Abstract
In contrast to gut, the oral microbiome of MS patients has not been characterized. Deep sequencing of saliva DNA from a pair of monozygotic twins (MSF1 with relapsing remitting MS; MSF2 with clinically isolated syndrome) identified 2,036 bacterial species. Relative abundances of 3 phyla were higher, and 3 lower in MSF1 versus MSF2. Species diversity was greater in MSF2, and 20 abundant species differed at least 2-fold. Pathway analysis identified 116 functional hierarchies differing 50% or more. Although limited to one pair of twins, our data suggests that oral microbiome analysis may be useful for diagnosis or monitoring therapeutic efficacy.
Keywords: oral microbiome, saliva, monozygotic, bacteroidetes, firmicutes
Introduction
The precise etiology of multiple sclerosis (MS) remains to be determined, however recent progress in characterizing genetic diversity helps to distinguish genetic and environmental determinants. The first genetic factor identified for MS susceptibility was major histocompatibility locus HLA, with the main risk factor identified as HLA-DRB1*15:01, later found to be associated with earlier onset and faster conversion from clinically isolated syndrome (CIS) to definite MS (Isobe et al., 2016). The first genetic variants associated with MS disease course were found by whole exome sequencing that located variants in 3 non-HLA genes (Gil-Varea et al., 2018). More recent genome wide association studies (GWAS) (Beecham et al., 2013, George et al., 2016) led to identification of hundreds of risk factors for MS, but each having only a small effect on overall risk, and in total thought to account for up to 25% of total genetic inheritability (Patsopoulos, 2018).
In contrast to human genetic variants, it is now accepted that microorganism levels and diversity in human tissues are associated with disease progression and severity. Numerous factors induce changes in the microbiome including diet, social interactions, stress, and medications. Knowledge of microbiome structure could help guide therapeutic approaches to increase levels of beneficial taxa, while reducing levels of detrimental microorganisms. Differences in biomes could also help account for the fact that identical twins have only 30% concordance of MS (Kuusisto et al., 2008). Although fecal samples are most often used as a proxy for the gastrointestinal microbiome, saliva has a similar degree of microbial diversity, and the relative ease of obtaining samples has led to it being used to examine microflora changes in neurodegenerative diseases (Boaden et al., 2017, Liu et al., 2019, Panza et al., 2019, Singhrao and Olsen, 2019).
The aim of the current study was to investigate the oral microbiome of MS patients and determine how that differs between MS patients with different disease severity. To minimize the contribution of human genetic differences, we analyzed saliva from a pair of identical twins discordant for MS disease severity cohabitating with similar diets, one with a diagnosis of CIS, the other with RRMS. We report on the differences between twins at phyla, genera, and species levels; discuss similarities to the gut microbiota dysbiosis reported for MS patients; and the pathways found enriched in either twin.
Material & Methods
Patients
Identical female twins MSF1 and MSF2 of Hispanic descent were identified at the UIC MS clinic as part of our previous study to characterize a small nucleotide polymorphism in the STK11 gene encoding liver kinase B1 (Boullerne et al., 2015). MSF1 was diagnosed at age 25 with relapsing remitting MS (RRMS) based on clinical, imaging, and laboratory data; MSF2 was diagnosed at age 26 with clinically isolated syndrome (CIS) based on clinical and MRI data (Supplement 1). At the time of sample collection both were 32 years-old, MSF1 had an expanded disability status scale (EDSS) score of 1.0 and MSF2 had an EDSS score of 0 according to the 2017 revised McDonald’s criteria (Thompson et al., 2018). Significant co-morbidities were uterine fibroids and ovarian cysts in both twins. Both twins self-reported to not taking any disease modifying therapies for at least 18 months before samples were collected, and neither reported any gum disease or other oral disease. Short Tandem Repeat (STR) profiling (Promega PowerPlex® Fusion Systems) carried out by the Molecular Pathology laboratory at UIC gave a random match probability of 2.3×10−33 across 24 loci, confirming monozygosity. MSF1 and MSF2 were consented and the study carried out under UIC IRB protocol 2001–0721.
Samples
Saliva samples were collected from MSF1 (Omnigene-oral-501 kit, DNA Genotek, Ottawa, Canada) and MSF2 (Saliva kit RU49000, Norgen Biotek, Ontario, Canada). At the time DNA collection, both twins had been living together, sharing a similar life style, were following identical low-fat, vegetable-rich diet, and were off medication for 18 months. Neither were tobacco users nor had antibiotic exposure over the month before sample collection. Genomic DNA (gDNA) was extracted using QIAGEN Allprep kit (Cat # 80204) per manufacturer’s recommendations, quality determined using TapeStation2200, and quantified using a Qubit 2.0 (Invitrogen) fluorimeter. Samples were stored at 80°C until use. Saliva samples (n=80) from a cohort of healthy individuals of Hispanic descent living in the Chicago area were collected, genomic DNA isolated, and microbial taxa determined by 16S rRNA sequencing as part of a larger study examining the diversity of the oral microbiome in dentate versus edentulous individuals (Gazdeck et al., 2019).
