Diet can influence the adult gut microbiome (the community of bacteria) and health outcomes, but the ability to make changes persisting beyond feeding of a particular diet is poorly understood. We investigated whether feeding highly purified diets to adult dogs for 36 weeks would alter bacterial populations sufficiently to result in a persistent change following the dogs’ return to a commercial diet. As expected, the microbiome changed when the purified diet was fed, but the original microbiome was reconstituted within weeks of the dogs returning to the commercial diet. The significance of these findings is in identifying an intrinsic stability of the host microbiome in healthy dogs, suggesting that dietary changes to support adult dog health through modifying the gut microbiome may be achieved only through maintenance on a specified diet, rather than through feeding transitionary diets.
KEYWORDS: canine, diet, fecal metagenome, fecal microbiota
ABSTRACT
The gut microbiome has an important role in health, and diet represents a key lever for shaping the gut microbiome across all stages of life. Maternal milk consumption in neonates leads to long-term health effects, indicating that pliability in the infant gut microbiome in response to diet can drive enduring change. The ability of diet to drive lasting changes in the adult gut microbiome is less understood. We studied the effect of an extreme dietary shift on the fecal microbiome of 46 Labrador retriever dogs (mean age, 4.6 years) over 11 months. Dogs were fed a nutritionally complete, commercially available complex diet (CD) for a minimum of 5 weeks, followed by highly purified diets (PDs) for 36 weeks, and the initial CD for at least a further 4 weeks. Fecal samples were collected at regular intervals for DNA extraction. By analyzing 16S rRNA genes and the metagenomes, we observed minor effects on microbial diversity but significant changes in bacterial taxa and genetic potential when a PD was fed. Specifically, metagenomics identified an enrichment of quinone- and GABA-related pathways on PD, providing insights into dietary effects on cross-feeding strategies impacting community structure. When dogs returned to the CD, no significant differences were found with the initial time point. These findings are consistent with the gut microbiome being rapidly adaptable but capable of being reconstituted when provided with similar diets. These data highlight that long-term changes in the adult dog gut microbiome may only be achieved through long-term maintenance on a specified diet, rather than through feeding a transitionary diet.
IMPORTANCE Diet can influence the adult gut microbiome (the community of bacteria) and health outcomes, but the ability to make changes persisting beyond feeding of a particular diet is poorly understood. We investigated whether feeding highly purified diets to adult dogs for 36 weeks would alter bacterial populations sufficiently to result in a persistent change following the dogs’ return to a commercial diet. As expected, the microbiome changed when the purified diet was fed, but the original microbiome was reconstituted within weeks of the dogs returning to the commercial diet. The significance of these findings is in identifying an intrinsic stability of the host microbiome in healthy dogs, suggesting that dietary changes to support adult dog health through modifying the gut microbiome may be achieved only through maintenance on a specified diet, rather than through feeding transitionary diets.
INTRODUCTION
In humans, gut bacterial communities and their genetic potential (the gut microbiome) begin to establish immediately after birth and are influenced by mode of birth (caesarean versus natural birth), environment, and early life nutrition (1, 2). Postweaning, divergent diets in different geographical communities can drive the establishment of distinct adult gut microbiome profiles (3, 4). Microbial perturbations following antibiotic exposure also shape the gut microbiome and can lead to lasting changes (5, 6). These changes may have important consequences for the host, as the gut microbiome has associations with a number of conditions, including obesity, cardiovascular disease, inflammatory bowel disease, and diabetes (7–9).
Improving health through dietary management of the microbiome is an attractive proposition. While temporary change of the adult gut microbiome through diet is well established (10), the ability of diet interventions to induce significant, long-term change in the adult microbiota remain relatively unexplored. Research studies could provide insights into the development of transitionary diets to establish new microbiomes to improve health. However, controlled dietary intervention studies in humans are often hampered by an inability to exert sufficient dietary control to observe meaningful effects on health mediated through the microbiome. Rodent models have offered some insights into potential fecal microbial extinction during transitionary diet feeding (11), but dogs have a close nutritional relation to humans and experience similar environmental stimuli and disease pathologies. To date, canine research has shown that early life represents an important time for the shaping of the gut microbiome (12), that manipulation of dietary macronutrients plays key roles in controlling the fecal microbiome (13, 14), and that the gut microbiome has an association with health and disease in dogs and cats (14).
The purpose of this study was to investigate whether, through feeding of a diet substantially different in composition for 36 weeks, it may be possible to alter the gut microbiome of healthy adult dogs sufficiently to enable the establishment of new microbiome community structures following return to the initial diet. The study took advantage of a nutrition intervention study using a nutritionally complete, highly purified diet (PD) (15), fed between periods of feeding a nutritionally complete, commercially available complex diet (CD). PDs have been used since the 1920s (16, 17) to meet the needs of individual studies by offering complete control over nutrient compositions. Such diets are commonly used in research as background diets to investigate the addition of novel raw materials/functional foods (18, 19) because they are much less complex and less variable than CDs. The stepwise breakdown of fibrous components of CDs is expected to require complex cross-feeding strategies and the establishment of diverse niches for a range of gut bacteria compared with those that may exploit other environments where nutrients are supplied in their more basic or purified form. As such, we hypothesized that an extremely different, highly purified diet fed for 36 weeks could be offered as a transitionary diet to alter the gut microbiome sufficiently to disrupt the host microbiome and enable the establishment of different, stable microbiomes when returned to CD. We tested this hypothesis using both taxonomic and metagenomic analyses of fecal samples.
