Abstract
Japanese encephalitis virus (JEV) is an infectious pathogen spreading in a wide range of vertebrate species. Pigs are amplifying hosts of JEV and thought to be maintained in nature predominantly by avian-mosquito cycles. In the innate immune system, interferon-inducible transmembrane protein (IFITM) is a small transmembrane protein family and has been identified as the first line of defense against a broad range of RNA virus invasion. In this paper, we found that swine IFITM (sIFITM) could restrict the replication of both JEV vaccine strain and wild strain NJ-2008. The cysteine S-palmitoylation modification of sIFITM plays important roles in their anti-JEV effects and intracellular distributions. Our findings show the anti-JEV activities of swine interferon-inducible transmembrane proteins and broaden the antiviral spectrum of IFITM protein family. The preliminary exploration of S-palmitoylation modification of sIFITM may contribute to understanding of the antiviral molecular mechanism of sIFITM.
Keywords: Innate immune, Swine IFITMs, Antiviral protein, Japanese encephalitis virus, S-palmitoylation
1. Introduction
Host innate immunity orchestrated by the interferon (IFN) plays a pluripotent role in natural defense and the pathogenesis of related diseases through IFN production and action. Interferon-inducible transmembrane proteins (IFITMs) are small membrane-spanning proteins with high sequence conservation in different vertebrates. Human IFITMs mainly refers to IFITM1, IFITM2, IFITM3, IFITM5 and IFITM10 and these IFITM genes forms a locus located on chromosome 11 (Bassano et al., 2017). Among them, IFITM1, IFITM2 and IFITM3 were identified as innate antiviral factors against a broad spectrum of pathogenic viruses, especially enveloped RNA viral pathogens of 9 viral families e.g., Orthomyxoviruses (Influenza A virus, IAV), Filoviruses (Ebola virus, EBOV), Coronaviruses (severe acute respiratory syndrome coronavirus, SARS-CoV), Lentiviruses (human immunodeficiency virus, HIV), Marburg virus (MARV), Bunyaviruses (Rift Valley fever virus, RVFV) and Hantaan virus (HTNV), and relative more Flaviviridae members including hepatitis C virus (HCV), dengue virus (DENV), Zika Virus (ZIKV), West Nile virus (WNV), Yellow Fever Virus(YFV), Omsk Hemorrhagic Fever Virus (OHFV) (Liao et al., 2019; Zhao et al., 2019). In mice models, IFITM3 demonstrated the critical role in inhibiting the infections of IAV, and three Flaviviruses members, WNV, Chikungunya virus and Venezuelan equine encephalitis virus (Poddar et al., 2016). From the mentioned antiviral effect of IFITMs from human and mouse, it seems to indicate that IFITM protein show the extensive antiviral activity against different flaviviruses. However, the effect of IFITMs on JEV infection, an important member of flaviviruses, has not been reported in vitro and in vivo.
Japanese encephalitis virus (JEV) is an epidemic virus in tropical regions that is transmitted through Culex mosquitoes among pigs, human and other animals. Different species of animals infected with JEV may exhibit different symptoms. Patients, especially children infected with JEV present clinically with encephalitis caused by central nervous system injury. Pigs have a high risk of JEV infection and are the most important domestic amplifying hosts (Rosen, 1986). When severe infection occurs, JEV infected pigs have the symptoms of boar testis or stillbirth. Recent years, the domestic pig comes to be thought of the central role in epidemiology of Japanese encephalitis, whether for virus amplification and maintenance, or transmission to humans (Ladreyt et al., 2019). Therefore, effective prevention and control of JEV spread in pigs is an important task for public health.
Many investigations manifested the subcellular distribution and topological structural function relationship of human and mouse IFITM proteins (Bailey et al., 2013; Ling et al., 2016; Weston et al., 2014; Smith et al., 2019; Jia et al, 2012, 2015; Foster et al., 2016). Post translational modification of human IFITM protein, especially S-palmitoylation of the N-terminal conserved cysteine residues are essential for the regulation of their antiviral function (McMichael et al., 2017; Narayana et al., 2015; Spence et al., 2019; Yount et al., 2010). While people have a deep understanding of the restriction on viral infection of human and mouse IFITMs, the investigation on the functions of IFITMs in other species especially domestic livestock closely related to human beings, is still seriously insufficient. Some scientists investigated the restriction of swine IFITM (sIFITM) on several kinds of viruses, such as foot-and-mouth disease virus (Xu et al., 2014; Zhang et al., 2016), swine influenza virus(SIV) (Benfield et al., 2015), porcine reproductive and respiratory syndrome virus (PRRSV) (Wang et al., 2014), classical swine fever virus (CSFV) (Li et al., 2019a), African swine fever virus (Munoz-Moreno et al., 2016), lyssa viruses (Benfield et al., 2015), and pseudorabies virus (Li et al., 2019b). However, most of these researches focused on demonstrate the antiviral role of IFITM3. So far, no one had published on whether swine interferon-inducible transmembrane proteins combat the infection caused by JEV.
The aim of study was to elucidate the anti-JEV activities of swine IFITM and revealed the important role of S-palmitoylation modification of swine IFITM1 from biochemistry. We also examined the proteins distribution when the S-palmitoylation of swine IFITM proteins changed by the inhibitor for palmitoylation or by the replacement of cysteine to serine.
2. Materials and methods
2.1. Gene cloning and plasmid constructions
The cDNAs of swine IFITM1, IFITM2, IFITM3 were synthesized from the isolated total RNA of porcine kidney epithelial PK15 cells and PCR amplified with a pair of specific primers (Table 1 ). The confirmed correct sequences were subcloned into the corresponding eukaryotic expression plasmids using DNA restriction endonucleases and ligases. Based on the aims of different experiments and convenience of detection, the fusion expression vectors with different tags, such as hemagglutinin (HA), FLAG, green fluorescent protein (GFP) or red fluorescent protein (RFP), were constructed respectively. The primary vectors were obtained from Invitrogen (Carlsbad, USA). Other molecular biological reagents were purchased from Takara (Shiga, Japan).
