Abstract
Prevalence of inflammatory bowel disease (IBD), a chronic inflammatory disorder of the gut, has been on the rise in recent years—not only in the adult population but also especially in pediatric patients. Despite the absence of curative treatments, current therapeutic options are able to achieve long-term remission in a significant proportion of cases. To this end, however, there is a need for biomarkers enabling accurate diagnosis, prognosis, and prediction of response to therapies to facilitate a more individualized approach to pediatric IBD patients. In recent years, evidence has continued to evolve concerning noncoding RNAs (ncRNAs) and their roles as integral factors in key immune-related cellular pathways. Specific deregulation patterns of ncRNAs have been linked to pathogenesis of various diseases, including pediatric IBD. In this article, we provide an overview of current knowledge on ncRNAs, their altered expression profiles in pediatric IBD patients, and how these are emerging as potentially valuable clinical biomarkers as we enter an era of personalized medicine.
Keywords: pediatrics, inflammatory bowel disease, Crohn’s disease, ulcerative colitis, noncoding RNA, microRNA
In this article, authors provide an overview of current knowledge on noncoding RNAs, their altered expression profiles in pediatric IBD patients, and how these are emerging as potentially valuable clinical biomarkers.
INTRODUCTION
Inflammatory bowel disease (IBD) is an umbrella term for ulcerative colitis (UC) and Crohn’s disease (CD). These chronic inflammatory disorders of the gastrointestinal tract often are diagnosed in adolescence and young adulthood. Some 8%–25% of IBD patients have early onset of the disease in childhood.1, 2 These cases are more severe,3 with many extraintestinal issues such as delayed growth and development.4 The prevalence of these diseases is steadily rising worldwide, and the increase is particularly rapid in the pediatric population.5–7 Current diagnostic routine includes symptom assessment, endoscopic examination and biopsy, histology, serology, and radiology.8, 9 No standard diagnostic routine and reliable direct biomarkers are currently available. The biomarkers we have now reflect only general inflammation rather than specific pathogenesis associated with ongoing IBD or a specific subtype of IBD. A time-consuming and often painful diagnostic process eventually leading to surgical intervention is a particularly traumatic experience for young children, but this could very well be avoided by the use of noninvasive or minimally invasive biomarkers for diagnostics and therapeutic disease monitoring.
Although novel therapeutic strategies are effective in managing symptoms and achieving long-term remission, these approaches are not curative, and in some patients, no or only poor response is observed.10, 11 Early identification of such patients by innovative diagnostic approaches and their redirection to other therapeutic options is therefore essential for improving therapeutic outcomes. Moreover, novel discoveries in IBD pathogenesis are necessary to identify the targets and to develop novel therapeutic strategies. Noncoding RNAs (ncRNAs) are currently being studied intensively in pediatric IBD patients because they constitute a promising, novel class of biomarkers and therapeutic targets.
NONCODING RNAs, THEIR CLASSIFICATION, FUNCTION, AND BIOGENESIS
After the Human Genome Project revealed that only 1.5% of the genome is protein-coding,12 it became clear that there is more to DNA than mere proteins. Even earlier, there had been some sparse information available about the existence of RNAs untranslated to the proteins, but this was long considered an exception rather than a rule. In addition to transfer and ribosomal RNA, the first observations of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) were made in the 1980s.13–16 Later, cancer-related deregulation of ncRNAs RNA H19,17 growth arrest-specific transcript 5 (GAS5),18 and prostate cancer antigen (PCA3/DD3)19 pointed to the now well-documented phenomena as to the involvement of noncoding genome in the development of many complex diseases. Only after the discovery of gene expression regulation through RNA interference facilitated by short noncoding RNAs,20–22 however, did exploration of noncoding genome really begin to take off.
Noncoding RNAs can be divided according to their function into 2 groups: housekeeping ncRNAs (e.g., tRNAs, rRNAs, snRNAs, snoRNAs) and regulatory ncRNAs. The latter of these is historically subdivided into 2 large groups according to the arbitrary dividing line of 200 nucleotides in length. Transcripts shorter than 200 nucleotides are termed short noncoding RNAs (sncRNAs), and transcripts exceeding 200 nucleotides are called long noncoding RNAs (lncRNAs). Both groups are involved in regulating gene expression and operate on several levels depending on the type of ncRNA. The sncRNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), are involved mostly in post-transcriptional regulation but also in many other specific processes such as transposon silencing or rRNA maturation.23 The lncRNAs are much longer by definition and comprise a more diverse group of transcripts. They are known to affect many cellular processes on transcriptional, post-transcriptional, and translational levels. Although both short and long noncoding transcripts usually possess no protein coding capacity, there has been some evidence of cryptic reading frames and translation in to shorter micropeptides in lncRNAs formerly regarded as noncoding.24–26
MicroRNAs
