Abstract
Identification of proteins that are synthesized de novo in response to specific microenvironmental cues is critical for understanding molecular mechanisms that underpin vital physiological processes and pathologies. Here, we report that a brief period of SILAM (Stable Isotope Labeling of Mammals) diet enables the determination of biological functions corresponding to actively translating proteins in the mouse brain. Our results demonstrate that the synapse, dendrite, and myelin sheath are highly active neuronal structures that display rapid protein synthesis, producing key mediators of chemical signaling as well as nutrient sensing, lipid metabolism, and amyloid precursor protein processing/stability. Together, these findings confirm that protein metabolic activity varies significantly between brain functional units in vivo. Our data indicate that pulsed SILAM approaches can unravel complex protein expression dynamics in the murine brain and identify active synthetic pathways and associated functions that are likely impaired in neurodegenerative diseases.
1. Introduction
Rapid changes in the cellular proteome are required to support critical biological responses to a diverse range of internal and external stimuli. Rates of protein synthesis are thus tightly regulated in vivo and vary significantly between the cell type and the molecule in question, even under normal physiological conditions. In contrast, disruption of steady-state proteome dynamics is thought to be a vital component of the aging process and is implicated in abnormal tissue development and various pathological disorders,1,2 including chronic inflammation,3 Alzheimer’s disease,4 skeletal dysplasia, fibrosis, and cancer.5 However, it is extremely challenging to unravel protein expression dynamics under normal physiological conditions in vivo; hence, we have only limited understanding of how translational activity is modified naturally over time or pathologically altered during progression to a disease.
In the brain, regulation of protein synthesis is critically required for a wide range of normal biological functions, including neuron growth, electrical impulses, signals transmission, learning, and memory formation.6,7 Previous studies have attempted to assess the brain protein turnover using approaches such as “stable isotope labeling by amino acids in cell culture” (SILAC) for in vitro analyses and “stable isotope labeling of mammals” (SILAM) to perform experiments in animal models.6,8−11 Using an innovative approach of feeding mice with N15-labeled blue-green algae prior to conducting mass spectrometry analysis of brain tissues, Pierce et al. were able to determine turnover rates for 1010 proteins at the whole organ level.8 Similarly, McClatchy et al. used SILAM diet in Rattus norvegicus to identify 3081 brain proteins that undergo significant changes during normal organ development and 1896 proteins that exhibited differential translation patterns between the cerebellum, cortex, and hippocampus.6 Subsequent work by Cohen et al. successfully discerned the half-life of 2802 different brain proteins via proteomic analysis of cortical cultures,9 while Dörrbaum et al. used mass spectrometry to assess the stability of >5100 proteins in cultures of rat hippocampus tissue.10 However, proteins generated in vivo are derived from at least two different amino acid sources: dietary intake and degradation of pre-existing proteins. Consequently, it remains unclear whether protein longevity as assessed in earlier studies is truly reflecting the protein lifespan in vivo. Recently, Fornasiero et al. made significant progress in this area by establishing a workflow for assessing the protein turnover in the mouse brain in vivo by using a combination of isotope labeling via SILAC diet with mass spectrometry and mathematical modeling.12 With this approach, they were able to determine the longevity of ∼3500 brain proteins in vivo, thus providing valuable new insights into protein stability in this organ. However, it remains unclear as to what extent specific cellular signals can trigger progressive changes in protein translational activity within different regions of the brain. Indeed, brain cells interact with their local environment and neighboring cells via a complex network of signaling pathways that can modify patterns of protein translation, modification, and degradation, leading to significant effects on overall organ functions. De novo protein synthesis in response to a specific stimulus is required to facilitate the rapid generation and transduction of appropriate signaling events that directly influence downstream physiological processes. It is therefore highly desirable to develop a method capable of profiling proteins that are being actively translated in vivo in order to better understand the molecular events that regulate essential functions under both normal and pathological conditions. Biorthogonal noncanonical amino acid tagging (BONCAT) is a novel method for newly synthesized protein profiling. It uses azidohomoalanine (AHA), an analog of methionine containing an azide moiety, to label proteins during ribosome protein synthesis.13,14 The AHA can be supplied through diet,15 injection,16 cell culture media,17 etc. and is accepted by the endogenous methionine tRNA in vivo without modulating on protein functions. AHA-modified proteins can then be isolated by azide-alkyne click-chemistry, avidin-based affinity purification, and on-resin trypsinization to enrich and identify newly synthesized proteins selectively (14). In recent years, SILAM-based labeling has emerged as a powerful method for mammalian protein labeling and quantification in vivo,6,8−11,18 but current protocols require that animals are fed with an expensive isotype-tagged diet for at least two generations prior to conducting experiments.19 The subsequent development of neutron-encoded (NeuCode) stable isotope metabolic labeling reagents has since reduced the minimum feeding period to 3–4 weeks before investigators can perform multiplex analyses of proteome dynamics. The NeuCode labeling protocol requires neuron-encoded amino acids, lysyl endopeptidase (Lys-C), and high-resolution MS1 spectra (≥240,000 resolving power @ m/z 400), while assuming equal efficiency of label incorporation across all the isotopologues present.20 Pulsed-SILAC approaches can trace active cellular translation events in response to a specific trigger at a particular time point, as demonstrated by previous in vitro studies of microbial resistance factors,21 oncoprotein expression in cancer cells,22,23 and host mediators of antiviral immunity.24,25 These stimulus-specific data can account for variable expression over time and provide essential information about normal cell physiology and potentially uncover novel targets for therapy in a range of diseases, but comparative analyses of different tissue compartments in vivo will be essential in realizing this potential.
In the current study, we hypothesized that proteins being actively translated under specific conditions in vivo correspond to the molecular mediators and pathways that underpin normal cellular physiology in a given tissue compartment. We therefore developed a new protocol termed “pulsed Stable Isotope in vivo Labeling of Mammals” (pSILAM) that facilitates analysis of actively translating proteins after just 2 days of mouse feeding with a diet containing 13C6-l-lysine. Proteomic profiling of the brain tissues from these animals by liqiud chromatography-tandem mass spectrometry (LC–MS/MS) enabled robust and reproducible identification of 7868 total protein groups among which 1223 proteins were being actively translated primarily within the synapse, dendrite, and myelin sheath. Our study demonstrates that pSILAM is both a cost-effective and time-saving new protocol that supports efficient analysis of actively translating protein profiles in vivo. This approach can reveal the active molecular events underlying critical biological processes within specific tissue compartments in living animals and will shed important new light on how protein expression profiles change in response to defined stimuli encountered in disparate organ systems.
2. Results and Discussion
2.1. Optimal SILAM Diet Feeding for Analysis of Active Protein Translation in the Murine Brain
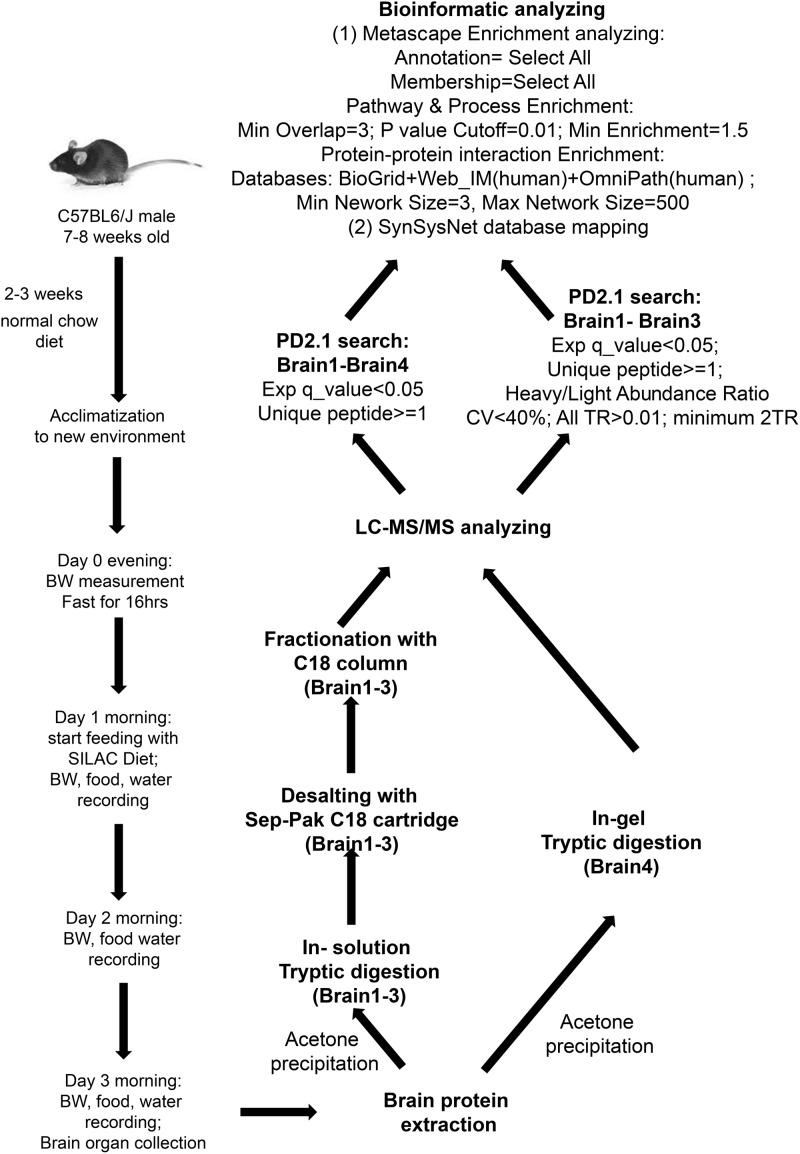
Measuring proteins that are actively synthesized under specific conditions in vivo provides valuable insights into the molecular events that underlie critical biological functions. We therefore developed a new protocol that uses the brief provision of a 13C6-l-lysine-labeled diet to enable “Stable Isotope in vivo labeling of Mammals” (pulsed SILAM) and facilitated tracing of proteins that are being actively translated in live animal tissues. We first determined the optimal pSILAM diet feeding time using previously published data reporting brain protein turnover rates (shown in Table S1 and Figure S1).8 In these experiments, the number of heavy-labeled peptides detected was found to increase exponentially over the first few days of the pSILAM diet, with >23% of total detectable peptides incorporating the label within just 2 days of feeding. We therefore concluded that 2 days of pSILAM diet is sufficient to measure the most active protein groups in the mouse brain that likely correspond to essential physiological and/or pathological processes. In our protocol (Figure 1), male C57BL6/J mice were fasted for 16 h then transferred onto SILAM diet with free access to drinking water for 48 h after that. Daily recording of the mouse body mass and food consumption indicated that the average SILAM dietary intake was 0.15 ± 0.01 g per gram body weight per day (Table S2 and Figure S2). After SILAM pulsing, the brain tissues were excised from euthanized mice for total protein extraction and in-solution digestion followed by high performance liquid chromatography (HPLC) tryptic peptide fractionation (samples Brain1, Brain2, and Brain3), or protein fractionation using SDS-PAGE followed by in-gel trypsin digestion (Brain4). The extracted and fractionated peptides obtained were then injected into a Q-Exactive LC–MS/MS system for further analysis. The LC–MS/MS raw data were subsequently searched using Proteome Discoverer version 2.1 (PD2.1) for protein identification and quantification.
