Abstract
The precise mechanism leading to profound immunodeficiency of HIV-infected patients is still only partially understood. Here, we show that more than 80% of CD4+ T cells from HIV-infected patients have morphological abnormalities. Their membranes exhibited numerous large abnormal membrane microdomains (aMMDs), which trap and inactivate physiological receptors, such as that for IL-7. In patient plasma, we identified phospholipase A2 group IB (PLA2G1B) as the key molecule responsible for the formation of aMMDs. At physiological concentrations, PLA2G1B synergized with the HIV gp41 envelope protein, which appears to be a driver that targets PLA2G1B to the CD4+ T cell surface. The PLA2G1B/gp41 pair induced CD4+ T cell unresponsiveness (anergy). At high concentrations in vitro, PLA2G1B acted alone, independently of gp41, and inhibited the IL-2, IL-4, and IL-7 responses, as well as TCR-mediated activation and proliferation, of CD4+ T cells. PLA2G1B also decreased CD4+ T cell survival in vitro, likely playing a role in CD4 lymphopenia in conjunction with its induced IL-7 receptor defects. The effects on CD4+ T cell anergy could be blocked by a PLA2G1B-specific neutralizing mAb in vitro and in vivo. The PLA2G1B/gp41 pair constitutes what we believe is a new mechanism of immune dysfunction and a compelling target for boosting immune responses in HIV-infected patients.
Keywords: AIDS/HIV, Immunology
Keywords: Cytokines, Lipid rafts, T cells
Introduction
CD4+ lymphocytes play a critical role in the severe immunodeficiency that characterizes HIV-infected patients. Although fewer than 0.5% of blood CD4+ T cells are infected, almost all are dysfunctional. Their progressive decline leads to CD4 lymphopenia. In addition, functional defects of the remaining CD4+ T cells lead to their unresponsiveness, or anergy, to certain antigens and cytokines (1). Major progress has been made in treatment of the viral infection. Antiretrovirals (ARVs) prevent acquired immunodeficiency syndrome (AIDS). However, further improvement in HIV therapy will require a better understanding of the mechanisms responsible for CD4+ T cell defects following HIV infection (2, 3).
The mechanism explaining CD4+ T cell loss during HIV infection is still debated (4, 5). Persistent immune activation plays a critical role in the induction of this CD4+ T cell decline (6–8). A major mechanism results from damage of the mucosal barrier and lymphoid tissues of the gastrointestinal (GI) tract that follows acute infection. HIV targets subpopulations of CCR5-expressing CD4+ T cells, which are dense in the GI. Following this damage, microbial products translocate across the GI barrier and cause general activation of the immune system (9–11). In this context, it is noteworthy that HIV controller patients, who maintain high CD4 counts and good control of viremia, show low inflammation (12).
Numerous investigations have described the impairment of CD4+ T cell function in HIV-infected individuals, in whom CD4+ T cells fail to proliferate after stimulation by antigens or mitogens (13, 14). A progressive loss of T helper function has also been reported (15–17). These results may be partially explained by changes in the T cell receptor (TCR) repertoire (18), but they may also result from a defect in the intrinsic capacity of the CD4+ T cells to respond to physiological signals. For example, a selective defect in IL-2 production, but not IFN-γ synthesis, has been reported after anti-CD3 stimulation (19). In this context, we previously analyzed CD4+ T cell responses to IL-2 and IL-7, 2 cytokines that are crucial for the control of the function, proliferation, and survival of CD4+ T cells. We showed that the β and γc chains of the IL-2 receptor (IL-2R) are underexpressed and nonfunctional, as measured by reduced entry into the S and G2/M phases of the cell cycle (20). Similarly, decreased expression of the IL-7Rα chain (CD127) on the surface of CD4+ T cells from HIV-infected patients has been described and their function was shown to be defective (21). Altered induction of the antiapoptotic molecule Bcl-2 and decreased expression of CD25 after in vitro exposure to IL-7 were measured (22). We subsequently showed that the Janus kinase (Jak)/signal transducer and activator of transcription 5 (STAT5) signaling pathway is involved in these defects (23, 24). Similar results involving γc have been published by other investigators (25–30).
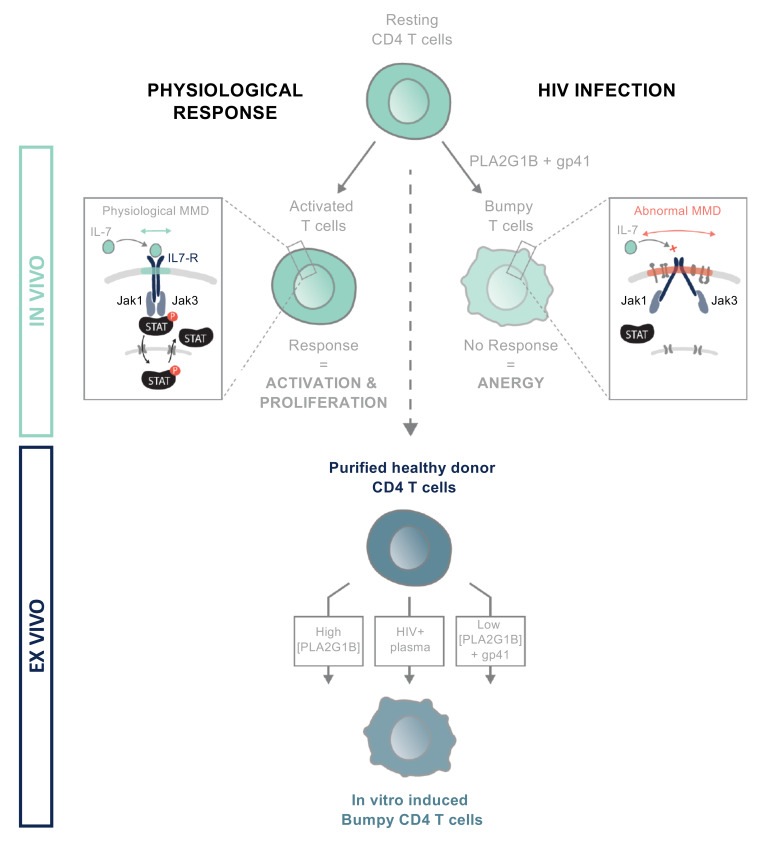
Most of the studies performed in lymphocytes from HIV-infected patients have used FACS analysis and in vitro functional immunological assays. Here, we reinvestigated this question by studying purified CD4+ T lymphocytes from viremic HIV-infected patients using imaging techniques (31) and molecular approaches rarely used in this field (32–34). We detected large, abnormal membrane microdomains (aMMDs) at the surface of CD4+ T cells purified from HIV-infected patients in the absence of any stimulation. The aMMD-bearing cells were named “Bumpy T cells,” due to their appearance after labeling. Their large aMMDs were shown to trap all IL-7R chains (α and γc). IL-7R chains lose their function when embedded in these aMMDs. Consequently, the Jak/STAT pathway was not functional and IL-7–induced phospho-STAT5 nuclear translocation (p-STAT5 NT) was inhibited. Bumpy T cells were recovered after exposure of CD4+ T cells from healthy donors (HDs) to plasma from HIV-infected patients. After characterization, we found that phospholipase A2 group IB (PLA2G1B) (35) was able to recapitulate the effects of plasma from HIV-infected patients. It induced aMMDs and consequently strongly affected numerous CD4 functions in vitro and in vivo (mouse model). However, PLA2G1B was found to synergize with HIV gp41 envelope protein in the blood of HIV-infected patients at physiological concentrations. Overall, our results provide insights into CD4+ T cell dysfunction and a mechanism for the CD4+ T cell anergy and lymphopenia observed in chronically HIV-infected patients.
Results
Membrane alterations and signaling defects in CD4+ T cells from HIV-infected patients.
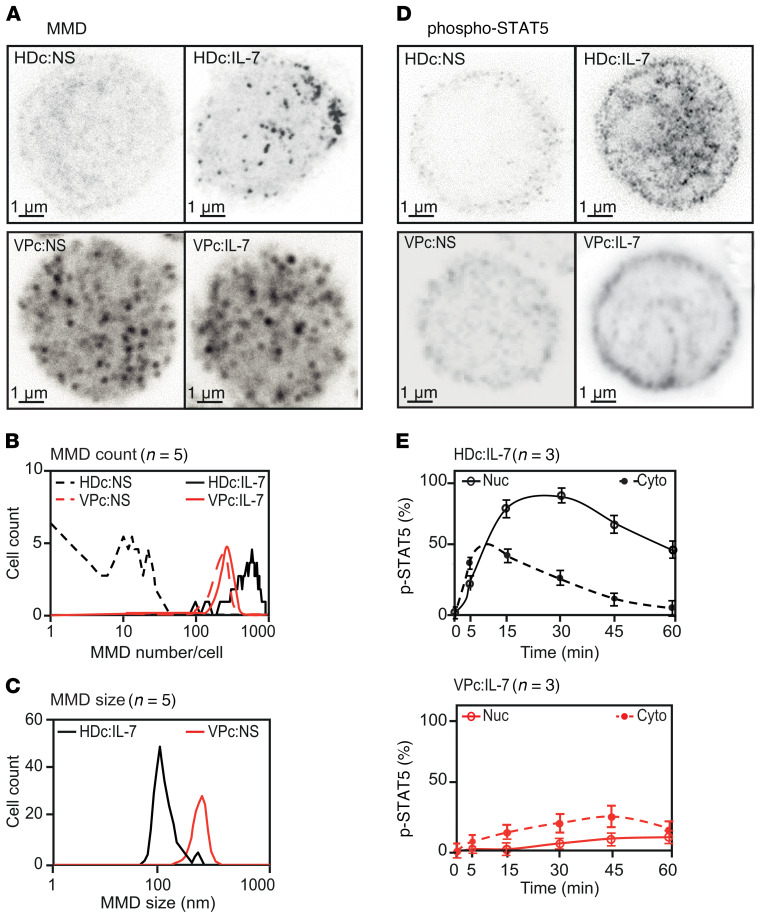
Stimulated emission depletion (STED) microscopy revealed numerous large aMMDs (up to 500/cell, with an average size >200 nm) at the surface of purified CD4+ T cells from viremic patients (VPs), in the absence of any activation. More than 80% of purified CD4+ T cells, also called Bumpy T cells, from all VPs exhibited aMMDs (Figure 1, A–C). Under the same conditions, resting CD4+ T cells from HDs did not spontaneously exhibit any aMMDs, whereas IL-7 stimulation promoted the formation of numerous physiological MMDs (pMMDs) of smaller size (approximately 800/cell, with an average size of 100 nm) (Figure 1, A–C). In contrast, IL-7 stimulation of CD4+ T cells from VPs did not induce any observable changes in their membranes (Figure 1, A and B).
Figure 1. Characterization of Bumpy T cells from HIV-infected patients.