Whole Genome Sequencing
Whole Genome Sequencing was carried out by Novagene (Beijing, China). Libraries were prepared from 1 μg gDNA (Illumina Truseq Nano DNA HT Sample Preparation Kit) following manufacturer’s recommendations. Briefly, gDNA samples were sheared by sonication to 350bp, fragments endpolished, poly(A) -tailed and amplified by PCR. Paired-end 150bp reads were sequenced on an Illumina HiSeq X System. High quality raw sequence reads provided a depth of 50x coverage for MSF1 and 54x for MSF2. Mapping was performed using the BWA-MEM assembler with soft clipping against the human reference genome version hg38 masked for low complexity regions. Resulting BAM files were cleaned of PCR duplicates using PICARD software.
Taxonomic profiling
Raw reads that were unmapped against reference genome hg38 were mapped to the NCBI noredundant nucleotide (nrNT) database using Centrifuge (Kim et al., 2016). MSF1 had 28.5M unmapped read pairs out of 640M total, and MSF2 had 6.2M unmapped read pairs out of 722M total. Taxonomic annotations for each read were obtained using the least common ancestor algorithm (MEGAN), then summarized across all reads to create counts per taxon.
Functional profiling
Raw reads were mapped to the Swissprot protein database using DIAMOND (Buchfink et al., 2015). Gene ortholog annotations were assigned using the consensus of aligned references and summarized across all reads to create counts per ortholog for each sample. Higher level summaries of orthologous functions were created using KEGG BRITE hierarchical annotations (Kanehisa et al., 2017).
Results
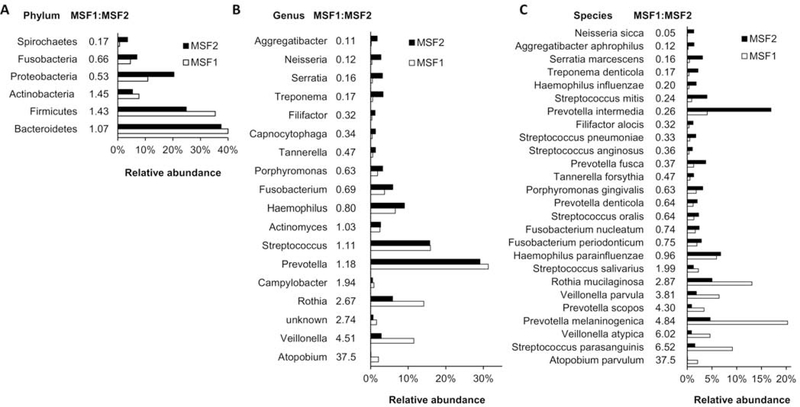
Unmapped read pairs from MSF1 and MSF2 were mapped to the NCBI non-redundant nucleotide (nrNT) database, which yielded 15.14M read pairs for a total of 2.32 Gbp for MSF1, and 2.83M read pairs for a total of 0.45 Gbp for MSF2. Based on this difference, for subsequent comparisons we normalized MSF2 reads by a factor of 5.184. The majority of reads mapped to Bacteria (97.8%), with a small percentage mapping to Archaea (0.04%), Virus and Phage (0.2%), and Eurkaryota (2.0%). Mapping identified 8,033 different annotated taxa (Table S1). This list was filtered to remove species annotations that were reflective of sample origin, rather than a curated species name – those containing ‘clone’, ‘symbiont’, ‘uncultured’, ‘unidentified’, ‘other’, ‘sp.’, or ‘bacterium’ were excluded, resulting in removal of 4,091 annotated taxa (Table S2). These constituted a small percentage of total reads (1.51M reads, or 10% for MSF1; 2.27M reads, or 15% for MSF2), with only 2 taxa having an abundance greater than 1%. The remaining 3,942 taxa were filtered for ones having <100 reads in both MSF1 and MSF2 which led to removal of 1,906 taxa (0.25% of all reads). The remaining taxa (Table S3) could be assigned to 35 different phyla (Table S4), 64 classes, 145 orders, 308 family, 829 genera (Table S5), and 2,036 species (Table S6). There were 6 phyla which accounted for >98.7% of all sequences in either MSF1 or MSF1 (Figure 1A). Of those, 3 (Firmicutes, Bacteroidetes, and Actinobacteria) were higher in MSF1 than in MSF2; while 3 (Proteobacteria, Fusobacteria, and Spirochaetes) were higher in MSF2 than in MSF1.
Figure 1.
Relative taxa abundance in monozygotic twins discordant for MS severity
Total reads were summed for each unique (A) phylum, (B) genus, and (C) species present in saliva DNA isolated from MSF1 and MSF2. For genus and species, only those present at 1% or greater abundance are shown. The ratio of reads in MSF1 compared to MSF2 is shown.