Figure 1.
Workflow for pulsed-SILAC in vivo labeling of mouse brain tissues and subsequent proteomic analysis. Male C57BL6/J mice aged 7–8 weeks were acclimatized to their environment for 2–3 weeks on a regular mouse chow diet containing 12C6-l-lysine. After 16 h fasting overnight, the animals were switched onto a 13C6-l-lysine-labeled diet for 2 days duration. The mice were then euthanized, and brain tissues were excised for protein extraction and mass spectrometry analysis. Subsequent proteome analysis allowed the identification of newly synthesized proteins (13C6-l-lysine-labeled) within whole mouse brain tissues. Mouse photograph courtesy of https://imgbin.com/ Copyright 2020.
We identified a total of 7868 unique protein groups (Table S3B) in the four mouse brain proteome samples, which displayed an average H/L ratio of 5.7% (calculated as the cumulative abundance of all heavy-labeled proteins divided by the cumulative abundance of all light proteins in our datasets). This protein labeling efficiency was comparable with previously reported protein turnover values for the mouse brain (Figure S1B).8 We then compared our protein list against three different expression data sets, (1) mouse brain proteome (http://www.mousebrainproteome.com/); (2) ProteinAtlas (https://www.proteinatlas.org/) for protein and RNA expression; and (3) Bgee for RNA expression (https://bgee.org/). These analyses confirmed that 7681 out of the 7868 protein groups detected are known to be expressed in the brain (97.6%), and 263 out of 303 protein groups correspond to the genes known to be transcribed in the brain. Among these 7868 protein groups, 4004 were detected in all four mouse brains analyzed with peptide false discovery rate (FDR) <1%, protein FDR <5%, and ≥1 unique high confidence peptide in each group. We then determined relative synthetic activity by measuring the heavy-to-light abundance ratio of each protein based on precursor ion quantification (using only unique tryptic peptides associated with each protein). To do this, the protein abundance ratio was calculated using the relative MS signal intensities of the heavy isotope-labeled protein over its unlabeled counterpart by using PD2.1 default settings. PD2.1 considers the extracted ion chromatogram (areas under the peak) of each precursor detected and adds the quantification channel values of the PSMs that meet the specified criteria (set to include all peptide groups and proteins, irrespective of charge and modification state). The minimum and maximum fold change values were set to 0.01 and 100, respectively, then used to identify actively translating proteins in the murine brain samples (illustrative examples are provided in Figure S3). With this approach, we discovered a total of 729 quantifiable protein groups at high confidence across all the four data sets, with the heavy-to-light abundance ratio ranging between 0.05 and 1.57. These data confirmed that pSILAM allows efficient identification of brain proteins that are being actively translated in vivo after only a short duration of the isotope-labeled diet.
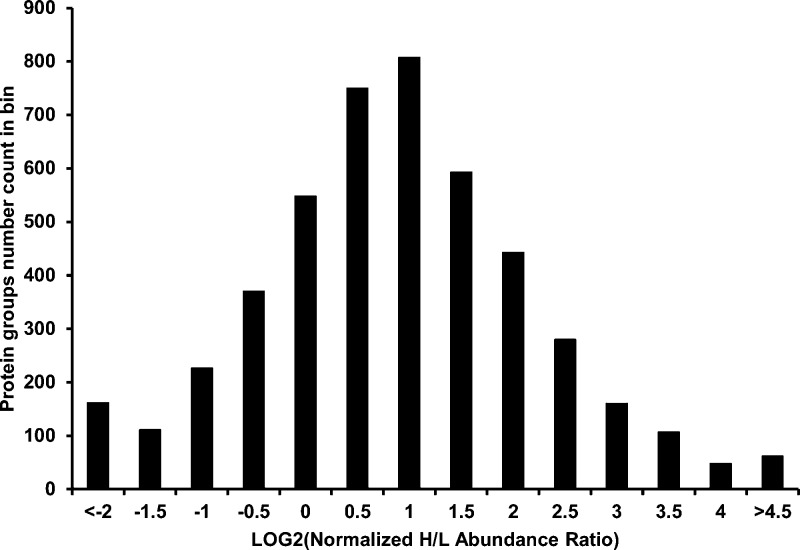
We next focused on brain samples 1–3 that were prepared by in-solution digestion, which yielded a total of 7679 unique protein groups (master proteins) (Table S4A-Master Proteins). Actively translating proteins are defined here as proteins with the H/L ratio > proteome-wide average H/L ratio. The average brain protein heavy lysine labeling efficiency in 2 days as determined by the average (H/L) ratio of the three brain proteomes was estimated at 5.6% (Table S4B-Average H/L). This value was then used to normalize the abundance ratio of each brain protein detected (i.e., the H/L ratio of each protein is divided by 0.056). Among the 7679 unique protein groups identified, 3011 proteins have no H/L ratio quantified by PD2.1. The H/L ratios exhibited by 1418 protein groups were less than the proteome-wide average of 0.056, and 3250 protein groups displayed a higher-than-average H/L ratio. As shown in Figure 2, we observed that the 4668 H/L quantifiable protein groups displaying translational activity followed a normal distribution when assessed by LOG2(normalized H/L abundance ratio) (Tables S4A4 and S4A5-Bin/Histogram). A total of 2331 protein groups were found to be both heavy-labeled and quantifiable with a biological correlation ≥0.79 (FDR <1% for peptides, <1% for proteins; Table S4A, Selected proteins). We further refined the candidate list by applying two additional criteria; the average H/L abundance ratio between biological samples (cutoff: 0.05) and the respective technical H/L abundance ratio coefficient variance (Abundance Ratio H/L CV(%):40% cutoff). After filtering, a total of 1222 protein groups were selected as high confidence candidates (biological correlations ≥ 0.82, technical replicate correlations ≥ 0.93; Table 1).
Figure 2.
Distribution of the average H/L abundance ratio of master protein groups. Brain protein heavy lysine labeling efficiency was determined as 5.6% (based on the average H/L ratio of the three brain proteomes after 2 days pSILAM diet). This value was then used to normalize the abundance ratio, i.e., average H/L of each protein from three biological replicates was divided by 0.056. Among the 4668 protein groups that have been quantified in all the three biological replicates, with LOG2(normalized H/L abundance ratio) adopting a normal distribution, a total of 1418 protein groups displayed H/L ratios less than the proteome-wide average of 0.056, while 3250 protein groups displayed a H/L ratio greater than the average.
Table 1. Biological and Technical Replicate Correlation Values between Brain Samples 1–3a.
| (A) biological replicates | correlation |
|---|---|
| Brain1 vs Brain2 | 0.82 |
| Brain1 vs Brain3 | 0.82 |
| Brain2 vs Brain3 | 0.90 |
| (B) technical replicates | correlation |
|---|---|
| Brain1_TR1 vs Brain1_TR2 | 0.94 |
| Brain1_TR1 vs Brain1_TR3 | 0.95 |
| Brain1_TR2 vs Brain1_TR3 | 0.94 |
| Brain2_TR1 vs Brain2_TR2 | 0.94 |
| Brain2_TR1 vs Brain2_TR3 | 0.93 |
| Brain2_TR2 vs Brain2_TR3 | 0.94 |
| Brain3_TR1 vs Brain3_TR2 | 0.96 |
| Brain3_TR1 vs Brain3_TR3 | 0.96 |
| Brain3_TR2 vs Brain3_TR3 | 0.96 |
The protein groups listed in Table S4B were used to calculate correlation values via filtering using two additional parameters: (1) heavy-to-light abundance ratio coefficient variance between the technical triplicate <40%, and (2) heavy-to-light abundance ratio > 0.05 (n = 1223).
2.2. Synapse, Dendrite, and Myelin Sheath are Active Neuronal Structures in the Murine Brain
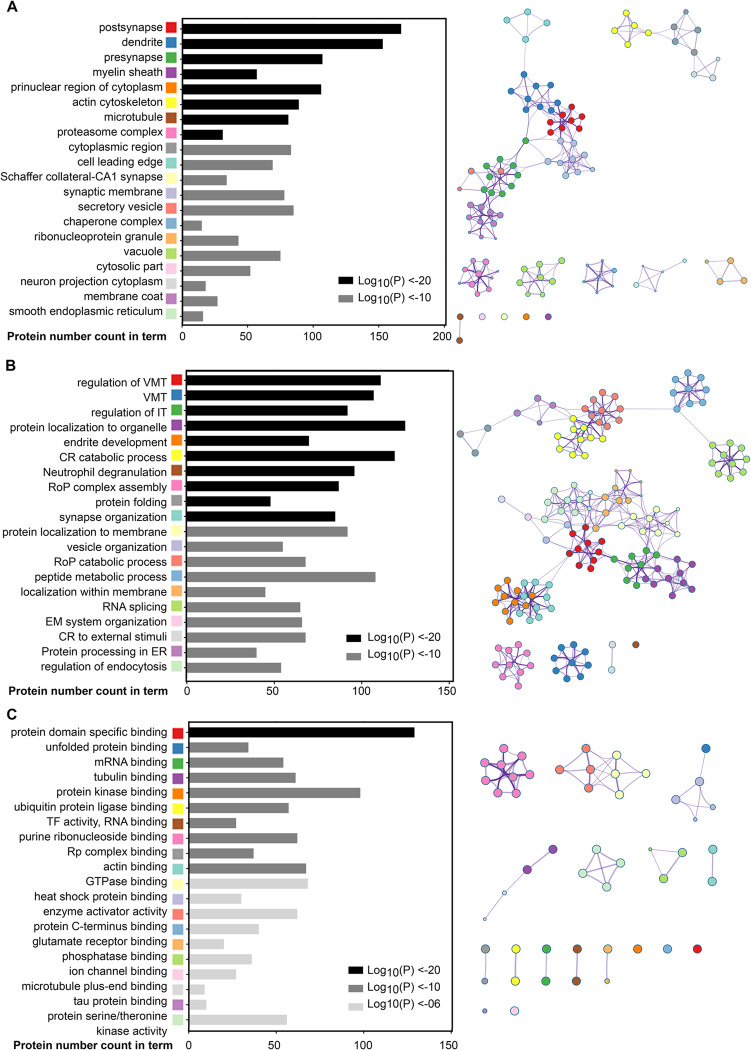
To understand the biological functions of proteins being actively translated in the murine brain, we next performed Metascape gene enrichment clustering analysis on the 1218 corresponding genes across four distinct categories, (1) structure, (2) pathway, (3) function and (4) protein–protein interaction (Table S5). As shown in Figure 3 and Table S5A, the structures found to be most actively translating proteins in the murine brain were postsynapse, presynapse, dendrite, and myelin sheath. Figure 3A indicates the top 20 nonredundant enrichment clusters identified, while Figure 3B shows the network of intracluster and intercluster similarities between enrichment terms. We next compared our candidate list with the synaptic protein database on SynSysNet (http://bioinformatics.charite.de/synsys/), which contains 1028 proteins that define pre- and postsynaptic proteins, as well as those present in subdomains of the synapse (such as synaptic vesicles and associated proteins, lipid rafts and postsynaptic density). A total 298 out of the 1218 highly synthesized proteins (24.5%) were matched to synaptic proteins (listed in Table S6A), with 29% of these being actively translated in the murine brain, while 883 out of the 7245 genes found in Brain1–3 whole data set matched to the provided synaptic protein list (Table S6B). This observation was further verified when we performed pathway and function clustering in Metascape, which returned a nonredundant cluster named “synapse organization” among several other members of the top 20 nonredundant cluster terms. For example, a total of 12 term clusters that are involved in a variety of synapse vesicle biological processes under the top one nonredundant enrichment cluster named regulation of vesicle-mediated transport (Figure 4, Table S5A).
