Abstract
BACKGROUND
An outbreak of listeriosis was identified in South Africa in 2017. The source was unknown.
METHODS
We conducted epidemiologic, trace-back, and environmental investigations and used whole-genome sequencing to type Listeria monocytogenes isolates. A case was defined as laboratory-confirmed L. monocytogenes infection during the period from June 11, 2017, to April 7, 2018.
RESULTS
A total of 937 cases were identified, of which 465 (50%) were associated with pregnancy; 406 of the pregnancy-associated cases (87%) occurred in neonates. Of the 937 cases, 229 (24%) occurred in patients 15 to 49 years of age (excluding those who were pregnant). Among the patients in whom human immunodeficiency virus (HIV) status was known, 38% of those with pregnancy-associated cases (77 of 204) and 46% of the remaining patients (97 of 211) were infected with HIV. Among 728 patients with a known outcome, 193 (27%) died. Clinical isolates from 609 patients were sequenced, and 567 (93%) were identified as sequence type 6 (ST6). In a case–control analysis, patients with ST6 infections were more likely to have eaten polony (a ready-to-eat processed meat) than those with non-ST6 infections (odds ratio, 8.55; 95% confidence interval, 1.66 to 43.35). Polony and environmental samples also yielded ST6 isolates, which, together with the isolates from the patients, belonged to the same core-genome multilocus sequence typing cluster with no more than 4 allelic differences; these findings showed that polony produced at a single facility was the outbreak source. A recall of ready-to-eat processed meat products from this facility was associated with a rapid decline in the incidence of L. monocytogenes ST6 infections.
CONCLUSIONS
This investigation showed that in a middle-income country with a high prevalence of HIV infection, L. monocytogenes caused disproportionate illness among pregnant girls and women and HIV-infected persons. Whole-genome sequencing facilitated the detection of the outbreak and guided the trace-back investigations that led to the identification of the source.
LISTERIOSIS, A SEVERE FOODBORNE Disease that has substantial mortality (20 to 30%), primarily affects persons with impaired cell-mediated immunity associated with pregnancy, extremes of age, underlying malignant conditions, human immunodeficiency virus (HIV) infection, chronic disease, or immunosuppressive therapy.1–5 Outbreaks are increasingly recognized,6,7 predominantly in upper-income countries where infection is more readily diagnosed,8 where existing surveillance programs facilitate early recognition,9 and where strain typing by whole-genome sequencing, which allows for identification of outbreak-linked cases and definitive attribution of the source, is accessible.10–14
An increase in the number of cases of listeriosis at two public hospitals in Gauteng Province, South Africa, during July and August 2017 prompted an investigation. Case numbers rapidly increased nationwide, and whole-genome multilocus sequence typing15 of Listeria monocytogenes isolates from patients identified a single sequence type (sequence type 6 [ST6]) in 93% of the cases. We used whole-genome sequencing and intensive epidemiologic and trace-back investigations to pursue the source of the outbreak. This report describes the key findings from the investigation.
METHODS
CASE DEFINITION
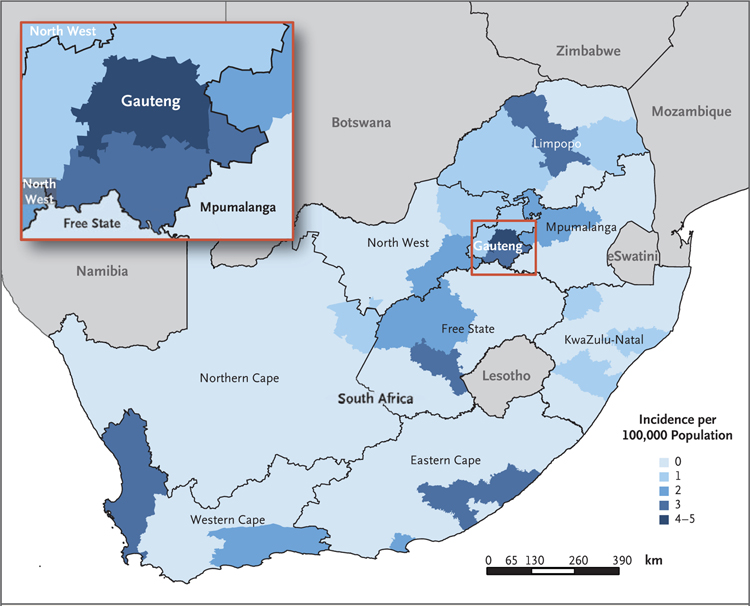
We defined an outbreak-associated case as laboratory-confirmed infection with L. monocytogenes, as determined by means of bacterial culture or polymerase-chain-reaction (PCR) assay of any clinical sample, during the outbreak period (June 11, 2017 [epidemiologic week 24 of that year], to April 7, 2018 [epidemiologic week 14]). This period was defined as the interval during which the case numbers at the national level exceeded and remained above a weekly threshold of five cases per week. The threshold of five cases per week was determined with the use of baseline laboratory data from January 1, 2013, to December 31, 2016. All L. monocytogenes infections were initially included in the case definition, because it was not possible to definitively exclude non-ST6 cases from the outbreak event; however, for the case–control analysis, the case definition was later refined to include ST6 cases only. Pregnancy-associated cases included illness with an onset during pregnancy or within the first 2 weeks of the postpartum period and illness in the neonate. The mothers of infected neonates were not counted among those who had cases if they did not have symptomatic laboratory-confirmed listeriosis; infection in a maternal–neonatal pair was defined as laboratory-confirmed infection in both the mother and neonate and was counted as a single case. Neonatal cases were classified as early onset (diagnosed between birth and day 6) or late onset (diagnosed between days 7 and 28). A map showing the incidence of infections according to district was generated (Fig. 1).