RESULTS
Detection of fecal microbiota composition by 16S rRNA gene sequence analysis.
Of the 276 samples proposed for collection pretrial (Fig. 1), a total of 230 fecal samples from 46 dogs were successfully collected and analyzed. Before rare count (see Materials and Methods) and noise removal, the mean read count per sample was 54,175 (median read count per sample of 51,940). After rare count and noise removal, the mean read count per sample was reduced to 52,160 (median read count per sample, 50,057). A total of 35,398 operational taxonomic units (OTUs) were detected from 16S rRNA gene sequence analysis, which reduced to 238 OTUs after rare count and noise removal. There was no significant difference in read depth between the 6 sample time points.
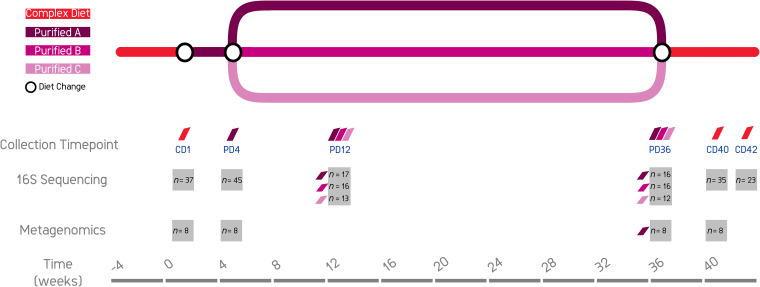
FIG 1.
Study design showing timelines, diet changes, and fecal sample collection points, with numbers analyzed for each group, for 16S sequencing and metagenomics.
Methionine:cystine ratio had no observable effect on microbial community structure.
The three PDs differed in the methionine:cystine ratio. Previous reports indicated that methionine may affect discrete populations of bacteria (20, 21). However, principal-component analysis (PCA) and heatmap analysis comparing PD and CD indicated that the three PDs fostered similar fecal microbial community composition with the methionine:cystine ratio and did not induce a detectable impact on fecal bacterial representation (Fig. 2A). Based on a lack of observed influence of PDs on bacterial phyla, microbiota data from the 3 diets at the 4-week (PD4), 12-week (PD12), and 36-week (PD36) time points (i.e., the initial PD time point and 8 and 32 weeks thereafter, respectively) were classified as PD for statistical analyses to compare the effects of long-term PD exposure across the cohort.
FIG 2.
Principal-component analysis (PCA) score plots of OTU data. Following Illumina sequencing analysis of fecal DNA, PCA was used to assess the variance of samples when fed one of three purified diets (PDs) only, where all dogs were fed diet A at PD4 (A), and variance when also including phases when fed complete diets (CDs) (B). Numbers represent week of study.
Feeding of complex versus purified diets differentiates fecal microbial communities at the taxonomic level.
Principal-component analysis of the OTU data identified variance between CD and PD samples, observable in PC1 (Fig. 2B), indicating that the microbiota composition was affected by diet within 4 weeks of diet transition. We had hypothesized that a prolonged period of consuming PD would not only alter the microbial community structure but also reduce the diversity and species richness. Using observed diversity, the mean values were significantly lower on PD (Fig. 3); however, using the Shannon index as our primary measure of diversity, no significant difference between microbial community diversity across sample points was observed (Fig. 3). The data were analyzed using four additional indices, and except for Shannon, a significant reduction in diversity/richness of the fecal bacterial community composition was detected when dogs consumed PDs compared with those observed at the initial baseline fecal sample (CD1) time point (Fig. 3).
FIG 3.
The diversity of fecal microbiota was determined at each sampling phase using 6 different approaches. Box plots represent quartiles. Time points that do not share a letter are significantly different by Tukey tests (P < 0.05).
When considering the distribution of bacteria at different taxonomic levels, distinct shifts in abundance were also observed. Bacteroidetes, Fusobacteria, and Firmicutes were the dominant phyla across the entire sample cohort (Fig. 4A). Fusobacteria showed the largest changes in proportional representation, increasing by around 2.5-fold during PD feeding. This phylum was represented by a single genus, Fusobacterium. Bacteroidetes decreased in proportion ∼2-fold (Fig. 4A), and at the family level, an increased proportion of Bacteroidaceae and a decreased proportion of Prevotellaceae were observed (Fig. 4B). Within the Firmicutes family, large proportional increases in Lachnospiraceae and decreases in Lactobacillaceae were observed when dogs transitioned from CD to PD (Fig. 4C). When considering changes within individual phyla, Deferribacteres showed the most consistent and statistically significant shift with diet. This phylum, represented by a single genus, Mucisprillium, was consistently present on CD and largely absent on the PD diet (see Fig. S1 in the supplemental material). Although overall representation of the Proteobacteria remained relatively unchanged, large proportional increases in Enterobacteriaceae and concurrent decreases in Alcaligenaceae were observed following the transition from CD to PD (Fig. 4D). Similar observations were also made within the Actinobacteria phylum where large proportional changes at the family level (increases in Coriobacteriaceae and decreases in Bifidobacteriaceae) following the transition from CD to PD were observed (Fig. 4E). Despite these changes, PCA analysis and taxonomic profiles pre- and post-PD showed that CD1, CD40, and CD42 communities are similar (Fig. 2B, Fig. 4A to E), suggesting the reconstitution of the pretrial microbial communities occurred within 4 to 6 weeks of changing the diet.