Figure 3.
Synapse, dendrite, and myelin sheath are the most active protein translating structures in the murine brain. The left panel bar graph shows the top 20 nonredundant enrichment clusters across input protein lists (n = 1218) according to structure (A), pathway (B), and function (C). The lowest p-value cluster term and corresponding protein hit count in each group have been used to represent the respective clusters, with statistical significance indicated by color coding. The right panel shows the enrichment network visualization of intracluster and intercluster similarities among the top 20 enriched terms, including up to 15 terms per cluster (no more than 250 terms in total). Cluster annotations are shown by color code and cluster ID, where each node represents an enriched term, whereas terms with similarity >0.3 are connected by edges. The lowest p-value cluster term was used to determine ID and represent each nonredundant enrichment cluster. VMT: vesicle-mediated transport; IT: intracellular transport; CP: cellular protein; CR: cellular response; RoP: regulation of protein; ER: endoplasmic reticulum; EM: endomembrane; TF: translation factor; and Rp: ribonucleoprotein.
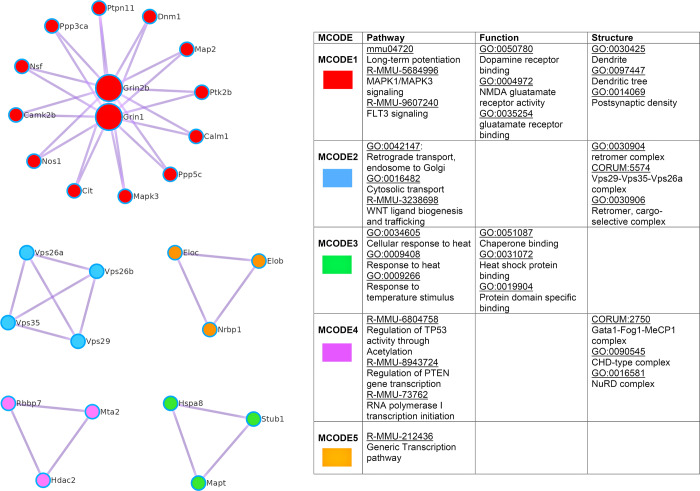
Figure 4.
Five protein complexes identified by the molecular complex detection (MCODE) method in Metascape. Protein–protein interaction enrichment analysis as conducted by Metascape with the BioGrid database (a subanalysis of structure, pathway, and function enrichment clustering). The MCODE algorithm was applied to the protein–protein interaction networks in order to identify neighborhoods where proteins are densely connected. The left panel shows the MCODE network color-coded according to identities. The right panel table shows the corresponding functional labels of the network plots. The top three highest-scoring terms based on p-values derived from GO-enrichment analysis have been retained as the functional labels and are described next to the corresponding components.
2.3. Murine Brain Actively Translates Proteins Involved in Neuronal Development and Function
An additional subset of highly active proteins in our data set was related to the processes of neuronal development and function (Table S5B). These proteins included several N-myc downstream-regulated gene family members (Ndrg1, Ndrg2, Ndrg3, and Ndrg4), RNA-binding protein Qki/Qk (KH domain containing RNA binding), Ataxin10 (Atxn10), Ras protein-specific guanine nucleotide-releasing factor1 (Rasgrf1), and E3 ubiquitin ligase enzyme Nedd4 (Neuronal precursor cell expressed developmentally downregulated 4). The Ndrg family is known to protect brain tissues from ischemic/hypoxic injury and has also been implicated in various nervous system disorders, including dementia and various neuronal cancers such as glioma, neuroblastoma, and meningioma.26,27 In particular, Ndrg1 is well known to be responsible for Charcot–Marie–Tooth disease type 4D (CMT4D, also known as hereditary motor and sensory neuropathy-Lom).28 Previous data have suggested that Ndrg2 may function to maintain blood–brain barrier permeability after ischemic strokes,29 while Ndrg3 has been reported to regulate the hypoxic response to cerebral ischemia.30 Similarly, Ndrg4 appears to mediate resistance to neuronal cell death during ischemia but also contributes to steady-state maintenance of the intracerebral BDNF (brain-derived neurotrophic factor), which is critical for spatial learning.31 These proteins therefore fulfill a range of critical roles in normal brain physiology and responses to injury, consistent with reports that both Ndrg2 and Ndrg4 are downregulated in glioblastoma and correlate with poor patient survival32 (Tables S4B and S5). Other vital proteins uncovered by this analysis included Qki, which influences the glial cell fate and development in addition to playing an essential role in myelinization processes, such that spontaneous mutations in this protein result in hypomyelinization of the central and peripheral nervous systems.33−35 Rasgrf1 instead contributes to long-term memory formation in mouse model systems by promoting dissociation of GDP from RAS protein in response to Ca2+ influx, muscarinic receptor signaling, or activation of the G protein beta–gamma subunit.36,37 Atxn10 is required for neuron survival, differentiation, and neuritogenesis via activation of the mitogen-activated protein kinase cascade.38,39 Nedd4 promotes endocytosis and proteasomal degradation of various ion channels and membrane transporters, thereby contributing to the formation of neuronal dendrites, neuromuscular junctions, cranial neural crest cells, motor neurons, and axons.40 Like several other highly translated proteins in this group, the critical roles played by Nedd4 are not limited to neurological functions alone, since this mediator has also been implicated in tumorigenesis.41
2.4. Glutamate Receptor Interactors and Cholesterol Homeostasis Mediators are Actively Expressed in Mouse Brain Tissues
Two highly active protein clusters identified by our functional enrichment analysis were “glutamate receptor binding” and “tau protein binding” , which are strongly associated with neurodegenerative diseases (Figure 3C, Tables S4B and S5C). Both glutamate receptors and tau proteins contribute to essential neuronal functions, such that their dysregulation can trigger neurological disorders such as Alzheimer’s disease.42−44 A total of 20 proteins were identified within the glutamate receptor cluster, including both Grin1 (glutamate ionotropic receptor NMDA type subunit 1) and Grin2b (glutamate ionotropic receptor NMDA type subunit 2b), which are critical subunits of N-methyl-d-aspartate (NMDA) receptors and acted as the two main seeds of the largest molecular complex detected by our protein–protein interaction network analysis (Figure 4, Table S5D). NMDA receptors are a specific type of ionotropic glutamate receptor that is concentrated at postsynaptic sites on dendrites and participates in rapid excitatory synaptic transmission.42 Regulation of NMDA receptor activity and downstream signal transduction is therefore essential for controlling synaptic plasticity and memory functions.45 Several enzymes involved in brain cholesterol homeostasis are highly active even under normal physiological conditions. These include critical mediators of cholesterol synthesis (3-hydroxy-3-methylglutaryl-CoA synthase 1; Hmgcs1) cholesterol ester processing (acyl-CoA cholesterol acyltransferase [ACAT]1 and ACAT2), cholesterol excretion (cholesterol-24-hydroxylase; Cyp46a1), and transmembrane lipid transport (ATP-binding cassette transporter subfamily A member 2; Abca2). In particular, Cyp46a1 is a crucial enzyme involved in brain-specific cholesterol export and generation of 24-HC (24(S)-hydroxycholesterol), which plays important roles in the trafficking of the amyloid precursor protein (APP). In addition, we observed active synthesis of multiple intercellular cholesterol trafficking proteins and apolipoproteins including ApoA4, ApoA1, ApoE, ApoJ/Clusterin, and Lrp1 (LDL receptor-related protein 1), each of which displayed synthetic rates ∼10 times faster than average (proteome-wide mean H/L was 0.057, whereas the mean H/L for this apolipoprotein subset was >0.5) (Table S4B).
2.5. Amyloid-β Precursor/A4 Protein Processing and Stability
App (Amyloid-β precursor/A4 protein) is a well-established central player in Alzheimer’s disease pathology.46 Brain-expressed β- and γ-secretases mediate App cleavage at the amino-terminus and carboxyl-terminus, respectively, thereby generating the amyloid-intracellular domain (AICD) as well as soluble ectodomains sAPPβ and β-amyloid peptides (Aβ) that enable extracellular deposition of Aβ, which is a critical event in the formation of β-amyloid/senile plaques.47 Consequently, App protein expression is highly dynamic and subject to tight control in healthy brain tissues. Accordingly, we observed that both App (Figure S3D) and Alzheimer peptide protein precursor family members Aplp1 and Aplp2 were being translated ∼10 times faster than average synthesis rates in the brain. Intriguingly, multiple other β-amyloid binding proteins were also prevalent in our data sets, including mediators of App processing and protein stability such as Apbb1 (amyloid beta A4 precursor protein-binding family B member 1; average H/L 0.473 ± 0.054) and Apba2/Xl1l (amyloid beta A4 precursor protein-binding family A member 2; average H/L 0.394 ± 0.031) (Table S4B). Adaptor protein Apbb1 is localized in the nucleus and can directly interact with App, as well as with LDL receptor and various transcription factors. Apbb1 can also form a complex with the γ-secretase-derived App intracellular domain to modulate App turnover and processing,48 suggesting a potential role in the pathogenesis of Alzheimer’s disease. However, our analyses also identified active synthesis of Apba2/Mint2/X11, which instead stabilizes App and inhibits the production of proteolytic fragments (including the Aβ peptide that is characteristically deposited in the brain tissues from Alzheimer’s disease patients).49 Also, prominent in our data set were the heavy-labeled retromer complex members Vps26a and Vps35 (Table S4B, Figure 4), which have been reported to alter the β-secretase cleavage of App via effects on SorLA(Sorl1)-mediated recycling between endosomes and Golgi.50 Our findings thus confirm that both App and production of Aβ peptides are subject to tight regulation under normal physiological conditions in the murine brain.