Of the 829 different genera identified, 645 were present in MSF1 with abundance >100, and 812 in MSF2 having >100 reads. Of those, 628 were shared, with 17 genera unique to MSF1 and 184 unique to MSF2. The unique genera were all present at 0.01% or less abundance. In MSF1 10 genera were present at 1% or greater abundance, which together comprised 91.1% of all genera (Figure 1B). In MSF2, 15 genera were present at 1% or greater, making up 88.7% of all genera. Amongst those, 5 Campylobacter, Rothia, Veillonella, Atopobium, and unknown) were approximately 2-fold or more higher in MSF1 than MSF2, while 7 (Aggregatibacter, Neisseria, Serratia, Treponema, Filifactor, and Capnocytophaga) were 2-fold or more higher in MSF2. The distribution and evenness of genera is higher in MSF2 (Shannon H-value of 2.85; 43% of the maximum of 6.7) compared to MSF1 (Shannon H-value of 2.42; 37% of the maximum of 6.5).
Overall species diversity was lower in MSF1 with 1,515 different species (>100 reads) compared to MSF2 having 1,975 different species (>100 reads). In MSF1, there were 17 species with a relative abundance of >1%, which together comprised 81% of all sequences annotated to the species level. In MSF2, there were 23 species with a relative abundance of >1%, and together these taxa comprised 73% of all sequences annotated to the level of species. 8 species were at least 2-fold higher in MSF1 versus MSF2 (Figure 1C) and 12 were at least 2-fold higher in MSF2 than in MSF1. There were 1,454 species shared between MSF1 and MSF2, with 61 unique to MSF1 and 521 unique to MSF2 resulting in 8.5-fold more unique species in MSF2 (Table 1). The distribution and evenness of species represented in the Shannon H-value was 3.81 (57% of the maximum of 6.72) for MSF2 compared to 3.26 in MSF1 (49% of the maximum of 6.72).
Table 1:
Genus and Species distribution in MSF1 and MSF2
| Total | Uique* | Unique MSF2:MSF1 | Shared | Shannon Diversity | Shannon Evenness | ||
|---|---|---|---|---|---|---|---|
| Genera in nrNT |
MSF1 MSF2 |
645 812 |
17 184 |
10.8 | 628 | 2.42 2.85 |
0.37 0.43 |
| Genera in HOMD |
MSF1
MSF2 |
115 121 |
0 6 |
n/a | 115 | 2.13 2.47 |
0.45 0.52 |
| Species in nrNT |
MSF1
MSF2 |
1,515 1,975 |
61 521 |
8.5 | 1,454 | 3.26 3.81 |
0.49 0.57 |
| Species in HOMD |
MSF1
MSF2 |
200 211 |
14 25 |
1.9 | 186 | 2.87 3.27 |
0.53 0.60 |
Unique is defined as having less than 100 reads in one twin but not the other
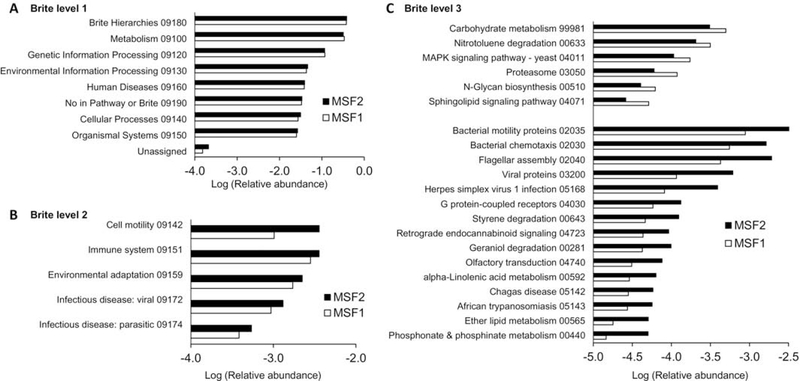
Pathway analysis identified 7 BRITE level 1 hierarchies present at roughly equal abundance in both patients (Figure 2A). Differences between the twins could be observed upon classification into level 2 BRITE hierarchies, which identified 5 pathways present at 0.05% abundance or greater that differed by at least 20% between MSF1 and MSF2 (Figure 2B). Classification into level 3 BRITE hierarchies identified 116 that differed by at least 50% between twins, of which 21 were present at 0.005% abundance or greater (Figure 2C). Of those 15 features were present at higher relative abundance in MSF2 relative to MSF1, while 6 features were lower in MSF2. Features with the largest differences were related to proteasome function (2-fold higher in MSF1) and phosphonate and phosphinate metabolism (3-fold higher in MSF2).
Figure 2.
Pathway analysis of encoded proteins
Total reads for MSF1 and MSF2 were translated to proteins, then analyzed for KEGG BRITE hierarchical annotations. (A) Level 1; (B) Level 2; (C) Level 3 hierarchies. The KEGG identifiers are provided for all hierarchies.