(A) MMD analysis by CW-STED microscopy. From top to bottom, purified HD CD4+ T cells (HDc) and VP CD4+ T cells (VPc). For each group, the top half of a representative nonstimulated (NS) CD4+ T cell, or after IL-7 stimulation, is shown from Z-stack images. (B) Quantification of MMDs on the surface of HD CD4+ cells (HDc) and VP CD4+ cells (VPc) before (NS) and after IL-7 stimulation. (C) Size of MMDs at the surface of IL-7–stimulated HD cells (HDc:IL-7) and VP cells before stimulation (VPc:NS). Lines represent the mean values. (D) Analysis of IL-7–induced phosphorylation and nuclear translocation of STAT5 by pulsed-STED microscopy (0.5 μm slices) in nonstimulated and IL-7–stimulated HD CD4+ T cells (top) or VP CD4+ T cells (bottom). In A–D, an average of 50 cells from each HD and 15 to 50 cells from each VP (HIV RNA/mL = 49,144 ± 33,689) were examined from 5 donors in each group and representative images are shown in A and D. (E) The kinetics of p-STAT5 in the nucleus (Nuc) and cytoplasm (Cyto) of HD and VP CD4+ T cells after IL-7 stimulation was measured using ImageJ and represented as the mean ± SD for 3 donors.
We then examined the functional consequences of these morphological changes in the membrane using the IL-7/IL-7R system as a readout. The function of IL-7Rs of VP CD4+ T cells was altered, as the IL-7–induced phosphorylation of STAT5 differed between CD4+ T cells of HDs and VPs (Figure 1D); p-STAT5 NT was almost completely abolished in the CD4+ T cells from VPs after IL-7 stimulation. This resulted from the difference in the kinetics of cytoplasmic phosphorylation of STAT5 and p-STAT5 NT between CD4+ T cells of VPs and HDs (Figure 1E).
We previously showed that IL-7–induced cytoskeletal organization is required for efficient p-STAT5 NT in CD4+ T cells of HDs and that colchicine and cytochalasin D treatment abolishes p-STAT5 NT (34). These results are comparable to those obtained for nontreated VP CD4+ T cells (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI131842DS1). Microfilaments and microtubules can be observed after IL-7 stimulation. Because of the very small size of the cytoplasm of lymphocytes, these structures were observed by pulsed-STED microscopy. After staining by anti-tubulin antibodies, microtubules could be observed in HD CD4+ T cells but not VP CD4+ T cells (Supplemental Figure 1C). Similarly, microfilaments were visible in HD CD4+ T cells after staining by anti-actin antibodies but not in VP CD4+ T cells (Supplemental Figure 1D). This further supports the idea that the IL-7/IL-7R system is nonfunctional in VP CD4+ T cells.
Overall, these results confirm and structurally characterize the activation status of CD4+ T lymphocytes from VPs and provide insight into the mechanism of unresponsiveness of these CD4+ T cells.
Biochemical analysis of aMMDs from the CD4+ T lymphocytes of VPs.
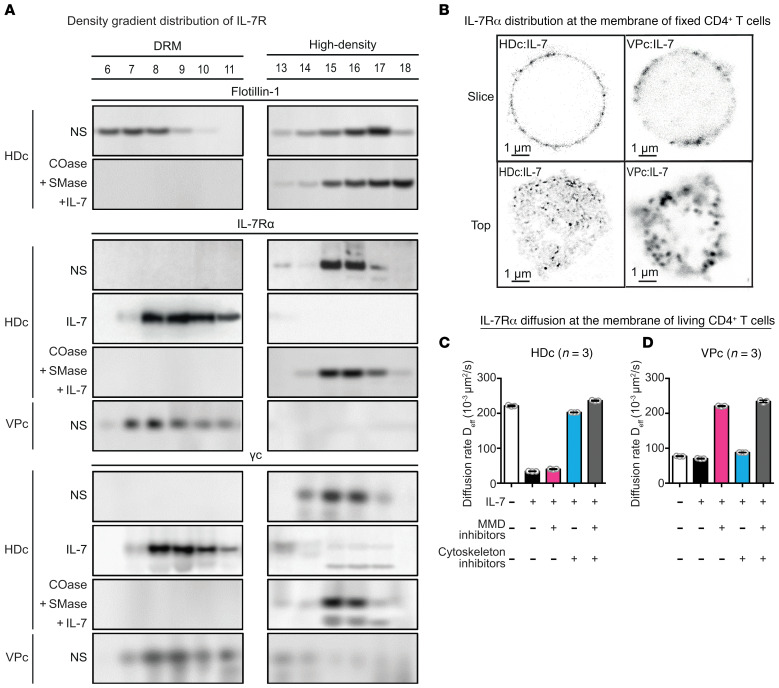
We further examined the mechanism linking the presence of aMMDs at the surface of the CD4+ T lymphocytes from VPs and the loss of function of the IL-7/IL-7R system by performing a biochemical analysis of their membranes. Cell lysates obtained after moderate detergent treatment were examined on sucrose gradients. This technique allowed us to separate free molecules that migrate in high-density fractions and the detergent-resistant microdomains (DRMs), structurally related to MMDs, which are recovered in the low-density fractions. Flotillin-1 was found in both fractions and was used as a marker in these experiments (Figure 2A and Supplemental Methods). IL-7Rα and γc were found as free molecules in the high-density fractions of HD CD4+ T cell lysates. They were found in low-density DRMs only after IL-7 stimulation (Figure 2A). In contrast, IL-7Rα and γc were spontaneously found associated with the low-density DRMs in lysates prepared from the Bumpy T cells of VPs, in the absence of any stimulation (Figure 2A). IL-7Rα appeared as clusters in STED images of the CD4+ T cell membranes from VPs (Figure 2B), further supporting the presence of low-density DRMs containing IL-7R chains. We further verified that IL-7Rα is included in aMMDs by studying its diffusion rate at the membrane surface. IL-7Rα was included in the aMMDs of VP CD4+ T cells, as their diffusion was limited and could be restored after disruption of the aMMDs by cholesterol oxidase and sphingomyelinase treatment (Figure 2, C and D).
Figure 2. Analysis of membrane domains and IL-7R distribution on the surface of HD and VP CD4+ T cells.
(A) Purified CD4+ T lymphocytes were lysed (0.5% Triton X-100) and the lysates loaded onto a 5% to 40% sucrose gradient. After 16 hours of centrifugation (280,000 g) at 4°C, 18 fractions were collected (no. 1 left = tube top = 5% sucrose; no. 18 right = tube bottom = 40% sucrose). Each fraction was analyzed by SDS-PAGE (2 gels). Flotillin-1 was used as a marker to indicate low-density fractions corresponding to DRMs. IL-7R α and γ chains were revealed by Western blotting. Results are shown for purified nonstimulated HD CD4+ T cells (HDc:NS), IL-7–stimulated HD CD4+ T cells (HDc:IL-7), or HD CD4+ T cells pretreated with cholesterol oxidase (COase: 31 μM, 25 minutes) and sphingomyelinase (SMase: 2.7 μM, 5 minutes), and IL-7–stimulated (HDc:COase+SMase/IL-7), as well as nonstimulated VP CD4+ T cells (VPc:NS) (n = 3 donors). (B) IL-7Rα chain localization at the membrane of CD4+ T cells from HDs and VPs was analyzed by CW-STED. Images of a section (slice) and the top view (top) of representative cells are shown among 50 cells per donor for HD (n = 3 donors) and 15 to 50 cells for VP CD4+ T cells (n = 3 donors). (C and D) The effect of cytoskeletal reorganization and MMD inhibition on IL-7R compartmentalization was evaluated by measuring the 2-dimensional effective diffusion rates (Deff) of the IL-7Rα chain by fluorescence correlation spectroscopy (FCS), as described in Tamarit et al. (34). Histograms represent the mean ± SD of the effective diffusion rate Deff in each condition at the surface of (C) HD (n = 3 donors) and (D) VP CD4+ T cells (n = 3 donors).
Overall, these results demonstrate that IL-7Rα and γc are spontaneously embedded in specific macrostructures of the membranes of CD4+ T cells from VPs, measured as aMMDs or DRMs. These data also show that the receptors lose their function when trapped in this abnormal structure of Bumpy CD4+ T cells.
Plasma from VPs induces the Bumpy T cell phenotype in HD CD4+ T cells.
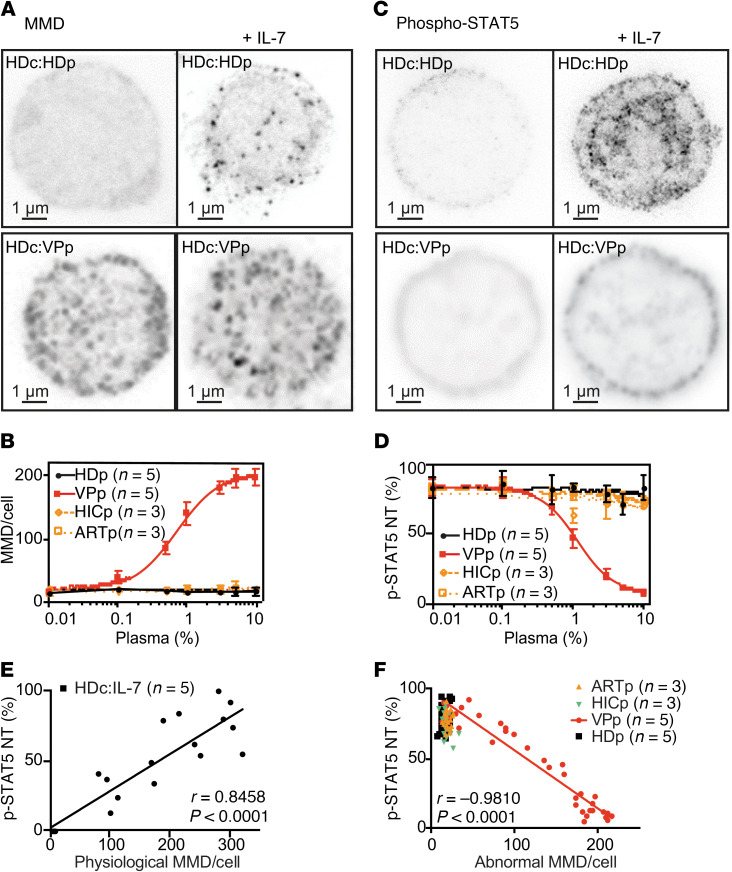
We investigated the molecular mechanism leading to the Bumpy T cell phenotype. The addition of plasma from VPs (Figure 3A) to HD CD4+ T cells was sufficient to induce the Bumpy T cell phenotype. Titration showed the phenotype to be induced in 50% of the cells at approximately 1% VP plasma (Figure 3B). HD CD4+ lymphocytes treated with VP plasma and Bumpy T cells were microscopically indistinguishable, and the number and size of aMMDs at their surface were not influenced by IL-7. In addition, plasma from elite controllers (HICp) (36, 37) and patients with suppressed viremia after 10 years of ARV (ARTp) could not induce this phenotype (Figure 3B).
Figure 3. Induction of Bumpy CD4+ T cells by plasma from HIV-infected patients.