Our findings demonstrate that a short period of pSILAM diet enables the identification of actively translating proteins that mediate vital biological functions in the mouse brain. In particular, we observed that the synapse, dendrite, and myelin sheath are highly active sites of protein synthesis, rapidly expressing mediators of chemical signaling, nutrient sensing, and lipid metabolism, as well as regulators of synaptic functions and axon guidance. However, in addition to proteins associated with high energy utilization, the brain is also a lipid-rich organ that employs distinct lipid/lipoprotein metabolic pathways to maintain normal functions behind the impermeable blood–brain barrier.51,52 Brain lipids consist primarily of glycerophospholipids, sphingolipids, and cholesterol, with previous studies suggesting that almost all CNS cholesterol is synthesized de novo (where this lipid species exhibits a half-life of 0.5–5 years, compared with just a few days for blood plasma cholesterol). The adult brain contains ∼20 to 25% of total cholesterol in the body,51,53 with the majority of this being nonesterified within the myelin sheaths and comprising the plasma membranes of the astrocytes and neurons.54 Steady-state maintenance of cholesterol levels is therefore essential for normal brain tissue morphology and function. Our results show that enzymes involved in cholesterol homeostasis and transport are among the most actively translated proteins in the brain. Accordingly, impairing the function of these enzymes leads to disruption of steady-state cholesterol homeostasis and has been linked to neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Niemann-Pick type C disease.55
Apolipoproteins play pivotal roles in the transport and metabolism of lipids within the CNS, where trafficking is mediated by specialized “high density lipoprotein (HDL)-like particles” enriched in ApoE/ApoA1.56,57 Among the apolipoproteins identified to date, nine out of 22 have previously been detected at the mRNA and/or protein level in the CNS (ApoC1, ApoC2, ApoD, ApoE, Clu/ApoJ, ApoL2, ApoL3, and ApoA4).58 Intriguingly, ApoA4 has previously been detected at lower levels than other apolipoproteins in the Sprague–Dawley rat brain,59,60 although here we detected higher levels of heavy labeling in this protein than were observed for other family members (average H/L abundance ratio = 1.549 ± 0.138 in three biological replicates; 27 times higher than the average brain proteome H/L). ApoA4 synthesis is typically thought to be confined to the intestine, although low-level expression has also been reported in the hypothalamus and prefrontal cortex.59,60 The primary function of these molecules in lipid metabolism remains somewhat unclear but roles in satiety and appetite regulation, as well as antioxidant and antiatherogenic properties, have been identified in rodent models.61−63 Polymorphisms in the ApoA4 gene have also been reported to enhance activation of LCAT (lecithin: cholesterol acyltransferase) and potentially increase the Alzheimer’s disease risk.64 Our data now suggest that ApoA4 likely plays an important role in the CNS that depends on the active synthesis and rapid degradation to maintain low-level expression in the healthy brain. It will be very interesting to compare the endogenous ApoA4 protein synthesis rate (reflected by heavy-to-light abundance ratio), its stability (emPAI values that reflect relative protein expression levels), and modulation on LCAT activity in different brain disease models.
Another key member of the apolipoprotein family is ApoA1, which is the major protein constituent of plasma HDL. In addition to high expression in several peripheral tissues, including the liver and intestine, ApoA1 is also one of the most abundant apolipoproteins in CSF (cerebrospinal fluid) and also serves as an essential cofactor for LCAT activation.65 Indeed, our data indicated that ApoA1 is far more abundant than ApoA4 in the murine brain (emPAI value 34.062 ± 16.639) while still displaying a high level of heavy labeling by pSILAM analysis (average H/L abundance ratio 0.890 ± 0.026, 15 times higher than the brain average; Table S4B). An earlier study has linked early-onset Alzheimer’s disease with a polymorphism (−75A/G) in the promoter region of the ApoA1 gene, which conferred a modest increase in plasma levels of this protein. However, it remains controversial whether ApoA1 levels are indeed increased in CSF from patients with Alzheimer’s disease and dementia.57,66−69 Our approach provides a possibility to measure whether there is an increase of the ApoA1 newly synthesized protein pool or a relative increase in the protein level (based on the emPAI value) in CSF under interesting pathological conditions through the available transgenic mouse models of AD. These findings suggest that analysis of proteome dynamics in the brain, and apolipoprotein biology in particular, could provide a new insight into the molecular basis of major neurological disorders. Indeed, our study also detected high CNS expression of ApoE, which serves as the primary transport protein for extracellular cholesterol and other lipids in this compartment. Since, there is known to be no exchange of ApoE between the brain and peripheral pools,70 we can be confident that the heavy-labeled protein detected here was newly synthesized locally in the brain. In the healthy adult brain, nascent ApoE lipoprotein synthesis occurs mainly in astrocytes, and cholesterol is then transferred to ApoE to form a mature lipidated particle that can be acquired by surrounding neurons.71,72 Under physiological conditions, ApoE protein levels are relatively stable and mediate dynamic transfer of lipids between the brain cells in the CNS, whereas detrimental events such as tissue injury can lead to dramatic increases in glial/neuronal levels of ApoE (up to 150-fold increase).73,74 In our study, ApoE displayed similar abundance levels and synthesis rates to ApoA1 (average H/L = 0.674 ± 0.074, emPAI value 31.060 ± 6.713; Figure S3B), confirming that multiple apolipoproteins are rapidly expressed in the murine brain. Indeed, while present at markedly lower levels, the alternative family member ApoJ/Clu also displayed a heavy labeling profile consistent with active synthesis (average H/L = 0.566 ± 0.100 with emPAI value 4.171 ± 0.807). Comparatively, ApoJ/Clu is more widely distributed throughout the body than ApoA1, ApoA4, and ApoE, being expressed in multiple peripheral organs as well as the brain. Within the CNS, ApoJ is produced primarily by astrocytes, but this protein can also be detected in pyramidal neurons of the hippocampus and Purkinje neurons in the cerebellum.75 Previous studies have identified that stresses such as cytotoxic insult and cellular injury can significantly upregulate ApoJ/Clu expression levels.76,77 Together, these data confirm that the dynamics and distribution of apolipoprotein expression are critical components of a healthy brain function and that dysregulation of this biology is likely to confer diseases. Indeed, both ApoE and ApoJ have been identified as genetic risk factors for the development of late-onset Alzheimer’s disease78−81 due to their crucial role in regulating App and Aβ metabolism in the brain.81,82 Consistent with these data, App also displayed marked heavy labeling/active synthesis in healthy mouse brain tissues subjected to pSILAM analysis. These findings convinced the vital Alzheimer’s disease molecular ApoA1, ApoA4, ApoE, ApoJ, and App proteins belong to very dynamic and tightly regulating protein pools in the brain. It would be fascinating to investigate the immediate crosstalk between these proteins in Alzheimer’s disease mouse models. Multiple studies have now reported that different human isoforms of ApoE (E2, E3, and E4) exhibit not only the differential binding affinity for Aβ,80,83 but also their relative expression levels can confer the increased risk of neurodegenerative disorders and stroke.84,85 Expression of a mutated form of the human App precursor protein in a transgenic mouse model leads to significant App deposition in the brain, but these deposits are substantially reduced by performing the same experiment in mice with an ApoE knockout background.86 ApoE is the single most significant genetic risk factor for sporadic Alzheimer’s disease. While ApoE promotes disease pathology by seeding Aβ aggregation in the brain, recent data indicate that Alzheimer’s disease risk can be reversed by loss of the neuronal receptor Lrp1.87 Intriguingly, Lrp1 was also identified in our experiments as undergoing active synthesis in the murine brain (average H/L: 0.263 ± 0.003, average emPAI value: 1.787 ± 0.135; Figure S3C). These data indicate that pSILAM can provide a novel insight into the biology of ApoE isoforms that are central to the Alzheimer’s disease pathology in human patients. Future studies may be able to use human ApoE knock-in approaches together with mouse brain proteomic methodologies to understand better how apolipoproteins interact with Aβ biology to promote human dementia.
Proteasome-mediated proteolysis is known to be crucial for synaptic plasticity in both mice and humans,88 and our analyses identified an extensive range of different subunits and numerous interacting partners of this complex. In particular, the microtubule-associated protein tau (Mapt) and associated molecules play a central role in this protein in normal brain physiology, which is frequently disrupted in Alzheimer’s and Parkinson’s diseases. The tau protein displayed an average heavy-to-light ratio of 0.115 ± 0.033, an average emPAI value of 485.049 ± 291.870, and was clustered together with adaptor protein Apbb1, immunophilin Fkbp4, apolipoproteins (ApoE and Clu), protein kinases (Fyn, Gsk3β), heat-shock proteins (Hsp90ab1 and Hsp90aa1), and serine/theronine phosphatase subunits (Ppp2ca and Ppp2r2a). Together, these data identify multiple actively translating proteins involved in the maintenance of brain proteostasis, thus confirming that pSILAM can be used to uncover mechanisms or protection against brain pathology in addition to those that promote neurodegeneration and tumor formation in the brain.
3. Conclusions
Our study presents a novel optimized protocol for studying protein dynamics in vivo that facilitates investigation of the molecular mechanisms underlying various pathologies in a cost-effective and time-efficient manner. We applied this approach to determine relative rates of protein turnover in different regions of the mouse brain, thus paving the way for future studies into how these dynamics are influenced by site-specific stimuli. It is now possible to use pSILAM feeding for a short time and then sample specific tissue regions to generate a highly detailed picture of active proteomic regulation in vivo. Improving our understanding of protein physiology in the healthy brain will subsequently lead to advances in our knowledge of the mechanisms underpinning different dementia syndromes. For example, pSILAM could potentially be used to perform direct comparisons of different age groups of male and female mice to help clarify why the clinicopathologic features of dementia vary between genders in humans, eventually leading to tailored therapies for each cohort. Provided that variation in the food intake can be controlled to ensure uniform labeling efficiency between animals (e.g., by daily monitoring of individual body weight and food consumption), there are a number of potential applications for pSILAM across multiple different fields. Indeed, there is also scope to combine pSILAM methodology with transgenic mice to increase the insight into human proteinopathy in various organs and facilitate preclinical testing of novel therapies in these systems.