Figure 1. Incidence of Laboratory-Confirmed Cases of Listeriosis in South Africa during the Outbreak Period, According to District.
A total of 937 cases were detected during the outbreak period (June 11, 2017, to April 7, 2018). Incidence was calculated with the use of mid-year population data from 2017.16
EPIDEMIOLOGIC CASE INVESTIGATION
Clinical and demographic details and underlying medical conditions were ascertained through patient interviews or abstracted from medical records or laboratory reports with the use of a standardized case-investigation form. This investigation was reviewed in accordance with local and Centers for Disease Control and Prevention procedures for protection of human research participants and was considered nonresearch disease-control activity in a public health emergency. From November 1, 2017, all patients with newly reported cases were contacted to assess food exposures during the 4 weeks preceding the onset of illness with the use of a semistructured questionnaire. In cases in which the patient was a child, had died, or was too ill to respond, the next of kin were interviewed as proxies. In neonatal cases, history of food consumption by the mother during pregnancy was obtained.
Among the subgroup of patients with a detailed food history and available whole-genome sequencing results, a case–control analysis was performed to estimate the odds ratios for the association between specific food exposures and outbreak-associated illness. In this analysis, a case patient was defined as a person with L. monocytogenes ST6 infection and a control patient as a person with non-ST6 listeriosis during the outbreak period.
ENVIRONMENTAL AND TRACE-BACK INVESTIGATIONS
Health authorities initiated the collection of food samples from the homes of patients in mid-November 2017. When L. monocytogenes was isolated from a food sample, a trace-back investigation was conducted.
CHARACTERIZATION OF THE OUTBREAK STRAIN
L. monocytogenes isolates were sent to a national reference laboratory, where genomic bacterial DNA was isolated and whole-genome sequencing analysis performed as described previously.17 Genome assemblies were analyzed with the use of the multilocus sequence typing analysis pipeline at the Center for Genomic Epidemiology (www.genomicepidemiology.org). Data from multilocus sequence typing were used to determine clonal complexes and sequence types.15 Raw sequencing data were analyzed with the use of the Bacterial Isolate Genome Sequence Database for L. monocytogenes (BIGSdb-Lm, http://bigsdb.pasteur.fr/listeria/listeria.html) to determine sublineages and core-genome multilocus sequence types.18 The data exported from the BIGSdb-Lm were analyzed with BioNumerics Software, version 7.6.2 (bioMérieux) in order to perform a core-genome multilocus sequence typing–based phylogenetic analysis with the use of a single-linkage clustering algorithm.
The virulence of the ST6 strain was assessed in 7-to-10-week-old E16P KI C57BL/6 female mice, as previously described19; approval was obtained from the Institut Pasteur ethics committee. We assessed the virulence of the ST6/CT4148 YA00061615 CLIP2018/00699 human isolate (L1-SL6-ST6-CT4148, in which L denotes phylogenetic lineage, SL sublineage, ST sequence type, and CT core-genome multilocus sequence type) as the South African strain, and compared it with that of the EGDe ST9 reference strain (L2-SL9-ST35-CT637; National Center for Biotechnology Information (NCBI) GenBank accession number, NC_003210)20 and the CLIP2009/01092 ST6 isolate (L1-SL6-ST6-CT451; NCBI accession number, PRJEB10792).21 Overnight culture of L. monocytogenes was diluted in brain–heart infusion medium to reach mid-log growth phase. The mice were inoculated intragastrically through a feeding needle with 2×108 colony-forming units. The infected animals were killed 4 days after inoculation, and the organs were dissected and homogenized. Serial dilutions of ground-tissue suspensions in phosphate-buffered saline were inoculated on brain–heart infusion agar plates. After 24 hours of incubation at 37°C, the colony-forming units were counted.
RESULTS
OUTBREAK CASES
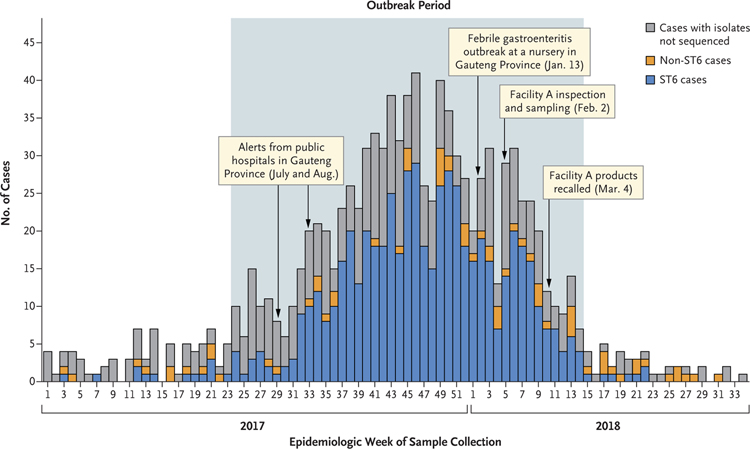
A total of 937 cases were reported during the outbreak period, with case numbers peaking at 41 per week in mid-November 2017 (epidemiologic week 46) (Fig. 2). ST6 was identified in 567 of 609 sequenced clinical isolates (93%). Although ST6 cases predominated during the outbreak period, smaller peaks of non-ST6 cases were noted. The number of cases decreased dramatically after recall of the implicated products on March 4, 2018. By mid-April 2018 (6 weeks after recall), fewer than 5 cases were reported weekly. Although cases were reported in all provinces, 543 of the 937 cases (58%) occurred in Gauteng Province, where the incidence reached 5 cases per 100,000 population in several districts (Fig. 1).
Figure 2. Number of Laboratory-Confirmed Cases of Listeriosis, According to Epidemiologic Week and Major Events (January 1, 2017, to August 21, 2018).