(A) CW-STED images of MMDs on HD CD4+ T cells treated with 10% HD plasma (HDc:HDp) or VP plasma (HDc:VPp) before and after IL-7 stimulation. (B) Dose effect of plasma from HD (HDp, n = 5), VP (VPp, n = 5, HIV RNA/mL = 49,144 ± 33,689), HIV-controllers (HICp, n = 3), and ART-treated donors (ARTp, n = 3) on the number of MMDs per HD CD4+ T cell. (C) Pulsed-STED images of p-STAT5 of HD CD4+ T cells pretreated with 10% plasma from HDs or VPs. (D) Plasma dose effects as in B on p-STAT5 NT in IL-7–treated HD CD4+ T cells. Data are represented as the mean ± SD. In A–D, for each condition, an average of 50 HD CD4+ T cells were analyzed from 5 donors and representative images are shown in A and C. (E) Pearson’s correlation between the kinetics of p-STAT5 NT and the number of physiological MMDs throughout IL-7 activation of HD cells (up to 60 minutes). Linear regression for the mean of the 5 HD plasma samples is shown. (F) Pearson’s correlation between p-STAT5 NT and abnormal MMDs per HD CD4+ T cell treated with various amounts of HDp (n = 5), VPp (n = 5, HIV RNA/mL = 49,144 ± 33,689), HICp (n = 3), and ARTp (n = 3). Linear regression for the mean of the 5 VP plasma samples is shown.
We then studied the responsiveness of VP plasma–induced Bumpy T cells. IL-7–induced p-STAT5 NT was inhibited by VP plasma, with a half-maximum dose of 1% (Figure 3, C and D). These results suggest a direct link between the induction of aMMDs and the mechanism leading to the inhibition of p-STAT5 NT. We found a positive correlation between the number of pMMDs and the frequency of cells with translocated p-STAT5 during IL-7 responses in HD CD4+ T cells (Figure 3E). Conversely, we found a negative correlation between the number of aMMDs per CD4+ T cell and the percentage of cells with nuclear p-STAT5 in IL-7–stimulated plasma–induced Bumpy T cells (Figure 3F). These correlations further support the idea that plasma-induced aMMDs are responsible for the loss of IL-7 response.
PLA2G1B is a unique inducer of Bumpy T cells.
We biochemically characterized the plasma molecule involved in these morphological and functional changes. Size-exclusion and ion-exchange chromatography were used to determine the apparent MW and pI of the bioactive molecule(s) from the plasma of 3 VPs, using microscopy as a readout (Supplemental Figure 2, A–C). A list of one hundred and three 10- to 15-kDa proteins with a pI between 6.5 and 7.5 and a secretory signal peptide was determined. Differential mass spectrometry analysis identified PLA2G1B, also known as pancreatic phospholipase A2 (35), as the top candidate (PA21B in Supplemental Figure 2D). Active PLA2G1B is produced after the cleavage of 7 N-terminal residues from inactive proPLA2G1B (38). Recombinant active PLA2G1B was produced, purified, crystallized, and structurally characterized. The position of residues H48 and D99 and the Ca2+-binding loop, critical for the activity of the enzyme, are shown in Figure 4A.
Figure 4. Cloned plasma PLA2G1B induces the Bumpy T cell phenotype.
(A) Crystal structure of PLA2G1B (PDB: 6Q42). (B) PLA2G1B effect on MMD formation followed by STED (representative of 2 experiments at 250 nM and verified at 500 nM and 1 μM). (C) Dose effect of PLA2G1B on IL-7–induced p-STAT5 NT in HD purified CD4+ T cells after analysis of confocal images. Results are shown as the mean ± SD from 4 donors. (D–G) The effects on aMMD formation (D and E) and p-STAT5 NT (F and G) in CD4+ T cells of 250 nM WT PLA2G1B were compared to those of the inactive mutant H48Q (D and F) and other PLA2s (PLA2GIIA, PLA2GIID, or PLA2GX) (E and G). Results are shown as the mean ± SD from 5 (D–F) or 7 donors (G). (H) VP plasma (3%, from 5 donors) was depleted with anti-PLA2G1B, anti-PLA2GIIA, or anti-PLA2GIID rabbit polyclonal antibodies (100 μg/mL). The effect of depletion was analyzed by following p-STAT5 NT in IL-7–stimulated CD4+ T cells (n = 3 donors) incubated with depleted plasma. Results were normalized to the response obtained with HD plasma and are shown as the mean ± SD. (I) Effect of VP plasma treated with various doses of neutralizing anti-PLA2G1B mAb 14G9 on p-STAT5 NT in CD4+ T cells from 1 donor and the effect of 100 μg/mL of 14G9 mAb on p-STAT5 NT in CD4+ T cells from 5 donors. **P < 0.01; ***P < 0.001 by Mann-Whitney t test (D, F, and I) and by the Kruskal-Wallis test followed by the Mann-Whitney test with P values adjusted for multiple comparisons between groups (E and G) or 1-way ANOVA (H) with Tukey’s correction for multiple comparisons.
Recombinant PLA2G1B alone was able to induce aMMDs (Figure 4B) and inhibit p-STAT5 NT in HD CD4+ T cells (Figure 4C). This property was catalytic-site dependent, as the inactive H48Q mutant (39) was unable to induce aMMDs or inhibit p-STAT5 NT on human CD4+ T cells (Figure 4, D and F). These effects were specific to PLA2G1B; indeed, other members of the PLA2 family such as PLA2GIIA, PLA2GIID, or PLA2GX showed no significant effect on either aMMD formation or p-STAT5 NT inhibition (Figure 4, E and G). Similarly, only polyclonal antibodies specific for PLA2G1B decreased plasma-induced p-STAT5 NT inhibition, whereas polyclonal antibodies specific for PLA2GIIA or PLA2GIID had no effect (Figure 4H).
We developed mouse monoclonal antibodies (mAbs) specific for PLA2G1B. Among them, mAb 14G9 efficiently inhibited the enzymatic activity of PLA2G1B and abrogated VP plasma inhibition of p-STAT5 NT in a dose-dependent manner (Figure 4I). These experiments show that, at physiological concentrations, PLA2G1B is involved in the phenotypic and functional changes induced by VP plasma in HD CD4+ T cells and the Bumpy T cell phenotype observed in VPs.
PLA2G1B induces anergy of CD4+ T cells: specificity and reversal of the effects.
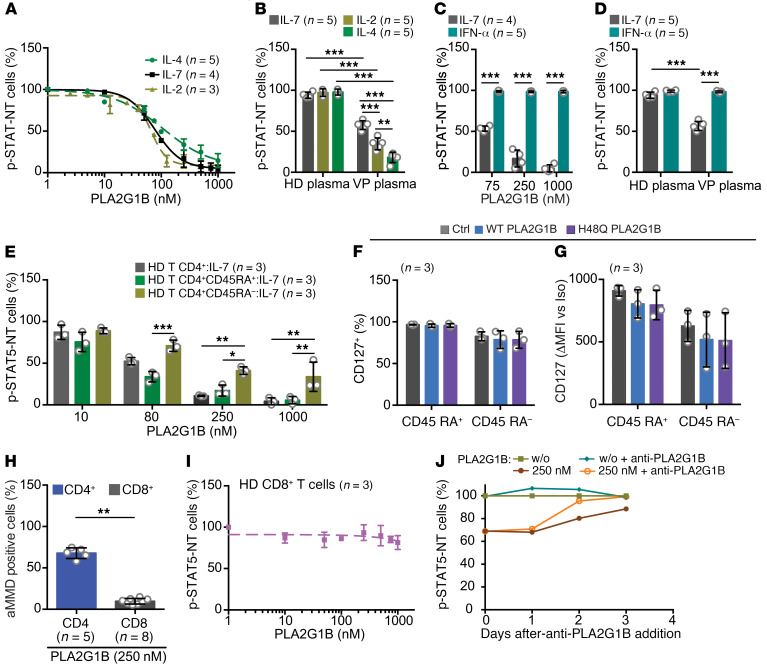
The unresponsiveness of CD4+ T cells to IL-7 induced by PLA2G1B was also observed for IL-2 and IL-4, 2 other γc cytokines. Similarly to IL-7, p-STAT NT induced by these 2 cytokines was inhibited by PLA2G1B in a dose-dependent manner and with a comparable IC50 (Figure 5A). These observations were verified using VP plasma–induced Bumpy T cells (Figure 5B). Unlike IL-2, IL-4, and IL-7, IFN-α–induced p-STAT1 NT was not inhibited by PLA2G1B or the plasma of VPs (Figure 5, C and D). IFN-α is known to signal by a mechanism independent of MMDs (40, 41). Thus, these results suggest that PLA2G1B only affects signaling pathways that involve compartmentalization into pMMDs. We then studied the effects of PLA2G1B first observed on total unseparated CD4+ T cells, and on naive (CD45RA+) and memory (CD45RA–) CD4+ T cells. PLA2G1B was slightly more active on CD45RA+CD4+ T cells than CD45RA–CD4+ T cells (Figure 5E). Such differential sensitivity is not the consequence of selective modulation of IL-7R (CD127) expression at the cell surface by PLA2G1B (Figure 5, F and G, and Supplemental Figure 3). As previously described, the percentage of CD127+ cells was slightly higher in CD45RA+ than CD45RA–CD4+ T cells (Figure 5F and refs. 42, 43). In addition, MFI analysis of CD127 expression (Figure 5G and Supplemental Figure 3) showed a slight reduction in CD45RA– cells, as previously reported (44). Overall, these analyses establish that PLA2G1B does not influence CD127 expression and support our hypothesis that PLA2G1B acts on signal transduction (as described above) and not by decreasing receptor expression.
Figure 5. Effect of PLA2G1B on CD4+ T cell subpopulations, specificity, and reversion.
(A) Dose effect of PLA2G1B (IL-7, n = 4; IL-2, n = 3; IL-4, n = 5) and (B) of 1% HD plasma (IL-7, n = 4; IL-2 and IL-4, n = 3) and VP plasma (n = 5) on IL-2–, IL-4–, and IL-7–induced p-STAT NT in CD4+ T cells. (C) Effects of PLA2G1B (IL-7, n = 4; IFN-α, n = 5) and (D) plasma (HD [n = 4] or VP [n = 5], 1%) on IL-7–induced p-STAT5 NT and IFN-α–induced p-STAT1 NT in CD4+ T cells (n = 5 donors). (E) The effect of PLA2G1B (30 minutes) on IL-7–induced p-STAT5 NT was analyzed in total (HD T CD4+:IL-7), naive (HD T CD4+ CD45RA+:IL-7), and memory (HD T CD4+ CD45RA–:IL-7) CD4+ T cells from the same donor in response to IL-7 (n = 3 donors). (F) Percentage of CD127+ cells among and (G) CD127 expression (Δ anti-CD127 MFI minus isotype control MFI) on CD45RA+ and CD45RA– CD4+ T cells after treatment with 1 μM WT or H48Q PLA2G1B (see gating strategy in Supplemental Figure 3A, n = 3 donors). (H) Effect of PLA2G1B (250 nM) on aMMD induction in CD4+ T cells (n = 5) and CD8+ T cells (n = 8) and (I) on IL-7–induced p-STAT5 NT in CD8+ T cells (dose effect, n = 3). In A–I, results are shown as the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA (B–D) and 2-way ANOVA (E) with Tukey’s correction for multiple comparisons or by the Mann-Whitney 2-tailed unpaired t test (H). (J) Anti-PLA2G1B treatment accelerates the recovery of a functional p-STAT5 NT response of PLA2G1B-treated CD4+ T cells to IL-7. The results of 1 representative experiment of 3 are presented.