4. Experimental Section
4.1. Animal Housing and In Vivo Protein Labeling
A total of six C57BL6/J male mice aged 7–8 weeks were obtained from InVivos Pte Ltd (Singapore) and used for two independent studies. For each study, the animals were randomly housed in groups of three animals per plastic cage and maintained under controlled temperature, humidity, and 12 h light/dark cycles (lights on from 7 a.m. to 7 p.m.) in a specific pathogen-free (SPF) room with free access to food and water. After 3 weeks of adaptation to their new environment with the provision of standard mouse chow (Altromin), the mice were fasted for 16 h starting in the evening and then transferred onto a scaled SILAM diet (13C6-l-lysine, SILANTET) the next morning with sterilized drinking water provided ad libitum for 2 days after that. Food intake, drinking water consumption, and body weight were monitored and recorded daily each morning around 10 a.m. Animal facilities were AAALAC-approved, and all the experiments were performed according to the established guidelines and protocols approved by the NTU Institutional Animal Care and Use Committee (NTU-IACUC) (IACUC protocol # ARF-SBS/NIE-A18018).
4.2. Brain Tissue Processing
Mice were euthanized with CO2 and immediately subjected to blood collection by cardiac puncture before brain tissues were excised, dissected, and immediately snap-frozen in liquid nitrogen. For total protein extraction, the brain tissues were disaggregated using a liquid nitrogen-cooled pulverizer (BioSpec), and 100 mg of the resultant sample was resuspended in 100 mM ammonium bicarbonate (ABB, Sigma) a lysing buffer containing 2% SDS together with a protease inhibitor cocktail (Merck). The tissue suspension was then further homogenized using 1 mm magnetic beads (Next Advance) in a bullet blender homogenizer (BioFrontier Technology) under high intensity at 4 °C for 5 min. The tissue homogenates were subsequently centrifuged at 10,000 × g, 4 °C for 10 min and the supernatants collected. Further rounds of homogenization were performed as required until no visible pellet remained. The collected supernatants were then combined, quantified, and purified using cold acetone precipitation (4 h at −20 °C), then processed for in-gel digestion or in-solution digestion. The protein pellets were collected by centrifugation at 5000 × g, 4 °C for 5 min. The pellets were then air-dried and redissolved in 100 mM ABB buffer containing 8 M urea and a protease inhibitor cocktail for in-solution tryptic digestion. For in-gel tryptic digestion, the protein gel-loading buffer for SDS-PAGE was used to dissolve the protein pellet.
4.3. In-Gel Tryptic Digestion
A total of 200 μg brain protein per mouse was separated with 10% SDS-PAGE gels before the lanes were cut into eight separate bands and subjected to in-gel digestion. Each gel band was further divided into approximately 1–2 mm2 pieces and washed several times with 25 mM ABB followed by 25 mM ABB containing 50% acetonitrile (ACN, Fisher Chemical) until the gel pieces were completely destained. The destained gel pieces were then dehydrated with ACN, SpeedVaced for 5–10 min, and further reduced by incubation for 1 h at 60 °C in freshly made 10 mM dithiothreitol (Sigma) prepared in 25 mM ABB. The resultant gel pieces were then alkylated with 55 mM Iodocetamide (IAA, Sigma) prepared in 25 mM ABB and left in the dark at room temperature (RT) for 1 h. Next, the gel pieces were dehydrated using ACN and subjected to overnight digestion at 37 °C with sequencing-grade modified trypsin (Promega). Peptides were extracted with 50% ACN, 5% acetic acid (Merck), and dried using SpeedVac (Eppendorf) then stored at −20 °C until use.
4.4. In-Solution Tryptic Digestion and HPLC Fractionation
A total of 600 μg brain protein was subjected to reduction with 10 mM Tris(2-carboxyethyl) phosphine (TCEP, Sigma) at 30 °C for 2 h followed by alkylation with 20 mM IAA in 100 mM ABB in the dark at RT for 30 min. Sequencing-grade modified trypsin was added immediately and incubated at 37 °C overnight. The tryptic peptides were then desalted with Sep-Pak C18 cartridges (Waters) and dried in a SpeedVac. Peptides were then dissolved with buffer A (0.02% NH4OH) and subjected to high pH reverse-phase liquid chromatography fractionation with buffer B (0.02% NH4OH, 80% ACN) on a C18 column (4.6 × 200 mm, 5 μm, 300 Å, Waters, USA) at a flow rate of 1.0 mL/min using HPLC. The established 60 min gradient was set as 3–10% buffer B for 5 min, 10–35% buffer B for 40 min, 35–70% buffer B for 5 min, and 100% buffer B for 10 min. A total of 60 individual fractions were collected and then combined into 15 separate pools according to the concatenation order. All fractions were SpeedVac-dried and stored at −20 °C until use.
4.5. LC–MS/MS Analysis
A total of four independent biological replicates were performed; brain samples 1–3 were prepared by in-solution digestion and HPLC separation into 15 fractions, while brain sample 4 was separated into eight individual fractions by SDS-PAGE prior to in-gel digestion. Tryptic peptides were resuspended in 0.1% formic acid (FA, Fisher Chemical), and each fraction was injected three times (TR1, TR2, and TR3 for in-solution digested samples) or twice (in-gel digestion sample) as technical replicates in LC–MS/MS. The peptides were separated and analyzed on a Dionex Ultimate 3000 RSLCnano system coupled to a Q-Exactive tandem mass spectrometer (Thermo Fisher). Approximately 2 μg peptide from each fraction was injected into an Acclaim peptide trap column (Thermo Fisher) via a Dionex RSLCnano autosampler. Peptides were separated in a Dionex EASY-Spray 75 μm × 10 m column packed with PepMap C18 3 μm, 100 Å (PepMap C18) at 35 °C. The flow rate was maintained at 300 nL/min. Mobile phase A (0.1% FA) and mobile phase B (0.1% FA in 100% acetonitrile) were used to establish a 60 min gradient. The peptides were then analyzed with a Q-Exactive MS with EASY nanospray source (Thermo Fisher) at an electrospray potential of 1.5 kV. A full MS scan (350–1600 m/z range) was acquired at a resolution of 70,000 at m/z 200 and a maximum ion accumulation time of 100 ms. The dynamic exclusion was set to 30s. The resolution of the HCD spectra was set to 35,000 at m/z 200. The AGC settings of the full MS scan and the MS2 scan were 3E6 and 2E5, respectively. The 10 most intense ions above the 2000 count threshold were selected for fragmentation in HCD with a maximum ion accumulation time of 120 ms. An isolation width of two was used for MS2. Single and unassigned charged ions were excluded from MS/MS. For HCD, the normalized collision energy was set to 28%. The Underfill ratio was defined as 0.2%.
4.6. Mass Spectrometric Data Analysis
Raw data files from 11 replicates (three injections per biological replicate 1–3, plus two injections of sample 4) were analyzed as four independent experiments using Proteome Discoverer (PD) v2.1 (Thermo Scientific, San Jose, CA) with the Uniprot mouse protein database (downloaded on 16 March 2017, 91,089 sequences, 38,788,886 residues) using a SILAC (K6) protein quantitation workflow. Briefly, this workflow includes eight processing nodes numbered from 0 to 7. Node 0 labeled “Spectrum Files” allows selection of raw files, and Node 1 labeled “Spectrum Selector” extracts spectra within a retention time window and precursor ion mass window. Node 2 labeled “IMP MS2 Spectrum Processor” deisotopes and deconvolutes isotopic clusters. Node 3 selected search engine SequestHT, and Node 4 selected Mascot with database search parameters as follows: enzyme: trypsin, maximum miss cleavage: 2, minimum peptide length: 6, maximum peptide length: 144, maximum number of peptides reported: 100, precursor mass tolerance: 10 ppm, fragment mass tolerance: 0.02 Da, dynamic modification: Label:13C(6)(K), deamidation of N and Q, and methionine oxidation. Static modification: carbamidomethyl(C), Node 5 labeled “Percolator” where target FDR(strict) was set as 0.01, and target FDR(relaxed) was set as 0.05. Node 6 labeled as “Event Detector”, mass precursor set as 4 ppm, S/N threshold set as 1, and Node 7 labeled “Precursor ions Quantifier”, where RT tolerance of isotope pattern multiplets(min): 0.2 and single peak/missing channels allowed: 1. Designed consensus workflow was used to analyze further the peptide/protein list obtained with PD2.1. In short, total nine nodes, Node 0 named as MSF Files, which the “spectra to store” set as both identified or quantified, “merging of identified” is set globally by search engine type, and the “reported FASTA Title lines: Best match, Node 1 is “PSM Grouper”, which “peptide group modification the site probability threshold set as 75 and ‘modification sites” show only the best position; Node 2 is “Peptide Validator”, “Validation mode set as “automatic and the target FDR for both PSMs and Peptides set as Strict”:0.01 and “Relaxed”: 0.05; Node 3 is “Peptide and protein filter” peptide confidence: high, minimum peptide length: 6, minimum peptide sequence: 1; Node 4 was “Protein scorer”, which including Node 5 named as “Protein grouping”: the strict parsimony principle was applied; Node 6 labeled as “Protein FDR Validation”: 0.01 as strict and 0.05 as relaxed; Node 7 named as “Peptide in protein annotation”; and Node 8 was “Peptide and Protein quantifier”: use Unique+Razor peptide, consider protein groups for unique peptides, normalized mode: none, scaling mode: on channels average(per file), quantification: abundance (ion intensity) based on: intensity, coisolation threshold: 50, average reporter S/N threshold: 100. The results were exported from PD2.1 as a text file that was subsequently processed in Excel. The abundance (intensities) of all the detected master proteins (unique protein groups) in individual heavy or light labeled channels were combined to obtain the total values and determine the average in vivo labeling efficiency (H/L) of the pSILAM experiment. The emPAI value reported by PD2.1 was used to estimate the abundance of the identified protein in the brain.