Shown are the whole-genome sequence types of 1055 isolates from the patients. The outbreak period was defined as the interval during which the case numbers exceeded and remained above a weekly threshold of five cases per epidemiologic week (June 11, 2017, to April 7, 2018). ST6 denotes sequence type 6.
CLINICAL INFORMATION
A total of 465 of the 937 cases (50%) were associated with pregnancy: 406 cases (43%) occurred in neonates and 59 (6%) in pregnant girls and women. Nine maternal–neonatal pairs were identified. Early-onset disease occurred in 95% of the neonatal cases. Of the 937 cases, 229 (24%) occurred in patients 15 to 49 years of age (excluding those who were pregnant) (Table 1). With the exclusion of the 59 girls and women known to be pregnant, female patients were overrepresented in the age group of 15 to 49 years (140 of 229 [61%]). A total of 83 cases (9%) occurred in persons 65 years of age or older. Overall, all but 2 patients were hospitalized, and no health care–associated infections were documented.
Table 1.
Characteristics of Patients with Laboratory-Confirmed Listeriosis during the Outbreak Period (June 11, 2017, to April 7, 2018).*
| Characteristic | No. of Cases (N = 937) | Pregnancy-Associated Cases | Cases According to Age Group | |||||
|---|---|---|---|---|---|---|---|---|
| Neonates (N = 406) | Pregnant Patients (N = 59) | 1 Mo to 14 Yr (N = 64) | 15 to 49 Yr (N = 229)† | 50 to 64 Yr (N = 90) | ≥65 Yr (N = 83) | Age Unknown (N = 6)‡ | ||
| Female sex — no./total no. of patients with known sex (%) | 516/918 (56) | 208/392 (53) | 59/59 (100) | 25/64 (39) | 140/229 (61) | 43/90 (48) | 39/83 (47) | 2/6 (33) |
| Risk factor — no./total no. of patients with known risk factor (%) | ||||||||
| HIV§ | 174/415 (42) | 60/158 (38) | 17/46 (37) | 2/33 (6) | 69/109 (63) | 21/43 (49) | 4/25 (16) | 1/1 (100) |
| Malignant condition | 20/188 (11) | NA | 0/42 | 2/15 (13) | 1/50 (2) | 10/42 (24) | 7/39 (18) | — |
| Diabetes mellitus | 20/187 (11) | NA | 1/42 (2) | 0/14 | 5/50 (10) | 8/42 (19) | 6/39 (15) | — |
| Renal failure | 10/187 (5) | NA | 0/42 | 0/14 | 3/50 (6) | 2/42 (5) | 5/39 (13) | — |
| Immunocompromising condition other than HIV¶ | 10/186 (5) | NA | 0/42 | 1/14 (7) | 1/50 (2) | 6/41 (15) | 2/39 (5) | — |
| Alcohol abuse | 10/187 (5) | NA | 0/42 | 0/14 | 2/50 (4) | 5/42 (12) | 3/39 (8) | — |
| Liver failure | 7/187 (4) | NA | 1/42 (2) | 0/14 | 2/50 (4) | 3/42 (7) | 1/39 (3) | — |
| Died — no./total no. of patients with known outcome (%) | 193/728 (27) | 86/308 (28) | 4/51 (8) | 7/49 (14) | 40/175 (23) | 27/73 (37) | 28/70 (40) | 1/2 (50) |
| Culture site — no./total no. of patients (%) | ||||||||
| Blood | 677/937 (72) | 376/406 (93) | 35/59 (59) | 39/64 (61) | 120/229 (52) | 50/90 (56) | 53/83 (64) | 4/6 (67) |
| Cerebrospinal fluid | 197/937 (21) | 19/406 (5) | 1/59 (2) | 22/64 (34) | 96/229 (42) | 36/90 (40) | 22/83 (27) | 1/6 (17) |
| Otherǁ | 63/937 (7) | 11/406 (3) | 23/59 (39) | 3/64 (5) | 13/229 (6) | 4/90 (4) | 8/83 (10) | 1/6 (17) |
The completeness of responses on the case-investigation forms varied among the patients; therefore, the patient denominators vary among the characteristics assessed. HIV denotes human immunodeficiency virus, and NA not applicable.
This age group excludes patients who were known to be pregnant.
A dash indicates that data were not available.
HIV-exposure status (i.e., maternal HIV status) was reported for neonates.
Immunocompromising conditions other than HIV included the receipt of glucocorticoid therapy or chemotherapeutic agents.
Other sample types included stool, urine, synovial fluid, tissue-biopsy specimen, pus, umbilical cord, placenta, amniotic fluid, cervical swab, and gastric aspirate.
HIV status was known in 415 of the 937 cases (44%). In 204 pregnancy-associated cases with known HIV status, 77 patients (38%) had positive HIV status, which included HIV exposure in 60 of 158 neonates (38%) and HIV infection in 17 of 46 pregnant girls and women (37%). Among the remaining 211 patients, 97 (46%) were infected with HIV. Among the 114 patients (excluding neonates) who were infected with HIV, 82 (72%) had available data on the CD4 T-lymphocyte count; the median count was 194 cells per cubic millimeter (interquartile range, 91 to 387). Maternal CD4 T-lymphocyte counts were known for 12 HIV-exposed neonates (20%); the median count was 479 cells per cubic millimeter (interquartile range, 322 to 575). After adjusting for age and sex, we found that the odds of ST6 infection were 48% lower among HIV-infected patients than among those without HIV infection, although odds lower than 78% or higher than 25% are also compatible with our data (odds ratio for ST6 infection, 0.52; 95% confidence interval [CI], 0.22 to 1.25). HIV-infected patients older than 1 month of age were 2.6 times as likely to have meningitis (confirmed by means of PCR assay of cerebrospinal fluid or cerebrospinal fluid culture) as HIV-negative patients (odds ratio, 2.55; 95% CI, 1.38 to 4.72). Predisposing medical conditions other than HIV infection were more common among patients 50 years of age or older.
