Abstract
The emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a global public health emergency. SARS-CoV-2 employs the host cell receptor ACE2 for cellular entry. Nonetheless, the differences in ACE2 expression pattern in lung versus other normal and solid tumor tissues remain incompletely characterized. Here, we analyze a large data set comprising ACE2 mRNA expression for 7592 tissue samples across 22 types of primary solid tumor and 4461 samples across matched 18 non-diseased tissues. Our results unravel eight normal tissues and 10 primary solid tumors, which might be at high risk of SARS-CoV-2 infection. These findings may provide additional insight into the prevention and treatment of SARS-CoV-2 infection, in particular for patients with these 10 vulnerable cancer types.
Keywords: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), ACE2, Expression, Solid tumors, Non-diseased tissues
To the Editor,
In December 2019, a novel pneumonia disease, now termed coronavirus disease 2019 (COVID-19), emerged in Wuhan, Hubei, China (Huang et al., 2020). A previously unknown coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in lower respiratory tract specimens from patients and serves as etiological agent responsible for COVID-19 (Zhu et al., 2019), which is threatening public health worldwide. As of April 4, 2020, a total of 1,051,635 laboratory-confirmed cases and 56,985 deaths caused by COVID-19 have been reported globally according to World Health Organization (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports).
Angiotensin-converting enzyme 2 (ACE2) is known to be a host cell receptor for severe acute respiratory syndrome coronavirus (SARS-CoV) (Kuhn et al., 2004). It has been confirmed that SARS-CoV-2 like SARS-CoV utilizes ACE2 as cellular entry receptor (Hoffmann et al., 2020; Zhou et al., 2020). Notably, elevated ACE2 expression has previously promoted susceptibility to SARS-CoV spike protein-driven infection in vitro (Hofmann et al., 2004; Li et al., 2007), implying potential positive correlation between ACE2 expression level and SARS-CoV-2 infection. Although ACE2 shows a widespread distribution in various human tissues (Harmer et al., 2002), a statistically robust comparison of expression levels in lung, the main target of SARS-CoV-2 infection (Wu and McGoogan, 2020), versus other tissues based on a large sample size is still lacking.
SARS-CoV-2 infection commonly presents with fever and cough, which frequently elicits lower respiratory tract disease (Wu and McGoogan, 2020). Nonetheless, extrapulmonary clinical manifestations have been observed (Chen et al., 2020a; Guan et al., 2020; Huang et al., 2020; Wang et al., 2020),such as diarrhea, nausea or vomiting, liver abnormality, acute cardiac injury, and acute kidney injury. It is reported that cancer patients might harbor a higher risk of SARS-CoV-2 infection and inferior prognosis than those in infection without cancer (Liang et al., 2020). However, whether a heterogeneity of risk for infection exists among various cancer types remains unclear.
Here, we retrieved ACE2 mRNA expression data of 7592 tissue samples across 22 primary solid tumor types in The Cancer Genome Atlas (TCGA) and 4461 samples across 18 matched non-diseased tissues in Genotype-Tissue Expression (GTEx) from UCSC Xena (https://xena.ucsc.edu), where TCGA and GTEx data were co-analyzed by the same Toil RNA-seq pipeline to eliminate computational batch effects (Vivian et al., 2017). The expression values of ACE2 were quantified by RNA-Seq by Expectation-Maximization algorithm (Li and Dewey, 2011) and then normalized using the upper quartile method. The normalized values were log2-transformed after adding an offset of 1 to avoid taking log of zero before analysis. Further, virus abundances for TCGA tumors, quantified by numbers of virus-supporting reads per hundred million reads processed (RPHM), were obtained (Cao et al., 2016). We defined a tumor sample with RPHM ≥100 for a given virus as virus-positive and examined ACE2 expression across seven tumor types with frequent viral presence according to the previous study (Cao et al., 2016) (Supplementary Table). Mann-Whitney U test was used to compare the expression level between two groups. This study is exempt from ethical review because its data are publicly available and deidentified.