Table 1.
The primers for the cDNAs synthesis of swine IFITMs, RT-PCR and gene knockdown.
| Primer purpose | Primer name | Forward: 5′–3′ | Reverse: 5′–3′ |
|---|---|---|---|
| Gene clone | sifitm1 | CGGAATTCATGATCAAGAGCCAGCAC | CGGGATCCCGGTAGCCTCTGTTACTCTTTG |
| sifitm2 | CGGAATTCATGAACTGCGCTTCCCAGCC | CGGGATCCCGGTAGCCTCTGTTACTCTTTG | |
| sifitm3 | CGGAATTCATGAACTGCGCTTCCCAGCC | CGGGATCCCG TAGCCTCTGTAATCCTTTA | |
| RT-PCR | JEV-E | ACTGACATCTCGACGGTGGC | CTCCCAATCGCTTTACTGGT |
| Gene knockdown | shRNA | CCGGGCAAAGAGTAACAGAGGCTACTTCAAGAGAGTAGCCTCTGTTACTCTTTGCTTTTTTGGTACC | AATTGGTACCAAAAAAGCAAAGAGTAACAGAGGCTACTCTCTTGAAGTAGCCTCTGTTACTCTTTGC |
2.2. Cell culture and transfection
Porcine kidney PK15 cells, baby hamster kidney BHK-21 cells, Homo sapiens embryonic kidney HEK293 cells were maintained in DMEM containing 10% FBS with at 37 °C/5% CO2. Cells were seeded into plates approximately 5–6 × 104 cells/well of 24-well plates and 2 × 105 cells/well of 6-well and cultured for 18–24 h before transfection. After cells adhered to the well for 18–24 h, the plasmids with objective genes and corresponding controls were introduced into cells using the X-tremeGENE DNA transfection reagent. The efficiency of cell transfection was checked respectively through fluorescence microscopic observation GFP or RFP tag, or using Western blotting to detected tag specific primary antibody.
2.3. Infection and antiviral assay
Two JEV strains, wild-type NJ-2008 and live vaccine strain SA14-14-2 were retrieved and propagated once in BHK-21 cells. The virus titer was determined through quantitative real-time PCR (qPCR) using a pair of specific primers targeted at conserved regions of the envelope protein (E) gene of JEV (Table 1). The viruses at 0.1–10 MOI (multiplicity of infection) were added into the medium of cells (transfected or untransfected) for 1 h of virus attachment. Supernatant medium was discarded, and the cells were washed with 1 × PBS twice to remove the uninfected viruses. The infected cells were maintained in medium containing 2% FCS for 24, 48, 72 h. At corresponding time points, supernatants containing viruses and cells were collected, the virus RNA in the media and the total cellular RNA were extracted and quantitated using qPCR. The cellular proteins were extracted and detected by Western blotting.
2.4. Stable sIFITM1-knockdown cell lines construction
The corresponding DNA sequences of shRNA targeting IFITM1 were amplified by PCR with a pair of specific primers (Table 1). The amplified DNA sequences verified by sequencing were inserted into a shRNA knockdown vector, pLKO.1-sh (Novagen, USA). HEK293T cells were co-transfected with the recombinant shRNA-encoding plasmids pLKO.1-shRNAifitm, and other two vectors of lentivirus system, pCMV-Gag-Pol and pCMV-VSV-G (Novagen, USA), using X-tremeGENE DNA transfection reagent as producer's protocol. The ratio of plasmids used were pCMV-Gag-Pol: pCMV-VSV-G: pLKO.1-shRNAifitm = 7: 2: 1. The supernatants containing viruses were collected at 48 and 72 h after transfection, filtered with a 0.45 μm filter. The virus titer of supernatant was detected by real time PCR with specific primers with specific primers (Table 1). The collected supernatant containing recombinant lentiviral virus infected PK15 and ST respectively at 1 MOI, and after 1 h post infection, the cultural supernatants were replaced with fresh medium. The positive cells were selected by puromycin under a working concentration of 5 μg/mL in the culture medium containing 10% FCS for 7 days. Then the clonogenic cells were transferred to 96-well plates by limiting dilution method. The knockdown of sifitm1 gene was validated by RT-PCR and Western blotting.
2.5. Construction of sIFITM1 mutants
To investigate whether the anti-JEV activity of swine IFITM was involved in S-palmitoylation, series mutants of sIFITM1 in which three conserved cysteines were substituted for serine were generated with PCR site directed mutagenesis technology. Then the mutant DNA sequences were inserted the eukaryotic expression vectors with the FLAG or RFP tag via restriction sites and transfected into cells according to different tests. The antiviral effects of sIFITM1 mutants were evaluated through transient overexpressing the mutants and determining the JEV copies with RT-PCR as mentioned above.
2.6. Confocal microscopy
To investigate the effect of S-palmitoylation on sIFITMs distribution in cell, different sIFITMs and sIFITM1 mutants with RFP tag at C terminal were introduced into cells by transfection. An analogue of palmitic acid, 2-bromopalmitic acid (2BP, Wako) was used as the inhibitors of palmitoylase. After 24 h post-transfection, 2-BP, at a final concentration of 100 μM, was added into supernatant cell culture medium. After 6 h 2-BP treatment, the medium was removed and washed 3 times with cold PBS, and the cells were covered with cold imaging buffer (containing 2 μM hoechst 33342). Then, the cells were immediately analyzed by the fluorescence of RFP using a Leica TCS SP8 laser scanning confocal microscope.