Among all ncRNAs, miRNAs have been studied thoroughly and have claimed the most attention in recent decades. Along with the discovery of RNA interference in the early 2000s, miRNAs were observed first in Caenorhabditis elegans as master regulators of developmental timing20, 21, 27–30 and later in many other species, including humans.30, 31 Their distinct length of 18 to 25 nucleotides makes them a very specific group of transcripts, currently encompassing about 2000 different mature miRNAs.32 Naturally produced endogenously, miRNAs constitute a pivotal cellular mechanism for regulating expression in as many as 60% of human genes31 by complementary binding to their target messenger RNAs (mRNAs). Of the 18 to 25 nucleotides, 8 are essential and make up what is termed the “seed” region, which binds to the 3′ untranslated region of the target, thereby leading to repressed translation of the target mRNA, either by its destabilization or degradation.33, 34 As the seed region is fairly short, many different mRNAs can contain a complementary sequence and be affected by a single miRNA, thus making it a pleiotropic regulator of several targets.33 Stemming from either individual miRNA genes or intergenic and intragenic regions of protein-coding genes,35 miRNAs are canonically transcribed by RNA polymerase II,36 thereby creating polyadenylated and capped pri-miRNAs.37, 38 This primary transcript is usually several hundred nucleotides long and contains a future mature miRNA sequence in the stem of the secondary hairpin structures of pri-miRNA. Next, splicing of the pri-miRNA is facilitated by a microprocessor complex consisting of RNase III Drosha39 and a dimer of DiGeorge critical region 8 (DGCR8).40, 41 The microprocessor cleaves the pri-miRNA transcript, creating 1 or several hairpin structures—pre-miRNAs—that each contain one future miRNA. Pre-miRNAs are transported by nuclear transporter protein exportin 542 to the cytoplasm, where they are processed further. The RNase III–type enzyme Dicer,43 together with other cooperating proteins (dependant on the species; in humans it is trans-activation-responsive RNA binding protein [TRBP]),44 cleaves pre-miRNA close to the terminal loop and creates a double-stranded RNA intermediate. One of the strands is recruited into an RNA-induced silencing complex (RISC) with proteins from the Argonaute family (AGO).44 The strand that is recruited is termed “leading,” and the other one, called the “passenger strand,” is usually degraded; although in some cases, it also can be recruited into the RISC.45
The canonical pathway of miRNA biogenesis can be overcome, as some miRNAs have been observed to be produced alternatively in noncanonical ways that exclude certain steps and also give rise to other types of sncRNAs.46–49
LONG NONCODING RNAs
Long ncRNAs were first regarded as nonfunctional because their roles in the cells have been unknown and their sequences are less conserved than are those of protein-coding genes.50, 51 In comparison with miRNAs, lncRNAs encompass a much broader group due to their definition by length. Though miRNAs encompass only a specific 18 to 25 nucleotides in length of the spectrum, everything from 200 nucleotides and larger is considered an lncRNA unless it is a coding sequence.52–56 Next-generation sequencing revealed that lncRNAs originate from more than 59,000 genes.57 That number was expanded even further by the NONCODE database to more than 96,000 genes producing over 172,000 transcripts.58 Not many of these, however, have been experimentally validated to date.59 Nevertheless, structural and functional variability makes it difficult to create a meaningful and useful classification system; 60 currently, several systems are being used based upon localization in the genome in relation to the protein-coding genes, according to their function or depending upon the means of their origin.
Although sharing many similarities with mRNAs, lncRNAs are more tissue- and time-specific and operate in much lower concentrations.52, 60–62 They are localized both in cell nucleus and in the cytoplasm in 1 or more copies, but nuclear localization, especially close to the chromatin, is their preferential one.52, 63 Close to the chromatin, they affect gene expression by facilitating chromatin interactions and guiding chromatin-remodeling complexes,64, 65 thus activating or repressing transcription. Other ways of transcriptional regulation include cooperation with transcription factors,66, 67 binding to regulatory sequences68–70 and promoting splicing of mRNAs in complexes with other splicing molecules.71, 72
When translocated to the cytoplasm, lncRNAs are involved in post-transcriptional and post-translational regulation of gene expression while acting in cooperation with RNA-binding proteins, or they affect the stability and degradation of such proteins and thus facilitate protein turnover.73 The mRNA stability is affected by the binding of RNA–protein complexes containing lncRNAs as guiding molecules, which causes either degradation or enhanced translation of the target mRNA.74, 75 The RNA–protein complexes are also involved in various signaling pathways,76 fulfill certain roles in cellular organelles, or help transport other molecules into organelles.77 A separate category of lncRNAs, so-called “competing endogenous RNAs” (ceRNAs), serve as decoys or sponges for miRNAs and so alter the relative availability of miRNAs.78 Similarly, lncRNAs serve also as protein decoys, averting proteins from binding to other transcripts.79