We compared the relative abundancies of the twins to those determined in a cohort of healthy individuals (HCs) of Hispanic descent living in the Chicago area. At the phyla level (Table S4) the relative abundance of bacteroidetes and proteobacteria were higher in both twins compared to the HCs, while levels of firmicutes and actinobacteria were lower. At the genus level (Table S5), relative levels of prevotella, rothia, and haemophilus were higher in both twins compared to HCs, while levels of streptococcus and actinomyces were lower. In addition, Shannon alpha diversities of genera in the twins (2.42 and 2.85) are comparable to the Shannon diversity of the HCs (2.84 ± 0.40, mean ± sd).
Discussion
Several studies have reported gut microbiome dysbiosis in MS patients compared to healthy controls, however to our knowledge the current study represents a first characterization of the MS oral microbiome. We evaluated microbiota in saliva samples from a pair of identical twins discordant for MS severity, where one (MSF2) had an ongoing CIS diagnosis 10 years after initial diagnosis, and the other (MSF1) a diagnosis of RRMS. At the time of DNA collection, the twins followed identical low-fat vegetable-rich diets, had no oral abscesses or mucosal lesions, and were off medication for 18 months. While small differences in relative microbiota abundance were apparent at higher taxonomic levels, for genera and species present at 1% abundance or greater we observed differences as large as 36-fold. In contrast, a comparison of the gut microbiome analyzed using 16S rRNA gene amplicon sequencing in identical twins discordant for MS severity showed few differences (Berer et al., 2017). In that study 34 twin pairs were examined in which one of the twins was unaffected; however, no differences were found in genera richness or abundance. Differences were observed when patients were stratified for use of disease modifying therapies, with the relative abundance of Akkermansia muciniphila higher in untreated patients compared to healthy controls. In contrast we found that the relative abundance of A. muciniphila was about 2-fold higher in MSF2 versus MSF1 (0.0036% versus 0.0018%).
Our approach of using whole genome sequencing (WGS) combined with alignment to the non-redundant nucleotide (nrNT) database allowed us to identify 8,033 species or species-level taxonomic features. Filtering of that data to remove taxa with unclear annotations (clone, symbiont, uncultured, unidentified, other, sp., bacterium) and any species detected at <100 reads (equivalent to 0.001% abundance) resulted in 2,036 species, 829 distinct genera, and 35 distinct phyla. In contrast, the current human oral microbiome database (HOMD) (Escapa et al., 2018) contains 15 different phyla, 211 genera, and 771 species. Of the 2,026 species we identified by shot-gun approach, 225 are listed in HOMD comprising 93.0% of all species; of the 829 genera 121 are in HOMD and comprise 95.8% of all genera, and of the 135 phyla only 10 are in HOMD comprising 99.3% of all phyla (Tables S4–6, highlighted entries). Re-analysis using only genera and species found in HOMD (Table 1) yields lower Shannon diversities and higher evenness in both MSF1 and MSF2; however, both diversity and evenness remain higher in MSF2 than MSF1.
There are several reasons that can account for the large difference in our results compared to the HOMD data. First, alignment to the nrNT identifies taxa whose annotations are not clearly associated with a defined species, but instead with sample origin. There are also distinct annotations in nrNT assigned to the same species, or to species at the strain level; and there are sequences for non −16S rRNA bacterial genes detected in nrNT not yet assigned to well annotated species and hence are not listed in HOMD. These differences are also consistent with a study in which 16S rRNA amplicon sequencing was directly compared to deep WGS using the same fecal sample (Ranjan et al., 2016), and which showed that WGS identified approximately twice as many species as the 16S method; and overall diversity was much greater. Identification of about 2,000 species in the current study is therefore in line with their finding of over 4,000 species in the fecal microbiome by WGS.
A comparison of differences between MSF1 and MSF2 to differences reported between gut microbiomes of MS patients and healthy controls (HC), or between patients with active versus nonactive disease) shows some similarities (Table 2). At the phylum level, Actinobacteria and Proteobacteria were decreased (Chen et al., 2016) in the gut biome of RRMS patients with active disease compared to HCs; in the current study Proteobacteria was lower, although Actinobacteria was higher in MSF1. Several genera and species were present at lower levels in the gut microbiome of MS patients compared to HCs, and lower levels of some of these were also seen comparing MSF1 to MSF2. While this suggests that a subset of relative abundance changes in oral taxa may parallel changes in the gut, only few studies have directly compared oral to gut (using fecal samples) microbiomes, and show that these microbiomes are highly distinct, with only weak correlations observed (Lokmer et al., 2020, Russo et al., 2017, Stewart et al., 2018).