The outcome was known for 728 patients (78%), among whom 193 deaths were reported (case-fatality ratio, 27%). HIV infection was associated with a 53% increased odds of death among patients older than 1 month, after adjustment for age and sex (odds ratio, 1.53; 95% CI, 0.75 to 3.15). Of the 4 maternal deaths reported, the underlying risk factors were known in 1 patient (diabetes mellitus and HIV infection). Fetal loss occurred in 27 of the 59 pregnant girls and women (46%).
CASE–CONTROL ANALYSIS
A total of 109 patients were interviewed. Consumption of polony (a ready-to-eat processed meat containing chicken, pork, beef, or any combination of these, similar to bologna) was reported by 93 patients (85%), and the brands produced by Facility A were the most commonly reported. Sequence data were available for 76 of the 109 patients: 65 had ST6 infections (case patients) and 11 had non-ST6 infections (control patients). The food items most strongly associated with ST6 infection included polony (odds ratio, 8.55; 95% CI, 1.66 to 43.35) and frozen chicken (odds ratio, 4.90; 95% CI, 1.04 to 25.55) (Table 2). Of the 57 case patients who reported eating polony, 50 (88%) reported eating brands manufactured at Facility A, although several patients reported eating several brands or did not specify a particular brand.
Table 2.
Case–Control Analysis of the Association between Specific Food Exposures and Listeria monocytogenes ST6 Infection.*
| Food Consumed | Case Patients (N = 65) | Control Patients (N = 11) | Odds Ratio (95% CI)† |
|---|---|---|---|
| no. (%) | |||
| Polony | 57 (88) | 5 (45) | 8.55 (1.66–43.35) |
| Kota‡ | 9 (14) | 0 | ND |
| Frozen chicken | 42 (65) | 4 (36) | 4.90 (1.04–25.55) |
| Any delicatessen-style ready-to-eat meat | 57 (88) | 8 (73) | 2.67 (0.37–14.38) |
| Apples | 51 (78) | 6 (55) | 3.86 (0.77–18.16) |
A total of 109 patients were interviewed, among whom sequence data were available for 76 patients (65 [86%] had L. monocytogenes ST6 infections [case patients] and 11 [14%] had non-ST6 listeriosis [control patients]). ND denotes not determined (unable to calculate odds ratio because of zero value in cell).
The odds ratio is for the association between the specific food item and L. monocytogenes ST6 infection.
Kota is a sandwich-type fast food containing polony.
TRACE-BACK AND ENVIRONMENTAL INVESTIGATIONS
On January 13, 2018, febrile gastroenteritis developed in 10 children from a nursery in Gauteng Province. Several stool samples were collected, and one yielded L. monocytogenes ST6. Sandwiches prepared and eaten at the nursery were the only common food exposure, and polony was the common ingredient. Polony was recovered from the nursery refrigerator, and L. monocytogenes ST6 was identified in the polony produced at Facility A.
On February 2, 2018, an environmental investigation was conducted at Facility A, located in Limpopo Province. Production of the polony entailed grinding and mixing raw ingredients, stuffing the emulsion into clipped nylon casings, cooking the polony loaves in hot water, and cooling the loaves in a brine chiller. Several areas were in poor repair, and many opportunities for cross-contamination of food products were identified, including condensation, unrestricted movement of workers, and prolonged reuse of brine for chilling.
L. monocytogenes was isolated from 47 of the 317 environmental samples (15%) collected at Facility A. A total of 34 of the 47 typed isolates (72%) were identified as ST6. These isolates originated from samples collected at several facility sections (precooking and postcooking), including from food-contact surfaces, non–food-contact surfaces, and chilling brine. L. monocytogenes ST6 was detected in 2 of 13 samples of unopened polony loaves collected at the facility and subsequently from polony loaves sold in retail stores.
CHARACTERIZATION OF THE OUTBREAK STRAIN
Whole-genome sequencing was performed in 710 human L. monocytogenes isolates (8 isolates collected in 2015; 37 in 2016; 455 in 2017; and 210 in 2018) and in 1061 food and environmental L. monocytogenes isolates collected between September 1, 2017, and March 31, 2018. On the basis of multilocus sequence typing, 567 of 609 isolates (93%) from the outbreak cases were identified as ST6, and the remainder represented 14 other sequence types. A total of 34 environmental isolates from Facility A and 19 isolates from food produced at Facility A were identified as ST6.