In contrast to the well-described canonical pathway of miRNAs, a general biogenetic pathway for lncRNAs is difficult to trace, as lncRNAs present a diverse group of transcripts produced in several ways. Nevertheless, an initial part of the biogenesis is shared not only among lncRNAs but also by all transcripts. This consists of transcription by polymerase II, polyadenylation, 5′ capping, and chromatin modifications typical for protein-coding sequences.52, 80 However, lncRNA genes usually contain fewer but longer exons, and their expression is more time- and tissue-specific. Enormous variability exists on the post-transcriptional level, which includes such specific modifications as cleaving of the 3′ end by RNAse P or back-splicing to creating a circular lncRNA (circRNA).81, 82 There is also some evidence that miRNA transcriptional apparatus is somewhat involved in lncRNA biogenesis. After all, sncRNAs, including for example miRNAs themselves, arise from formerly long primary transcripts classifiable as lncRNAs and only later are processed by specific biogenetic pathways.83, 84
NONCODING RNAs AS BIOMARKERS
Great demand exists for a precise, possibly noninvasive biomarker that can provide a faster, simpler, and more efficient way of characterizing patients and personalizing management of the disease. The ncRNAs have emerged as potential biomarkers for several diseases, as these are generally stable and abundantly present in a variety of clinical specimens, including tissues and bodily fluids; are highly tissue-specific, cell type-specific, and condition-specific; and can be readily detected by routine and inexpensive laboratory techniques.85, 86
The majority of the human genome encodes RNAs that do not code for proteins.20, 21, 87 These ncRNAs affect normal expression of the genes, including genes involved in the immune system, inflammation, and IBD pathogenesis. Although miRNAs are the most studied regulatory ncRNAs to date and miRNA-targeted diagnostics and therapeutics have already reached clinical development,28, 85, 86, 88 the importance of lncRNAs as potential biomarkers and therapeutic targets is increasingly recognized.85, 86, 89, 90
Both short and long ncRNAs function mostly as regulators and fine-tuners of gene expression. Although miRNAs share a simple structure and, in the majority of cases, bind to their target mRNAs through their 8-nucleotide long seed region to post-transcriptionally regulate gene expression,27 lncRNAs use many different molecular mechanisms depending on the length and structure of a given transcript. This enables a wide variety of functions, spanning from transcription regulation and acting as miRNA sponges to orchestrating epigenetic modifications.82 Several miRNAs89, 90 and specific miRNA signatures91, 92 have been identified in IBD-associated tissues. It has been shown that among many other cellular processes, miRNAs play a significant role in intestinal immunity.93 Nevertheless, there exists only sparse information on ncRNA profiles and their diagnostic potential in pediatric IBD patients (summarized in Tables 1 and 2). Given that adult and pediatric IBD have some differences in manifestation, etiology, and genetic background,4 it is expected that ncRNA profiles may reflect these differences. To examine these aspects thoroughly, we searched the PubMed database for relevant studies according to the following strategy: “miRNA” and “pediatric” and (“ulcerative” and “colitis”) or (“crohn” and “disease”) or “IBD.” When we excluded nonclinical studies and chose only studies carried out on pediatric patients, the remaining 11 articles (Tables 1 and 2) were relevant for our discussion.
TABLE 1.
Summary of Tissue ncRNAs Associated With Various Aspects of Pediatric IBD
| Study | ncRNA | Change in expression (patients) | Compared groups | Number of patients, sample type | Statistical parameters | Technological platform | ||
|---|---|---|---|---|---|---|---|---|
| Best P- achieved | AUC | Sensitivity/ Specificity (%) | ||||||
| Koukos et al., 2013 40 | miR-101 | Down | IBD vs. non-IBD | 45 biopsies | — | — | — | MicroRNA-library screen, RT-qPCR |
| miR-26 | Down | — | ||||||
| miR-124 | Down | pUC vs. pCD/non-IBD | <0.0001/<0.01 | |||||
| Koukos et al., 2015 42 | miR-4284 | Down | pIBD vs. non-IBD | 37 biopsies | <0.05 | — | — | mirCURY microRNA array, RT-qPCR |
| Zahm et al., 2014 43 | miR-192 miR-194 miR-200b miR-375 | Down | pUC vs. controls | 50 biopsies | 0.0006 0.0019 0.0056 0.0001 | — | — | nCounter, TaqMan low density array, RT-qPCR |
| miR-142-3p miR-146a miR-21 let-7i | Up | 0.0048 0.0027 0.0003 0.0007 | — | — | ||||
| miR-24 | — | pUC vs. pCD | — | 0.83 | 83.3/85.7 | |||
| Béres et al., 2016 44 | miR-122 miR-146a miR-155 | Up Up UP | ipCD vs. C/pUC pUC vs. C ipCD vs. C | 28 FF samples, 71 FFPE samples | <0.01 <0.001 <0.001 | — | — | RT-qPCR |
| Szűcs et al., 2016 45 | miR-146a miR-155 | Up | ipCD vs. inpCD vs. C | 30 FFPE samples | <0.001 | — | — | RT-qPCR |
| Béres et al., 2017 33 | miR-185 miR-223 | Up | ipCD vs. C | 44 FF samples | <0.05 <0.001 | 0.81 1 | 62.5/100 100/100 | NGS, RT-qPCR |
| miR-146a miR-142–3p | — | pCD vs. pUC | <0.01 <0.01 | 0.838 0.888 | 80/76.92 77.78/90.31 | |||
| Tang et al., 2018 48 | miR-15a | Down | CDre vs. CDac | 54 FF samples | <0.05 | — | — | RT-qPCR |
Abbreviations: AUC, area under the curve; C, control; FFPE, formalin-fixed paraffin-embedded; CDac, active Crohn’s disease; CDre, Crohn’s disease in remission; FF, fresh frozen tissue; ipCD, histologically intact tissue of pediatric patients with Crohn’s disease; inpCD, histologically inflamed tissue of pediatric patients with Crohn’s disease; NGS, next-generation sequencing; pCD, pediatric patients with Crohn’s disease; pIBD, pediatric patients with inflammatory bowel disease; pUC, pediatric patients with ulcerative colitis; non-IBD, control patients without inflammation typical for IBD; RT-qPCR, real-time quantitative polymerase chain reaction.
TABLE 2.