Table 2:
Taxa differences shared in gut and oral microbiomes
| Taxa Level | MSF1:MSF2 | MS:HC | Reference |
|---|---|---|---|
| Phylum | |||
| Proteobacteria | Down | Down | (Chen et al., 2016) |
| Genus | |||
| Anaerostipes | Down | Down | (Miyake et al., 2015) |
| Faecalibacterium | Down | Down | (Miyake et al., 2015) |
| Slackia | Down | Down | (Jangi et al., 2016) |
| Haemophilus | Down | Down | (Chen et al., 2016) |
| Lactobacillus | Down | Down | (Chen et al., 2016) |
| Parabacteroides | Down | Down | (Chen et al., 2016) |
| Atopobium | Up | Up | (Tremlett et al., 2016) |
| Bifidobacterium | Up | Up | (Tremlett et al., 2016) |
| Megamonas | Up | Up | (Tremlett et al., 2016) |
| Megasphaera | Up | Up | (Tremlett et al., 2016) |
| Prevotella | Up | Up | (Tremlett et al., 2016) |
| Species | |||
| [Eubacterium] rectale | Down | Down | (Miyake et al., 2015) |
| Anaerostipes hadrus | Down | Down | (Miyake et al., 2015) |
| Prevotella stercorea | Down | Down | (Jangi et al., 2016) |
| Faecalibacterium prausnitzii | Down | Down | (Tremlett et al., 2016) |
| Eggerthella lenta | Up | Up | (Miyake et al., 2015) |
| Streptococcus thermophilus | Up | Up | (Miyake et al., 2015) |
In humans, two phyla dominate in the gut: Firmicutes (associated with a Western, carnivore diet) and Bacteroidetes (associated with a vegetarian and omnivore diet). MS patients with greater disease activity have a higher ratio of Firmicutes to Bacteroidetes in the gut (Cosorich et al., 2017). It was also reported that RRMS patients have increased Actinobacteria; and decreased Bacteroides compared to healthy controls (Furusawa et al., 2015); and that levels of Bacteroides were inversely correlated with Treg numbers in MS patients, while Fusobacteria correlated with Tregs in healthy controls (Tremlett et al., 2016). Consistent with these findings, in MSF2 the relative level of Firmicutes to Bacteroides (24.9% versus 37.5%) is less than in MSF1 (35.4% versus 40.0%); levels of Actinobacteria were higher in MSF1 than MSF2 (7.7% versus 5.3%); and levels of Fusobacteria were lower in MSF1 than MSF2 (4.6% versus 7.0%). Correlation of these differences with MS disease may be due in part to reduced bacterial production of metabolites such as short chain fatty acids (SCFAs, primarily due to Bacteroides) which have well-characterized anti-inflammatory effects and can promote regulatory Tcell numbers and function (Furusawa et al., 2015, Melbye et al., 2019, Smith et al., 2013). In this regard, it was reported that in RRMS patients (Jangi et al., 2016), the relative abundance of several bacterial genus involved in the metabolism of the SCFA butyrate were lower as compared to healthy controls. Butyrate is a bacterial-derived metabolite that can increase the population of regulatory T cells (Furusawa et al., 2015), which suppress Th1 Tcell activation and production of inflammatory factors. Inspection of our data (Table S7) shows 11 different bacterial species associated with butyrate metabolism, all 1.5 to 3-fold higher in MSF2 than in MSF1.
Amongst the most abundant species in both MSF1 and MSF2 is Porphyromonas gingivalis (P. gingivalis), which accounted for 1.8% of all species in MSF1 and 3.1% in MSF2. This is in contrast with abundance levels reported in 16s rRNA analysis of salivary samples from HCs of 0.0036% (Damgaard et al., 2019) and 0.09% (Chen et al., 2019), this difference may be due in part to the method of measurement (shot-gun versus 16s RNA amplicon sequencing). P. gingivalis is an established biomarker for periodontitis (Szafranski et al., 2015), and produces proinflammatory serine dipeptide lipids, and lipopolysaccharide-G which can activate cytokine production from monocytes (Ballerini et al., 2017). Moreover, both oral infection and subcutaneous injection with P. gingivalis worsened EAE (Polak et al., 2018, Shapira et al., 2002). These data suggest that P. gingivalis in the oral cavity may contribute to inflammatory activation in these patients.