2.7. Quantitative real-time PCR
Viral RNA and cell total RNA were extracted respectively from the culture supernatant and cells using TRIzol (Invitrogen, USA) and reverse transcribed with SuperScriptase (Invitrogen, USA). Quantitative real-time PCR (qPCR) measurement of JEV or target genes was conducted with specific primers described in Table 1 using the SYBR Green qPCR kit (Vazyme Biotech, China). The standard curve method was used to determine the copy number of JEV according to the cycle threshold conversion based on a positive control plasmid with E gene. The expression level of ifitm1 gene was relatively quantified using the comparative threshold cycle (C T) method. The housekeeping gene hprt1 was used as a reference control.
2.8. Western blotting
Cultured cells were harvested and washed with 1 × PBS twice and then lysed with 1 mL lysis buffer (Solarbio Life Science, Beijing, China) for 30 min on ice. Protein concentration was quantified by the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, California, USA). The proteins in lytic supernatants were separated by SDS-PAGE and then transferred onto a PVDF membrane. The membrane was blocked with 5% nonfat dried milk in TBST buffer and then incubated with specific or tag antibodies followed by HRP-conjugated goat anti-mouse IgG or goat anti-rabbit IgG. Primary antibodies were as follows: rabbit anti-IFITM1 polyclonal antibody (Cusabio, China), monoclonal mouse anti-actin (Proteintech, Portland), mouse monoclonal anti-FLAG M2 antibody (Sigma-aldrich, USA). Pierce ECL Plus Substrate (Thermo Scientific, USA) was used to detect blots. Digital images were obtained by Amersham Imager 600 (GE, USA).
2.9. Acyl-PEGyl exchange gel shift (APEGS)
The constructed different sIFITMs expressing plasmids were transfected into HEK293T cells in 6 well plates, followed by TEA buffer with 4% SDS for lysate. The protein concentration of cell lysate was determined by BCA. The acyl-PEGyl exchange gel shift (APEGS) assay was carried out in the way as Yokoiet al. described (Yokoi et al., 2016) and modified appropriately according to Kanadome's method (Kanadome et al., 2019). In brief, tris-(2-carboxyethyl) phosphine (TCEP, Thermo) and N-ethyl maleimide (NEM, Wako) was added into the cell lysis successively and incubated with at 25 °C and then recovered twice using Chloroform/Methanol Precipitation (CMppt). Then the recovered protein was dissolved in TEA buffer containing 4 mM EDTA, 4% SDS, and divided into 2 portions, one portion was added 0.75 M neutralized NH2OH dissolved in TEA buffer and 0.2% Triton X-100, another one portion, as controls, containing TEA buffer and 0.2% Triton X-100. The mixtures above were incubated at 25 °C for 2 h. Then the 5 KDa maleimide-conjugated PEGs (mPEG-5k; NOF) were used for the replacement of S-palmitate of cysteines and incubated at 25 °C for 3 h. Finally, all treated samples including controls and untreated input and internal reference were measured by Western blotting and visualized by fluorescence gel scanning. Image-Pro Plus image software was used to calculate the band intensity for quantitative analysis of S-palmitoylated protein.
2.10. Statistical analysis
Graphs were presented as means and standard deviations of normalized data points. Levels of significance were determined using student's t-test.
3. Results
3.1. Swine IFITMs overexpression inhibited JEV in vitro
To investigate whether swine IFITM proteins could inhibit JEV infection of cells, we first studied the effects of sIFITM1, sIFITM2, and sIFITM3 on the replication of JEV strain SA14-14-2 in PK15. As shown in Fig. 1 a, the RT-PCR results of JEV genomic RNA in the supernatants indicated that transiently overexpression of sIFITM1, sIFITM2, and sIFITM3 drastically decreased the copies of JEV compared to the control group. In addition, sIFITM1 demonstrated more significant and persistent restriction. Although SA14-14-2 has been often used in the research field of JEV, the attenuated strain might not fully present the actual infection process. Therefore, we further investigated the viral proliferation of a JEV wild strain, NJ-2008, in sIFITMs-overexpressing PK15 cells. JEV genomic RNA was harvested for RT-quantitative PCR. As shown in Fig. 1b, genomic RNAs of NJ-2008 strain greatly reduced in IFITM-overexpressing cells, compared with the control vector. Together, our data demonstrated that extrinsic IFITM proteins from pig were restriction factors for JEV and sIFITM1 may play a more prominent role in their inhibitory actions.
Fig. 1.
Swine IFITMs inhibit JEV replication. Different cells were transfected by the plasmids inserted with swine IFITMs gene and 24 h later, infected JEV strains. At 24h, 48h and 72h post-infection, thelture supernatants were collected to detect virus genome copies number. (a) and (b) PK15 cells infected by SA14-14-2 and NJ-2008 respectively. The anti-JEV activity of sIFITM1 was further verified in PK15 (c), HEK293(d) and BHK-21 (e) cells. Bar charts represent mean ± SEM of three experiments and each experiment included triplicate repeats. *p<0.05; **p <0.01; ***p <0.001, Student's t-test.
Then, we further evaluated the anti-JEV activity of swine IFITM1 in PK15, and other two non-porcine cell lines, HEK293 and BHK-21 through similar transiently overexpression method. In three different cell lines, the exogenous sIFITM1 gene expression presented significant affection on viral genomic amplification (Fig. 1c, d and e). And that, in BHK-21, a cell line commonly used for JEV multiplication, the restriction on JEV by sIFITM1 was stronger than that in the other two cell lines (Fig. 1e). The observed anti-viral effects were not due to the tag sequences attached to sIFITM1 though transient expression of protein without tags (date not shown). Thus, these results demonstrated that porcine IFITM proteins could restrict JEV attenuated SA14-14-2 strain and wild strain NJ-2008 infection in vitro and sIFITM1 displayed the strongest resistance to JEV among three objective sIFITMs. These data suggested that swine interferon-inducible transmembrane protein were restriction factors that inhibited JEV infection.