Serum ncRNAs With Successfully Validated Biomarker Potential for Various Aspects of Pediatric IBD
| Study | ncRNA | Change in expression (patients) | Compared groups | Number of patients, sample type | Statistical parameters | Technological platform | |||
|---|---|---|---|---|---|---|---|---|---|
| Best P- achieved | AUC | Sensitivity/ Specificity (%) | |||||||
| Zahm et al., 201149 | miR-484 miR-16 | Up | pCD vs. control vs. celiac | 102 blood serum samples | <0.0001 | 0.917 0.912 | 82.61/84.38 73.91/100 | TaqMan human microRNA array, RT-qPCR | |
| Zahm et al., 201443 | miR-192 miR-142–3p miR-21 | Up | pUC vs. control | 47 blood serum samples | 0.0045 0.0078 — | 0.757 0.723 0.718 | 79.31/77.78 75.86/66.67 75.86/66.67 | TaqMan low density array human microRNA panel, RT-qPCR | |
| Heier et al., 201650 | miR-146a miR-146b miR-320 miR-486 | Down | pIBD pharmaco-dynamics | 19 PBMC samples | <0.05 <0.01 <0.01 <0.01 | — | — | RT-qPCR | |
| De Iucidibus et al., 201851 | miR-29c-3p | Up | pIBD pharmaco-dynamics | 10 PBMC samples | <0.01 | — | — | NGS, RT-qPCR | |
| Lucafò et al., 201852 | GAS5 | Up | pIBD pharmaco-dynamics | 19 PBMC samples | <0.05 | — | — | RT-qPCR |
Abbreviations: AUC, area under the curve; C, control; GAS5, growth arrest-specific transcript 5; NGS, next-generation sequencing; PBMC, peripheral blood mononuclear cells; pCD, pediatric patients with Crohn’s disease; pIBD, pediatric patients with inflammatory bowel disease; pUC, pediatric patients with ulcerative colitis; RT-qPCR, real-time quantitative polymerase chain reaction.
NONCODING RNAs IN TISSUES OF PEDIATRIC IBD PATIENTS
The research group of Koukos et al focused on differences in ncRNA expression profiles in pediatric and adult IBD patients and published its study in 2013.94 In addition to showing that the IL6/STAT3 pathway is a critical factor in the development and progression of IBD, those authors identified 5 miRNAs suppressing activity of STAT3. These are miR-125, miR-101, miR-26, miR-124, and let-7, with miR-124 probably being a central regulator of STAT3 due to its greater than 90% inhibitory effect on STAT3 in human colonocytes.95 Further investigation using real-time quantitative polymerase chain reaction (RT-qPCR) on adult and pediatric samples revealed that let-7 and miR-125 were downregulated specifically in adult patients, miR-101 and miR-26 in pediatric and adult patients, and miR-124 only in pediatric patients with active UC. Thus, these are potential diagnostic biomarkers depicting disease activity. Downregulation of miR-124 was due to methylation of miR-124 promoter, which provided the first evidence as to a role of epigenetic regulation in pediatric IBDs.94 The Koukos team continued its research efforts and 2 years later published another study on pediatric IBD patients,96 again comparing active and inactive disease vs healthy controls and adult UC patients. They discovered a 24-miRNA signature that was deregulated in colonic tissue, with miR-4284 being the most downregulated ncRNA in pediatric UC patients. Its expression was also downregulated in patients with active vs inactive UC. Further in vitro experiments showed that miR-4284 is present in colonic epithelial cells and regulates expression of C-X-C motif chemokine 5 (CXCL5) by binding to its 3′UTR. Correspondingly, CXCL5 levels are increased in pediatric patients with UC due to miR-4284 downregulation.96 The CXCL5 is known for its expression in colonic epithelial cells, and as a facilitator of neutrophil recruitment, it is one of the major players in the development of UC.93, 95
In the study by Zahm et al,97 tissue expression profiles from rectal biopsies revealed specific miRNA patterns associated with pediatric UC and CD compared with controls. Four miRNAs that were enriched in epithelial cells (miR-192, miR-194, miR-200b, and miR-375) were significantly downregulated in UC patients compared with controls. Contrarily, 4 miRNAs that were overexpressed in inflammatory cells (miR-142–3p, miR-146a, miR-21, and let-7i) were upregulated in UC patients compared with controls. Only miR-375 and miR-21 were significantly altered in pediatric CD patients in comparison with controls. In UC patients receiving the immunomodulator 6-mercaptopurine or methotrexate, significant elevation was observed of miR-375 and miR-192 compared with in UC patients not receiving immunomodulators. A single miRNA, miR-24, was differentially expressed between UC and CD patients and enabled correct classification of 84% of patients, with a sensitivity of 83% and specificity of 86%.97
Another study focusing on both pediatric CD and UC came from the group of Béres et al.98 They selected for their study miR-146a, miR-122, and miR-155, which previously had been shown to play an important role in immune processes and immune-mediated diseases. MiR-146a and miR-155 levels were significantly higher in the inflamed mucosa of children with CD and UC compared with the intact mucosa.98 Moreover, the authors demonstrated induction of miR-146a and miR-155 after treatment with TNF-α (a potent inflammatory cytokine and effective therapeutic target in IBD)—and hence, their potential involvement in TNF-α pro-inflammatory signaling. The same team achieved similar results when comparing expression of miR-146a, miR-155, and miR-122 in inflamed duodenal tissue of CD patients, intact duodenal tissue of CD patients, and healthy controls.99
Their follow-up publication describes the most robust biomarker study to date concerning ncRNAs in pediatric IBD patients.89 Using small RNA sequencing of fresh-frozen tissue biopsies, the authors obtained specific miRNA profiles of CD patients with inflamed and intact histology and also those of healthy controls. The validation phase of the study by Béres et al89 was conducted not only in CD but also in pediatric UC patients, thereby providing additional information on the discovered miRNAs. The most interesting results from a diagnostic perspective are summarized in Table 1. There was significant overlap between the miRNA expression profiles differentiating CD and UC from healthy patients (upregulation of miR-18a, miR-21, miR-31, miR-99a, miR-99b, miR-125a, miR-126, miR-142-5p, miR-146a, and miR-223 and downregulation of miR-141 and miR-204 in diseased tissue). Nevertheless, there were some miRNAs upregulated in UC compared with CD, namely miR-21, miR-31, miR-125, miR-142-3p, and miR-146a; on the other hand, the expression levels of miR-100, miR-150, and miR-185 were increased in CD patients compared with UC patients. Through combined pathway analysis of miRNAs and their target mRNAs identified in CD, those authors revealed a strong association of these miRNAs and mRNAs with inflammation, fibrosis, and response to microbiome, in addition to immune and inflammatory response. Five miRNAs differentially expressed in UC patients (miR-20a, miR-126, miR-141, miR-142, and miR-223) were connected to the ABCG2 and ABCB1 efflux transport proteins important in intestinal barrier protection against external stimuli.89
It seems that some of these miRNAs are probably not specific for pediatric IBD patients but more likely are important for IBD in general or inflammation as such. Similar profiles have been detected in studies performed on samples from adult IBD patients,90 and some miRNAs are well-known players in inflammatory processes (eg, miR-146a, miR-155, and miR-21),100 regardless of whether these are in adult or pediatric patients.