The action of PLA2G1B appears to be specific to CD4+ T cells. Indeed, PLA2G1B did not induce aMMD formation in purified CD8+ T cells from HDs (Figure 5H). Similarly, p-STAT5 NT was not affected in CD8+ T cells by PLA2G1B, even at high concentrations (Figure 5I). These results are consistent with ex vivo observations of VP CD8+ T cells in which aMMDs were not detectable and p-STAT5 NT continued to occur (Supplemental Figure 4, A–C). In addition, physiological concentrations of PLA2G1B present in VP plasma inhibited p-STAT5 NT on CD4+ T cells but had no functional effects in CD8+ lymphocytes purified from HDs (Supplemental Figure 4D).
PLA2G1B is known to digest lipids, we thus further explored the difference between the response of CD4+ and CD8+ T cells to PLA2G1B by lipidomics analysis. There were significant differences in the proportions of the ganglioside GM3, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), sphingomyelin (SM), and triacylglycerol (TG) between HD CD4+ and CD8+ T cells (Supplemental Figure 4E). Similarly, differences in the lipid proportions have been reported between murine CD4+ and CD8+ T cells (45). It is possible that the differential effects of PLA2G1B on CD4+ and CD8+ T cells are associated with differences in lipid composition, but direct evidence will require extensive studies.
We next investigated the reversal of the induced Bumpy T cell phenotype in vitro and show the results of 1 of 3 representative experiments (Figure 5J). Purified CD4+ T cells were first treated in vitro with PLA2G1B and then cultured for various periods of time up to 3 days. Inhibition of p-STAT5 NT was examined every day. Under our experimental conditions, 30% of the cells were anergized and did not respond to IL-7. After 3 days in culture, p-STAT5 NT returned to normal, clearly showing that the Bumpy T cell phenotype is reversible. Furthermore, neutralizing mAb 14G9 accelerated the reversion (Figure 5J).
PLA2G1B affects CD4+ T cell survival in vitro: inhibition by neutralizing mAb 14G9.
Aside from the unresponsiveness of CD4+ T cells to physiological signals (anergy), HIV-infected patients suffer from CD4+ lymphopenia. We thus tested the effects of PLA2G1B on the half-life of CD4+ T cells in vitro. Purified CD4+ lymphocytes were cultured and the number of live cells counted over time as described in Methods. The number of surviving CD4+ T cells varied between donors in control cultures. The effects of PLA2G1B on the CD4+ lymphocytes are thus expressed as the percentage of the surviving cells relative to that in control cultures in the absence of PLA2G1B at each time point. The effect of PLA2G1B on CD4+ T cell survival was first tested at various concentrations up to 100 μM (Figure 6A). We then verified that 50% of the cells died after 18 days of culture in the presence of 1 μM PLA2G1B (Supplemental Figure 5A) and 40% of the cells died after 24 days of culture in the presence of 250 nM PLA2G1B (Figure 6B).
Figure 6. PLA2G1B acts on dying CD4+ T cells and reduces CD4+ T cell survival.
(A and B) PLA2G1B reduces the survival of human CD4+ T cells. (A) Cells were treated with PBS (Ctrl) or various amounts of PLA2G1B (1, 10, 100 μM) for 1 experiment. Results are shown as the percentage of CD4+ T cell counts normalized to the number of Ctrl cells at each time point. (B) Cells were treated with PBS (Ctrl) or 250 nM PLA2G1B (n = 6 donors). Results are shown as the mean ± SD of the percentage of CD4+ T cell counts normalized to the number of Ctrl cells at each time point. (A and B) The lines show the linear regression and the P values indicate the significance of the difference from control. (C–E) PLA2G1B acts on dying CD4+ T cells and digests phosphatidylserine. FACS analysis of CD4+ T cells for annexin V–APC on Live/Dead marker (Zombie-Violet) positive cells after treatment with (C) 250 nM PLA2G1B WT or H48Q or (D) 250 nM PLA2G1B with anti-PLA2G1B (14G9) or not (without Ab). (C and D) Annexin V–APC labeling (MFI) at various time points after treatment are presented (1 representative experiment of 2 in C and 3 in D is presented). (E) Results are shown as the mean ± SD of the percentage of annexin V–negative Zombie-positive CD4+ T cells after treatment with PBS (Ctrl), PLA2G1B alone (without Ab), or anti-PLA2G1B (14G9) (n = 3 donors). (F) Anti-PLA2G1B treatment inhibits the effect of PLA2G1B on the survival of CD4+ T cells. Results are shown as the mean ± SD of the percentage of CD4+ T cell counts normalized to the number of Ctrl cells at each time point (n = 3 donors). Lines show the linear regression and P values indicate the significance of the difference between experimental conditions. ***P < 0.001.
During these experiments, we further analyzed the CD4+ T cells. As expected, numerous dying cells became annexin V+Zombie+. However, we also detected annexin V–Zombie– cells (Figure 6, C and D). Their percentage increased during culture, reaching more than 70% of the recovered lymphocytes (Figure 6E). This was a specific consequence of PLA2G1B treatment, as such cells were not detected when CD4+ T cells were cultured in the presence of the inactive PLA2G1B mutant H48Q (Figure 6C and Supplemental Figure 5B). Similarly, their percentage was lower when cultures were performed in the presence of mAb 14G9, which neutralizes the enzymatic activity of PLA2G1B (Figure 6, D and E). This can be explained by the fact that PLA2G1B digested one of its substrates during culture, PS, which is also the binding site of annexin V. This confirms that the action of PLA2G1B on CD4+ T cells is mediated by its enzymatic activity.
We then tested the effect of mAb 14G9 on CD4+ T cell survival. The mAb significantly increased the survival of CD4+ T cells exposed to PLA2G1B (up to >50%) relative to cultures in the presence of the control isotype (Figure 6F and Supplemental Figure 5C).
In vitro and in vivo effects of human PLA2G1B in a mouse model.
We studied the effects of human PLA2G1B in mice after verifying its activity in vitro on mouse CD4+ T cells to extend our data in vivo. Upon exposure to PLA2G1B we also observed annexin V–negative Zombie-positive mouse CD4+ T cells (Supplemental Figure 6). CD4+ T cells from C57BL/6 mice were purified from the spleen and stimulated by anti-CD3 plus anti-CD28 beads, in the presence of IL-2. Human PLA2G1B was active on mouse CD4+ T cells and induction of CD25 (IL-2Rα) was inhibited by PLA2G1B by day 5 in a dose-dependent manner (Figure 7, A and B). Similarly, survival and proliferation were profoundly altered (Figure 7, C–E). These effects depended on the catalytic activity of PLA2G1B, as the H48Q mutant was ineffective (Figure 7, A–E). Furthermore, neutralizing mAb 14G9 blocked CD25 induction and decreased survival of the CD4+ cells (Figure 7, F and G). These data demonstrate that PLA2G1B can also inhibit TCR responses. In addition to the human experiments, these data establish that the effects of PLA2G1B can be measured after several days in culture.
Figure 7. Immunological effects of hPLA2G1B on mouse CD4+ T cells in vitro and in vivo.
(A–G) FACS analysis of the effect of hPLA2G1B on mouse CD4+ T cells after anti-CD3/CD28 and IL-2 stimulation (5 days, see gating strategy on Supplemental Figure 6C). (A–E) mCD4+ T cells were pretreated with WT or H48Q hPLA2G1B. (A) CD25 expression after treatment with 125 nM hPLA2G1B. (B) CD25 expression (MFI) and (C) cell survival (n = 3, 10 mice). (D) mCD4+ T cell proliferation profile after treatment with 125 nM hPLA2G1B. (E) Percentage of live mCD4+ T cells per cell generation (Go to G5; n = 3, 9 mice). (F and G) Effects of mAb anti-PLA2G1B 14G9 in vitro treatment on 125 nM hPLA2G1B action on CD4+ T cell survival and CD25 expression (n = 4, 11 mice). (H–L) In vivo effects of hPLA2G1B on CD4+ T cell response to IL-7. Spleen CD4+ T cells were isolated after intraperitoneal injection into C57BL/6 mice and the ex vivo p-STAT5 NT response to IL-7 was evaluated by confocal microscopy, with an average of 200 cells examined for each condition. Effect of hPLA2G1B injection at several doses of PLA2G1B for 3 hours (H, 6 mice, 2 experiments) and at several times after injection (I, 3 mice, 1 experiment; J, 8 mice, 2 experiments). (K) Effects of mAb anti-hPLA2G1B 14G9 injected in vivo on the hPLA2G1B (100 μg, 3 hours) response (5 mice, 1 experiment). (L) Inhibition of the effects of hPLA2G1B after injection into hPLA2G1B/BSA–immunized mice (5 mice, 1 experiment). Results are shown as the mean ± SEM (B, C, and E–G) or mean ± SD (H–L). *P < 0.05; **P < 0.01; ***P < 0.001 adjusted for multiple comparisons by Kruskal-Wallis test P < 0.001, followed by the Mann-Whitney test (B) and 2-way ANOVA with correction for multiple comparisons by Tukey’s (C, H, and J–L), Dunnett’s for the condition without PLA2G1B as a control group (E), or Sidak’s (F, G, and I) post hoc test.
In vivo, PLA2G1B showed activity on mouse CD4+ lymphocytes in a dose-dependent manner (Figure 7H). Injection of 100 μg of PLA2G1B induced a long-lasting effect, which persisted for up to 72 hours and began to diminish after 168 hours (Figure 7I). The effect of PLA2G1B was maximal 3 hours after injection (Figure 7J). We tested the effects of pretreatment with the anti-PLA2G1B neutralizing mAb 14G9 under the same experimental conditions (Figure 7K). The blockade was close to 100%. We obtained a comparable result in a group of mice that was preimmunized against human PLA2G1B (Figure 7L).
The cell specificity of the effects of PLA2G1B was further verified in this experimental model. Injection of PLA2G1B did not result in any loss of IL-7–induced p-STAT5 NT in mouse CD8+ lymphocytes, as measured ex vivo 3 hours after injection (Supplemental Figure 4F). Overall, these results open the possibility of using mouse models to evaluate anti-PLA2G1B neutralizing mAbs as an immunotherapeutic strategy.
Synergy between PLA2G1B and plasma HIV gp41 protein.