3.2. Interference of sIFITM1 expression facilitated JEV replication
To further verify the anti-JEV role of swine IFITMs, we knocked down sIFITM1 expression in PK15 and ST cells by RNA-interference technology. The results of quantitative RT-PCR clearly showed that sIFITM1mRNA was significantly reduced both in shIFITM1-PK15 cells and shIFITM1-ST cells (Fig. 2 b and e). And that, Western blot showed that the bands of sIFITM1 expression were almost undetectable (Fig. 2c and f). Two shIFITM1 cells were infected with SA14-14-2 and after 48 h post-infection, viruses in medium were collected for genomic RNA analysis by RT-PCR. The virus infection results showed that sIFITM1 knockdown could make cells produced more viruses in both tested cell lines (Fig. 2a and d). Although JEV DNA replication did not augment in a time-dependent manner as expected, there was still significant increase of virus copies in the culture medium of the experimental cells at 24, 48, and 72 h compared to control empty vector lentivirus group. These knockdown studies indicated that partially reduction of the expression of endogenous sIFITM1 resulted in the increase of the cellular susceptibility to JEV.
Fig. 2.
Swine IFITM1 knockdown enhances JEV infection. (a) At 24h, 48h and 72h post-infection, the JEV genome copies number of culture supernatants of IFITM1 shRNA PK15 cells. (b) Quantitive RT-PCR analysis of sIFITM1 mRNA expression in control and IFITM1 shRNA PK15 cells. (c) Western blot analysis of whole cell extracts from control and IFITM1 shRNA PK15 cells infected with JEV. (d) At 24h, 48h and 72h post-infection, the JEV genome copies number of culture supernatants of IFITM1 shRNA ST cells. (e) Quantitive RT-PCR analysis of sIFITM1 mRNA in control and IFITM1 shRNA ST cells. (f) Western blot analysis of whole cell extracts from control and IFITM1 shRNA ST cells infected with JEV. Bar charts represent mean ± SEM of three experiments and each experiment included triplicate repeats. *p<0.05; **p <0.01; ***p <0.001, Student's t-test.
3.3. The conserved cysteines in sIFITM1 are critical for its anti-JEV activity
We analyzed the amino acid sequences of sIFITM1, sIFITM2, and sIFITM3 and found all of three sIFITMs had transmembrane domain–proximal cysteine residues as potential sites of S-palmitoylation: at positions 50, 51 and 84 in sIFITM1; 71, 72 and 105 in sIFITM2 and sIFITM3 (Fig. 3 a and b). In order to investigate whether these cysteines in sIFITMs play critical roles in the resistance to viruses, we constructed a series of sIFITM1 mutants in which cysteine was replaced by serine (Fig. 3c) and analyzed their anti-JEV effects. Results showed that the C84S single site mutation of sIFITM1 entirely lost its anti-JEV activity, while the single site mutation C51S, C84S and double mutation C5051S still could restrict the virus production (Fig. 3d and e). And surprisingly, the triple mutants C505184S exhibited slightly elevated viral copies. These results indicated that potential palmitoylated C84 residue localized at the C-terminus of sIFITM1 proteins played a significant role in preventing JEV infection. Hence, the C-terminal cysteine at 84 of sIFITM1 was critical for the restriction of viruses. And our results implied that the antiviral activities of swine IFITM could be related to its cysteine S-palmitoylation.
Fig. 3.
The predicted conserved palmitoylated cysteines of sIFITM1 are required for anti-JEV activity. (a) Sequence and domain structure analysis and of three swine IFITMs. The conserved residues are marked with the asterisk "*". Three conserved S-palmitoylated cysteines are highlighted with in transparent orange and markerd. NTD: N-terminal domain; IMD: intramembrane domain; CIL: intracellular loop; TMD: transmembrane domain; CTD: C-terminal domain. The IMD and CIL domain comprise the canonical CD225 domain. (b) The putative topologic structure of sIFITMs. The location of S-palmitoylated cysteine residues (red font) in transmembrane domains were predicted based on human IFITM from previous study. (c) In PK15 cells, the effects of three conserved S-palmitoylated cysteines of sIFITM1 on viral production. (d) JEV infection of mutants of sIFITM1 transduced PK15 and HEK293 cells. Data are expressed as mean ± SEM of three experiments and each experiment included triplicate repeats with a significance of *p<0.05; **p<0.01; ***p <0.001. Student's t-test.