However, there are some examples showing just the opposite. Particularly noteworthy is that miR-223, identified in the Béres study as one of the most significantly upregulated miRNAs in UC and CD (in both inflamed and intact tissue),89 has been described in adult IBD as a negative regulator of inflammation.101 Results from Neudecker et al relating to adult IBD patients and an animal model showed that overexpression of miR-223 can attenuate inflammation. Moreover, they observed the release of proinflammatory cytokines and chemokines in myeloid-derived cells through the miR-223–NOD-like receptor 3 (NLRP3)–interleukin-1β (IL-1β) regulatory circuit as a critical component of intestinal inflammation and homeostasis.101 MiR-223 is probably one of the examples indicating the differences between the underlying IBD pathogenesis in adult and pediatric patients.
The most recent work from Tang et al102 was focused on miR-15 as a regulator of Cdc4, a potent regulator of the cell cycle. The miR-15 level was quantified in 33 pediatric IBD patients and 21 controls, and moderate increase in miR-15 expression was observed in CD patients. Unfortunately, the variability of miR-15 expression was too high, thus precluding its use as a reliable biomarker of CD. Testing for potential correlation between miR-15 expression and PCDAI score also was unsuccessful.102
NONCODING RNAs IN BODILY FLUIDS OF PEDIATRIC IBD PATIENTS
Concerning ncRNAs in bodily fluids (Table 2), Zahm et al provided initial promising findings.97, 103 In their early work, these authors revealed that levels of miR-484 and miR-16 were significantly deregulated in blood serum of CD patients compared with healthy controls. Clinical testing achieved 83% sensitivity and 84% specificity for miR-484 and 74% sensitivity and 100% specificity for miR-16; these levels are indisputably higher than those for such laboratory markers currently used, such as C-reactive protein or anti-Saccharomyces cerevisiae antibody.103 In addition to the tissue miRNA profiles from rectal biopsies of pediatric UC and CD patients described previously, they identified in their further work miRNA biomarkers in blood serum. Circulating miR-192, miR-142-3p, and miR-21 were confirmed to be elevated in both UC and CD samples relative to controls, and they correctly classified 79%, 72%, and 72% of IBD patients, respectively. In patients from whom both serum and rectum miRNAs were measured, circulating miRNA levels did not correlate with those of the tissue. There were also no differences in circulating miRNAs that would enable differentiating between CD and UC patients.97
Heier et al performed expression profiling of 24 circulating miRNAs involved in inflammation or steroid response to examine their responsiveness to anti-inflammatory treatments (eg, prednisone, infliximab).104 They identified that 3 miRNAs (miR-146a, miR-146b, and miR-320a) known to be induced by inflammatory signaling were responsive to—or downregulated by—both drugs. A fourth miRNA, miR-486, showed a significant change in response to prednisone but not to infliximab. Together, measuring levels of these miRNAs could potentially help in assessing inflammatory disease and therapeutic response.104 A similar study evaluated differentially expressed miRNAs during glucocorticoid treatment in blood cells (specifically peripheral blood mononuclear cells [PBMCs]) of pediatric patients with IBD (8 UC, 2 CD) enrolled at diagnosis and followed for the first weeks of steroid therapy.105 Peripheral blood was obtained at diagnosis (T0) and after 4 weeks of prednisone treatment (T4). Among the 18 miRNAs differentially expressed from T0 to T4, 16 were upregulated and 2 were downregulated. Three miRNAs (miR-144, miR-142, and miR-96) could putatively recognize the 3′UTR of the glucocorticoid receptor gene, and 3 miRNAs (miR-363, miR-96, miR-142) contained glucocorticoid responsive element sequences, thereby potentially enabling direct regulation by the glucocorticoid receptor.105