FIG 4.
Taxonomic relative abundance distribution of fecal microbiota at each sampling phase, at the phylum level (A), with number of OTUs in each phylum listed in the legend, and at the family level for Bacteroidetes (B), Firmicutes (C), Proteobacteria (D), and Actinobacteria (E).
Metagenomics identifies significant changes in genetic potential of the gut microbiome associated with diet.
Metagenomic analysis of a subset of the fecal samples was used to interrogate the data set. Prior to filtering for quality and host DNA contamination, there was a median and mean read count per sample of 56,045,949 × 2 and 55,601,432 × 2, respectively. After trimming and host contaminant DNA removal the median read count per sample was 41,864,532 × 2 and the mean read count per sample was 42,534,357 × 2.
PCA analysis of the functional pathway relative abundances showed separation between PD and CD samples in PC1/PC2 (Fig. 5). Subsequent univariate analysis showed no significant differences in any of the pathways comparing CD1 and CD40 time points (Fig. 6). Together, these data suggested that feeding dogs PDs for 36 weeks did not result in long-lasting alterations to the fecal microbial composition or its genetic potential.
FIG 5.
Multigroup PCA scores of metagenome data at the pathway level, for four time points (data for 8 dogs fed PD “A”).
FIG 6.
A comparison of significant changes in pathway abundance between different time points. Following the transition to PD, 178 pathways increased and 31 decreased in abundance (bar chart, left-hand side; CD1 to PD4). Following the transition back to CD, 142 pathways decreased and 6 increased on the return to CD (bar chart, right-hand side; PD36 to CD40). Of the 178 pathways that increased from CD to PD, 124 decreased following the return to CD (top Venn diagram), while 5 of the 31 that decreased from CD to PD increased on return to CD (bottom Venn diagram). The total number of significant changes between time points (described in the time point bar at the bottom of the figure) shows that no pathway had a significant difference between PD4 and PD36 or between CD1 and CD40.
Diet-induced changes in the metagenome are stable over 32 weeks but are reversible within 4 weeks.
To compare metagenomics data, abundance differences in the genetic potential from a total of 354 pathways were analyzed. Following dietary transition from CD1 to PD4, the relative abundance (RA) of 209 pathways differed significantly (P < 0.05). Of these, 178 pathways increased in RA and 31 decreased in RA after transition to a purified diet. Conversely, dietary transition back from PD36 to CD40 resulted in 148 pathways that significantly differed in RA (P < 0.05). Of these, 142 pathways decreased in RA, while only 6 increased on return to CD. Of the 178 pathways that increased from CD to PD, 124 decreased following return to CD, while 5 of the 31 that decreased from CD to PD increased on return to CD. No pathway differed significantly in the same direction at the two transitions (e.g., was elevated following transition at both time points). In addition, no pathway had a significant difference between PD4 and PD36 nor between CD1 and CD40, suggesting that the genetic potential was stable on these diets within 4 weeks of feeding a diet and was restored within 4 weeks of feeding CD (Fig. 6).
An analysis of the 129 pathways identified as significantly different following both diet transitions identified pathways related to metabolic functions, such as energy metabolism, vitamin B and other cofactor syntheses, nucleotide syntheses, and bioenergetics, as well as bacterial membrane component synthesis (see Table S2 in the supplemental material).
Pathway enrichment analysis was used to aid context and provide an objective analysis of the 129 pathways that changed significantly following both dietary transitions. The 129 metabolic pathways are members of 28 superpathways, of which four superpathways met our enrichment inclusion criteria for further interpretation, namely, cofactor biosynthesis (33 pathways), tricarboxylic acid cycle (TCA) variants (6 pathways), amine degradation (5 pathways), and metabolic regulators (2 pathways) (Table 1).
TABLE 1.