3.4. APEGS revealed sIFITM1 was S-palmitoylated
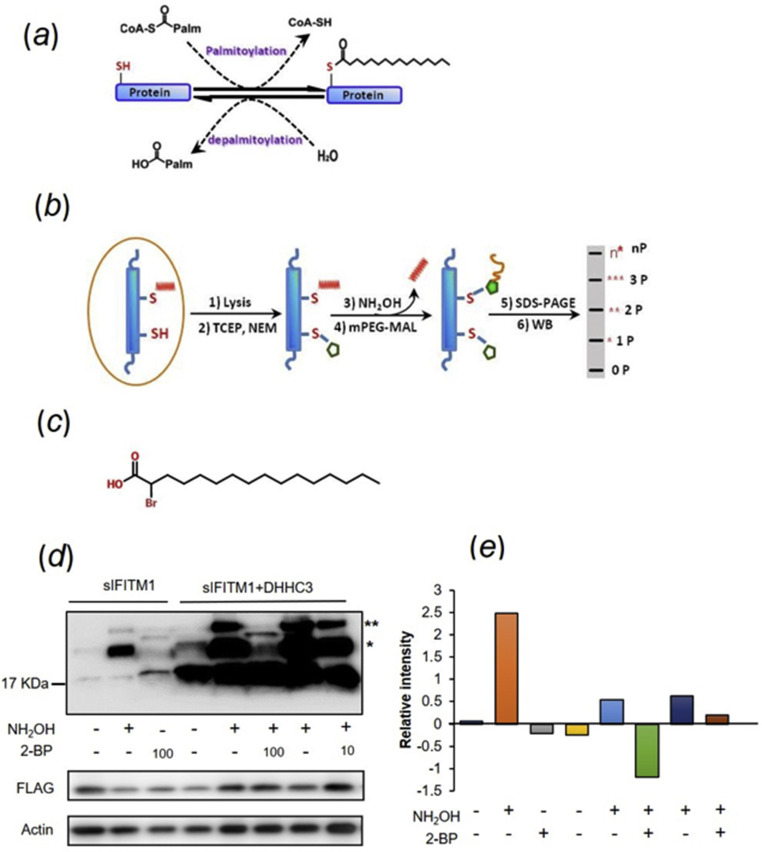
S-palmitoylation/depalmitoylation of membrane protein is a crucial cycle for signaling pathway and for their transferring and locating in eukaryotic cells (Fig. 4 a). In this dynamic cycle, one or more palmitates are covalently attached to the cysteines residues of the target protein and removed through enzymatic reactions in cells, which regulates the protein stability through modification of their hydrophobic properties and are associated with the occurrence and development of a variety of diseases (De and Sadhukhan, 2018). To further verify the S-palmitoylation of swine IFITMs, we investigated the state of the cysteine sulfydryl using the method of acyl-PEGyl exchange gel shift (APEGS) assay (Fig. 4b). Furthermore, for comparative analysis, a recombinant expression plasmid inserted with DHHC-type containing 3(DHHC3) (an identified human palmitoyl transferase gene) were co-transfected with sIFITM1 to HEK 293 and 2-bromopalmitate (2-BP), an inhibitor of palmitoyl acyl-transferase, was used to interfere with the palmitoylation of interest protein (Fig. 4c). Fig. 4d and e showed Western blot analysis and quantitative results of the ratio of intensity of total palmitoylated bands to the unpalmitoylated. From the results of APEGS, there were more than one mPEG-MAL labeled proteins bands (marked with asterisks), which indicated the robust S-palmitoylation of sIFITM1 at more than one cysteine site whether with or without DHHC3 coexpression (Fig. 4d). As shown in Fig. 4d, 2-BP administration weakened the mPEG-MAL and DHHC enhanced the intensity of the signal of mPEG-MAL. The intensity of mPEG-MAL-linked protein bands receded apparently when the cells suffered with lower concentration of 100 μM 2-BP. These results indicated 100 μM 2-BP treatment significantly inhibited the palmitoylation of sIFITM1 and 10 μM 2-BP, a lower concentration, did not have significant effect on palmitoylation (Fig. 4d and e). Quantitative analysis of palmitoylation based on Western blotting indicated the S-palmitoylation of sIFITM1 was very common, especially one or two cysteine residues modified (Fig. 4d and e). We found when cotransfection with DHHC3, the total amount of IFITM1 detected was increased distinctly compared single sIFITM1 overexpression (Fig. 4d) while it still needs to take further work to study whether DHHC3 enhance sIFITM1 stability or upregulate its expression.
Fig. 4.
Biochemical analyses for S-palmitoylation on swine IFITM1. (a) The palmitoylation/depalmitoylation is a dynamic reversible reaction in which the palmitoyl thioester group is covalently connected to coenzyme A(CoA), is transferred to cysteine by palmitoyl acyl transferase (PAT). (b) The principle of acyl-PEGyl exchange gel shift. (c) Molecular structure of 2-bromopalmitic acid (2BP). (d) S-palmitoylated sIFITM1 was monitored through APEGS method mass-shift based detection. The cell lysates are incubated with TCEP, and free cysteine residues are protected with NEM. S-fatty acid groups of sIFITM1 are replaced by 5 KDa mPEG-Mal through two subsequent reactions, a reduction mediated by NH2OH results to cysteines exposure and then a ligation with mPEG-Mal. Proteins are subjected to SDS-PAGE and then measured by Western blot. The number 10 and 100 represented the μM concentration of 2-BP. The number of PEGylation exchange is indicated by asterisks (*). The lower two panels were SDS-PAGE of whole-cell lysate input fraction from cells used in upper APEGS analysis. Immunoblotting was performed with anti-FLAG and anti-actin. (e) The relative intensity of the palmitoylated and unpalmitolated sIFITM1. The relative intensity was a ratio of average intensity of estimates (n = 5) worked out according to the intensity of different mass-shift bands of sIFITM1 measured by Image Pro Plus software. The expression of actin and sIFITM1 in whole lysate were taken as important consideration in data processing.