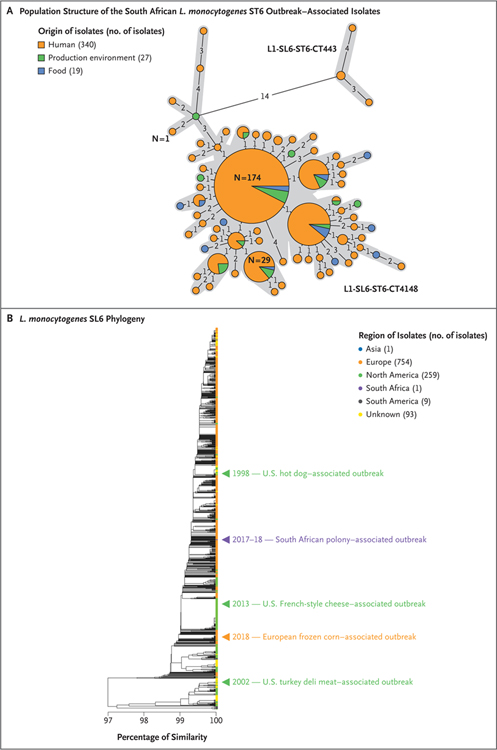
Whole-genome sequencing data for 386 ST6 isolates were analyzed with the use of core-genome multilocus sequence typing. Four human ST6 isolates that were recovered in 2017 and 2018 differed by at least 14 alleles, but the remaining 382 differed by no more than 4 alleles (out of 1748 loci included in the scheme) (Fig. 3A).18 This maximum 4-allelic difference is within the 7-allelic difference threshold that defines potentially epidemiologically linked isolates, as described by Moura et al.18 These 382 isolates, including 336 human isolates and 19 food and 27 environmental isolates from Facility A, were assigned to the same core-genome multilocus sequence type (CT4148; complete genotype, L1-SL6-ST6-CT4148). The oldest South African CT4148 isolates date from September 2015 and are related to a cluster of three cases of listeriosis in Western Cape Province28; this finding suggests a potential epidemiologic link to the 2017–2018 outbreak (Fig. 3A).
Figure 3. Population Structure of the South African Listeria monocytogenes ST6 Outbreak–Associated Isolates and of L. monocytogenes ST6 Isolates Collected Worldwide.
Panel A shows the diversity of the South African ST6 outbreak isolates based on core-genome multilocus sequence typing profiles. A total of 386 South African isolates are represented. The minimum spanning tree was constructed with the use of the minimum-spanning-tree algorithm available in the software tool BioNumerics, version 7.6.2 (bioMérieux). Each circle represents isolates that have a single core-genome multilocus sequence typing profile. Gray zones surround the isolates that have no more than 7 allelic differences (out of 1748 L. monocytogenes core genes),18 thus delineating the core-genome multilocus sequence types. The genotypes of the isolates are indicated next to the corresponding gray zone and provide information on the phylogenetic lineage (L), sublineage (SL), sequence type (ST), and core-genome multilocus sequence type (CT).18 The numbers shown on the black lines linking the circles correspond to the numbers of allelic differences between the isolates. These links are only indicative, because alternative links with equal weight might exist. The size of the circle reflects the number of isolates having similar core-genome multilocus sequence typing profiles; the number on or next to the circles indicates the number of isolates they contain. Panel B shows the genotypic relatedness of the South African ST6 outbreak isolates to the global ST6 population. The position of one isolate representative of the South African outbreak (HM00108598, CT4148, food origin) is indicated by the purple arrowhead. The positions of isolates representative of past outbreaks involving ST6 are also shown.22–27 Single-linkage–based cluster analysis was performed with the use of the core-genome multilocus sequence typing allelic profiles of isolates, as described previously.18 The scale bar indicates the percentage of allelic similarity among the core-genome multilocus sequence typing profiles of the isolates.
Whole-genome sequencing data were compared with information from curated databases through international networks and with isolates representative of previous major ST6 outbreaks to detect possible matches, and none were found (Fig. 3B, and Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org); genome sequences for 10 L. monocytogenes ST6 isolates associated with this South African outbreak have been deposited at the NCBI GenBank repository under the accession numbers QEXB00000000 to QEXK00000000 (Bio-Project number, PRJNA451422; and BioSample numbers, SAMN08970424 to SAMN08970415). Outbreak ST6 isolates had a hypervirulent phenotype, as described previously for L. monocytogenes ST6,21 but were not more virulent than a strain representative of ST6 (Fig. S2).
CONTROL MEASURES
On March 4, 2018, the Minister of Health announced the outbreak source. Facility A products were traced and recalled from distributors and retailers, and the public was advised to return products for reimbursement. Facility A was closed immediately. The World Health Organization assisted in recalling the products that had been exported to 15 African countries (Angola, Botswana, Democratic Republic of Congo, Ghana, Lesotho, Madagascar, Malawi, Mauritius, Mozambique, Namibia, Nigeria, eSwatini, Uganda, Zambia, and Zimbabwe). A single case of listeriosis was reported in Namibia during March 2018, but the patient’s isolate was confirmed as non-ST6; no other countries reported cases during the period from January 1, 2017, to September 3, 2018, when the outbreak was declared over.
DISCUSSION
Contaminated polony that was produced at a single facility was the cause of a large national outbreak of listeriosis predominantly associated with an L. monocytogenes ST6 strain in South Africa. Evidence supporting the cause includes a strong association between eating polony and ST6 infection, the detection of ST6 isolates in polony recovered from the refrigerator in the nursery, the detection of ST6 isolates in unopened polony loaves collected at the production facility, the detection of ST6 isolates in environmental samples collected from the production facility, a decline in ST6 cases after a recall of the products and closure of the facility, and the fact that outbreak-associated ST6 isolates from patients as well as food and environmental isolates from the facility belonged to a single core-genome multilocus sequence type.
Ready-to-eat processed meats are a well-known vehicle for listeriosis outbreaks.29,30 Polony is a low-cost, readily available food popular across all socioeconomic groups in South Africa and is used in kota, a fast food favored in urban areas. Polony has a shelf life of 5 months and is produced in large quantities by several manufacturers for local consumption and export.