Superpathway members meeting enrichment set analysis criteria
| Superpathway | Pathway name | Pathway code |
|---|---|---|
| Amine degradation | 4-Aminobutanoate degradation V | PWY-5022 |
| Allantoin degradation to glyoxylate III | PWY-5705 | |
| GABA shunt | GLUDEG-I-PWY | |
| Superpathway of N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminate degradation | GLCMANNANAUT-PWY | |
| Superpathway of ornithine degradation | ORNDEG-PWY | |
| Cofactor biosynthesis | 2-Carboxy-1,4-naphthoquinol biosynthesis | PWY-5837 |
| Molybdenum cofactor biosynthesis | PWY-6823 | |
| NAD de novo biosynthesis I (from aspartate) | PYRIDNUCSYN-PWY | |
| NAD salvage pathway I (PNC VI cycle) | PYRIDNUCSAL-PWY | |
| NAD salvage pathway III (to nicotinamide riboside) | NAD-BIOSYNTHESIS-II | |
| NAD/NADH phosphorylation and dephosphorylation | PWY-5083 | |
| NAD/NADP-NADH/NADPH mitochondrial interconversion (yeast) | PWY-7269 | |
| Polyisoprenoid biosynthesis (Escherichia coli) | POLYISOPRENSYN-PWY | |
| Pyridoxal 5′-phosphate biosynthesis I | PYRIDOXSYN-PWY | |
| Pyridoxal 5′-phosphate salvage II (plants) | PWY-7204 | |
| Superpathway of demethylmenaquinol-9 biosynthesis | PWY-5862 | |
| Superpathway of menaquinol-10 biosynthesis | PWY-5896 | |
| Superpathway of menaquinol-11 biosynthesis | PWY-5897 | |
| Superpathway of menaquinol-12 biosynthesis | PWY-5898 | |
| Superpathway of menaquinol-13 biosynthesis | PWY-5899 | |
| Superpathway of menaquinol-6 biosynthesis I | PWY-5850 | |
| Superpathway of menaquinol-7 biosynthesis | PWY-5840 | |
| Superpathway of menaquinol-8 biosynthesis I | PWY-5838 | |
| Superpathway of menaquinol-9 biosynthesis | PWY-5845 | |
| Superpathway of phylloquinol biosynthesis | PWY-5863 | |
| Superpathway of tetrahydrofolate biosynthesis | PWY-6612 | |
| Superpathway of tetrahydrofolate biosynthesis and salvage | FOLSYN-PWY | |
| Superpathway of thiamine diphosphate biosynthesis I | THISYN-PWY | |
| Superpathway of thiamine diphosphate biosynthesis II | PWY-6895 | |
| Superpathway of ubiquinol-8 biosynthesis (prokaryotic) | UBISYN-PWY | |
| Tetrapyrrole biosynthesis I (from glutamate) | PWY-5188 | |
| Tetrapyrrole biosynthesis II (from glycine) | PWY-5189 | |
| Ubiquinol-10 biosynthesis (prokaryotic) | PWY-5857 | |
| Ubiquinol-7 biosynthesis (prokaryotic) | PWY-5855 | |
| Ubiquinol-8 biosynthesis (prokaryotic) | PWY-6708 | |
| Ubiquinol-9 biosynthesis (prokaryotic) | PWY-5856 | |
| Pantothenate and coenzyme A biosynthesis III | PWY-4242 | |
| 1,4-Dihydroxy-2-naphthoate biosynthesis II (plants) | PWY-5791 | |
| Metabolic regulators | Mannosylglycerate biosynthesis I | PWY-5656 |
| ppGpp biosynthesis | PPGPPMET-PWY | |
| TCA variants | Superpathway of glyoxylate bypass and TCA | TCA-GLYOX-BYPASS |
| TCA cycle I (prokaryotic) | TCA | |
| TCA cycle IV (2-oxoglutarate decarboxylase) | P105-PWY | |
| TCA cycle V (2-oxoglutarate:ferredoxin oxidoreductase) | PWY-6969 | |
| TCA cycle VII (acetate producers) | PWY-7254 | |
| TCA cycle VIII (Helicobacter) | REDCITCYC |
DISCUSSION
The objective was to investigate whether long-term (36-week) feeding of PD to adult dogs would alter the gut microbial community sufficiently to enable new, stable bacterial community compositions to develop following a return to the original diet. If so, transitionary diets might be an approach to modify the gut microbiome to support host health in dogs with, or predisposed to, chronic conditions. Our observations were consistent with reports that diet change significantly alters the taxonomic structure of the gut microbial communities (13, 22). However, we also demonstrated that changes induced by long-term feeding were transitory because fecal microbial communities pre- and posttrial showed similar microbial taxonomic structure and genetic potential. Considering the extreme nature of the diet manipulation used, we suggest that long-term manipulation of the adult microbiome may be achieved only by maintaining feeding of a specified diet and not by deployment of transitionary diets.
We surmised that the CD microbiome would be more diverse, with different microbes specializing in niches required for the stepwise breakdown and utilization of complex substrates (including polysaccharides and fermentable fibers present in complex diets), than the PD microbiome, with limited nutrient diversity (21). This would be consistent with observations in human populations differing in fiber content with fecal microbiota (4) and gene richness (23). It was, therefore, unexpected when long-term feeding on PD had no detectable impact on our primary measure of fecal microbial diversity, the Shannon index. Further analysis at the taxonomic level identified significant changes in the fecal microbial community profiles between CD to PD (Fig. 3), consistent with reports that diet, particularly those differing macronutrient profiles, can have a profound impact on fecal microbial composition in dogs (13, 22). Notably, 4 weeks after an extreme diet transition, it was possible to have a distinct fecal microbiome community structure, but with a similar diversity, that remained stable over a further 32 weeks.