3.5. Three conserved cysteine residues of sIFITM1 were S-palmitoylation sites
Swine IFITM1 amino acid sequence analysis showed cysteine residues at positions 50, 51 and 84 were high conservative (Fig. 3a). Therefore, we presumed that these cysteines in swine IFITM1 protein may be involved in S-palmitoylation just as in the case of human IFITMs. The APEGS results showed sIFITM1 mutant displayed that, both the number of bands mPEG-MAL and the intensity significantly changed compared with that of WT (Fig. 5 a and b). Single cysteine mutation of C50S, C51S and C84S displayed two mPEG-MAL-linking bands, which indicated that the serine substitution at 50, 51 and 84 resulted in these positions lost the ability to be palmitoylated. And in PK15 cells, in terms of the band intensity, the C84S was much weaker than C50S, C51S. Dual mutation C5051S (cysteine 84 still exist) demonstrated one band with strong palmitoylation signal, for this band intensity alone, there is no significant difference with that of single mutation C50S, C51S. But only a weak fluorescence was detected in the dual mutants C5184S. These results suggested that cys84 was more important in S-palmitoylation, similar with antiviral analysis of mutants above (3d, e). The triple mutant (C505184S) completely lost the S-palmitoylation because all three cysteines were substituted. And there was no band detected at the position of lower molecular-mass un-modified sIFITM1, which suggested that the unpalmitoylated sIFITM1 was instable and degraded easily. In summary, sIFITM1 was a S-palmitoylated protein and all the three conserved cysteines were potential modified sites and the cys84 was more important site compared to cys50 and cys51.
Fig. 5.
Identification of S-palmitoylated sites of sIFITM1. (a) The palmitoylated state of different mutants of sIFITM1. S-palmitoylated sIFITM1 was monitored through APEGS method mass-shift based detection. The number of PEGylation exchange are indicated by asterisks (*). The lower two panels were SDS-PAGE of whole-cell lysate input fraction from cells used in upper APEGS analysis. Immunoblotting was performed with anti-FLAG and anti-actin. (b) The relative intensity of different palmitoylated state of sIFITM1 mutants. The relative intensity was a ratio of average intensity of estimates (n = 5) worked out according to the intensity of different mass-shift bands of sIFITM1 measured by Image Pro Plus software.
3.6. Cellular distribution of sIFITMs
Confocal microscopy imaging demonstrated that the intracellular distribution of each sIFITM presented divergent characteristic (Fig. 6 ). The fluorescence emitted by fusion sIFITM1-RFP localized in the whole cytoplasmic region out of nucleus like small star dots, which corroborated the known human IFITM1 localized predominantly at the plasma membrane and some endolysosome. But upon 2-BP treatment, IFITM1 mostly concentrated in the areas close to the nucleus. By contrast, IFITM2-RFP and IFITM3-RFP were seen mostly in intracellular compartments with distinct distributions. IFITM2-RFP localized in a tight cluster of punctate close to the nucleus, while IFITM3-RFP had a more dispersed, punctate distribution. Therefore, the distinctively different distribution of palmitoylated and unpalmitoylated sIFITMs indicated that S-palmitoylation was critical to subcellular localization of swine-origin IFITMs.
Fig. 6.
Localization changes of swine sIFITMs caused by palmitoylation inhibitor. Confocal microscopy imaging of sIFITM1-RFP in cells. Left 3 panels were the images of cells 2-BP untreatment. Right 3 panels were the images of cells treatment with 2-BP. Hoechst 33342 staining was used to visualize nuclei. Red channel, rhodamine. Scale bar represents 10 μm. PK15 cells transduced with the empty vector control (Vector) or different IFITMs with RFP tag. The depalmitoylation process of IFITMs was monitored by treatment with 100 μM 2-BP for 6 h before microscopy. All images are representative of 3 independent experiments.
We further determined the effect on distribution of three potential S-palmitoylated cysteine residues. Results showed that all mutants, whether single mutation, dual mutation or triple mutation, presented difference from WT sIFITM1 (Fig. 7 ). In the absence of 2-BP, among the single site mutants, cysteine 84 mutating displayed a more remarkable change than the other two, just similar as that of WT sIFITM1 treatment with 2-BP. Dual mutant C5051S in the first membrane-associated domain of sIFITM1 brought obvious location alteration in spite of still with significant antiviral activity, which indicated the S-palmitoylation played conflicting roles in IFITM localization compared with its anti-JEV effect (Fig. 3d and e). Whether one-point mutation or dual mutation prompted sIFITM1 gathering in the area near the nucleus. Triple mutation completely overthrew intracellular distribution compared with WT and other mutants, which suggest that the protein, completely incapable of palmitoylation, possibly have been transported to the digestive organelles for degradation. After treatment with 2-BP, both one-point mutation or dual mutation displayed distinct changes, which manifested that S-palmitoylation of each conservative cysteine residue played a role in the distribution of sIFITM1 in cell. Unchanged distribution of triple mutation upon 2-BP treatment indicated that incapability of palmitoylation had lost its sensitive signals to inhibitor for palmitoylation, which was consist with the results of antiviral experiment.
Fig. 7.
Confocal microscopy of sIFITM1 mutants in PK15 cells. Swine IFTIM1 and its different mutational sIFITM1 fusion with RFP were transduced into PK15 cells. The depalmitoylation process of IFITMs was monitored by treatment with 100 μM 2-BP for 6 h before microscopy. Left 3 panels were the images of cells 2-BP untreatment. Right 3 panels were the images of cells treatment with 2-BP. Hoechst 33342 staining was used to visualize nuclei. Red channel, rhodamine. Scale bar represents 10 μm. All images are representative of 3 independent experiments.
4. Discussion
The data reported in the present study demonstrated sIFITMs were restriction factors for JEV in vitro. S-palmitoylation was crucial modification for antiviral activity and cellular distribution.
