestral population contributions in the Continental United States. Hum Biol. 2003;75(1):1–11. [DOI] [PubMed] [Google Scholar]
- 18. Ziv E, John EM, Choudhry S, et al. Genetic ancestry and risk factors for breast cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bryc K, Velez C, Karafet T, et al. Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci. 2010;107(Suppl 2):8954–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fejerman L, Ahmadiyeh N, Hu D, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun. 2014;5(1):5260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffman J, Fejerman L, Hu D, et al. Identification of novel common breast cancer risk variants at the 6q25 locus among Latinas. Breast Cancer Res. 2019;21(1):3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. John EM, Horn-Ross PL, Koo J.. Lifetime physical activity and breast cancer risk in a multiethnic population: The San Francisco Bay Area Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2003;12(11 pt 1):1143–1152. [PubMed] [Google Scholar]
- 23. John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6(4):R375–R389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kvale MN, Hesselson S, Hoffmann TJ, et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200(4):1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angeles-Llerenas A, Ortega-Olvera C, Pérez-Rodríguez E, et al. Moderate physical activity and breast cancer risk: the effect of menopausal status. Cancer Causes Control. 2010;21(4):577–586. [DOI] [PubMed] [Google Scholar]
- 27. The Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacDonald DJ, Blazer KR, Weitzel JN.. Extending comprehensive cancer center expertise in clinical cancer genetics and genomics to diverse communities: the power of partnership. J Natl Compr Canc Netw. 2010;8(5):615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander DH, Novembre J, Lange K.. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galanter JM, Fernandez-Lopez JC, Gignoux CR, et al. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8(3):e1002554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drake KA, Torgerson DG, Gignoux CR, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133(2):370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ziv E, Tice JA, Sprague B, et al. Using breast cancer risk associated polymorphisms to identify women for breast cancer chemoprevention. PLoS One. 2017;12(1):e0168601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willer CJ, Li Y, Abecasis GR.. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeLong ER, DeLong DM, Clarke-Pearson DL.. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1998;44(3):837–845. [PubMed] [Google Scholar]
- 36. Fejerman L, Romieu I, John EM, et al. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol Biomarkers Prev. 2010;19(4):1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fejerman L, John EM, Huntsman S, et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res. 2008;68(23):9723–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ziv E, Burchard EG.. Human population structure and genetic association studies. Pharmacogenomics. 2003;4(4):431–441. [DOI] [PubMed] [Google Scholar]
- 40. Conomos MP, Laurie CA, Stilp AM, et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2016;98(1):165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuchenbaecker KB, McGuffog L, Barrowdale D, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109(7):djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sirugo G, Williams SM, Tishkoff SA.. The missing diversity in human genetic studies. Cell. 2019;177(1):26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin AR, Kanai M, Kamatani Y, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gravel S, Henn BM, Gutenkunst RN, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011;108(29):11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.