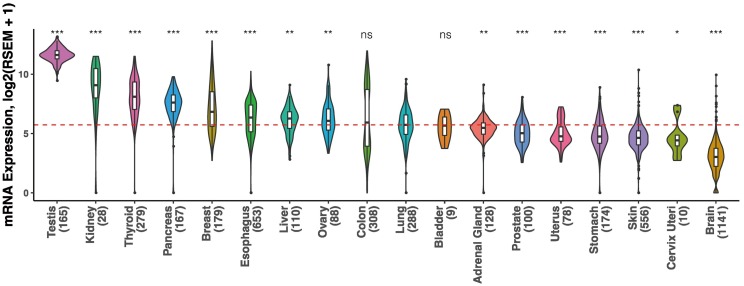
As shown in Fig. 1 , we observed a widespread distribution of ACE2 in these normal tissues, which is consistent with previous reports (Harmer et al., 2002). Notably, eight normal tissues, including testis, kidney, thyroid, pancreas, breast, esophagus, liver, and ovary, had significantly higher ACE2 levels than lung as a reference (all P < .05), and expression levels in colon and bladder were similar to that in lung (both P > .05). The differences in the expression abundance between these tissues and lung indicate possible SARS-CoV-2 infection in extrapulmonary organs. For instance, highest ACE2 abundance in testis may implicate its great possibility of SARS-CoV-2 exposure. Of note, a pathological analysis of testes from six patients who died of SARS showed that orchitis is a complication of SARS (Xu et al., 2006). Thus, we propose strengthening follow-ups for reproductive functions of recovered SARS-CoV-2 male patients. Additionally, comparably high expression observed in kidney, liver, and colon may partially contribute to acute kidney injury, liver impairment, and diarrhea at onset of COVID-19, respectively (Chen et al., 2020a; Guan et al., 2020; Huang et al., 2020; Wang et al., 2020). Interestingly, our finding that ACE2 was highly expressed in breast appears to be in contrast to a retrospective study on nine pregnant women with COVID-19 in the third trimester, in which the colostrum from six patients tested negative for SARS-CoV-2 (Chen et al., 2020b). However, considering the small sample size and short duration of the study period, the risk of vertical transmission via breastfeeding deserves further investigations.
Fig. 1.
Distribution of ACE2 expression abundance across 18 non-diseased tissues.
The vertical axis depicts the expression level shown as log2 (RSEM normalized count +1), whereas non-diseased tissues (No.) are ordered on the horizontal axis according to their median ACE2 expression values. The dashed red line represents median value (5.73) of ACE2 expression in lung. P values are calculated for comparison of expression levels in lung versus other tissues. The widths of curved shapes indicate the probability density of expression values. Box plots display the median and interquartile range, whiskers extend to 1.5 times the interquartile range, and outlier data are shown as dots. *, P < .05; **, P < .01; ***, P < .001; ns, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
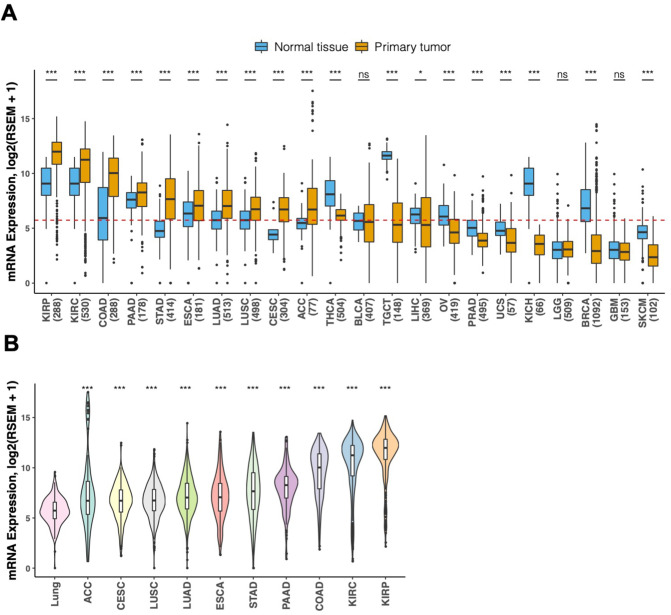
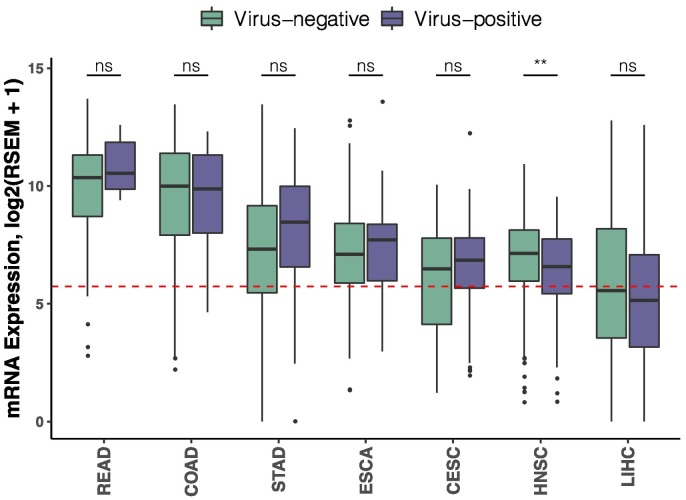