JEV is widely distributed around Asia and has begun to spread to other geographic areas, including previously unaffected Australia (Mackenzie et al., 2002) and Pakistan (Erlanger et al., 2009). The high mortality, residual neurological complications, and the wide epidemic characteristics of JEV have made it a serious public health threat. Because the infectious mechanism of JEV is not yet clear, there is no specific therapy available for JE except early vaccination. The fight between JEV and the immune system decides whether a successful infection occurs. However, the research in innate immune signaling against JEV is insufficient and fewer anti-viral host factors have been identified. It has been proved that IFITMs, especially human, murine or avian IFITM3, could limit the infection of some other members of Flaviviridae, such as hepatitis C virus (Narayana et al., 2015; Wilkins et al., 2013), WNV (Brass et al., 2009), DENV (Brass et al., 2009), Zika virus (Savidis et al., 2016), avian Tembusu virus (Chen et al., 2017). In this study, we found that sIFITM1, sIFITM2, and sIFITM3 could restrain the replication of JEV and identified S-palmitoylation of sIFITM1 and its key sites. Here, our results show IFITM1 presented a stronger anti-JEV effect compared with IFITM2 and IFITM3, which is similar to the known anti-HCV effect of IFITM1 (Narayana et al., 2015). Many previous reports that IFITM3 showed stronger restriction effects against variety of viruses and its antiviral activity manifested at the early entry stage, mainly at the level of the late endosome, particularly for viruses that require the low pH endosomal environment to trigger viral fusion. As transmembrane protein, most of previous reports demonstrated IFITMs were located at plasma membrane or endosomes. Considering the different distribution of three swine IFITMs (Fig. 6) and that of human and mouse IFITMs, it suggests that stronger inhibition on JEV from sIFITM1 could concerning the cellular entry mechanism. Zhu and co-workers reported that JEV fusion occurred at the level of the early endosome because successful infection required passage through Rab5-positive early endosomes but is independent of Rab7 both in neuroblastoma cells and in non-neuronal cells (Zhu et al., 2012). Microscopy of swine IFITM1 displayed similar distribution character with early endosomes (Fig. 6, Fig. 7). And a previous report highlighting an essential role for membrane cholesterol (Kalia et al., 2013). In this work, we carried out our research mainly in non-neuronal cells, such as PK15, HEK293, BHK-21 and ST. Two groups demonstrated that IFITMs act as organizers of cell membranous structure through interaction with other cellular proteins (Wilkins et al., 2013). The subcellular localization of sIFITM1 in the membrane area is basically consistent with that of IFITMs of humans and mice reported before (Narayana et al., 2015; Huang et al., 2011; Feeley et al., 2011). We speculate that overexpression of sIFITM1 might also form a tight barrier difficult for JEV virions entry.
Our results also showed S-palmitoylation of swine IFITM1 and identified three conserved key cysteine residues as potential palmitoylation sites which are crucial for its anti-JEV activity (Fig. 4, Fig. 5). It can be speculated that endogenous palmitoylase increases the antiviral effects of IFITM1, IFITM2, and IFITM3 because of the conserved cysteines, although the specific palmitoylase directly modifying sIFITMs remains to be proved in detail. IFITMs plays a role in regulating cytokine production important for resistance against virus infection or other abnormal conditions (Narayana et al., 2015; Alteber, 2018; Trepanier et al., 2016; Wee et al., 2015). For other neurotropic viruses of Flaviviridae, such as CHIKV (Poddar et al., 2016) and WNV (Gorman et al., 2016), IFITM3 prevents excessive adverse effects that lead to exacerbation or even death. Another piece of supporting evidence is that the mice deficient for IFITMs present metabolic dysregulation, typically age-related obesity (Wee et al., 2015). And IFITM proteins play important roles in adaptive immunity and can drive differentiation of type 2 T helper cell (Yanez et al., 2019). Therefore, a conjecture that palmitoylase related to IFITM could participate in this kind of metabolic disorder does not seem to be groundless. Two research groups demonstrated their interesting findings that excessive IFITMs may cause the pregnancy complications during innate infections or other pathologies caused by IFN. And mutation display suggested that the inhibition of trophoblast cell fusion was possibly directly related to the three potential palmitoylation cysteines of IFITM (Buchrieser et al., 2019; Zani et al., 2019). Our latest results support this hypothesis because excessive amounts of palmitate gathered in the ifitm -/- −/− cells (data not shown), which probably contributes to the abnormal lipid metabolism. Therefore, follow-up explorations of the exact molecular mechanism will be required.
The pathogenesis of severe cases caused by JEV infection remains poorly understood. It is believed that the powerful strategies of JEV to evade or suppress the host innate immune response may be one of the molecular mechanisms contributing to JEV virulence (Ye et al., 2017; Garcia-Sastre, 2002; Katze et al., 2002). Type I IFN is the crucial part of the earliest innate immune response against viral infection (Takeuchi and Akira, 2009). Therefore, in the future, it would be very interesting to carry out some related studies to explore how JEV interferes with the signaling pathway of IFITM1 and to identify the specific palmitoylase for IFITM protein.
Declaration of competing interest
All authors declare no competing interests.
Acknowledgements
This project was financially supported by the National Natural Science Foundation of China of China (U1804108), the Foundation of Henan Educational Committee (19A230005) and the Preferred Foundation for Returned Scholar from Overseas of Ministry of Human Resources and Social Security of China (Study abroad personnel and expert service center of Henan Province) (2017–02).