Functional analysis identified several BRITE hierarchies which target a specific metabolite, or metabolite class. This includes pathways involved in phosphonate, ether lipids, alpha-linolenic acid, and geraniol metabolism. Although these were present at relatively low abundance (from 0.005% to 0.01%), they were present at 2.5 or greater abundance in MSF2 than MSF1. Interestingly, these have been suggested to play protective roles in EAE or to reduce inflammatory responses. The ether phospholipid edelfosine reduces EAE disease (Abramowski et al., 2014b, Chabannes et al., 1992) and Tcell activation (Abramowski et al., 2014a). Linolenic acid delays onset and severity in EAE (Adkins et al., 2019) and is associated with lower MS risk (Bjornevik et al., 2017, Bjornevik et al., 2019). Geraniol has anti-inflammatory properties (Huang et al., 2018, Jiang et al., 2017). Bisphosphonates have been tested for efficacy in a variety of neurological diseases acting through inhibition of mevalonate pathways; inhibition of isoprenoid synthesis; and regulation of cholesterol synthesis (Zameer et al., 2018). Two other pathways at higher levels in MSF2 are African trypanosomiasis and Chagas disease. Mice with Trypanosoma bruci infection developed less severe and delayed EAE (Wallberg and Harris, 2005), while infection with Trypanosoma cruzi led to immunosuppression and reduced EAE (Tadokoro et al., 2004). In contrast the only metabolite pathway enriched in MSF1 was nitrotoluene degradation, a chemical used in manufacture of pigments, photographic chemicals, and agricultural chemicals. As there are no reports of nitrotoluene in EAE or MS, it is not known if alterations in genes involved in its degradation could influence disease.
Comparison of relative taxa abundancies in MSF1 and MSF2 to HCs shows several differences. While these may reflect the limited nature of the current study, increased levels of proteobacteria (Qiao et al., 2018) and bacteroidetes, and reduced levels of firmicutes were also been found when comparing the saliva microbiome of patients with autism to HCs (Kong et al., 2019); and differences at the genus level when comparing the buccal microbiome of Parkinson’s disease patients to HCs (Pereira et al., 2017).
Our study has several limitations, the most important that a single pair of monozygotic twins was analyzed, preventing statistical comparisons to be made. Second, although neither twin reported gum disease or oral infection, we did not carry out oral examinations before sample collection which could have influenced the microbiome. As this was a pilot study, we die not compare the oral microbiome of the twins to matched healthy controls. Finally, we obtained saliva samples from MSF1 and MSF2 using 2 different saliva collection kits, which could introduce variance. However, both kits are reported by the manufacturers to provide over a year stability, with no significant degradation of high molecular weight DNA or reduction in qPCR analysis for specific targets; and both include reagents to prevent growth of gram-negative and gram-positive bacteria and fungi, and to inactivate viruses. While a direct comparison of these kits has not been reported, the respective versions sold to isolate DNA from stool have been compared directly and shown to produce no differences in sample richness, alpha or beta diversity, or phylum or genera level composition when tested on the same samples (Iizumi et al., 2017, Watson et al., 2019). This suggests that any differences in these kits would be unlikely to have contributed to observed oral microbiome differences.
In summary, comparison of the oral microbiome in saliva samples from monozygotic twins discordant for MS severity revealed differences at all taxonomic levels. Several are consistent with ones reported in the gut microbiome between MS patients and HCs, or patients with active versus inactive disease. This may reflect displaced gut bacteria present at low levels in saliva or bacteria that reside on oral surfaces in addition to the gut. Although we characterized the salivary biome of monozygotic twins living under similar conditions including diet and overall health status, other environmental influences likely contribute to the differences observed (Gomez and Nelson, 2017) and it has been reported that environment is the dominant factor that dictates the oral microbiome rather than host genetics (Shaw et al., 2017). Further studies using large group sizes will be needed to dissect these relative contributions. Since MSF1 and MSF2 reflect differences in MS clinical presentation we suggest differences in oral microbiome diversity and speciation may reflect loss of tolerance which may not parallel similar gut interactions because of unique oral immunologic activity (e.g., variation in oral mucosa site specific keratinization and physiology, number and type of immune effectors, and presence of γ/δ TCR vs α/β TCR). Our findings are limited in scope to analysis of a single pair of twins, but due to the increased depth of shot-gun sequencing, supplies a long list of taxa and bacterial genes that can be found in the saliva of MS patients that may normally go undetected. The data thus provide groundwork for further studies to validate analysis of the oral biome as an easily accessible sample source as a prognostic or diagnostic tool.
Supplementary Material
Highlights.
In contrast to the gut, the oral microbiome of MS patients has not been examined
Shot-gun sequencing of saliva DNA from monozygotic twins discordant for MS identified over 2000 species
Differences between the twins were present at all taxonomic levels
Several differences parallel those reported for the gut microbiome between MS and controls
Examination of the oral microbiome could help inform as to MS diagnosis, severity, and treatment.
Acknowledgements
We thank Yash Patel for assistance with data analysis.