Notable changes in bacterial communities between the two diets were observed, particularly in the two most dominant phyla. Fusobacteria showed approximately 2.5-fold increased prevalence during PD feeding. These OTUs were, in general, represented by Fusobacterium at the genus level. Fusobacteria are non-spore-forming, Gram-negative, obligate anaerobes which are asaccharolytic (incapable of breaking down carbohydrates for energy). Fusobacteria have been reported in dog feces and were higher when dogs were fed a “natural” diet (bone, raw meat, and vegetable) than two commercial feeds (24). Reduced incidence of members of the Bacteroidetes phylum (2-fold decrease) was observed on PD (Fig. 3A). Bacteroidetes are composed of three large classes of Gram-negative, non-spore-forming, anaerobic or aerobic, rod-shaped bacteria, described to be prevalent in dogs (13, 22). We also observed significant reductions in Deferribacteres on PD, although the proportional representation of this group was small. Deferribacteres are a group of fast-growing opportunists, many of which do not have a sequenced genome, but which have been described in dogs (25). Deferribacteres were represented by Mucispirillum at the genus level, which are bacteria inhabiting the mucous layer and the prevalence of which has correlated with the thickness of the intestinal mucous barrier (26).
To determine whether long-term PD feeding altered the gut microbiome sufficiently to result in a novel fecal microbial community after dogs were offered a complex diet again, we compared CD1 to CD40/CD42 time points. Dogs returned to a bacterial community structure similar to that seen at CD1, suggesting that the intrinsic microbial composition in these dogs was resilient. In other words, bacteria unable to flourish in the lumen on the PD regime were able to reestablish within weeks of hosts being fed a diet similar to the original one. Individual taxonomic changes can be difficult to interpret, because at various taxonomic levels, bacteria are metabolically diverse (27). Thus, qualitative descriptions of these diet-induced alterations are of limited value and further insights were sought through comparing changes to the metagenome.
To investigate the genetic potential induced by long-term feeding of PD, a subset of samples were analyzed using metagenomic sequencing. A group of 32 samples from 8 dogs provided a representative perspective of the microbiome pretrial (CD1) and posttrial (CD40) alongside those from early (PD4) and late (PD36) stages during the dietary intervention. Analysis of the metagenomic data showed a broad effect of diet on the pathway level (357 pathways representing 28 of 40 superpathways affected). This effect was stable between 4 and 36 weeks of feeding the PD but did not persist, with a significant but transitory effect for many pathways reverting within 4 weeks of transitioning back to CD. These data support the findings of the 16S rRNA composition analysis regarding a broad-ranging and stable effect of diet that was transient, with no significant change between CD1 and CD40, returning to a similar diversity profile within 4 weeks. However, unlike taxonomic diversity that reduced on PD, the genetic potential was elevated when dogs were fed PD compared with being fed CD. This observation is based on the number of pathways that increased on PD and decreased on CD (124 pathways) compared with those that decreased on PD and then increased on CD (5 pathways). This highlights that bacterial diversity did not reflect the metabolic functional potential of the microbiome, nor did it conform to our expectation that those genes required to exploit the complex raw materials would represent increased genetic potential on CD.
An interpretation of the areas of metabolism most significantly changing with diet highlighted four superpathways related to energy metabolism, specifically related to amine degradation, TCA cycle, and electron transfer systems. As PDs containing purified amino acids are readily absorbed by the host (reflected in the high fecal crude protein apparent digestibility of 96% [15]) we suggest that enrichment of the N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminate degradation pathway reflects the ability of bacteria to scavenge from intestinal sources, including host cell mucus and cell membranes, and bacterial cell wall structures (28). The other amine degradation pathways relate to carbon and nitrogen metabolism via the GABA shunt. The GABA shunt provides a mechanism for generating energy from glutamate without losing nitrogen as ammonia (29), which may be particularly important when amino acid availability is low. In addition, GABA has a role in cross-feeding, with transcription of GABA-producing pathways reported in Bacteroides, Parabacteroides, and Escherichia species in stool samples and GABA being identified as the sole growth factor produced by Bacteroides fragilis required to support growth of a previously only coculturable bacterium KLE1738 (30). We suggest that, under conditions of sufficient nutrient supply, the glutamate decarboxylase (GAD) system, induced under anaerobic conditions, provides a mechanism for some bacteria to manage acid stress through a glutamate/GABA antiporter, releasing extracellular GABA with an additional proton from glutamate decarboxylation (29). GABA export then supports the growth of other, luminal bacteria able to utilize the GABA as C and N sources. However, the relatively nutrient-deprived digesta provided by highly digestible dietary PD resulted in donor bacteria using strategies other than GABA export. Subsequently, those bacteria catabolizing GABA had fewer resources and were less able to replicate. This resulted in their decreased relative abundance in the fecal microbiome, while bacteria scavenging closer to the host mucosa under more microaerobic conditions increased in relative abundance.