References
- Alteber Z. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol. Cell Biol. 2018;96(3):284–297. doi: 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- Bailey C.C. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J. Biol. Chem. 2013;288(45):32184–32193. doi: 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassano I. Accurate characterization of the IFITM locus using MiSeq and PacBio sequencing shows genetic variation in Galliformes. BMC Genom. 2017;18 doi: 10.1186/s12864-017-3801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield C.T. Bat and pig IFN-induced transmembrane protein 3 restrict cell entry by influenza virus and lyssaviruses. J. Gen. Virol. 2015;96(Pt 5):991–1005. doi: 10.1099/vir.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019;365(6449):176–+. doi: 10.1126/science.aaw7733. [DOI] [PubMed] [Google Scholar]
- Chen S. Avian interferon-inducible transmembrane protein family effectively restricts avian Tembusu virus infection. Front. Microbiol. 2017;8:672. doi: 10.3389/fmicb.2017.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De I., Sadhukhan S. Emerging roles of DHHC-mediated protein S-palmitoylation in physiological and pathophysiological context. Eur. J. Cell Biol. 2018;97(5):319–338. doi: 10.1016/j.ejcb.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Erlanger T.E. Past, present, and future of Japanese encephalitis. Emerg. Infect. Dis. 2009;15(1):1–7. doi: 10.3201/eid1501.080311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E.M. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T.L. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;20(4):429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microb. Infect. 2002;4(6):647–655. doi: 10.1016/s1286-4579(02)01583-6. [DOI] [PubMed] [Google Scholar]
- Gorman M.J. The interferon-stimulated gene Ifitm3 restricts West Nile virus infection and pathogenesis. J. Virol. 2016;90(18):8212–8225. doi: 10.1128/JVI.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1) doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86(24):13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R. The C-terminal sequence of IFITM1 regulates its anti-HIV-1 activity. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia M. Japanese encephalitis virus infects neuronal cells through a clathrin-independent endocytic mechanism. J. Virol. 2013;87(1):148–162. doi: 10.1128/JVI.01399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadome T. Systematic screening of depalmitoylating enzymes and evaluation of their activities by the acyl-PEGyl exchange gel-shift (APEGS) assay. Methods Mol. Biol. 2019;2009:83–98. doi: 10.1007/978-1-4939-9532-5_7. [DOI] [PubMed] [Google Scholar]
- Katze M.G., He Y., Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2(9):675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Ladreyt H. How central is the domestic pig in the epidemiological cycle of Japanese encephalitis virus? A review of scientific evidence and implications for disease control. Viruses-Basel. 2019;11(10) doi: 10.3390/v11100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Antiviral role of IFITM proteins in classical swine fever virus infection. Viruses. 2019;11(2) doi: 10.3390/v11020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Antiviral role of IFITM proteins in classical swine fever virus infection. Viruses-Basel. 2019;11(2) doi: 10.3390/v11020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front. Microbiol. 2019;10:1097. doi: 10.3389/fmicb.2019.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci. Rep. 2016;6:24029. doi: 10.1038/srep24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr. Top. Microbiol. Immunol. 2002;267:49–73. doi: 10.1007/978-3-642-59403-8_3. [DOI] [PubMed] [Google Scholar]
- McMichael T.M. The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity. J. Biol. Chem. 2017;292(52):21517–21526. doi: 10.1074/jbc.M117.800482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Moreno R. Antiviral role of IFITM proteins in african swine fever virus infection. PloS One. 2016;11(4) doi: 10.1371/journal.pone.0154366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana S.K. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015;290(43):25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar S. The interferon-stimulated gene IFITM3 restricts infection and pathogenesis of arthritogenic and encephalitic alphaviruses. J. Virol. 2016;90(19):8780–8794. doi: 10.1128/JVI.00655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L. The natural history of Japanese encephalitis virus. Annu. Rev. Microbiol. 1986;40:395–414. doi: 10.1146/annurev.mi.40.100186.002143. [DOI] [PubMed] [Google Scholar]
- Savidis G. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15(11):2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- Smith S.E. Interferon-Induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019;93(6) doi: 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.S. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019;15(3):259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepanier M.O. Postmortem evidence of cerebral inflammation in schizophrenia: a systematic review. Mol. Psychiatr. 2016;21(8):1009–1026. doi: 10.1038/mp.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Porcine reproductive and respiratory syndrome virus counteracts the porcine intrinsic virus restriction factors-IFITM1 and Tetherin in MARC-145 cells. Virus Res. 2014;191:92–100. doi: 10.1016/j.virusres.2014.07.025. [DOI] [PubMed] [Google Scholar]
- Wee Y.S. Age-related onset of obesity corresponds with metabolic dysregulation and altered microglia morphology in mice deficient for Ifitm proteins. PloS One. 2015;10(4) doi: 10.1371/journal.pone.0123218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S. A membrane topology model for human interferon inducible transmembrane protein 1. PloS One. 2014;9(8) doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57(2):461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Swine interferon-induced transmembrane protein, sIFITM3, inhibits foot-and-mouth disease virus infection in vitro and in vivo. Antivir. Res. 2014;109:22–29. doi: 10.1016/j.antiviral.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez D.C. IFITM proteins drive type 2 T helper cell differentiation and exacerbate allergic airway inflammation. Eur. J. Immunol. 2019;49(1):66–78. doi: 10.1002/eji.201847692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. Japanese encephalitis virus NS5 inhibits type I interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-kappaB. J. Virol. 2017;91(8) doi: 10.1128/JVI.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi N. Identification of PSD-95 depalmitoylating enzymes. J. Neurosci. 2016;36(24):6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount J.S. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6(8):610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A. Interferon-induced transmembrane proteins inhibit cell fusion mediated by trophoblast syncytins. J. Biol. Chem. 2019;294(52):19844–19851. doi: 10.1074/jbc.AC119.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Induction of systemic IFITM3 expression does not effectively control foot-and-mouth disease viral infection in transgenic pigs. Vet. Microbiol. 2016;191:20–26. doi: 10.1016/j.vetmic.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.S. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front. Microbiol. 2019;9 doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.Z. Japanese encephalitis virus enters rat neuroblastoma cells via a pH-dependent, dynamin and caveola-mediated endocytosis pathway. J. Virol. 2012;86(24):13407–13422. doi: 10.1128/JVI.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]