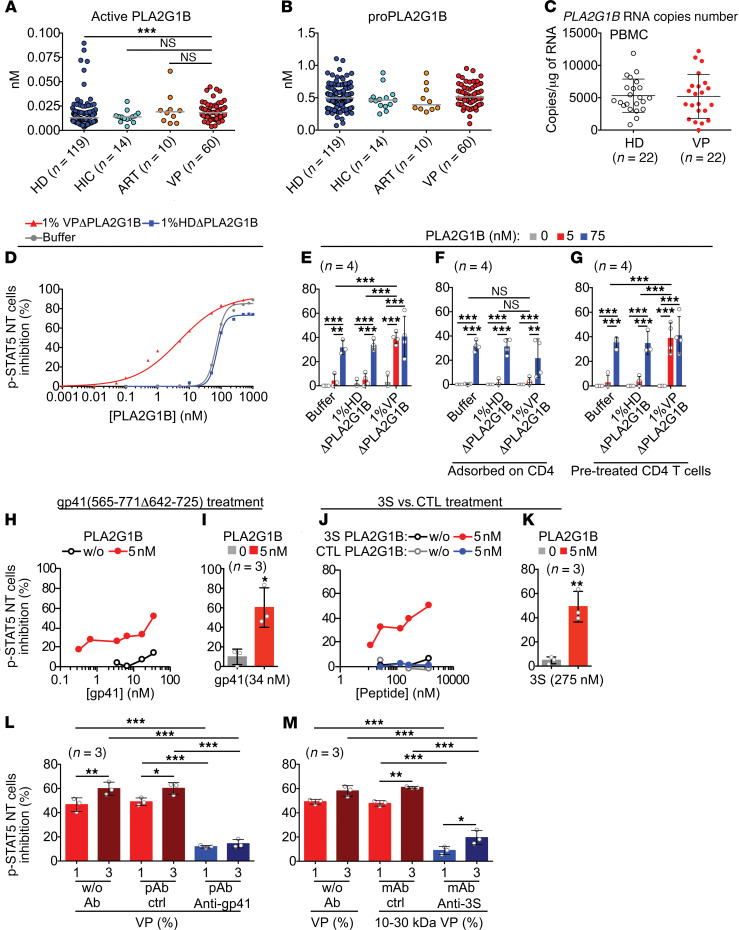
We then measured active and proPLA2G1B in HD plasma, HICp, ARVp, and VP plasma using PLA2G1B ELISAs (Figure 8, A and B). VPs had similar levels of active PLA2G1B as HIC and ART patients but slightly more active PLA2G1B than HDs (median increase of 1.4). In addition, comparable copy numbers of PLA2G1B RNA were found in the PBMCs of HDs and VPs by qPCR (Figure 8C). We thought that these results cannot explain the difference in PLA2G1B activity observed with the functional assays (Figure 3, B and D). This observation led us to consider that one or more cofactors are present in the plasma of VPs and are required for the induction of aMMDs and blockade of p-STAT5 NT. Indeed, the dose-response curve (from 0.001 to 1,000 nM) of PLA2G1B diluted in HD or VP plasma previously depleted of endogenous PLA2G1B showed striking differences in the IC50 values (Figure 8D), supporting the presence of a cofactor. The IC50 of PLA2G1B was 75 nM when diluted in PBS or HD plasma, but decreased to 5 nM when diluted in PLA2G1B-depleted VP plasma. We concluded that PLA2G1B was not acting alone but in synergy with another factor present in VP plasma. A bioassay was developed, using a limiting amount of PLA2G1B inactive by itself (5 nM, Figure 8D) and PLA2G1B-depleted VP plasma to detect the potential cofactor (Figure 8E), and used it to show that the enhancement was lost after plasma was incubated with purified CD4+ T cells (Figure 8F), suggesting that the cofactor was adsorbed on CD4+ T cells. After such pretreatment, the addition of 5 nM PLA2G1B to the CD4+ T cells, without further addition of VP plasma, resulted in the inhibition of IL-7–driven p-STAT5 NT (Figure 8G). This cofactor activity was sensitive to trypsin treatment and could be fractionated with an apparent MW between 10 and 30 kDa. The search then focused on HIV peptides possibly released into the plasma of infected patients. A gp41 fragment (MN strain, 565–771Δ642–725), and its corresponding 3S peptide (46), the sequence of which is highly conserved among various HIV isolates, exerted strong cofactor activity (Figure 8, H–K). In addition, the depletion of VP plasma with anti-gp41 antibodies that do not bind to gp120 (Supplemental Figure 7) resulted in the loss of cofactor activity (Figure 8L). This critical point was definitively established after depletion by a mAb characterized in the laboratory. This mAb, 1C5, was raised against the 3S peptide and shown to also recognize gp41 protein but not gp120 (Supplemental Figure 7). It was also able to completely deplete various VP plasma samples of cofactor activity (Figure 8M). These results support the hypothesis that fragments of gp41 containing the 3S sequence may act as cofactors that target PLA2G1B to the surface of CD4+ lymphocytes to exert its enzymatic activity.
Figure 8. Synergy between PLA2G1B activity and gp41.
(A and B) ELISA quantification of PLA2G1B in plasma from HD, VP, HIV-controllers (HIC), and ART-treated donors (ART) (median is shown). The Kruskal-Wallis test P value was 0.0025 in A, and then multiple comparisons were performed using the Mann-Whitney test. ***P < 0.001. (C) Level of PLA2G1B RNA in PBMCs from HDs and VPs. Results are shown as the mean ± SD of the number of copies of PLA2G1B/μg of total RNA. (D) Inhibitory activity of PLA2G1B diluted in PBS or plasma from HDs or VPs previously depleted (Δ) of endogenous PLA2G1B (P < 0.0001, nonlinear regression in VP plasma relative to HD plasma or buffer). (E) The same experiment as in D with 1% plasma and 5 or 75 nM PLA2G1B. (F) VP plasma previously adsorbed on CD4+ T cells. Adsorbed plasma or buffer was collected and used to treat other CD4+ T cells together with PLA2G1B or not. (G) PLA2G1B activity on VP plasma–pretreated CD4+ T cells. CD4+ T cells were pretreated with plasma or buffer, and then plasma or buffer was removed and PLA2G1B was added, or not, to the pretreated CD4+ T cells. (H–K) PLA2G1B inhibitory activity in the presence of the gp41 fragment 565–771Δ642–725 (H and I) or 3S or control (CTL) peptides (J and K). (L and M) Inhibitory activity of 1% or 3% VP plasma depleted with anti-gp41 (gp41) polyclonal antibody (pAb) (L) or anti–gp41 3S monoclonal antibody (anti-3S) (M), control (ctrl) or not depleted (without Ab) on CD4+ T cells. D, H, and J show 1 representative dose-response experiment among 2 to 3. In E–G, I, and K–M, results are shown as the mean ± SD of the percentage of p-STAT5 NT cells inhibition on 3 or 4 donors, as indicated. *P < 0.05; **P < 0.01; ***P < 0.001 by 2-tailed unpaired t test (I and K) or by ANOVA with Tukey’s correction for multiple comparisons (E–G, L, and M).
Discussion
Here, we further characterize the activation status of the CD4+ T cell compartment in HIV-infected patients. Aside from currently used cell-surface activation markers, such as CD38 and HLA-DR, we show structural alterations of CD4+ T cell membranes, consisting of large aMMDs observed at the cell surface. They arise due to the activity of the PLA2G1B enzyme, acting in synergy with gp41. These findings support a mechanism of immunosuppression in HIV-infected patients. Here, we show that this mechanism is involved in CD4+ T cell anergy to γc cytokine and TCR responses. Furthermore, its action in the decreased survival of CD4+ T cells suggests a role in CD4 lymphopenia.
Bumpy T cells represent a new phenotype characterizing activated CD4+ T cells found in HIV-infected patients. It results from remodeling of the CD4+ T cell membrane under the influence of the enzymatic activity of PLA2G1B. This exposes GM1 gangliosides, which are enriched in aMMDs and recognized by labeled cholera toxin B using the STED imaging technique. Bumpy T cells expressing numerous aMMDs up to 500/cell can be recovered from patient blood and easily identified. In the blood of these individuals, aMMDs are spontaneously expressed by CD4+ T cells and their characteristic pattern is not modified by a strong stimulus, such as IL-7. Given that this phenotype is observed in more than 80% of the CD4+ T cells of HIV-infected patients, a correlation with CD38 and HLA-DR activation markers should be investigated. In any case, this observation offers insights into our understanding of the dysfunction of the CD4 compartment of HIV-infected patients.
At low concentrations, such as those found in the blood, PLA2G1B cannot act by itself. In HIV-infected patients, it requires a cofactor, which we identified as a soluble fragment of gp41. Gp41 appears to be a driver that targets PLA2G1B to the CD4+ T cell surface. Molecular dissection of gp41 demonstrated that the 3S peptide of gp41 is the active moiety. Plasma gp41 is most likely a degradation product of circulating HIV or HIV-infected dead cells. We analyzed the spatiotemporal mode of action of these 2 molecules when in the plasma. Our results suggest that they act in 2 separate steps. Gp41 acts first and possibly modifies the CD4+ T cell membrane. Gp41 may bind to CD4+ T cell membranes by binding to gC1qR through its 3S sequence (47, 48). This may change the membrane composition, possibly through induction of the fusion of exocytic vesicles with the plasma membrane, as previously shown for NKp44L induction (47). In the second step, these changes in lipid composition of the outer leaflet of the CD4+ T cell may increase the binding and activity of PLA2G1B (49), making the modified membrane a target of PLA2G1B.
PLA2G1B is the active moiety of the PLA2G1B/gp41 pair. At high concentrations, it appears to act, in the absence of gp41, as a newly discovered immune modulator that acts on CD4+ T cells. Its unique properties, under these conditions, were defined in vitro or in vivo in mouse experiments. PLA2G1B induces numerous aMMDs, trapping and inactivating the function of receptors (γc family, TCR, etc.), thus decreasing physiological activation, proliferation, and survival. The results obtained with IFN-α show that PLA2G1B works selectively on systems that use pMMDs for physiological signaling. It is possible that, by digesting phospholipids, PLA2G1B modifies membrane composition and changes its fluidity, allowing pMMDs to fuse and form receptor-inactivating aMMDs.
Despite this broad activity, the action of PLA2G1B remains specific and it does not act directly on CD8+ T cells. However, this mechanism may indirectly affect CD8 responses in HIV-infected patients, which are mostly highly CD4+ T cell dependent (50). It will be of interest to extend our studies to other immune cells, such as NK cells, which are highly dependent on the γc cytokine IL-15 for their activity (51).
In this context, we sought the origins of active PLA2G1B in the blood. We found the pancreas to be a major source of PLA2G1B, followed by the duodenum, jejunum, and ileum (Supplemental Figure 8, A–C). We used 2 different mAbs, 1C11 recognizing both proPLA2G1B and active PLA2G1B and 14G9 specific for the active form, and observed that proPLA2G1B is expressed in the endocrine pancreas, while active PLA2G1B is mainly expressed in the exocrine pancreas and intestinal tissues. We also observed low amounts of PLA2G1B transcripts in lymphoid cells (CD4+ or CD8+ T cells and NK cells) by qPCR, whereas they were almost undetectable in myeloid cells (lung macrophages, myeloid DCs, and plasmacytoid DCs) (Supplemental Figure 8D). The mechanism leading to the presence of active and proPLA2G1B in the blood is still unknown. They may leak from the intestinal tract, where proPLA2G1B is probably cleaved to form active PLA2G1B by proteolytic enzymes. Conversion of proPLA2G1B into the active molecule may also take place at immune sites where inflammatory cells produce proteolytic enzymes. Initially, PLA2G1B was thought to play solely a digestive role (35). Because of its function as an immunomodulator, demonstrated here, it may also play a crucial role in the regulation of the lymphoid compartment of the gut immune system. It may be involved in tolerance to food and microbiota-derived components (52). There is a high level of PLA2G1B RNA in the duodenum (Supplemental Figure 8A) and a high concentration of active PLA2G1B protein in the intestinal lumen (Supplemental Figure 8B). Thus, it may act directly on CD4+ T cells or, alternatively, cofactors derived from bacteria or viruses of the microbiota may be involved. These hypotheses may open new challenging areas of investigation.