Other evidence supports the increased relative abundance of mucosa-associated bacteria, with enrichment within the superpathway representing the largest number of pathways (n = 33), namely, that of cofactor biosynthesis. Of 33 pathways, at least 24 were related to the synthesis of specific menaquinones and ubiquinones, which provide the electron transfer link to cytochrome oxidases and are known to have different redox optima (31). Increased abundance suggests that bacteria capable of adapting to more varied microaerobic conditions, such as in close proximity to host cells, were more prevalent, an interpretation that is also consistent with enrichment of the TCA-variant superpathway. It is also notable that, similar to GABA, quinone biosynthesis by donor bacteria was identified as an essential growth factors for unculturable gut microbiota (32). Menaquinone biosynthetic pathways are missing from many unculturable OTUs from human stool samples (32, 33), and anaerobic respiration by some bacteria require donor-derived menaquinone biosynthesis (e.g., members of the Faecalibacterium genus [32, 34]). The loss of quinone biosynthesis has occurred independently across lineages and been suggested as a “cheater” strategy, taking advantage of quinones produced and released by other organisms into the environment (32). Although speculative, enrichment for quinone biosynthesis pathways in the metagenomes on PD may simply reflect a reduced relative abundance of those bacteria reliant on quinone extracellular transport from donor bacteria, which is reduced under these conditions. Future work could include in vitro cross-feeding studies to determine the relative abundance of GABA and quinone utilizers with conditions suitable for GABA and quinone production.
Other cofactor pathways enriched on PD are those for biosynthesis and salvage of folate, vitamin B6, and NAD. Other nutrient-related pathways (in purine and pyrimidine synthesis) were also increased on the PD, consistent with the very restricted provision of nucleotides from the ingredients used. We suggest that these pathways reflect the increased demand for bacterial biosynthesis of nucleotides for DNA and RNA synthesis rather than scavenging them from the diet. The other superpathway, metabolic regulators, included the guanosine pentaphosphate (ppGppp) pathway, representing that the stringent stress response can be induced by amino acid starvation, due to stalled ribosomes. This is consistent with PD resulting in a nutrient-deprived lumen, where bacteria living in closer proximity to the host were capable of tolerating stress or scavenging from fewer resources and were relatively more abundant than those that benefit from a more nutrient-diverse environment provided by CD. An important consideration from this is the confounding role complex diets have in driving population changes in the lumen, resulting in a relatively low representation of those members of the gut microbiome more commonly inhabiting the microhabitats of the inner and outer intestinal mucus layers and crypts (35) that may better reflect host-microbe interactions.
Restoration of the CD fecal microbiome within 4 weeks suggests a stable resident gut microbiome that is capable of recolonization of the lumen from reservoirs in response to diet change. Some bacteria, for example B. fragilis strains, routinely exchange between the intestinal crypts and the lumen (36), while regional structural variations, such as the cecum or appendix, may similarly serve as microbial reservoirs (37). Together, these reservoirs may explain the transitory effect of feeding a purified diet over such an extended period of time and explain why transitionary diets may not provide an effective approach to modify the adult microbiome. Our findings support the view that intentional manipulation of the microbiome may be more effective in early life, when perturbations of the microbiota may shape the adult microbiota (38), and may establish desired microbial species and/or community structures to support health benefits (6, 39, 40). As many dogs obtain nutrition predominantly from nutritionally complete, commercial diets throughout life, there are opportunities to support appropriate host-microbe interactions that are unmanageable in normal, healthy human populations. Dogs experience similar stimuli and disease pathologies as humans (41), and a recent diet intervention study involving dogs, humans, mice, and pigs observed the greatest phylogenetic similarly and genetic potential between the dog and human microbiome (42). Therefore, canine-based studies may also highlight the potential for dietary influences in supporting beneficial microbe-human interactions.
In conclusion, our work has provided detailed insights into the changes that occur in the adult dog fecal microbial communities following long-term diet change and suggests that they are dynamic and fully reversible. Consequently, our findings are consistent with the proposition that nutritional interventions aiming to modulate health through the microbiome are likely to require maintained feeding to persist.
MATERIALS AND METHODS
Animals.
Adult Labrador retriever dogs (n = 46) with an average age of 4.6 years (range, 2.0 to 7.8 years) were enrolled. Veterinary checks (physical and blood analysis) prior to and during the course of the study indicated they were healthy throughout. Dogs were housed and exercised with other dogs in their dietary group. Studies were approved by the Waltham Animal Welfare and Ethical Review Body and performed under the UK Regulation of Animals (Scientific Procedures) Act of 1986.
Diets.
Diets included a standard, commercially available, nutritionally complete, and balanced kibble-type diet (Pedigree Adult Complete, dry; referred to as CD) and three highly purified, nutritionally complete diets (Ssniff Speziäldiaten Gmbh; referred to as PD). PDs contained crystalline amino acids instead of intact protein and no complex carbohydrates, except for pure corn starch, maltodextrin, and refined cellulose powder. The three PDs differed in the ratio of methionine to cystine (0.98, 0.34, and 0.24), with total sulfur-containing amino acids being maintained at 2.68 g/1,000 kcal. PDs were isocaloric and isonitrogenous (via alanine). The complete dietary analysis of PDs is described in Table S1 in the supplemental material. CD was offered in specific amounts to maintain an individuals’ ideal body condition score (43). PDs were initially offered at individually determined maintenance energy requirements (MERs), estimated by energy intake equivalent to the CD phase, but intakes were adjusted as required to maintain individuals’ ideal body condition score (43).
Study design.