4.7. Statistical Analysis
Each of the PD2.1 search peptide/protein lists generated was exported to Microsoft Excel and then subjected to cutoff filtering according to the following parameters: Exp q_value < 0.05, unique peptide ≥ 1, protein false discovery rate (FDR) < 0.05, and unique protein groups. However, to reduce the false positive candidate list, only proteins identified with high confidence FDR < 0.01 were used for further functional analysis (Table S4B). The labeled protein group lists were further cutoff-filtered using a coefficient variance of the heavy/light abundance ratio between technical triplicates <40%, minimum two technical replicate detection, and all heavy/light abundance ratios >0.01. All statistical analyses were performed using PD2.1 default settings. Pearson correlation coefficients (default Excel Correl function) were used to assess relationships between biological and technical triplicates; correlation coefficient values >+0.8 were considered significantly positively correlated. Standard deviation (SD) was used to measure variance between biological triplicates of candidate proteins. The distribution figure was generated and analyzed using Excel.
4.8. Bioinformatic Analysis
Bioinformatic analysis was performed using Metascape. The parameter settings were ″Annotation″ set as check all terms; ″Membership″ set as select all membership; ″Enrichment″ background genes set as all genes; and other parameters set as (1) Pathway & Pathway Enrichment: min overlap is 3, P-value cutoff is 0.01 and min enrichment is 1.5. (2) Protein–protein interaction enrichment: min and max network size are set as 3 and 500. The molecular complex detection (MCODE) method was applied to identify closely related proteins in the protein–protein interaction (PPI) network using the BioGrid+InWeb_IM(human) + OmniPath(human) PPI databases.89 The synaptic protein/gene database was downloaded from SynSysNet.90
4.9. Data Availability
LC–MS/MS raw data from the 11 replicates and results for protein and peptide identification and quantification from PD2.1 have been deposited with the ProteomeXchange Consortium via the PRIDE91 partner repository under the data set identifier PXD013502. The raw data and search results can be accessed using the following login to the PRIDE data depository:
Username: reviewer36496@ebi.ac.uk
Password: poIjhLxu
Acknowledgments
This work is in part supported by the Singapore Ministry of Education (MOE2018-T1-001) and Singapore National Medical Research Council (NMRC/OFIRG/0003/2016). R. N. K. is supported by a grant from Alzheimer’s Research UK (ARUK).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04439.
Record of mouse body weight and average 13C6-l-Lysine-labeled diet consumption; dynamic changes in murine brain proteome; mouse body weight change and average daily SILAM food intake; and distribution of ApoA1, ApoE, Lrp1, and App protein precursor ion quantification ratios (PDF)
Dynamic change in murine brain proteins with the SILAM diet1 (XLS)
Proteomic results of the 4 biological replicates of pSILAM mouse brains (XLSX)
Proteomic results of the 3 biological replicates of pSILAM mouse brains processed by in-solution digestion (XLSX)
Brain protein groups identified with high confidence within the 13C6-l-Lysine-labeled fraction (XLSX)
Metascape enrichment analysis of selected candidate genes (XLSX)
List of synaptic protein candidates referenced in the SynSysNet synaptic database (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Medina-Cano D.; Ucuncu E.; Nguyen L. S.; Nicouleau M.; Lipecka J.; Bizot J. C.; Thiel C.; Foulquier F.; Lefort N.; Faivre-Sarrailh C.; Colleaux L.; Guerrera I. C.; Cantagrel V. High N-glycan multiplicity is critical for neuronal adhesion and sensitizes the developing cerebellum to N-glycosylation defect. Elife 2018, 7, e38309 10.7554/eLife.38309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steklov M.; Pandolfi S.; Baietti M. F.; Batiuk A.; Carai P.; Najm P.; Zhang M.; Jang H.; Renzi F.; Cai Y.; Abbasi Asbagh L.; Pastor T.; De Troyer M.; Simicek M.; Radaelli E.; Brems H.; Legius E.; Tavernier J.; Gevaert K.; Impens F.; Messiaen L.; Nussinov R.; Heymans S.; Eyckerman S.; Sablina A. A. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 2018, 362, 1177–1182. 10.1126/science.aap7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C.; Ackermann S.; Ma Z.; Mohanta S. K.; Zhang C.; Li Y.; Nietzsche S.; Westermann M.; Peng L.; Hu D.; Bontha S. V.; Srikakulapu P.; Beer M.; Megens R. T. A.; Steffens S.; Hildner M.; Halder L. D.; Eckstein H. H.; Pelisek J.; Herms J.; Roeber S.; Arzberger T.; Borodovsky A.; Habenicht L.; Binder C. J.; Weber C.; Zipfel P. F.; Skerka C.; Habenicht A. J. R. ApoE attenuates unresolvable inflammation by complex formation with activated C1q. Nat. Med. 2019, 25, 496–506. 10.1038/s41591-018-0336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W. γ-Secretase and its modulators: Twenty years and beyond. Neurosci. Lett. 2019, 701, 162–169. 10.1016/j.neulet.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen S.; Laperre K.; Eelen G.; Rinaldi G.; Fraisl P.; Torrekens S.; van Looveren R.; Loopmans S.; Bultynck G.; Vinckier S.; Meersman F.; Maxwell P. H.; Rai J.; Weis M.; Eyre D. R.; Ghesquière B.; Fendt S.-M.; Carmeliet P.; Carmeliet G. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature 2019, 565, 511–515. 10.1038/s41586-019-0874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy D. B.; Liao L.; Lee J. H.; Park S. K.; Yates J. R. III Dynamics of subcellular proteomes during brain development. J. Proteome Res. 2012, 11, 2467–2479. 10.1021/pr201176v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome T. J.; Helmstetter F. J. Protein degradation and protein synthesis in long-term memory formation. Front. Mol. Neurosci. 2014, 7, 61. 10.3389/fnmol.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. C.; Guan S.; Burlingame A.; Prusiner S. B.; Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 14508–14513. 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L. D.; Zuchman R.; Sorokina O.; Müller A.; Dieterich D. C.; Armstrong J. D.; Ziv T.; Ziv N. E. Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS One 2013, 8, e63191 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörrbaum A. R.; Kochen L.; Langer J. D.; Schuman E. M. Local and global influences on protein turnover in neurons and glia. Elife 2018, 7, e34202 10.7554/eLife.34202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauniyar N.; McClatchy D. B.; Yates J. R. III Stable isotope labeling of mammals (SILAM) for in vivo quantitative proteomic analysis. Methods 2013, 61, 260–268. 10.1016/j.ymeth.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Fornasiero E. F.; Mandad S.; Wildhagen H.; Alevra M.; Rammner B.; Keihani S.; Opazo F.; Urban I.; Ischebeck T.; Sakib M. S.; Fard M. K.; Kirli K.; Centeno T. P.; Vidal R. O.; Rahman R. U.; Benito E.; Fischer A.; Dennerlein S.; Rehling P.; Feussner I.; Bonn S.; Simons M.; Urlaub H.; Rizzoli S. O. Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 2018, 9, 4230. 10.1038/s41467-018-06519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiick K. L.; Saxon E.; Tirrell D. A.; Bertozzi C. R. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 19–24. 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C.; Lee J. J.; Link A. J.; Graumann J.; Tirrell D. A.; Schuman E. M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007, 2, 532–540. 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- McClatchy D. B.; Ma Y.; Liu C.; Stein B. D.; Martínez-Bartolomé S.; Vasquez D.; Hellberg K.; Shaw R. J.; Yates J. R. III Pulsed Azidohomoalanine Labeling in Mammals (PALM) Detects Changes in Liver-Specific LKB1 Knockout Mice. J. Proteome Res. 2015, 14, 4815–4822. 10.1021/acs.jproteome.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S.; Witten A. J.; Ocken A. R.; Kinzer-Ursem T. L. Incorporation of non-canonical amino acids into the developing murine proteome. Sci. Rep. 2016, 6, 32377. 10.1038/srep32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C.; Link A. J.; Graumann J.; Tirrell D. A.; Schuman E. M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 9482–9487. 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M.; Moser M.; Ussar S.; Thievessen I.; Luber C. A.; Forner F.; Schmidt S.; Zanivan S.; Fässler R.; Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008, 134, 353–364. 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Zanivan S.; Krueger M.; Mann M. In vivo quantitative proteomics: the SILAC mouse. Methods Mol. Biol. 2011, 757, 435–450. 10.1007/978-1-61779-166-6_25. [DOI] [PubMed] [Google Scholar]