Our results also have other consequences for our understanding of the immunodeficiency of HIV-infected patients. They show PLA2G1B to be the active component of the PLA2G1B/gp41 pair and to contribute to the processes that render most of the major conventional CD4+ T cell subpopulations anergic. First, we have clearly demonstrated dysfunction of the IL-7R/IL-7–induced microtubule and microfilament reorganization, thereby blocking p-STAT5 NT. Furthermore, all γc cytokines lost their function because of sequestering of the γc chains in macro-MMDs (aMMDs). This was verified by studying the blockade of IL-2– and IL-4–induced p-STAT NT after PLA2G1B treatment. Furthermore, the TCR responses induced by anti-CD3/anti-CD28 were also inhibited after PLA2G1B treatment. Inhibition of the function of γc cytokines and TCR responses leads to the blockade of antigen-specific responses. Overall, our results show that Bumpy T cells obtained in vitro are anergic and strongly suggest that the same is true for Bumpy T cells recovered from HIV-infected patients.
Furthermore, our data suggest that PLA2G1B is also involved in the CD4 lymphopenia observed in HIV-infected patients. IL-7 is the main cytokine that controls homeostasis of the CD4 compartment (53, 54). Thus, the PLA2G1B-induced defect in the IL-7R signaling mechanism described here should significantly contribute to CD4 lymphopenia. Furthermore, the number of surviving CD4+ lymphocytes decreased after exposure to PLA2G1B in culture (Figure 6, A and B). During these in vitro studies, we found numerous annexin V–Zombie+ CD4+ T cells, which can be considered to be the direct consequence of PLA2G1B activity, as they indicate the degradation of PS at the surface of the dying cells (Figure 6, C–E). The activity of PLA2G1B on the membrane of dying cells led us to hypothesize an additional role of PLA2G1B in the removal of damaged cells, as previously described for sPLA2 (35, 49). This may also contribute to the decrease in CD4+ T cell number. In the future, annexin V–negative dying cells could be used as a signature to study the effects of PLA2G1B on the CD4+ T cells of HIV-infected patients. This could be used to verify the activity of PLA2G1B in vivo and to follow its neutralization after anti-PLA2G1B immunotherapy.
PLA2G1B may represent a compelling therapeutic target for boosting immune responses in people contaminated by HIV. In this context, the potential of the specific mAb 14G9 has to be considered. It completely neutralized the effects of PLA2G1B in vitro, as measured by the inhibition of p-STAT5 NT. This property was also verified in vivo, using a mouse model. Furthermore, 14G9 accelerated the reversion of Bumpy T cells in vitro, as measured by their capacity to recover an IL-7 response. It also significantly reduced (up to >50%) the capacity of PLA2G1B to decrease cell survival in vitro. Thus, neutralization of the deleterious effects of the PLA2G1B/gp41 pair may be considered as a new therapeutic tool. It should be able to boost the immune system early in infection, at a time when PLA2G1B is pathogenic as a consequence of its synergy with gp41. Treatment during the beginning of ARV therapy, at a stage when the viral load remains detectable, may increase the CD4+ T cell–dependent immune defense and improve the control of HIV infection. In addition, anti-PLA2G1B therapy could have positive effects in patients infected by ARV-resistant HIV strains. After humanization, 14G9 could be a drug candidate that could be used to boost the functions of the CD4 compartment and the CD4-dependent immune responses of HIV-infected patients. By restoring IL-7 responses, decreasing anergy, and contributing to an increase in CD4+ T cell counts, neutralization of PLA2G1B may be one of the critical parameters toward remission of HIV infection (55–57).
The in vivo relevance of our data needs to be further underscored. The most significant data come from the analysis of the role of plasma from VPs. PLA2G1B in VP plasma is at physiological concentrations and, in the presence of gp41, induces unresponsiveness of CD4+ lymphocytes. The activity of plasma from VPs is well established. At the morphological level, Bumpy T cells directly purified from the blood of VPs are indistinguishable from in vitro VP plasma–induced Bumpy T cells. At the functional level, we found that aMMDs characterized from purified CD4+ T cells trap all γc and subsequently showed that HD CD4+ T cells treated with VP plasma become unresponsiveness to γc cytokines, such as IL-2, IL-4, and IL-7 (Figure 5, A and B). Inhibition or depletion of either PLA2G1B or gp41 abolishes the activity of VP plasma (Figure 4, H and I, and Figure 8, L and M). We thus propose a mechanism whereby PLA2G1B is the active moiety and gp41 a cofactor or driver that targets PLA2G1B to the surface of CD4+ T cells. The activity of the VP plasma results from synergy between the 2 molecules.
We then analyzed PLA2G1B and gp41 separately in vitro to understand the respective roles of the 2 molecules. At high concentrations, PLA2G1B is active alone. High concentrations appear to compensate for the absence of gp41. This allows characterization of the biochemical and immunological properties of the active molecule. More significantly, at low concentrations, cloned PLA2G1B was not active and did not induce unresponsiveness of the CD4+ lymphocytes in the absence of gp41. Under these experimental conditions, we clearly show that gp41 boosts PLA2G1B activity (Figure 8, H–K). PLA2G1B becomes pathogenic only after CD4+ lymphocytes have interacted with the gp41 3S peptide (Figure 8). Therefore, this in vitro analysis further supports the model in which synergy with viral gp41 is required to observe the effects of low concentrations of PLA2G1B.
In addition, studies of 3 clinical settings confirm the relevance of our observations. PLA2G1B activity strictly correlated with the presence of HIV particles. We only found PLA2G1B activity in VPs who had HIV particles in the circulating blood and therefore gp41 in the plasma. In contrast, HIV controllers and patients treated for more than 10 years with ARV, with an undetectable viral load in their plasma, had no detectable PLA2G1B activity. Furthermore, we observed a negative correlation between the ability of plasma to induce aMMDs in vitro and the CD4+ T cell counts of the patient source of plasma in a preliminary analysis (Supplemental Figure 9A). Similarly, plasma from patients with CD4+ T cell counts below 300/mm3 more strongly inhibited p-STAT5 NT than plasma from patients with CD4+ counts above 300/mm3 (Supplemental Figure 9B). Overall, these results corroborate the notion of a viral cofactor/driver that synergizes with PLA2G1B to induce CD4+ T lymphocyte unresponsiveness in vivo.
It is noteworthy that the PLA2G1B/gp41 pair appears to be a new mechanism of immunopathology, in which a physiological enzyme becomes pathogenic in the presence of molecules derived from the pathogen. This system may play a role in diseases in which immunodeficiency contributes to the emergence or progression of the disease. Preliminary data obtained with hepatitis C, Staphylococcus aureus, and Porphyromonas gingivalis support this concept.
Methods
Characterization of MMDs in primary human CD4+and CD8+T cells. Cell preparation and labeling of specific proteins were performed as previously described (34). Briefly, purified cells were equilibrated in RPMI with 5% FBS for 2 hours at 37°C and 5% CO2 before plating them onto poly-L-lysine–coated coverslips for 20 minutes at 37°C. Cells were treated with IL-7 (2 nM, 15 minutes, 37°C) and then fixed with 1.5% paraformaldehyde (PFA) (Electron Microscopy Sciences) and rehydrated for 15 minutes in PBS/5% FBS. GM1 gangliosides were labeled with Alexa Fluor–coupled cholera toxin subunit B (CtxB–Alexa Fluor 488, C22841; CtxB–Alexa Fluor 633, C34778; or CTxB-biotin, C34779; and streptavidin–Alexa Fluor 647, S32357; Life Technologies).
MMDs were analyzed at the surface of fixed CD4+ T cells and CD8+ T cells from VPs and HDs in response to IL-7 stimulation or not (Figure 1, A–C, and Supplemental Figure 4) or purified HD CD4+ or CD8+ T cells upon treatment with plasma samples (from HD, VP, ART, or HIC), WT or H48Q PLA2G1B, PLA2GIIA, PLA2GIID, or PLA2GX recombinant proteins for 30 minutes and stimulation with the cytokine for 15 minutes (Figures 3–5).
Images were acquired below the diffraction limit using a Leica TCS STED CW (ref. 31, Figures 1 and 3, and Supplemental Figure 1) or Leica TCS SP8 STED 3X (Figure 4B), or above the diffraction limit using an inverted laser-scanning confocal microscope (LSM700, Zeiss or LSM780 ELYRA PS.1, Zeiss), as previously described (34). Deconvolution was performed using Huygens Pro software (Scientific Volume Imaging). For each condition, the top half of a representative CD4+ T cell is shown from Z-stack images. MMDs were counted on the entire surface of the purified CD4+ T cells; an average of 50 cells were examined for HDs and between 15 to 50 for VPs. We determined the number of MMDs in Figures 1 and 3 or the percentage of cells with aMMDs on their surface in Figures 4 and 5.
p-STAT NT.
STAT phosphorylation and nuclear translocation in VP and HD CD4+ T cells were analyzed by microscopy after IL-7 stimulation (2 nM), or in HD CD4+ and CD8+ T cells incubated with plasma samples from HD, VP, ART, or HIC (30 minutes), WT or H48Q PLA2G1B, PLA2GIIA, PLA2GIID, or PLA2GX recombinant proteins, with or without neutralizing antibodies (30 minutes) before a 15-minute stimulation with 2 nM IL-7, IL-2, or IL-4 or 1 nM IFN-α2. WT and H48Q porcine PLA2G1B were used for the experiments shown in Figure 4, D and F. The effect of the anti-PLA2G1B 14G9 mAb on the recovery of a functional p-STAT5 NT response was studied by pretreating CD4+ T cells for 1 hour with 250 nM PLA2G1B before the addition of 667 nM anti-PLA2G1B mAb (14G9). All pretreatments and stimulations were performed at 37°C. Stimulation was stopped by addition of 4% PFA and incubation for 15 minutes at 37°C. Cells were then permeabilized overnight at –20°C in a 90% methanol/water solution.