The only study in pediatric IBD patients focusing on lncRNAs thus far was related to glucocorticoid therapy response and GAS5 levels in PBMCs. Clinical activity was assessed using the Pediatric Crohn’s Disease Activity Index (PCDAI) for patients with CD and the Pediatric Ulcerative Colitis Activity Index (PUCAI) for patients with UC. Clinical remission was defined as PCDAI <10 or PUCAI <10, whereas clinical improvement was defined as a reduction of at least 15 points from the baseline score for PCDAI and at least 20 points from baseline for PUCAI.106 Growth arrest-specific 5 levels were measured in PBMCs of 19 pediatric IBD patients at diagnosis and after the first cycle of glucocorticoids. This demonstrated upregulation of the lncRNA in patients with unfavorable steroid response, indicating that GAS5 can be considered a novel pharmacogenomic marker useful for personalizing glucocorticoid therapy.106
CONCLUSION AND FUTURE PERSPECTIVES
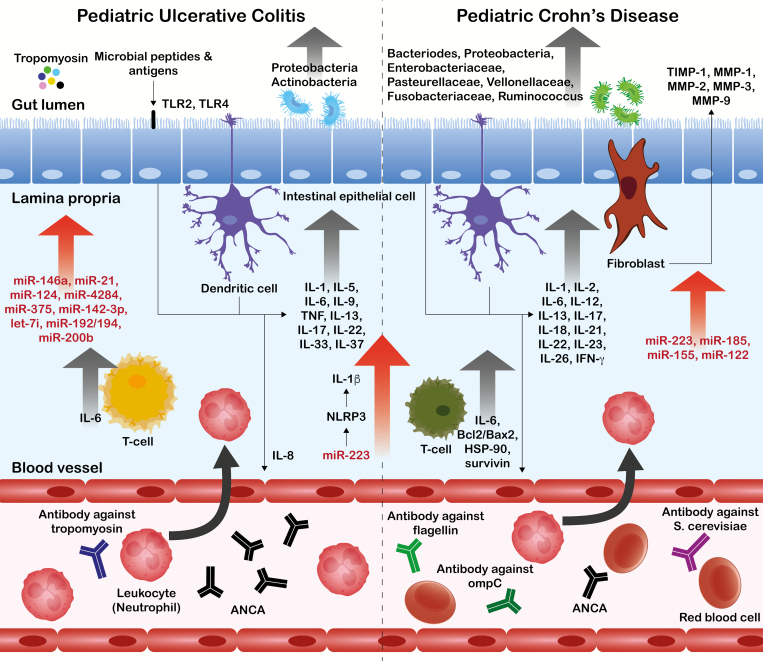
Not many studies to date have been focused on ncRNAs in pediatric IBD patients, very little knowledge exists as to the underlying biology of miRNAs involved in pediatric IBD patients (Fig. 1),107 and the current descriptive observations are often derived by extrapolation of discoveries from adult IBD experimental studies. Existing results show promise, however, as there is significant overlap in miRNA profiles across independent studies. Specifically, miR-146a, miR-142-3p, and miR-223 seem to be emerging as potential noninvasive biomarkers of pediatric IBD in the near future. Some of these miRNAs are specific for pediatric IBDs when compared with adult counterparts. There also are miRNA biomarkers (eg, miR-24), enabling accurate differentiation between UC and CD cases and tissue miRNA expression changes reflecting successful glucocorticoid treatment. In bodily fluids, miRNAs have been observed to differ by their levels in blood serum of IBD patients and controls. In PBMCs, miRNAs and lncRNA GAS5 have been shown responsive to the anti-inflammatory agents prednisone and infliximab. A variety of study designs are found in the current literature, however. These need to be unified and include independent validation cohorts of patients to provide solid and more convincing results. Also, high-throughput technologies for ncRNA profiling are not as common, and a majority of the studies are based on preselected groups of ncRNA candidates. A higher level of methodological standardization is necessary also to develop reliable clinical-level biomarkers.
FIGURE 1.
Tissue miRNAs involved in the development of pediatric IBD (modified from Park et al., 2017).107Abbreviations: TLR, toll-like receptor; TNF, tumor necrosis factor; ANCA, anti-neutrophil cytoplasmic antibodies; IFN, interferon; TIMP, tissue inhibitor of metalloproteinases; MMP, matrix metalloproteinase; Bcl2, B-cell lymphoma 2; BAX, BCL2 associated X; CCL, CC chemokine ligand; CCR, CC chemokine receptor; ompC, outer membrane protein C precursor.
Author contribution: PJ, JB, TP, and LK wrote the manuscript. OS and AG critically revised the manuscript. All authors reviewed and approved the final manuscript.
Supported by: This work was supported by the Ministry of Health, Czech Republic, Conceptual Development of Research Organization (FNBr, 65269705) and National Cancer Institute (CA184792 and CA202797).
Conflicts of interest: The authors declare that they have no conflict of interest.