- Overmyer K. A.; Tyanova S.; Hebert A. S.; Westphall M. S.; Cox J.; Coon J. J. Multiplexed proteome analysis with neutron-encoded stable isotope labeling in cells and mice. Nat. Protoc. 2018, 13, 293–306. 10.1038/nprot.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua S. L.; Yam J. K. H.; Hao P.; Adav S. S.; Salido M. M.; Liu Y.; Givskov M.; Sze S. K.; Tolker-Nielsen T.; Yang L. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat. Commun. 2016, 7, 10750. 10.1038/ncomms10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.; Park J. E.; Tse W.; Low J. K.; Kon O. L.; McCarthy N.; Sze S. K. ERO1α promotes hypoxic tumor progression and is associated with poor prognosis in pancreatic cancer. Oncotarget 2016, 10, 5970–5982. 10.18632/oncotarget.27235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. E.; Tse S. W.; Xue G.; Assisi C.; Maqueda A. S.; Ramon G. P. X.; Low J. K.; Kon O. L.; Tay C. Y.; Tam J. P.; Sze S. K. Pulsed SILAC-based proteomic analysis unveils hypoxia- and serum starvation-induced de novo protein synthesis with PHD finger protein 14 (PHF14) as a hypoxia sensitive epigenetic regulator in cell cycle progression. Oncotarget 2019, 10, 2136–2150. 10.18632/oncotarget.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K.; Lin K. M.; Ravenhill B. J.; Davies C.; Nobre L.; Fielding C. A.; Ruckova E.; Fletcher-Etherington A.; Soday L.; Nichols H.; Sugrue D.; Wang E. C. Y.; Moreno P.; Umrania Y.; Huttlin E. L.; Antrobus R.; Davison A. J.; Wilkinson G. W. G.; Stanton R. J.; Tomasec P.; Weekes M. P. High-Definition Analysis of Host Protein Stability during Human Cytomegalovirus Infection Reveals Antiviral Factors and Viral Evasion Mechanisms. Cell Host Microbe 2018, 24, 447–460.e11. 10.1016/j.chom.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood E. J. D.; Williamson J. C.; Sienkiewicz A.; Naamati A.; Matheson N. J.; Lehner P. J. Promiscuous Targeting of Cellular Proteins by Vpr Drives Systems-Level Proteomic Remodeling in HIV-1 Infection. Cell Rep. 2019, 27, 1579–1596.e7. 10.1016/j.celrep.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonkeren S. L.; Massen M.; van der Horst R.; Koch A.; Vaes N.; Melotte V. Nervous NDRGs: the N-myc downstream-regulated gene family in the central and peripheral nervous system. Neurogenetics 2019, 20, 173–186. 10.1007/s10048-019-00587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotte V.; Qu X.; Ongenaert M.; Criekinge W.; Bruïne A. P.; Baldwin H. S.; Engeland M. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. 2010, 24, 4153–4166. 10.1096/fj.09-151464. [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L.; Gresham D.; Gooding R.; Heather L.; Baas F.; de Jonge R.; Blechschmidt K.; Angelicheva D.; Chandler D.; Worsley P.; Rosenthal A.; King R. H. M.; Thomas P. K. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am. J. Hum. Genet. 2000, 67, 47–58. 10.1086/302978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takarada-Iemata M.; Yoshikawa A.; Ta H. M.; Okitani N.; Nishiuchi T.; Aida Y.; Kamide T.; Hattori T.; Ishii H.; Tamatani T.; Le T. M.; Roboon J.; Kitao Y.; Matsuyama T.; Nakada M.; Hori O. N-myc downstream-regulated gene 2 protects blood-brain barrier integrity following cerebral ischemia. Glia 2018, 66, 1432–1446. 10.1002/glia.23315. [DOI] [PubMed] [Google Scholar]
- Yao Y.; Wang W.; Jing L.; Wang Y.; Li M.; Hou X.; Wang J.; Peng T.; Teng J.; Jia Y. Let-7f Regulates the Hypoxic Response in Cerebral Ischemia by Targeting NDRG3. Neurochem. Res. 2017, 42, 446–454. 10.1007/s11064-016-2091-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto H.; Kokame K.; Okuda T.; Nakajo Y.; Yanamoto H.; Miyata T. NDRG4 protein-deficient mice exhibit spatial learning deficits and vulnerabilities to cerebral ischemia. J. Biol. Chem. 2011, 286, 26158–26165. 10.1074/jbc.M111.256446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej M. A.; Weischer C.; Reinges M. H.; Uhl E.; Weigand M. A.; Schwarm F. P.; Schanzer A.; Acker T.; Quint K.; Uhle F.; Stein M. NDRG2 and NDRG4 Expression Is Altered in Glioblastoma and Influences Survival in Patients with MGMT-methylated Tumors. Anticancer Res. 2016, 36, 887–897. [PubMed] [Google Scholar]
- Hardy R. J. QKI expression is regulated during neuron-glial cell fate decisions. J. Neurosci. Res. 1998, 54, 46–57. . [DOI] [PubMed] [Google Scholar]
- Larocque D.; Richard S. QUAKING KH domain proteins as regulators of glial cell fate and myelination. RNA Biol. 2014, 2, 37–40. 10.4161/rna.2.2.1603. [DOI] [PubMed] [Google Scholar]
- Lu Z.; Zhang Y.; Ku L.; Wang H.; Ahmadian A.; Feng Y. The quakingviable mutation affects qkI mRNA expression specifically in myelin-producing cells of the nervous system. Nucleic Acids Res. 2003, 31, 4616–4624. 10.1093/nar/gkg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman A.; Assmann A.; Richter S.; Soch J.; Schütze H.; Wüstenberg T.; Deibele A.; Klein M.; Richter A.; Behnisch G.; Düzel E.; Zenker M.; Seidenbecher C. I.; Schott B. H. Genetic variation of the RASGRF1 regulatory region affects human hippocampus-dependent memory. Front. Hum. Neurosci. 2014, 8, 260. 10.3389/fnhum.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyes L.; Holst S.; Lozano M.; Santos E.; Fernandez-Medarde A. Spatial learning and long-term memory impairments in RasGrf1 KO, Pttg1 KO, and double KO mice. Brain Behav. 2018, 8, e01089 10.1002/brb3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- März P.; Probst A.; Lang S.; Schwager M.; Rose-John S.; Otten U.; Özbek S. Ataxin-10, the spinocerebellar ataxia type 10 neurodegenerative disorder protein, is essential for survival of cerebellar neurons. J. Biol. Chem. 2004, 279, 35542–35550. 10.1074/jbc.M405865200. [DOI] [PubMed] [Google Scholar]
- Waragai M.; Nagamitsu S.; Xu W.; Li Y. J.; Lin X.; Ashizawa T. Ataxin 10 induces neuritogenesis via interaction with G-protein β2 subunit. J. Neurosci. Res. 2006, 83, 1170–1178. 10.1002/jnr.20807. [DOI] [PubMed] [Google Scholar]
- Donovan P.; Poronnik P. Nedd4 and Nedd4-2: ubiquitin ligases at work in the neuron. Int. J. Biochem. Cell Biol. 2013, 45, 706–710. 10.1016/j.biocel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Zou X.; Levy-Cohen G.; Blank M. Molecular functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim. Biophys. Acta 2015, 1856, 91–106. 10.1016/j.bbcan.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Wang R.; Reddy P. H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimers Dis. 2017, 57, 1041–1048. 10.3233/JAD-160763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V.; Tsai L.-H.; Götz J. Glutamate Receptors in Alzheimer’s Disease: Mechanisms and Therapies. Neural. Plast 2016, 2016, 8256196. 10.1155/2016/8256196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Tan L.; Yu J.-T.; Tan L. Tau in Alzheimer’s Disease: Mechanisms and Therapeutic Strategies. Curr. Alzheimer Res. 2018, 15, 283–300. 10.2174/1567205014666170417111859. [DOI] [PubMed] [Google Scholar]
- Hardingham G. NMDA receptor C-terminal signaling in development, plasticity, and disease. F1000Res. 2019, 8, F1000. 10.12688/f1000research.19925.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignante E. A.; Lorenzo A. APP signaling in Alzheimer’s disease. Aging 2018, 10, 3063–3064. 10.18632/aging.101641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão F. Jr.; Grokoski K. C.; da Silva B. B.; Lamers M. L.; Siqueira I. R. The amyloid precursor protein (APP) processing as a biological link between Alzheimer’s disease and cancer. Ageing Res. Rev. 2019, 49, 83–91. 10.1016/j.arr.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Chow W. N. V.; Ngo J. C. K.; Li W.; Chen Y. W.; Tam K. M. V.; Chan H. Y. E.; Miller C. C. J.; Lau K.-F. Phosphorylation of FE65 Ser610 by serum- and glucocorticoid-induced kinase 1 modulates Alzheimer’s disease amyloid precursor protein processing. Biochem. J. 2015, 470, 303–317. 10.1042/BJ20141485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y.; Sano Y.; Vassar R.; Gandy S.; Nakaya T.; Yamamoto T.; Suzuki T. X11 proteins regulate the translocation of amyloid β-protein precursor (APP) into detergent-resistant membrane and suppress the amyloidogenic cleavage of APP by β-site-cleaving enzyme in brain. J. Biol. Chem. 2008, 283, 35763–35771. 10.1074/jbc.M801353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett J.; Bugarcic A.; Collins B. M.; Teasdale R. D. Retromer’s Role in Endosomal Trafficking and Impaired Function in Neurodegenerative Diseases. Curr. Protein Pept. Sci. 2017, 18, 687–701. 10.2174/1389203717666160311121246. [DOI] [PubMed] [Google Scholar]
- Dietschy J. M. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biol. Chem. 2009, 390, 287–293. 10.1515/BC.2009.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B. V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008, 57, 178–201. 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Björkhem I.; Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 806–815. 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- Dietschy J. M.; Turley S. D. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J. Lipid Res. 2004, 45, 1375–1397. 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Martín M. G.; Pfrieger F.; Dotti C. G. Cholesterol in brain disease: sometimes determinant and frequently implicated. EMBO Rep. 2014, 15, 1036–1052. 10.15252/embr.201439225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S.; Donarski N.; Goetze K.; Kreckel M.; Stuerenburg H. J.; Buhmann C.; Beisiegel U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J. Lipid Res. 2001, 42, 1143–1151. [PubMed] [Google Scholar]
- Demeester N.; Castro G.; Desrumaux C.; De Geitere C.; Fruchart J. C.; Santens P.; Mulleners E.; Engelborghs S.; De Deyn P. P.; Vandekerckhove J.; Rosseneu M.; Labeur C. Characterization and functional studies of lipoproteins, lipid transfer proteins, and lecithin:cholesterol acyltransferase in CSF of normal individuals and patients with Alzheimer’s disease. J. Lipid Res. 2000, 41, 963–974. [PubMed] [Google Scholar]
- Elliott D. A.; Weickert C. S.; Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin. Lipidol. 2010, 51, 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Doi T.; Shen L.; Woods S. C.; Seeley R. J.; Zheng S.; Jackman A.; Tso P. Intestinal satiety protein apolipoprotein AIV is synthesized and regulated in rat hypothalamus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R1382–R1387. 10.1152/ajpregu.2001.280.5.R1382. [DOI] [PubMed] [Google Scholar]
- Shen L.; Pearson K. J.; Xiong Y.; Lo C. M.; Tso P.; Woods S. C.; Davidson W. S.; Liu M. Characterization of apolipoprotein A-IV in brain areas involved in energy homeostasis. Physiol. Behav. 2008, 95, 161–167. 10.1016/j.physbeh.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso P.; Chen Q.; Fujimoto K.; Fukagawa K.; Sakata T. Apolipoprotein A-IV: a circulating satiety signal produced by the small intestine. Obes. Res. 1995, 3, 689S–695S. 10.1002/j.1550-8528.1995.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Ostos M. A.; Conconi M.; Vergnes L.; Baroukh N.; Ribalta J.; Girona J.; Caillaud J. M.; Ochoa A.; Zakin M. M. Antioxidative and antiatherosclerotic effects of human apolipoprotein A-IV in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1023–1028. 10.1161/01.ATV.21.6.1023. [DOI] [PubMed] [Google Scholar]