Details of the experimental design and sample collection time points are shown in Fig. 1. Dogs were initially fed CD for a minimum of 5 weeks. Baseline fecal samples (CD1) were collected from dogs prior to transition onto the purified diet (PD, “A”). Fecal samples were collected again after 4 weeks (PD4) on PD A. The dogs, randomized into 3 groups, were then fed either PD A (n = 17), PD “B” (n = 16), or PD “C” (n = 13). Dogs in these dietary groups were balanced for age and sex. Diets were then fed for a further 32 weeks, and fecal samples were collected at 8 weeks (PD12) and 32 weeks (PD36) after entry onto the individual PD treatment arms. Dogs were transitioned back onto feeding the CD, and fecal samples were collected at 4 weeks (CD40) and 6 weeks (CD42) posttransfer onto this diet. Data are missing for two reasons, namely, either fecal samples were not collected or data were of low sequence data quality (see Fig. 1 for data sampled).
Fecal microbiota and microbiome analysis.
Samples of fecal material from the core of freshly evacuated feces were obtained using sterile metal spatulas by opening the sample longitudinally and subsampling from the core. Samples (200 mg wet weight) were stored in Eppendorf tubes at –80°C within 60 min of defecation.
DNA preparation.
DNA was extracted from fecal samples (100 mg) using the QIAamp 96 PowerFecal QIAcube high-throughput (HT) kit (Qiagen; according to the manufacturer’s instructions) and an epMotion 5075 robot (Eppendorf). In brief, material was homogenized using a tissue lyser (Qiagen) (5 minutes at 30 Hz, room temperature), followed by digestion with proteinase K (Qiagen) (10 minutes, room temperature). Lysates were applied to a QIAamp 96-well plate under vacuum at 90 kPa. Remaining bound impurities were removed by washing, and pure DNA was eluted under centrifugation in buffer ATE (100 μl). Purified DNA was analyzed by Nanodrop spectrophotometry. Only samples with a 260/280 nm ratio of >1.8 were used.
Dual indexing barcoding for microbiota analysis.
The 16S rRNA PCR amplicons were prepared for sequencing using the 319F/806R dual-indexed PCR approach (44). In brief, primers were arrayed into 96-well plates at a final concentration of 0.1 μM. Master mix containing the Phusion high-fidelity (HF) enzyme (New England BioLabs) and HF buffer including genomic DNA (10 ng) was then applied to each well and cycling performed as follows: 98°C for 30 s; 98°C for 15 s, 58°C for 15 s, and 72°C for 15 s (30 cycles); and 72°C for 60 s. Agarose gel electrophoresis confirmed successful amplification of the samples.
Sequencing and bioinformatic analyses of fecal bacterial community composition.
The 16S rRNA amplicons for sequencing on the Illumina MiSeq platform (Eurofins Genomics, Germany) were prepared according to standard protocols.
Forward and reverse reads were assembled into contiguous sequences spanning the entire 16S rRNA gene V3-V4 regions using FLASH assembler (45). Tags were removed using TagCleaner (46), and sequences were demultiplexed in QIIME using split_libraries_fastq.py. Chimeric sequences were removed using userarch61 (47). Sequences were clustered at >98% identity using uclust (48) to generate operational taxonomic units (OTUs), and the most abundant sequences were chosen as cluster representatives.
The relative abundance and distribution across samples was assessed for each OTU to separate noise from consistent but rare OTUs (latter defined as mean proportion of <0.05% in at least two dogs in any study group). Representative sequences of all OTUs that passed the filtering criteria were searched against the Silva small subunit (SSU) database release 128 (49) using blastall 2.2.25 (50). If the alignment did not meet the cutoff criteria of ≥98% sequence identity and ≥98% query sequence coverage, genus or higher level annotations were used.
Statistical analyses of bacterial diversity and community composition.
Diversity and richness indices (observed, Shannon, Simpson, abundance-based coverage estimator [ACE], Fisher, and Chao1) were calculated for all samples and modeled using a linear mixed effects model. The response of this model was the diversity, with phase included as a fixed effect, and a random effect of dog was included to account for the repeat measurements of the sequence composition within each dog. From this model, the average diversity of the microbiota at each phase was estimated for each diversity calculation, with 95% confidence intervals, and a Tukey test was performed to compare all pairs of phases while maintaining a false-positive rate of 5%. An R package (phyloseq v1.16.2) calculated the other alpha diversity metrics (see detailed methods in reference 32).
Multigroup principal-component analysis (PCA) was performed on the relative abundance of rare/noise-filtered OTUs with group set as dog to account for the repeat measurements, and OTUs were scaled to unit variance to allow changes in the smaller proportion OTUs to show. Scores and loadings were extracted from these results, and the scores were visualized by study phase to investigate changes over time and individual purified diet to assess whether this level of granularity was necessary.
All analyses were performed using R v3.3.3 using the lme4, multcomp, mixOmics, FactoMineR, and multigroup libraries (42).
Sequencing and bioinformatic approaches for fecal metagenome analysis.