Unique features of this outbreak include its recognition in a middle-income country with a high prevalence of HIV infection and a high fertility rate. HIV infection is a well-recognized risk factor for listeriosis.2,31–34 In 2017, the prevalence of HIV infection in South Africa was 12.6%, and an estimated 7.9 million people were living with HIV infection. The prevalence of HIV infection in the age group of 15 to 49 years was 26.3% among female persons and 14.8% among male persons.16 Of the nearly 1.2 million girls and women who gave birth in 2017, approximately 265,000 (22%) were infected with HIV.35 In this outbreak, HIV infection was the most common predisposing condition among patients younger than 65 years of age. We also found that a large percentage of pregnant girls and women were infected with HIV (37% [17 of 46]), and a similar percentage of neonates were exposed to HIV (38% [60 of 158]). The percentage of female patients was highest in the age group of 15 to 49 years, which suggests that possible unrecognized HIV infection or pregnancy were predisposing conditions. Culture-confirmed or PCR assay–confirmed L. monocytogenes infections were reported in more cerebrospinal fluid samples from HIV-positive patients than from HIV-negative patients. It is possible that HIV infection is associated with an increased mortality, but given the missing data and study design, the analyses were not powered to detect a difference between the HIV-infected group and the HIV-uninfected group.
The size and velocity of the outbreak were noteworthy and most likely resulted from the wide distribution of large volumes of contaminated products in a large population vulnerable to invasive listeriosis. Likely explanations for the predominance of cases in Gauteng Province include consumer behavior, food preferences, and higher socioeconomic status in this densely populated province, but increased reporting or differential health-seeking and physician-testing behavior may have been contributing factors. We found no evidence that this ST6 outbreak–associated strain was more virulent than a strain representative of ST6.21
No outbreak-associated cases were detected in the 15 low-to-middle-income countries that imported polony from Facility A. However, cases were probably missed because of nonspecific clinical presentation, physician-testing behavior, limited diagnostic capacity at the laboratory, and lack of surveillance for listeriosis. The burden of listeriosis is most likely higher than is currently recognized in low-income and middle-income countries,8 particularly those with large populations of people living with HIV.
Before this outbreak, listeriosis was not required to be reported and not under surveillance in South Africa. A national surveillance system has since been implemented, and all isolates from patients are analyzed by means of whole-genome sequencing. This outbreak catalyzed a revision of local food-safety regulations; certification through the Hazard Analysis and Critical Control Point system is now a legal requirement for ready-to-eat meat producers, and microbiologic criteria for L. monocytogenes in ready-to-eat foods are under review.
This outbreak investigation had several limitations. Not all clinical isolates were available for whole-genome sequence typing. Microbiologic investigations are not routinely conducted in pregnant women with mild, nonspecific febrile illness or in those who have had miscarriages or stillbirths; therefore, the number of patients with pregnancy-related listeriosis was most likely underestimated. With respect to the period under study, limited case-investigation forms were available, and data were of varying completeness. Insufficient clinical data prohibited a description of clinical syndromes. Sources of the data on HIV status included laboratory reports and reports from the patients themselves, which probably resulted in underreported positive status. On the basis of the findings from core-genome multilocus sequence typing, it is likely that the ST6 cluster in 2015 was associated with the 2017–2018 outbreak; however, incomplete histories of food consumption and trace-back data precluded a conclusive epidemiologic link.
The findings showcase the power of complementary epidemiologic data and whole-genome sequence typing for detecting and investigating foodborne disease outbreaks and show that whole-genome sequencing technology can be ably implemented and used in developing countries. As the global shift to whole-genome sequence typing for foodborne-pathogen surveillance accelerates,36–38 developing countries should build the capacity to leverage this technology in a rapidly evolving landscape of food-safety concerns. Targeted health communication for the prevention of listeriosis among pregnant girls and women and HIV-infected persons in developing countries may help mitigate the risk of disease in these vulnerable groups.
Supplementary Material
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
We thank the many people who assisted with this investigation, including laboratory colleagues, physicians, infection prevention and control practitioners, and nursing staff in public and private health sectors throughout South Africa; local, provincial, and national health departments; the Centre for Enteric Diseases, National Institute for Communicable Diseases (Masindi Ramudzulu, Dominic Ndlovu [in memoriam], Elias Khumalo, Jaime Macdonald, Sandrama Nadan, Rembuluwani Netshikweta, Tersia Kruger, Portia Mogale, Emily Dloboyi, Jack Kekana, Mzikazi Dickmolo, Sarah Gallichan, and Tina Duze); the personnel at the Sequencing Core Facility, National Institute for Communicable Diseases; the personnel at the Centre for Respiratory Diseases and Meningitis, National Institute for Communicable Diseases; Stefano Tempia and Cheryl Cohen for advice on statistical analyses; South African Field Epidemiology Training Programme residents (Poncho Bapela, Natasha Abraham, Mpho Sikhosana, Tracy Arendse, and Rudzani Mathebula); Katherine Calver; Vanessa Quan and surveillance officers of the Group for Enteric, Respiratory, and Meningeal Disease Surveillance in South Africa (GERMS-SA), National Institute for Communicable Diseases; Sue Candy and Jimmy Khoza, National Institute for Communicable Diseases; the personnel at the Infection Control Services Laboratory, National Health Laboratory Service; the acting chief executive officer and board of the National Health Laboratory Service; Mary-Anne Groepe and Rufaro Chatora, World Health Organization Country Office, South Africa; Yolandi Nel and Mohamed Cara at Food Consulting Services; colleagues from the Division of Foodborne, Waterborne, and Environmental Diseases at the Centers for Disease Control and Prevention in Atlanta for their input and guidance during the outbreak (especially Amanda Conrad, Matthew Wise, and Peter Gerner-Smidt); David Moore and Kathy-Anne Strydom for triggering the outbreak investigation by alerting the National Institute for Communicable Diseases of increased cases at their respective institutions; Preeteeben Vallabh at Chris Hani Baragwanath Academic Hospital for her recognition of the nursery outbreak; Wasingi Masingi and Gabisile Masingi for the nursery outbreak environmental investigation; the Minister of Health, Aaron Motsoaledi, for his support; Lynn Morris, executive director of the National Institute for Communicable Diseases, for her support and encouragement; and the National Institute for Communicable Diseases of the National Health Laboratory Service, Institut Pasteur, INSERM, Santé Publique France, and the Inception Program for providing internal funds to support the study.