CD4+ and CD8+ T cells were stained using respectively anti–human CD4 (mouse anti-CD4 clone RPA-T4, 555344, BD Biosciences; or goat anti-CD4, AF-379-NA, R&D Systems/Novus) and anti–human CD8 (mouse anti-CD8 clone RPA-T8, 555364, BD Biosciences), labeled with donkey anti-mouse–Alexa Fluor 488 (A21202, Thermo Fisher Scientific) or donkey anti-goat–Alexa Fluor 488 (A11055, Thermo Fisher Scientific). Phosphorylation of STAT5 in response to IL-2 or IL-7 stimulation was then revealed by staining with rabbit anti–p-STAT5 (9356, Cell Signaling Technology) labeled with goat anti-rabbit–Atto 647N (15068, Active Motif) or donkey anti-rabbit–Alexa Fluor 555 (A31572, Life Technologies); that of STAT6 in response to IL-4 stimulation by rabbit anti–p-STAT6 (9361, Cell Signaling Technology) labeled with anti-rabbit–Alexa Fluor 488 (A11034 or A21206, Life Technologies); and that of STAT1 in response to IFN-α2 stimulation by rabbit anti–p-STAT1 (9167, Cell Signaling Technology) labeled with anti-rabbit–Alexa Fluor 488 (A11034 or A21206, Life Technologies).
Images were acquired below the diffraction limit with a DM16000CS/SP5 inverted laser-scanning confocal microscope using pulsed-excitation STED (TCS STED, Leica) (58) or above the diffraction limit using an inverted laser-scanning confocal microscope (LSM700 or LSM780 ELYRA PS.1, Zeiss), as previously described (34). Deconvolution was performed using Huygens Pro software (Scientific Volume Imaging). The appearance of p-STAT was measured using ImageJ software (NIH). p-STAT5 was quantified in the cytoplasm and nucleus of the cells (Figures 1 and 3 and Supplemental Figure 1) where indicated. The number of cells positive for nuclear p-STAT among more than 200 in response to cytokines was analyzed by confocal microscopy in Figures 4, 5, 7, and 8, and Supplemental Figure 4.
Study of the effect of PLA2G1B on human CD4+ T cell survival.
Purified CD4+ T cells were cultured (7 × 106 cells/mL) in RPMI 1640 medium supplemented with 5% FBS (Life Sciences, Gibco). FBS was initially selected for its capacity to support efficient CD4+ T cell activation in response to anti-CD3/CD28 stimulation, as measured by CD69 cell-surface expression. The same FBS was also later found to support long-term survival of these lymphocytes. CD4+ T cells were treated with PBS, PLA2G1B, or the inactive mutant H48Q PLA2G1B alone or with the anti-PLA2G1B mAb 14G9 (Figure 6 and Supplemental Figure 5) or control isotype (mouse IgG1, 16-4714-85, Thermo Fisher Scientific; Figure 6 and Supplemental Figure 5). The effect of PLA2G1B on CD4+ T cell survival was evaluated by a Moxy Z Mini Automated Cell Counter (Moxy Z, Orflo Technologies). Moxy Z measures cell counts, cell size, and cell health. Cell heath is evaluated via the Moxy Viability Index (MVI) value (59). The results based on the MVI were analogous to those obtained by hemocytometer count of cells stained with trypan blue (0.1%).
For the annexin V experiments, CD4+ T cells were stained with Alexa Fluor 488–labeled antibodies against CD4 (300519, BioLegend) for 30 minutes at 4°C, the Zombie Violet Fixable Viability Kit (0.5 μL/test) (423114, BioLegend), and annexin V–APC (5 μL/test) (640941, BioLegend) for 15 minutes at room temperature. Cells were analyzed with a CytoFLEX cytometer (Beckman Coulter) and FlowJo software, version 10 (Tree Star).
Determination of gp41 and 3S plasma cofactor peptide activity on HD CD4+T cells. The effect of gp41 on PLA2G1B activity on CD4+ T cells was assessed by incubating purified CD4+ T cells in PBS/1% BSA containing peptides, recombinant proteins, VP or HD plasma, or the 10- to 30-kDa fraction previously depleted, or not, of PLA2G1B or gp41 (Supplemental Methods), together with recombinant PLA2G1B.
The binding of the viremic plasma cofactor to CD4+ T cells was tested by first incubating the PLA2G1B-depleted plasma with CD4+ T cells for 15 minutes. Then, the adsorbed plasma was collected and incubated with other CD4+ T cells from the same donor for 30 minutes, alone or together with PLA2G1B (Figure 8F).
The pretreatment effect of viremic plasma cofactor on CD4+ T cells was tested by first incubating the PLA2G1B-depleted plasma with CD4+ T cells bound onto poly-L-lysine–coated coverslips for 15 minutes. Then the supernatant was removed and the cells were washed and incubated for 30 minutes, with or without PLA2G1B (Figure 8G). p-STAT5 NT was analyzed in CD4+ T cells incubated with the adsorbed supernatants.
The effect of the recombinant gp41 and gp41 3S peptide on PLA2G1B activity was tested by pretreating the cell suspension for 15 minutes with 40 μL of the recombinant gp41 protein, peptides, with subsequent addition of 10 μL PLA2G1B for 30 minutes (Figure 8, H–K). The regulation of PLA2G1B by endogenous gp41 was tested by treating the cell suspension for 30 minutes with 50 μL of plasma dilutions or 10- to 30-kDa plasma fraction, previously depleted or not of gp41 (Figure 8, L and M). p-STAT5 NT inhibition was examined by microscopy as described above.
Additional reagents and procedures are detailed in the online Supplemental Methods (https://doi.org/10.1172/JCI131842DS1) which includes information on study design and human sample collection, recombinant proteins and peptides, cell purification and culture, DRM analysis, Western blot analyses, analysis of the IL-7R diffusion rate at the surface of living CD4+ T lymphocytes, the study of the effect of cytoskeleton inhibitors on p-STAT5 NT, analysis of cytoskeleton organization, identification of PLA2G1B as the active component in VP plasma, active human PLA2G1B structure determination, lipidomic analysis, ELISA, quantitative real-time PCR, immunodepletion experiments, flow cytometry analyses, in vitro experiments on mouse T cells, and in vivo experiments in mice.
Statistics.
Statistical parameters, including the exact value of n, precise measures (mean ± SD in all Figures, with the exception of the mean ± SEM in Figure 7, B–G), statistical significance, and tests used for each analysis are reported in the figures and figure legends. Analyses were performed using GraphPad Prism.
For experiments on human cells, 1 donor represents 1 experiment. For experiments on mice, the number of pooled mice from n independent experiments is shown.
Correlations between 2 variables were evaluated by Pearson’s correlation and linear regression.
Data were analyzed using the 2-tailed unpaired t test for 2 groups or ANOVA with correction for multiple comparisons (Tukey’s, Dunnett’s, or Sidak’s) when the distribution was Gaussian according to the D’Agostino and Pearson omnibus test. The effect of PLA2G1B on the survival of mouse CD4+ T cells in G0 to G5 was analyzed by 2-way ANOVA with Dunnett’s correction for multiple comparisons using the control condition as the control group. The anti-PLA2G1B effect on CD25 expression and the survival of PLA2G1B-treated mouse CD4+ T cells, as well as the kinetics of the effect of PLA2G1B injection on the percentage of cells showing p-STAT5 NT, were analyzed by 2-way ANOVA with Sidak’s correction for multiple comparisons. When data were not Gaussian, the Mann-Whitney nonparametric test was used to compare 2 groups and the Kruskal-Wallis test was used when more than 2 groups were compared. When the Kruskal-Wallis test showed significance, 2-by-2 comparisons were conducted to identify groups that differed by applying Bonferroni’s correction. The level of significance is indicated as *P < 0.05, **P < 0.01, and ***P < 0.001 in all figures.
Study approval.
The human study was supported by the ANRS and approved by the Comité des Personnes Ile-de-France VII under number 05-15. All participants were adults and provided written informed consent before inclusion in the study.
All animal experiments described in the present study were conducted at the Institut Pasteur according to European Union guidelines for the handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and were approved by the Institut Pasteur Animal Care and Use Committee (CETEA 89, Institut Pasteur de Paris) and the Direction Sanitaire et Vétérinaire de Paris under permit number 2016-0004 and APAFIS no. 6453-2016071912038344 v2. All experiments were subject to the 3 Rs of animal welfare (refine, reduce, and replace).
Author contributions
JP, TR, and FB designed and conducted the experiments on human and mouse cells. The study was initiated by TR and continued by JP, who participated in the writing of the manuscript. DG performed the TCS SP8 STED analysis. TR designed and conducted the plasma chromatography, biochemical analysis, microscopy image analysis, MS, and bioinformatics data analysis. LJ participated in the mAb characterization, ELISA development, and recombinant protein production. AM participated in the characterization of anti-gp41 mAb. AH and FS designed and conducted the structural analysis. JA and ERMC participated in the studies of plasma from HIV-infected patients. LT provided PLA2G1B protein. GL contributed to the design of certain experiments, provided ideas and models, and shared resources. JT was the lead senior author for this manuscript.
Supplementary Material
Acknowledgments
This work was part of the ANRS programs EP20, EP33, and EP36 (J.F. Delfraissy, O. Lambotte). It was initially supported by the Institut Pasteur (PTR 424) and the Pasteur-Weizmann Foundation. We are grateful to P. Pouletty for continuous interest and support. We wish to thank U. Schwarz (Leica Microsystems, Mannheim), E. Perret, P. Roux, A. Salles, and S. Shorte (Imagopole, Institut Pasteur) for their microscopy expertise, as well as A.H. Pillet for her expertise in biochemistry and P. Bochet for data processing and Benoit Colsch (Laboratoire d’Etude du Métabolisme des Médicaments (LEMM), CEA, INRA, Université Paris Saclay, MetaboHUB) for lipidomics analysis by mass spectrometry of CD4+ and CD8+ T cells. We thank Yoann Madec and Fredj Tekaia for their help and expertise in statistics. We acknowledge SOLEIL for the provision of synchrotron radiation facilities and thank the staff of the PROXIMA-1 beamline for their assistance. We benefited greatly from help and numerous discussions with C. Abrial, L. Touqui, B. Colsch, D. Troisvallet, M.L. Gougeon, P. Bruhns, and J. Tiollier. We also gratefully acknowledge J.P. Routy and B. Malissen for their critical review of the manuscript.
Version 1. 03/03/2020
In-Press Preview
Version 2. 04/27/2020
Electronic publication
Version 3. 04/29/2020
Updated to include graphical abstract.
Version 4. 06/01/2020
Print issue publication
Footnotes
Conflict of interest: JT is cofounder and CEO of DIACCURATE, a spin-off of the Institut Pasteur. JP, LJ, and AM are employees of DIACCURATE.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):2872–2887.https://doi.org/10.1172/JCI131842.
Contributor Information
Julien Pothlichet, Email: julien.pothlichet@diaccurate.com.
Thierry Rose, Email: thierry.rose@pasteur.fr.
Florence Bugault, Email: fbugault@pasteur.fr.
Louise Jeammet, Email: louise.jeammet@diaccurate.com.
Annalisa Meola, Email: annalisa.meola@diaccurate.com.
Ahmed Haouz, Email: ahmed.haouz@pasteur.fr.
Frederick Saul, Email: frederick.saul@pasteur.fr.
David Geny, Email: david.geny@inserm.fr.
José Alcami, Email: ppalcami@isciii.es.
Ezequiel Ruiz-Mateos, Email: ezequiel.ruizmateos@gmail.com.
Luc Teyton, Email: lteyton@scripps.edu.
Gérard Lambeau, Email: lambeau@ipmc.cnrs.fr.
Jacques Thèze, Email: jacques.theze@diaccurate.com.