Funding
This work was supported by grants BX002625 and 14S-RCS-003 from the Department of Veterans Affairs (DLF)
Footnotes
Declaration of Interests
The authors declare they have no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramowski P, Otto B, Martin R. The orally available, synthetic ether lipid edelfosine inhibits T cell proliferation and induces a type I interferon response. PloS one. 2014a;9:e91970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramowski P, Steinbach K, Zander AR, Martin R. Immunomodulatory effects of the ether phospholipid edelfosine in experimental autoimmune encephalomyelitis. Journal of neuroimmunology. 2014b;274:111–24. [DOI] [PubMed] [Google Scholar]
- Adkins Y, Soulika AM, Mackey B, Kelley DS. Docosahexaenoic acid (22:6n-3) Ameliorated the Onset and Severity of Experimental Autoimmune Encephalomyelitis in Mice. Lipids. 2019;54:13–23. [DOI] [PubMed] [Google Scholar]
- Ballerini P, Diomede F, Petragnani N, Cicchitti S, Merciaro I, Cavalcanti M, et al. Conditioned medium from relapsing-remitting multiple sclerosis patients reduces the expression and release of inflammatory cytokines induced by LPS-gingivalis in THP-1 and MO3.13 cell lines. Cytokine. 2017;96:261–72. [DOI] [PubMed] [Google Scholar]
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature genetics. 2013;45:1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:10719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornevik K, Chitnis T, Ascherio A, Munger KL. Polyunsaturated fatty acids and the risk of multiple sclerosis. Multiple sclerosis (Houndmills, Basingstoke, England). 2017;23:1830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornevik K, Myhr KM, Beiske A, Bjerve KS, Holmoy T, Hovdal H, et al. alpha-Linolenic acid is associated with MRI activity in a prospective cohort of multiple sclerosis patients. Multiple sclerosis (Houndmills, Basingstoke, England). 2019;25:987–93. [DOI] [PubMed] [Google Scholar]
- Boaden E, Lyons M, Singhrao SK, Dickinson H, Leathley M, Lightbody CE, et al. Oral flora in acute stroke patients: A prospective exploratory observational study. Gerodontology. 2017;34:343–56. [DOI] [PubMed] [Google Scholar]
- Boullerne AI, Skias D, Hartman EM, Testai FD, Kalinin S, Polak PE, et al. A single-nucle otide polymorphism in serine-threonine kinase 11, the gene encoding liver kinase B1, is a risk factor for multiple sclerosis. ASN neuro. 2015;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- Chabannes D, Ryffel B, Borel JF. SRI 62–834, a cyclic ether analogue of the phospholipid ET-18OCH3, displays long-lasting beneficial effect in chronic relapsing experimental allergic encephalomyelitis in the Lewis rat. Comparison with cyclosporin and (Val2)-dihydrocyclosporin effects in clinical, functional and histological studies. Journal of autoimmunity. 1992;5:199–211. [DOI] [PubMed] [Google Scholar]
- Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Scientific reports. 2016;6:28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu X, Zhu D, Xu M, Yu Y, Yu L, et al. Microbiota in Human Periodontal Abscess Revealed by 16S rDNA Sequencing. Frontiers in microbiology. 2019;10:1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Science advances. 2017;3:e1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C, Danielsen AK, Enevold C, Massarenti L, Nielsen CH, Holmstrup P, et al. Porphyromonas gingivalis in saliva associates with chronic and aggressive periodontitis. Journal of oral microbiology. 2019;11:1653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. mSystems. 2018;3:e00187–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Hase K. Commensal microbiota regulates T cell fate decision in the gut. Seminars in immunopathology. 2015;37:17–25. [DOI] [PubMed] [Google Scholar]
- Gazdeck RK, Fruscione SR, Adami GR, Zhou Y, Cooper LF, Schwartz JL. Diversity of the oral microbiome between dentate and edentulous individuals. Oral diseases. 2019;25:911–8. [DOI] [PubMed] [Google Scholar]
- George MF, Briggs FB, Shao X, Gianfrancesco MA, Kockum I, Harbo HF, et al. Multiple sclerosi risk loci and disease severity in 7,125 individuals from 10 studies. Neurology Genetics. 2016;2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Varea E, Urcelay E, Vilarino-Guell C, Costa C, Midaglia L, Matesanz F, et al. Exome sequencing study in patients with multiple sclerosis reveals variants associated with disease course. Journal of neuroinflammation. 2018;15:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Nelson KE. The Oral Microbiome of Children: Development, Disease, and Implications Beyond Oral Health. Microbial ecology. 2017;73:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang XL, Ni YH, Xu ZM. Geraniol suppresses proinflammatory mediators in phorbol 12myristate 13-acetate with A23187-induced HMC-1 cells. Drug design, development and therapy. 2018;12:2897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizumi T, Perez G,I, Blaser MJ. Evaluation of Methods for the Preservation of Human Fecal Samples, for Assessment of Microbiota Composition. Gastorenterology. 2017;152:S817. [Google Scholar]