APPENDIX
The authors’ full names and academic degrees are as follows: Juno Thomas, M.D., Nevashan Govender, M.Sc., M.P.H., Kerrigan M. McCarthy, M.D., Linda K. Erasmus, M.D., Timothy J. Doyle, Ph.D., Mushal Allam, Ph.D., Arshad Ismail, Ph.D., Ntsieni Ramalwa, M.P.H., Phuti Sekwadi, M.P.H., Genevie Ntshoe, M.P.H., Andronica Shonhiwa, M.P.H., Vivien Essel, M.D., Nomsa Tau, M.S., Shannon Smouse, M.S., Hlengiwe M. Ngomane, M.T., Bolele Disenyeng, M.T., Nicola A. Page, Ph.D., Nelesh P. Govender, M.D., Adriano G. Duse, M.D., Rob Stewart, M.T., Teena Thomas, M.D., Deon Mahoney, M.S., Mathieu Tourdjman, M.D., Olivier Disson, Ph.D., Pierre Thouvenot, B.S., Mylène M. Maury, Ph.D., Alexandre Leclercq, M.S., Marc Lecuit, M.D., Ph.D., Anthony M. Smith, Ph.D., and Lucille H. Blumberg, M.D.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
J. Thomas, Centre for Enteric Diseases, South Africa
N. Govender, Division of Public Health Surveillance and Response, South Africa
K.M. McCarthy, Division of Public Health Surveillance and Response, South Africa; National Institute for Communicable Diseases, National Health Laboratory Service, the University of the Witwatersrand, South Africa
L.K. Erasmus, Division of Public Health Surveillance and Response, South Africa
T.J. Doyle, Division of Global Health Protection, Center for Global Health, Centers for Disease Control and Prevention, Pretoria, South Africa
M. Allam, Sequencing Core Facility, South Africa
A Ismail, Sequencing Core Facility, South Africa.
N. Ramalwa, Centre for Enteric Diseases, South Africa; University of Pretoria, Tshwane, South Africa
P. Sekwadi, Centre for Enteric Diseases, South Africa
G. Ntshoe, Division of Public Health Surveillance and Response, South Africa; University of Pretoria, Tshwane, South Africa
A. Shonhiwa, Division of Public Health Surveillance and Response, South Africa
V. Essel, Division of Public Health Surveillance and Response, South Africa
N. Tau, Centre for Enteric Diseases, South Africa
S. Smouse, Centre for Enteric Diseases, South Africa
H.M. Ngomane, Centre for Enteric Diseases, South Africa
B. Disenyeng, Centre for Enteric Diseases, South Africa
N.A. Page, Centre for Enteric Diseases, South Africa
N.P. Govender, Centre for Healthcare-Associated Infections and Antimicrobial Resistance, South Africa; National Institute for Communicable Diseases, National Health Laboratory Service, the University of the Witwatersrand, South Africa; University of Pretoria, Tshwane, South Africa
A.G. Duse, National Institute for Communicable Diseases, National Health Laboratory Service, the University of the Witwatersrand, South Africa; School of Pathology, National Health Laboratory Service, Johannesburg, South Africa
R. Stewar, School of Pathology, National Health Laboratory Service, Johannesburg, South Africa
T. Thomas, National Institute for Communicable Diseases, National Health Laboratory Service, the University of the Witwatersrand, South Africa; School of Pathology, National Health Laboratory Service, Johannesburg, South Africa
D. Mahoney, Deon Mahoney Consulting, Melbourne, VIC, Australia
M. Tourdjman, Santé Publique France, the French Public Health Agency, Saint-Maurice, France
O. Disson, Institut Pasteur, Biology of Infection Unit, INSERM Unité 1117 and National Reference Center–WHO Collaborating Center for Listeria, France
P. Thouvenot, Institut Pasteur, Biology of Infection Unit, INSERM Unité 1117 and National Reference Center–WHO Collaborating Center for Listeria, France
M.M. Maury, Institut Pasteur, Biology of Infection Unit, INSERM Unité 1117 and National Reference Center–WHO Collaborating Center for Listeria, France
A. Leclercq, Institut Pasteur, Biology of Infection Unit, INSERM Unité 1117 and National Reference Center–WHO Collaborating Center for Listeria, France
M. Lecuit, Institut Pasteur, Biology of Infection Unit, INSERM Unité 1117 and National Reference Center–WHO Collaborating Center for Listeria, France; Université de Paris, Division of Infectious Diseases and Tropical Medicine, Necker–Enfants Malades University Hospital, Assistance Publique–Hôpitaux de Paris, Institut Imagine, France
A.M. Smith, Centre for Enteric Diseases, South Africa; National Institute for Communicable Diseases, National Health Laboratory Service, the University of the Witwatersrand, South Africa
L.H. Blumberg, Division of Public Health Surveillance and Response, South Africa; University of Stellenbosch, Stellenbosch, South Africa
REFERENCES
- 1.Gerner-Smidt P, Ethelberg S, Schiellerup P, et al. Invasive listeriosis in Denmark 1994–2003: a review of 299 cases with special emphasis on risk factors for mortality. Clin Microbiol Infect 2005; 11: 618–24. [DOI] [PubMed] [Google Scholar]
- 2.Goulet V, Hebert M, Hedberg C, et al. Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. Clin Infect Dis 2012;54: 652–60. [DOI] [PubMed] [Google Scholar]
- 3.Charlier C, Perrodeau É, Leclercq A, et al. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 2017;17:510–9. [DOI] [PubMed] [Google Scholar]
- 4.Listeria illnesses, deaths, and outbreaks — United States, 2009–2011. MMWR Morb Mortal Wkly Rep 2013; 62: 448–52. [PMC free article] [PubMed] [Google Scholar]
- 5.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect 2007;9:1236–43. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, Whiting RC. A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017; 75:1–13. [Google Scholar]
- 7.Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 2010;16:16–23. [DOI] [PubMed] [Google Scholar]
- 8.de Noordhout CM, Devleesschauwer B, Angulo FJ, et al. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14: 1073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Valk H, Jacquet C, Goulet V, et al. Surveillance of listeria infections in Europe. Euro Surveill 2005;10:251–5. [PubMed] [Google Scholar]