Comparison of ACE2 expression between cancer tissues and their respective normal tissues demonstrated significantly elevated ACE2 expression in 10 cancer types (all P < .001), including KIRP, KIRC, COAD, PAAD, STAD, ESCA, LUAD, LUSC, CESC, and ACC (Fig. 2A). Moreover, all the 10 cancer types exhibited significantly higher ACE2 abundance than normal lung tissue (all P < .001) (Fig. 2B). These findings suggest greater likelihood of SARS-CoV-2 infection for patients with these cancers. With regard to higher COVID-19 positive risk in cancer patients versus individuals without cancer reported by Liang et al. (2020), a possible explanation may be their immunosuppression from the malignance or anticancer treatment (Kamboj and Sepkowitz, 2009). Cancer cases analyzed in the study by Liang et al. (2020) is, however, limited in number (five lung cancers, four colorectal cancers, three breast cancers, two bladder cancers, and four other types of cancer). This restricts its ability to draw conclusions about the risk of SARS-CoV-2 infection in subgroups of specific cancer type. We hypothesize that another contributing mechanism may be increased likelihood of SARS-CoV-2 entry into certain cancer tissues due to aberrantly abundant ACE2 expression. Therefore, during this pandemic, we propose reinforcing personal protection, such as remote medical counselling, minimizing the number of hospital visits, and appropriate isolation procedures when admitted to hospitals, for cancer patients, especially patient subgroups with these 10 solid tumor types. Interestingly, our analysis for seven types of tumors related to viruses revealed that virus-positive HNSC samples showed significantly lower ACE2 abundance than virus-negative ones (P < .01) (Fig. 3 ), suggesting potential viral roles of ACE2 expression in HNSC.
Fig. 2.
Comparison of ACE2 expression abundance between solid tumor tissues and normal tissues.
(A) Comparison of ACE2 expression abundance in 22 types of solid tumors versus corresponding normal tissues. Cancer types (No.) are ordered on the horizontal axis according to their median ACE2 expression values. The dashed red line indicates median value (5.73) of ACE2 expression in normal lung tissue. (B) Violin plots depicting the differences in ACE2 expression abundance in 10 types of solid tumor, each of which exhibits significantly higher abundance than the respective matched normal tissue, versus normal lung tissue. The widths of curved shapes indicate the probability density of expression values. Box plots display the median and interquartile range, whiskers extend to 1.5 times the interquartile range, and outlier data are shown as dots. *, P < .05; ***, P < .001; ns, not significant. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; LGG, brain lower grade glioma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; UCS, uterine carcinosarcoma. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Comparison of ACE2 expression abundance in virus-positive versus negative solid tumor tissues.