REFERENCES
- 1. Benchimol EI, Fortinsky KJ, Gozdyra P, et al. . Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423–439. [DOI] [PubMed] [Google Scholar]
- 2. Ghione S, Sarter H, Fumery M, et al. ; Epimad Group Dramatic increase in incidence of ulcerative colitis and Crohn’s disease (1988-2011): a population-based study of French adolescents. Am J Gastroenterol. 2018;113:265–272. [DOI] [PubMed] [Google Scholar]
- 3. Van Limbergen J, Russell RK, Drummond HE, et al. . Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology. 2008;135:1114–1122. [DOI] [PubMed] [Google Scholar]
- 4. Kelsen J, Baldassano RN. Inflammatory bowel disease: the difference between children and adults. Inflamm Bowel Dis. 2008;14(Suppl 2):S9–11. [DOI] [PubMed] [Google Scholar]
- 5. Molodecky NA, Soon IS, Rabi DM, et al. . Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 6. Ng SC, Shi HY, Hamidi N, et al. . Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. [DOI] [PubMed] [Google Scholar]
- 7. Sýkora J, Pomahačová R, Kreslová M, et al. . Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol. 2018;24:2741–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. IBD Working Group of the European Society for Paediatric Gastroenterology HpaN. Inflammatory bowel disease in children and adolescents: recommendations for diagnosis--the Porto criteria. J Pediatr Gastroenterol Nutr. 2005;41:1–7. [DOI] [PubMed] [Google Scholar]
- 9. Levine A, Koletzko S, Turner D, et al. ; European Society of Pediatric Gastroenterology, Hepatology, and Nutrition ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr. 2014;58:795–806. [DOI] [PubMed] [Google Scholar]
- 10. De Iudicibus S, Stocco G, Martelossi S, et al. . Genetic predictors of glucocorticoid response in pediatric patients with inflammatory bowel diseases. J Clin Gastroenterol. 2011;45:e1–e7. [DOI] [PubMed] [Google Scholar]
- 11. Pastore S, Naviglio S, Canuto A, et al. . Serious adverse events associated with anti-tumor necrosis factor alpha agents in pediatric-onset inflammatory bowel disease and juvenile idiopathic arthritis in a real-life setting. Paediatr Drugs. 2018;20:165–171. [DOI] [PubMed] [Google Scholar]
- 12. Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. [DOI] [PubMed] [Google Scholar]
- 13. Wise JA, Weiner AM. Dictyostelium small nuclear RNA D2 is homologous to rat nucleolar RNA U3 and is encoded by a dispersed multigene family. Cell. 1980;22:109–118. [DOI] [PubMed] [Google Scholar]
- 14. Calvet JP, Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981;26:363–370. [DOI] [PubMed] [Google Scholar]
- 15. Calvet JP, Meyer LM, Pederson T. Small nuclear RNA U2 is base-paired to heterogeneous nuclear RNA. Science. 1982;217:456–458. [DOI] [PubMed] [Google Scholar]
- 16. Lacoste-Royal G, Simard R. Localization of small nuclear RNA by EM autoradiography in Chinese hamster ovary (CHO) cells. Exp Cell Res. 1983;149:311–323. [DOI] [PubMed] [Google Scholar]
- 17. Elkin M, Shevelev A, Schulze E, et al. . The expression of the imprinted H19 and IGF-2 genes in human bladder carcinoma. FEBS Lett. 1995;374:57–61. [DOI] [PubMed] [Google Scholar]
- 18. Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. . DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 20. Fire A, Albertson D, Harrison SW, et al. . Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development. 1991;113:503–514. [DOI] [PubMed] [Google Scholar]
- 21. Fire A, Xu S, Montgomery MK, et al. . Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [DOI] [PubMed] [Google Scholar]
- 22. Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. [DOI] [PubMed] [Google Scholar]
- 23. Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. [DOI] [PubMed] [Google Scholar]
- 24. Chooniedass-Kothari S, Emberley E, Hamedani MK, et al. . The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. [DOI] [PubMed] [Google Scholar]
- 25. Kondo T, Plaza S, Zanet J, et al. . Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. [DOI] [PubMed] [Google Scholar]
- 26. Bánfai B, Jia H, Khatun J, et al. . Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 28. Reinhart BJ, Slack FJ, Basson M, et al. . The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. [DOI] [PubMed] [Google Scholar]
- 29. Slack FJ, Basson M, Liu Z, et al. . The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. [DOI] [PubMed] [Google Scholar]
- 30. Pasquinelli AE, Reinhart BJ, Slack F, et al. . Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. [DOI] [PubMed] [Google Scholar]
- 31. Friedman RC, Farh KK, Burge CB, et al. . Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alles J, Fehlmann T, Fischer U, et al. . An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47:3353–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol. 2010;222:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee Y, Kim M, Han J, et al. . MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. [DOI] [PubMed] [Google Scholar]
- 39. Lee Y, Ahn C, Han J, et al. . The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. [DOI] [PubMed] [Google Scholar]
- 40. Han J, Lee Y, Yeom KH, et al. . The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 42. Yi R, Qin Y, Macara IG, et al. . Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee YS, Nakahara K, Pham JW, et al. . Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. [DOI] [PubMed] [Google Scholar]
- 44. Chendrimada TP, Gregory RI, Kumaraswamy E, et al. . TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A. 2016;113:E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herrera-Carrillo E, Berkhout B. Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res. 2017;45:10369–10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Babiarz JE, Ruby JG, Wang Y, et al. . Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okamura K, Hagen JW, Duan H, et al. . The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang J, Zhang J, Zheng H, et al. . Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:1 p following 757; discussion following 757. [PubMed] [Google Scholar]
- 51. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. [DOI] [PubMed] [Google Scholar]
- 52. Derrien T, Johnson R, Bussotti G, et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guttman M, Amit I, Garber M, et al. . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. [DOI] [PubMed] [Google Scholar]