- Duverger N.; Tremp G.; Caillaud J. M.; Emmanuel F.; Castro G.; Fruchart J. C.; Steinmetz A.; Denefle P. Protection against atherogenesis in mice mediated by human apolipoprotein A-IV. Science 1996, 273, 966–968. 10.1126/science.273.5277.966. [DOI] [PubMed] [Google Scholar]
- Császár A.; Kálmán J.; Szalai C.; Janka Z.; Romics L. Association of the apolipoprotein A-IV codon 360 mutation in patients with Alzheimer’s disease. Neurosci. Lett. 1997, 230, 151–154. 10.1016/S0304-3940(97)00500-4. [DOI] [PubMed] [Google Scholar]
- Cooke A. L.; Morris J.; Melchior J. T.; Street S. E.; Jerome W. G.; Huang R.; Herr A. B.; Smith L. E.; Segrest J. P.; Remaley A. T.; Shah A. S.; Thompson T. B.; Davidson W. S. A thumbwheel mechanism for APOA1 activation of LCAT activity in HDL. J. Lipid Res. 2018, 59, 1244–1255. 10.1194/jlr.M085332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbach H.; Heun R.; Morris C. M.; Edwardson J. A.; McKeith I. G.; Jessen F.; Schulz A.; Maier W.; Kölsch H. APOA1 polymorphism influences risk for early-onset nonfamiliar AD. Ann. Neurol. 2005, 58, 436–441. 10.1002/ana.20593. [DOI] [PubMed] [Google Scholar]
- Jeenah M.; Kessling A.; Miller N.; Humphries S. G to A substitution in the promoter region of the apolipoprotein AI gene is associated with elevated serum apolipoprotein AI and high density lipoprotein cholesterol concentrations. Mol. Biol. Med. 1990, 7, 233–241. [PubMed] [Google Scholar]
- Juo S.-H. H.; Wyszynski D. F.; Beaty T. H.; Huang H.-Y.; Bailey-Wilson J. E. Mild association between the A/G polymorphism in the promoter of the apolipoprotein A-I gene and apolipoprotein A-I levels: a meta-analysis. Am. J. Med. Genet. 1999, 82, 235–241. . [DOI] [PubMed] [Google Scholar]
- Harr S. D.; Uint L.; Hollister R.; Hyman B. T.; Mendez A. J. Brain expression of apolipoproteins E, J, and A-I in Alzheimer’s disease. J. Neurochem. 1996, 66, 2429–2435. 10.1046/j.1471-4159.1996.66062429.x. [DOI] [PubMed] [Google Scholar]
- Linton M. F.; Gish R.; Hubl S. T.; Bütler E.; Esquivel C.; Bry W. I.; Boyles J. K.; Wardell M. R.; Young S. G. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J. Clin. Invest. 1991, 88, 270–281. 10.1172/JCI115288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitas R. E.; Boyles J. K.; Lee S. H.; Foss D.; Mahley R. W. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim. Biophys. Acta 1987, 917, 148–161. 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- Kim W. S.; Rahmanto A. S.; Kamili A.; Rye K. A.; Guillemin G. J.; Gelissen I. C.; Jessup W.; Hill A. F.; Garner B. Role of ABCG1 and ABCA1 in regulation of neuronal cholesterol efflux to apolipoprotein E discs and suppression of amyloid-β peptide generation. J. Biol. Chem. 2007, 282, 2851–2861. 10.1074/jbc.M607831200. [DOI] [PubMed] [Google Scholar]
- Ignatius M. J.; Gebicke-Harter P. J.; Skene J. H.; Schilling J. W.; Weisgraber K. H.; Mahley R. W.; Shooter E. M. Expression of apolipoprotein E during nerve degeneration and regeneration. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 1125–1129. 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snipes G. J.; McGuire C. B.; Norden J. J.; Freeman J. A. Nerve injury stimulates the secretion of apolipoprotein E by nonneuronal cells. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 1130–1134. 10.1073/pnas.83.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinetti G. M.; Johnson S. A.; Oda T.; Rozovsky I.; Finch C. E. Clusterin (SGP-2): a multifunctional glycoprotein with regional expression in astrocytes and neurons of the adult rat brain. J. Comp. Neurol. 1994, 339, 387–400. 10.1002/cne.903390307. [DOI] [PubMed] [Google Scholar]
- Iwata A.; Browne K. D.; Chen X. H.; Yuguchi T.; Smith D. H. Traumatic brain injury induces biphasic upregulation of ApoE and ApoJ protein in rats. J. Neurosci. Res. 2005, 82, 103–114. 10.1002/jnr.20607. [DOI] [PubMed] [Google Scholar]
- Dragunow M.; Preston K.; Dodd J.; Young D.; Lawlor P.; Christie D. Clusterin accumulates in dying neurons following status epilepticus. Brain. Res. Mol. Brain Res. 1995, 32, 279–290. 10.1016/0169-328X(95)00088-A. [DOI] [PubMed] [Google Scholar]
- Harold D.; Abraham R.; Hollingworth P.; Sims R.; Gerrish A.; Hamshere M. L.; Pahwa J. S.; Moskvina V.; Dowzell K.; Williams A.; Jones N.; Thomas C.; Stretton A.; Morgan A. R.; Lovestone S.; Powell J.; Proitsi P.; Lupton M. K.; Brayne C.; Rubinsztein D. C.; Gill M.; Lawlor B.; Lynch A.; Morgan K.; Brown K. S.; Passmore P. A.; Craig D.; McGuinness B.; Todd S.; Holmes C.; Mann D.; Smith A. D.; Love S.; Kehoe P. G.; Hardy J.; Mead S.; Fox N.; Rossor M.; Collinge J.; Maier W.; Jessen F.; Schürmann B.; Heun R.; van den Bussche H.; Heuser I.; Kornhuber J.; Wiltfang J.; Dichgans M.; Frölich L.; Hampel H.; Hüll M.; Rujescu D.; Goate A. M.; Kauwe J. S. K.; Cruchaga C.; Nowotny P.; Morris J. C.; Mayo K.; Sleegers K.; Bettens K.; Engelborghs S.; De Deyn P. P.; Van Broeckhoven C.; Livingston G.; Bass N. J.; Gurling H.; McQuillin A.; Gwilliam R.; Deloukas P.; Al-Chalabi A.; Shaw C. E.; Tsolaki M.; Singleton A. B.; Guerreiro R.; Mühleisen T. W.; Nöthen M. M.; Moebus S.; Jöckel K.-H.; Klopp N.; Wichmann H.-E.; Carrasquillo M. M.; Pankratz V. S.; Younkin S. G.; Holmans P. A.; OˈDonovan M.; Owen M. J.; Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1088–1093. 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.-C.; Heath S.; Even G.; Campion D.; Sleegers K.; Hiltunen M.; Combarros O.; Zelenika D.; Bullido M. J.; Tavernier B.; Letenneur L.; Bettens K.; Berr C.; Pasquier F.; Fiévet N.; Barberger-Gateau P.; Engelborghs S.; De Deyn P.; Mateo I.; Franck A.; Helisalmi S.; Porcellini E.; Hanon O.; de Pancorbo M. M.; Lendon C.; Dufouil C.; Jaillard C.; Leveillard T.; Alvarez V.; Bosco P.; Mancuso M.; Panza F.; Nacmias B.; Bossù P.; Piccardi P.; Annoni G.; Seripa D.; Galimberti D.; Hannequin D.; Licastro F.; Soininen H.; Ritchie K.; Blanché H.; Dartigues J.-F.; Tzourio C.; Gut I.; Van Broeckhoven C.; Alpérovitch A.; Lathrop M.; Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1094–1099. 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Strittmatter W. J.; Saunders A. M.; Schmechel D.; Pericak-Vance M.; Enghild J.; Salvesen G. S.; Roses A. D. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 1977–1981. 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos R. B.; Cirrito J. R.; Parsadanian M.; May P. C.; O’Dell M. A.; Taylor J. W.; Harmony J. A. K.; Aronow B. J.; Bales K. R.; Paul S. M.; Holtzman D. M. ApoE and clusterin cooperatively suppress Aβ levels and deposition: evidence that ApoE regulates extracellular Aβ metabolism in vivo. Neuron 2004, 41, 193–202. 10.1016/S0896-6273(03)00850-X. [DOI] [PubMed] [Google Scholar]
- Huang Y.-W. A.; Zhou B.; Wernig M.; Südhof T. C. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell 2017, 168, 427–441.e21. e21 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D. M.; Bales K. R.; Tenkova T.; Fagan A. M.; Parsadanian M.; Sartorius L. J.; Mackey B.; Olney J.; McKeel D.; Wozniak D.; Paul S. M. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 2892–2897. 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A.; Lammertse D. P.; Coll J. R.; Charlifue S.; Coughlin C. T.; Whiteneck G. G.; Worley G. Apolipoprotein E s4 allele and outcomes of traumatic spinal cord injury. J. Spinal Cord Med. 2016, 31, 171–176. 10.1080/10790268.2008.11760708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slooter A. J. C.; Tang M. X.; van Duijn C. M.; Stern Y.; Ott A.; Bell K.; Breteler M. M.; Van Broeckhoven C.; Tatemichi T. K.; Tycko B.; Hofman A.; Mayeux R. Apolipoprotein E ε4 and the risk of dementia with stroke: A population-based investigation. JAMA 1997, 277, 818–821. 10.1001/jama.1997.03540340052032. [DOI] [PubMed] [Google Scholar]
- Fagan A. M.; Watson M.; Parsadanian M.; Bales K. R.; Paul S. M.; Holtzman D. M. Human and murine ApoE markedly alters Aβ metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 2002, 9, 305–318. 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- Tachibana M.; Holm M. L.; Liu C. C.; Shinohara M.; Aikawa T.; Oue H.; Yamazaki Y.; Martens Y. A.; Murray M. E.; Sullivan P. M.; Weyer K.; Glerup S.; Dickson D. W.; Bu G.; Kanekiyo T. APOE4-mediated amyloid-β pathology depends on its neuronal receptor LRP1. J. Clin. Invest. 2019, 129, 1272–1277. 10.1172/JCI124853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde A. N. The ubiquitin-proteasome pathway and synaptic plasticity. Learn. Mem. 2010, 17, 314–327. 10.1101/lm.1504010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Zhou B.; Pache L.; Chang M.; Khodabakhshi A. H.; Tanaseichuk O.; Benner C.; Chanda S. K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eichborn J.; Dunkel M.; Gohlke B. O.; Preissner S. C.; Hoffmann M. F.; Bauer J. M. J.; Armstrong J. D.; Schaefer M. H.; Andrade-Navarro M. A.; le Novere N.; Croning M. D. R.; Grant S. G. N.; van Nierop P.; Smit A. B.; Preissner R. SynSysNet: integration of experimental data on synaptic protein-protein interactions with drug-target relations. Nucleic Acids Res. 2012, 41, D834–D840. 10.1093/nar/gks1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Pérez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yılmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaíno J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC–MS/MS raw data from the 11 replicates and results for protein and peptide identification and quantification from PD2.1 have been deposited with the ProteomeXchange Consortium via the PRIDE91 partner repository under the data set identifier PXD013502. The raw data and search results can be accessed using the following login to the PRIDE data depository:
Username: reviewer36496@ebi.ac.uk
Password: poIjhLxu