References
- 1.Walker B, McMichael A. The T-cell response to HIV. Cold Spring Harb Perspect Med. 2012;2(11):a007054. doi: 10.1101/cshperspect.a007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev. 2013;254(1):326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitman MC, Lau JSY, McMahon JH, Lewin SR. Barriers and strategies to achieve a cure for HIV. Lancet HIV. 2018;5(6):e317–e328. doi: 10.1016/S2352-3018(18)30039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doitsh G, Greene WC. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe. 2016;19(3):280–291. doi: 10.1016/j.chom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39(4):633–645. doi: 10.1016/j.immuni.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Armas LR, et al. Reevaluation of immune activation in the era of cART and an aging HIV-infected population. JCI Insight. 2017;2(20):95726. doi: 10.1172/jci.insight.95726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sousa AE, Carneiro J, Meier-Schellersheim M, Grossman Z, Victorino RM. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J Immunol. 2002;169(6):3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 9.George V, et al. Associations of plasma cytokine and microbial translocation biomarkers with immune reconstitution inflammatory syndrome. J Infect Dis. 2017;216(9):1159–1163. doi: 10.1093/infdis/jix460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tincati C, Douek DC, Marchetti G. Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res Ther. 2016;13:19. doi: 10.1186/s12981-016-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 12.Hocini H, et al. HIV controllers have low inflammation associated with a strong HIV-specific immune response in blood. J Virol. 2019;93(10):e01690-18. doi: 10.1128/JVI.01690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer BE, Blyveis N, Fontenot AP, Wilson CC. Functional and phenotypic characterization of CD57+CD4+ T cells and their association with HIV-1-induced T cell dysfunction. J Immunol. 2005;175(12):8415–8423. doi: 10.4049/jimmunol.175.12.8415. [DOI] [PubMed] [Google Scholar]
- 14.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103(3):966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 15.Clerici M, et al. Detection of three distinct patterns of T helper cell dysfunction in asymptomatic, human immunodeficiency virus-seropositive patients. Independence of CD4+ cell numbers and clinical staging. J Clin Invest. 1989;84(6):1892–1899. doi: 10.1172/JCI114376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boswell KL, et al. Loss of circulating CD4 T cells with B cell helper function during chronic HIV infection. PLoS Pathog. 2014;10(1):e1003853. doi: 10.1371/journal.ppat.1003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallikkuth S, de Armas L, Rinaldi S, Pahwa S. T follicular helper cells and B cell dysfunction in aging and HIV-1 infection. Front Immunol. 2017;8:1380. doi: 10.3389/fimmu.2017.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, et al. Cycling memory CD4+ T cells in HIV disease have a diverse T cell receptor repertoire and a phenotype consistent with bystander activation. J Virol. 2014;88(10):5369–5380. doi: 10.1128/JVI.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieg SF, Bazdar DA, Harding CV, Lederman MM. Differential expression of interleukin-2 and gamma interferon in human immunodeficiency virus disease. J Virol. 2001;75(20):9983–9985. doi: 10.1128/JVI.75.20.9983-9985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David D, et al. Regulatory dysfunction of the interleukin-2 receptor during HIV infection and the impact of triple combination therapy. Proc Natl Acad Sci U S A. 1998;95(19):11348–11353. doi: 10.1073/pnas.95.19.11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colle JH, Moreau JL, Fontanet A, Lambotte O, Delfraissy JF, Thèze J. The correlation between levels of IL-7Ralpha expression and responsiveness to IL-7 is lost in CD4 lymphocytes from HIV-infected patients. AIDS. 2007;21(1):101–103. doi: 10.1097/QAD.0b013e3280115b6a. [DOI] [PubMed] [Google Scholar]
- 22.Colle JH, et al. Regulatory dysfunction of the interleukin-7 receptor in CD4 and CD8 lymphocytes from HIV-infected patients--effects of antiretroviral therapy. J Acquir Immune Defic Syndr. 2006;42(3):277–285. doi: 10.1097/01.qai.0000214823.11034.4e. [DOI] [PubMed] [Google Scholar]
- 23.Landires I, et al. HIV infection perturbs interleukin-7 signaling at the step of STAT5 nuclear relocalization. AIDS. 2011;25(15):1843–1853. doi: 10.1097/QAD.0b013e32834a3678. [DOI] [PubMed] [Google Scholar]
- 24.Juffroy O, et al. Dual mechanism of impairment of interleukin-7 (IL-7) responses in human immunodeficiency virus infection: decreased IL-7 binding and abnormal activation of the JAK/STAT5 pathway. J Virol. 2010;84(1):96–108. doi: 10.1128/JVI.01475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18(4):374–384. doi: 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JX, Leonard WJ. The common cytokine receptor γ chain family of cytokines. Cold Spring Harb Perspect Biol. 2018;10(9):a028449. doi: 10.1101/cshperspect.a028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman ML, Shive CL, Nguyen TP, Younes SA, Panigrahi S, Lederman MM. Cytokines and T-cell homeostasis in HIV infection. J Infect Dis. 2016;214 suppl 2:S51–S57. doi: 10.1093/infdis/jiw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaughlin D, Faller E, Sugden S, MacPherson P. Expression of the IL-7 receptor alpha-chain is down regulated on the surface of CD4 T-cells by the HIV-1 Tat protein. PLoS One. 2014;9(10):e111193. doi: 10.1371/journal.pone.0111193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Micci L, et al. Paucity of IL-21-producing CD4(+) T cells is associated with Th17 cell depletion in SIV infection of rhesus macaques. Blood. 2012;120(19):3925–3935. doi: 10.1182/blood-2012-04-420240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shive CL, et al. Inflammatory cytokines drive CD4+ T-cell cycling and impaired responsiveness to interleukin 7: implications for immune failure in HIV disease. J Infect Dis. 2014;210(4):619–629. doi: 10.1093/infdis/jiu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440(7086):935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 32.Dinic J, Riehl A, Adler J, Parmryd I. The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci Rep. 2015;5:10082. doi: 10.1038/srep10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13(4):257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 34.Tamarit B, et al. Membrane microdomains and cytoskeleton organization shape and regulate the IL-7 receptor signalosome in human CD4 T-cells. J Biol Chem. 2013;288(12):8691–8701. doi: 10.1074/jbc.M113.449918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambeau G, Gelb MH. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem. 2008;77:495–520. doi: 10.1146/annurev.biochem.76.062405.154007. [DOI] [PubMed] [Google Scholar]
- 36.Goulder P, Deeks SG. HIV control: Is getting there the same as staying there? PLoS Pathog. 2018;14(11):e1007222. doi: 10.1371/journal.ppat.1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thèze J, Chakrabarti LA, Vingert B, Porichis F, Kaufmann DE. HIV controllers: a multifactorial phenotype of spontaneous viral suppression. Clin Immunol. 2011;141(1):15–30. doi: 10.1016/j.clim.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W, Yi L, Feng Y, Chen L, Liu J. Structural insight into the activation mechanism of human pancreatic prophospholipase A2. J Biol Chem. 2009;284(24):16659–16666. doi: 10.1074/jbc.M808029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janssen MJ, et al. Catalytic role of the active site histidine of porcine pancreatic phospholipase A2 probed by the variants H48Q, H48N and H48K. Protein Eng. 1999;12(6):497–503. doi: 10.1093/protein/12.6.497. [DOI] [PubMed] [Google Scholar]
- 40.Blouin CM, Lamaze C. Interferon gamma receptor: the beginning of the journey. Front Immunol. 2013;4:267. doi: 10.3389/fimmu.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchetti M, et al. Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol Biol Cell. 2006;17(7):2896–2909. doi: 10.1091/mbc.e06-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercier F, et al. Persistent human immunodeficiency virus-1 antigenaemia affects the expression of interleukin-7Ralpha on central and effector memory CD4+ and CD8+ T cell subsets. Clin Exp Immunol. 2008;152(1):72–80. doi: 10.1111/j.1365-2249.2008.03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, et al. CD127 Expression in naive and memory T cells in HIV patients who have undergone long-term HAART. Lab Med. 2017;48(1):57–64. doi: 10.1093/labmed/lmw053. [DOI] [PubMed] [Google Scholar]
- 44.Booth NJ, et al. Different proliferative potential and migratory characteristics of human CD4+ regulatory T cells that express either CD45RA or CD45RO. J Immunol. 2010;184(8):4317–4326. doi: 10.4049/jimmunol.0903781. [DOI] [PubMed] [Google Scholar]
- 45.Nagafuku M, et al. CD4 and CD8 T cells require different membrane gangliosides for activation. Proc Natl Acad Sci U S A. 2012;109(6):E336–E342. doi: 10.1073/pnas.1114965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieillard V, Strominger JL, Debré P. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc Natl Acad Sci U S A. 2005;102(31):10981–10986. doi: 10.1073/pnas.0504315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fausther-Bovendo H, Vieillard V, Sagan S, Bismuth G, Debré P. HIV gp41 engages gC1qR on CD4+ T cells to induce the expression of an NK ligand through the PIP3/H2O2 pathway. PLoS Pathog. 2010;6:e1000975. doi: 10.1371/journal.ppat.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pednekar L, et al. Identification of the gC1qR sites for the HIV-1 viral envelope protein gp41 and the HCV core protein: Implications in viral-specific pathogenesis and therapy. Mol Immunol. 2016;74:18–26. doi: 10.1016/j.molimm.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brueseke TJ, Bell JD. A new hat for an old enzyme: waste management. Biochim Biophys Acta. 2006;1761(11):1270–1279. doi: 10.1016/j.bbalip.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Novy P, Quigley M, Huang X, Yang Y. CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. J Immunol. 2007;179(12):8243–8251. doi: 10.4049/jimmunol.179.12.8243. [DOI] [PubMed] [Google Scholar]
- 51.Becknell B, Caligiuri MA. Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv Immunol. 2005;86:209–239. doi: 10.1016/S0065-2776(04)86006-1. [DOI] [PubMed] [Google Scholar]
- 52.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279(1):70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. 2012;24(3):209–217. doi: 10.1016/j.smim.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kittipatarin C, Khaled AR. Interlinking interleukin-7. Cytokine. 2007;39(1):75–83. doi: 10.1016/j.cyto.2007.07.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davenport MP, Khoury DS, Cromer D, Lewin SR, Kelleher AD, Kent SJ. Functional cure of HIV: the scale of the challenge. Nat Rev Immunol. 2019;19(1):45–54. doi: 10.1038/s41577-018-0085-4. [DOI] [PubMed] [Google Scholar]
- 56.Deeks SG, et al. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med. 2016;22(8):839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lederman MM, et al. A cure for HIV infection: “Not in my lifetime” or “Just around the corner”? Pathog Immun. 2016;1(1):154–164. doi: 10.20411/pai.v1i1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–782. doi: 10.1364/OL.19.000780. [DOI] [PubMed] [Google Scholar]
- 59.Dittami GM, Sethi M, Rabbitt RD, Ayliffe HE. Determination of mammalian cell counts, cell size and cell health using the Moxi Z mini automated cell counter. J Vis Exp. 2012;(64):3842. doi: 10.3791/3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.