- 10.Schjørring S, Gillesberg Lassen S, Jensen T, et al. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Euro Surveill 2017;22(50):17–00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moura A, Tourdjman M, Leclercq A, et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg Infect Dis 2017; 23: 1462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson BR, Tarr C, Strain E, et al. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 2016; 63: 380–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong JC, Mercoulia K, Tomita T, et al. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 2016; 54: 333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid D, Allerberger F, Huhulescu S, et al. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 2014; 20: 431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragon M, Wirth T, Hollandt F, et al. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 2008; 4(9): e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Fifth South African National HIV Prevalence, Incidence, Behaviour and Communication Survey, 2017. Cape Town, South Africa: Human Sciences Research Council, 2018. (http://www.hsrc.ac.za/uploads/pageContent/9234/SABSSMV_Impact_Assessment_Summary_ZA_ADS_cleared_PDFA4.pdf). [Google Scholar]
- 17.Allam M, Tau N, Smouse SL, et al. Whole-genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc 2018;6:(25):e00538–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moura A, Criscuolo A, Pouseele H, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2016; 2: 16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Disson O, Nikitas G, Grayo S, Dussurget O, Cossart P, Lecuit M. Modeling human listeriosis in natural and genetically engineered animals. Nat Protoc 2009; 4: 799–810. [DOI] [PubMed] [Google Scholar]
- 20.Glaser P, Frangeul L, Buchrieser C, et al. Comparative genomics of Listeria species. Science 2001;294:849–52. [DOI] [PubMed] [Google Scholar]
- 21.Maury MM, Tsai YH, Charlier C, et al. Uncovering Listeria monocytogenes hyper-virulence by harnessing its biodiversity. Nat Genet 2016;48:308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Zhang W, Knabel SJ. Multi-virulence-locus sequence typing clarifies epidemiology of recent listeriosis outbreaks in the United States. J Clin Microbiol 2005;43:5291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Update: multistate outbreak of listeriosis — United States, 1998–1999. MMWR Morb Mortal Wkly Rep 1999; 47: 1117–8. [PubMed] [Google Scholar]
- 24.Outbreak of listeriosis — northeastern United States, 2002. MMWR Morb Mortal Wkly Rep 2002; 51: 950–1. [PubMed] [Google Scholar]
- 25.Kathariou S, Graves L, Buchrieser C, Glaser P, Siletzky RM, Swaminathan B. Involvement of closely related strains of a new clonal group of Listeria monocytogenes in the 1998–99 and 2002 multistate outbreaks of foodborne listeriosis in the United States. Foodborne Pathog Dis 2006; 3: 292–302. [DOI] [PubMed] [Google Scholar]
- 26.Multistate outbreak of listeriosis linked to Crave Brothers Farmstead cheeses (final update). Atlanta: Centers for Disease Control and Prevention, 2013. (https://www.cdc.gov/listeria/outbreaks/cheese-07−13/index.html). [Google Scholar]
- 27.European Food Safety Authority. Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections linked to frozen corn and possibly to other frozen vegetables — first update. Technical report. Stockholm: European Centre for Disease Prevention and Control, 2018. (https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/sp.efsa.2018.EN-1448). [Google Scholar]
- 28.Smith AM, Naicker P, Bamford C, et al. Genome sequences for a cluster of human isolates of Listeria monocytogenes identified in South Africa in 2015. Genome Announc 2016;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead PS, Dunne EF, Graves L, et al. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiol Infect 2006; 134:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacquet C, Catimel B, Brosch R, et al. Investigations related to the epidemic strain involved in the French listeriosis outbreak in 1992. Appl Environ Microbiol 1995;61:2242–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maertens De Noordhout C, Devleesschauwer B, Maertens De Noordhout A, et al. Comorbidities and factors associated with central nervous system infections and death in non-perinatal listeriosis: a clinical case series. BMC Infect Dis 2016; 16:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joint FAO/WHO expert consultation on risk assessment of microbiological hazards in foods. Rome: Food and Agriculture Organization of the United Nations, July 2000. (http://www.fao.org/3/x8124e/x8124e00.htm). [Google Scholar]
- 33.Kales CP, Holzman RS. Listeriosis in patients with HIV infection: clinical manifestations and response to therapy. J Acquir Immune Defic Syndr 1990; 3: 139–43. [PubMed] [Google Scholar]
- 34.Schuchat A, Swaminathan B, Broome CV. Epidemiology of human listeriosis. Clin Microbiol Rev 1991; 4: 169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mid-year population estimates, 2017. Pretoria: Statistics South Africa, 2017. (https://www.statssa.gov.za/publications/P0302/P03022017.pdf). [Google Scholar]
- 36.Jagadeesan B, Gerner-Smidt P, Allard MW, et al. The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol 2019; 79:96–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadon C, Van Walle I, Gerner-Smidt P, et al. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 2017; 22(23):30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whole genome sequencing for foodborne disease surveillance: landscape paper. Geneva: World Health Organization, 2018. (https://apps.who.int/iris/bitstream/handle/10665/272430/9789241513869-eng.pdf?ua=1). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.