Cancer types (No.) are ordered on the horizontal axis according to their median ACE2 expression values. The dashed red line indicates median value (5.73) of ACE2 expression in normal lung tissue. Box plots display the median and interquartile range, whiskers extend to 1.5 times the interquartile range, and outlier data are shown as dots. **, P < .01; ns, not significant. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In conclusion, we performed the first, to our knowledge, large-scale comparative analysis of ACE2 expression across multiple solid tumors and matched non-diseased tissues based on a consistently analyzed expression repository, which highlights eight normal tissues and 10 primary solid tumors with potentially similar or greater risk of SARS-CoV-2 exposure compared with lung, and identify a potential association between HNSC-related viruses and ACE2 expression. Given the large sample sizes for these 18 risky candidates, our results may be statistically robust and reliable; notably, we did not use normal tissue samples from TCGA (adjacent to the tumor), because they are typically limited in number and their proximity to tumor may introduce signals of tumor microenvironment in their ACE2 expression profile (Aran et al., 2017); moreover, we used expression data unified by a standardized bioinformatic pipeline (Toil RNA-seq), enabling the direct comparison of ACE2 expression level from two sources (TCGA and GTEx). Our findings may contribute additional insight into the prevention and treatment of COVID-19, especially in patient subgroups with certain vulnerable cancer types. However, further clinical and autopsy studies are required to validate these findings.
Declaration of Competing Interest
None.
Acknowledgments
This work was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001). The sponsor had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104428.
Appendix A. Supplementary data
Supplementary Table. Information on different viruses infection across seven cancer types.
References
- Aran D., Camarda R., Odegaard J., Paik H., Oskotsky B., Krings G., Goga A., Sirota M., Butte A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Wendl M.C., Wyczalkowski M.A., Wylie K., Ye K., Jayasinghe R., Xie M., Wu S., Niu B., Grubb R., 3rd, Johnson K.J., Gay H., Chen K., Rader J.S., Dipersio J.F., Chen F., Ding L. Divergent viral presentation among human tumors and adjacent normal tissues. Sci. Rep. 2016;6 doi: 10.1038/srep28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q., Liao J., Yang H., Hou W., Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Yu, Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang Jin-lin, Liang Z., Peng Y., Wei L., Liu Y., Hu Ya-hua, Peng P., Wang Jian-ming, Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj M., Sepkowitz K.A. Nosocomial infections in patients with cancer. Lancet. Oncol. 2009;10:589–597. doi: 10.1016/S1470-2045(09)70069-5. [DOI] [PubMed] [Google Scholar]
- Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. CMLS. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sui J., Huang I.-C., Kuhn J.H., Radoshitzky S.R., Marasco W.A., Choe H., Farzan M. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian J., Rao A.A., Nothaft F.A., Ketchum C., Armstrong J., Novak A., Pfeil J., Narkizian J., Deran A.D., Musselman-Brown A., Schmidt H., Amstutz P., Craft B., Goldman M., Rosenbloom K., Cline M., O’Connor B., Hanna M., Birger C., Kent W.J., Patterson D.A., Joseph A.D., Zhu J., Zaranek S., Getz G., Haussler D., Paten B. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu J., Qi L., Chi X., Yang J., Wei X., Gong E., Peh S., Gu J. Orchitis: a complication of severe acute respiratory syndrome (SARS) Biol. Reprod. 2006;74:410–416. doi: 10.1095/biolreprod.105.044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F. Tan, W., 2020. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table. Information on different viruses infection across seven cancer types.