- Isobe N, Keshavan A, Gourraud PA, Zhu AH, Datta E, Schlaeger R, et al. Association of HLA Genetic Risk Burden With Disease Phenotypes in Multiple Sclerosis. JAMA neurology. 2016;73:795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nature communications. 2016;7:12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Zhang T, Yin N, Ma X, Zhao G, Wu H, et al. Geraniol alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis. Oncotarget. 2017;8:71038–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2017;45:D353–d61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Liu J, Cetinbas M, Sadreyev R, Koh M, Huang H, et al. New and Preliminary Evidence on Altered Oral and Gut Microbiota in Individuals with Autism Spectrum Disorder (ASD): Implications for ASD Diagnosis and Subtyping Based on Microbial Biomarkers. Nutrients. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto H, Kaprio J, Kinnunen E, Luukkaala T, Koskenvuo M, Elovaara I. Concordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twins. European journal of neurology. 2008;15:1106–10. [DOI] [PubMed] [Google Scholar]
- Liu XX, Jiao B, Liao XX, Guo LN, Yuan ZH, Wang X, et al. Analysis of Salivary Microbiome in Patients with Alzheimer’s Disease. Journal of Alzheimer’s disease : JAD. 2019;72:633–40. [DOI] [PubMed] [Google Scholar]
- Lokmer A, Aflalo S, Amougou N, Lafosse S, Froment A, Tabe FE, et al. Response of the human gut and saliva microbiome to urbanization in Cameroon. Scientific reports. 2020;10:2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melbye P, Olsson A, Hansen TH, Sondergaard HB, Bang Oturai A. Short-chain fatty acids and gut microbiota in multiple sclerosis. Acta neurologica Scandinavica. 2019;139:208–19. [DOI] [PubMed] [Google Scholar]
- Panza F, Lozupone M, Solfrizzi V, Watling M, Imbimbo BP. Time to test antibacterial therapy in Alzheimer’s disease. Brain : a journal of neurology. 2019;142:2905–29. [DOI] [PubMed] [Google Scholar]
- Patsopoulos NA. Genetics of Multiple Sclerosis: An Overview and New Directions. Cold Spring Harbor perspectives in medicine. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PAB, Aho VTE, Paulin L, Pekkonen E, Auvinen P, Scheperjans F. Oral and nasal microbiota in Parkinson’s disease. Parkinsonism & related disorders. 2017;38:61–7. [DOI] [PubMed] [Google Scholar]
- Polak D, Shmueli A, Brenner T, Shapira L. Oral infection with P. gingivalis exacerbates autoimmune encephalomyelitis. Journal of periodontology. 2018;89:1461–6. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Wu M, Feng Y, Zhou Z, Chen L, Chen F. Alterations of oral microbiota distinguish children with autism spectrum disorders from healthy controls. Scientific reports. 2018;8:1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochemical and biophysical research communications. 2016;469:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo E, Bacci G, Chiellini C, Fagorzi C, Niccolai E, Taddei A, et al. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Frontiers in microbiology. 2017;8:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira L, Ayalon S, Brenner T. Effects of Porphyromonas gingivalis on the central nervous system: activation of glial cells and exacerbation of experimental autoimmune encephalomyelitis. Journal of periodontology. 2002;73:511–6. [DOI] [PubMed] [Google Scholar]
- Shaw L, Ribeiro ALR, Levine AP, Pontikos N, Balloux F, Segal AW, et al. The Human Salivary Microbiome Is Shaped by Shared Environment Rather than Genetics: Evidence from a Large Family of Closely Related Individuals. mBio. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhrao SK, Olsen I. Assessing the role of Porphyromonas gingivalis in periodontitis to determine a causative relationship with Alzheimer’s disease. Journal of oral microbiology. 2019;11:1563405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (New York, NY). 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CJ, Auchtung TA, Ajami NJ, Velasquez K, Smith DP, De La Garza R 2nd, et al. Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: a pilot study. PeerJ. 2018;6:e4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski SP, Wos-Oxley ML, Vilchez-Vargas R, Jauregui R, Plumeier I, Klawonn F, et al. Highresolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Applied and environmental microbiology. 2015;81:1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro CE, Vallochi AL, Rios LS, Martins GA, Schlesinger D, Mosca T, et al. Experimental autoimmune encephalomyelitis can be prevented and cured by infection with Trypanosoma cruzi. Journal of autoimmunity. 2004;23:103–15. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. The Lancet Neurology. 2018;17:162–73. [DOI] [PubMed] [Google Scholar]
- Tremlett H, Fadrosh DW, Faruqi AA, Hart J, Roalstad S, Graves J, et al. Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC neurology. 2016;16:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg M, Harris RA. Co-infection with Trypanosoma brucei brucei prevents experimental autoimmune encephalomyelitis in DBA/1 mice through induction of suppressor APCs. International immunology. 2005;17:721–8. [DOI] [PubMed] [Google Scholar]
- Watson EJ, Giles J, Scherer BL, Blatchford P. Human faecal collection methods demonstrate a bias in microbiome composition by cell wall structure. Scientific reports. 2019;9:16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zameer S, Najmi AK, Vohora D, Akhtar M. Bisphosphonates: Future perspective for neurological disorders. Pharmacological reports : PR. 2018;70:900–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.