- 55. Carninci P, Kasukawa T, Katayama S, et al. ; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 56. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Iyer MK, Niknafs YS, Malik R, et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fang S, Zhang L, Guo J, et al. . NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018;46:D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. An G, Sun J, Ren C, et al. . LIVE: a manually curated encyclopedia of experimentally validated interactions of lncRNAs. Database (Oxford). 2019;2019:bazz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cabili MN, Trapnell C, Goff L, et al. . Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ravasi T, Suzuki H, Pang KC, et al. . Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mercer TR, Dinger ME, Sunkin SM, et al. . Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Djebali S, Davis CA, Merkel A, et al. . Landscape of transcription in human cells. Nature. 2012;489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khalil AM, Guttman M, Huarte M, et al. . Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang KC, Yang YW, Liu B, et al. . A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feng J, Bi C, Clark BS, et al. . The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shamovsky I, Ivannikov M, Kandel ES, et al. . RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. [DOI] [PubMed] [Google Scholar]
- 68. Lai F, Orom UA, Cesaroni M, et al. . Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang L, Lin C, Jin C, et al. . lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang X, Arai S, Song X, et al. . Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ramos AD, Andersen RE, Liu SJ, et al. . The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gonzalez I, Munita R, Agirre E, et al. . A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yoon JH, Abdelmohsen K, Kim J, et al. . Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. Plos One. 2014;9:e107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carrieri C, Cimatti L, Biagioli M, et al. . Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. [DOI] [PubMed] [Google Scholar]
- 76. Willingham AT, Orth AP, Batalov S, et al. . A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. [DOI] [PubMed] [Google Scholar]
- 77. Noh JH, Kim KM, Abdelmohsen K, et al. . HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016;30:1224–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Salmena L, Poliseno L, Tay Y, et al. . A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kino T, Hurt DE, Ichijo T, et al. . Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Melé M, Mattioli K, Mallard W, et al. . Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- 82. Kunej T, Obsteter J, Pogacar Z, et al. . The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–357. [DOI] [PubMed] [Google Scholar]
- 83. Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett. 2018;592:2874–2883. [DOI] [PubMed] [Google Scholar]
- 84. Zheng GX, Do BT, Webster DE, et al. . Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol. 2014;21:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Salviano-Silva A, Lobo-Alves SC, Almeida RC, et al. . Besides pathology: long non-coding RNA in cell and tissue homeostasis. Noncoding RNA. 2018;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jia H, Osak M, Bogu GK, et al. . Genome-wide computational identification and manual annotation of human long noncoding RNA genes. Rna. 2010;16:1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lagos-Quintana M, Rauhut R, Meyer J, et al. . New microRNAs from mouse and human. Rna. 2003;9:175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Béres NJ, Kiss Z, Sztupinszki Z, et al. . Altered mucosal expression of microRNAs in pediatric patients with inflammatory bowel disease. Dig Liver Dis. 2017;49:378–387. [DOI] [PubMed] [Google Scholar]
- 90. Fasseu M, Tréton X, Guichard C, et al. . Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:e13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mirza AH, Kaur S, Brorsson CA, Pociot F. Effects of GWAS-associated genetic variants on lncRNAs within IBD and T1D candidate loci. Plos One. 2014;9:e105723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mirza AH, Berthelsen CH, Seemann SE, et al. . Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dalal SR, Kwon JH. The role of MicroRNA in inflammatory bowel disease. Gastroenterol Hepatol (N Y). 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 94. Koukos G, Polytarchou C, Kaplan JL, et al. . MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–52.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kwon JH, Keates AC, Anton PM, et al. . Topical antisense oligonucleotide therapy against LIX, an enterocyte-expressed CXC chemokine, reduces murine colitis. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1075–G1083. [DOI] [PubMed] [Google Scholar]
- 96. Koukos G, Polytarchou C, Kaplan JL, et al. . A microRNA signature in pediatric ulcerative colitis: deregulation of the miR-4284/CXCL5 pathway in the intestinal epithelium. Inflamm Bowel Dis. 2015;21:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zahm AM, Hand NJ, Tsoucas DM, et al. . Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J Crohns Colitis. 2014;8:1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Béres NJ, Szabó D, Kocsis D, et al. . Role of altered expression of miR-146a, miR-155, and miR-122 in pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016;22:327–335. [DOI] [PubMed] [Google Scholar]
- 99. Szűcs D, Béres NJ, Rokonay R, et al. . Increased duodenal expression of miR-146a and -155 in pediatric Crohn’s disease. World J Gastroenterol. 2016;22:6027–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sheedy FJ. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. 2015;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Neudecker V, Haneklaus M, Jensen O, et al. . Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tang WJ, Peng KY, Tang ZF, et al. . MicroRNA-15a - cell division cycle 42 signaling pathway in pathogenesis of pediatric inflammatory bowel disease. World J Gastroenterol. 2018;24:5234–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zahm AM, Thayu M, Hand NJ, et al. . Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Heier CR, Fiorillo AA, Chaisson E, et al. . Identification of pathway-specific serum biomarkers of response to glucocorticoid and infliximab treatment in children with inflammatory bowel disease. Clin Transl Gastroenterol. 2016;7:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. De Iudicibus S, Lucafò M, Vitulo N, et al. . High-throughput sequencing of microRNAs in glucocorticoid sensitive paediatric inflammatory bowel disease patients. Int J Mol Sci. 2018;19:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lucafò M, Di Silvestre A, Romano M, et al. . Role of the long non-coding RNA growth arrest-specific 5 in glucocorticoid response in children with inflammatory bowel disease. Basic Clin Pharmacol Toxicol. 2018;122:87–93. [DOI] [PubMed] [Google Scholar]
- 107. Park JH, Peyrin-Biroulet L, Eisenhut M, et al. . IBD immunopathogenesis: a comprehensive review of inflammatory molecules. Autoimmun Rev. 2017;16:416–426. [DOI] [PubMed] [Google Scholar]