DNA samples (n = 32) from eight dogs (5 female [F], 3 male [M]; average age of 5.0 years; range, 3.7 to 7.5 years) all receiving PD A were taken forward for a pilot study to assess metagenomic content. Only CD1, PD4, PD36, and CD40 time points were selected to provide representative profiles pre- and posttrial alongside short- and long-term PD exposure. Aliquots from the same purified DNA samples were used for both 16S rRNA and metagenomic sequencing. Standard genomic library preparation, DNA fragmentation, adapter ligation, amplification, and size selection were performed (Eurofins, Germany). Sequencing was completed using a 2 × 150-bp paired-end approach using the Illumina HiSeq 4000 platform to generate a total of 50 million reads per sample.
A dog reference genome database was created using bowtie2-build with the Canis lupus familiaris reference genome CanFam3.1 (NCBI accession number GCA_000002285.3) (51). Paired-end FASTQ files were processed using the dog reference genome database and KneadData (https://bitbucket.org/biobakery/kneaddata/wiki/Home) to perform quality control and remove contaminant host DNA. The composition of the microbial communities was profiled using MetaPhlAn2 (52). HUMAnN2 was used to profile the abundance of microbial pathways against the ChocoPhlAn nucleotide reference database and the Uniref90 protein reference database (53, 54). Relative abundances of MetaCyc pathways were calculated using humann2_renorm_table with the option “–units relab.”
Statistical analyses of fecal metagenome composition.
Comparisons of fecal metagenome composition were made between CD1 and PD4, CD1 and CD40, PD4 and PD36, and PD36 and CD40 for all pathway relative abundances using a Wilcoxon signed-rank test. This test was used as it makes no assumptions about the data distribution and is also unaffected by zero inflation. Within each contrast, Benjamini-Hochberg correction was applied to the P values to maintain a 5% false-positive rate.
To perform enrichment analysis for an assessment of diet effects on fecal metagenomes, each pathway was matched to all upstream level 2 pathways, referred to herein as superpathways, using the MetaCyc v23.0 (55) database. Level 1 pathways from the database were excluded, as they provided little discrimination in the subsequent analysis. Prior to enrichment analysis, each individual pathway was marked as being significant if it met the criteria of having significant differences between CD1 versus PD4 and PD36 versus CD40 in the univariate analysis by the Wilcoxon signed-rank test. Enrichment analysis was then performed by calculating the ratio of the total number of pathways to the number of significant pathways within each superpathway. This was then divided by the ratio of the total number of pathways to the total number of significant pathways in the whole data set to give an enrichment value. An enrichment value greater than 1 indicates that a superpathway contains more significant pathways than the data average. Permutation testing was also performed by randomly assigning the significance tag to the pathways 1,000 times to give a P value for each enrichment value.
Following the enrichment analysis, the following a priori criteria were implemented for inclusion: enrichment >1; at least 2 pathways detected, if 2 both must be significant; if 3 to 5 pathways are detected, then ≥50% must be significant; and if >5 are detected, then ≥25% must be significant and the overall P value is <0.05.
Data availability.
Metagenomics and 16S sequencing data were submitted to ENA under primary accession number PRJEB34360.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of our colleagues at the WALTHAM Centre for Pet Nutrition for their care and expertise in the training, welfare and sample collection of dogs used in the study and those in the research team who supported us, specifically Nicola Kirkwood for lab support and Matt Harrison who managed the purified diet study.
The work was funded by Mars Petcare UK, and the funders had no role in the design, analysis or writing of this article.
All authors were employed by Mars Petcare UK at the time of the study.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Molgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newburg DS, Morelli L. 2015. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res 77:115–120. doi: 10.1038/pr.2014.178. [DOI] [PubMed] [Google Scholar]
- 3.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jernberg C, Lofmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 6.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorburn AN, Macia L, Mackay CR. 2014. Diet, metabolites, and “Western-lifestyle” inflammatory diseases. Immunity 40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guard BC, Mila H, Steiner JM, Mariani C, Suchodolski JS, Chastant-Maillard S. 2017. Characterization of the fecal microbiome during neonatal and early pediatric development in puppies. PLoS One 12:e0175718. doi: 10.1371/journal.pone.0175718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Lauber CL, Czarnecki-Maulden G, Pan Y, Hannah SS. 2017. Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. mBio 8:e01703-16. doi: 10.1128/mBio.01703-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blake AB, Suchodolski JS. 2016. Importance of gut microbiota for the health and disease of dogs and cats. Anim Front 6:37–42. doi: 10.2527/af.2016-0032. [DOI] [Google Scholar]
- 15.Harrison M, Thomas G, Gilham M, Gray K, Colyer A, Allaway D. 2020. Short-term determination, and long-term evaluation of the dietary methionine requirement in adult dogs. Br J Nutr doi: 10.1017/S0007114520000690. [DOI] [PubMed] [Google Scholar]
- 16.McHargue JS. 1926. Further evidence that small quantities of copper, manganese and zinc are factors in the metabolism of animals. Am J Physiol 77:245–255. doi: 10.1152/ajplegacy.1926.77.2.245. [DOI] [Google Scholar]
- 17.Bertrand G, Benson R. 1922. Recherches sur l’importance du zinc dans l’alimentation des ainmaux. C R Hebd Seances Acad Sci 175:289. [Google Scholar]
- 18.Koleva P, Ketabi A, Valcheva R, Ganzle MG, Dieleman LA. 2014. Chemically defined diet alters the protective properties of fructo-oligosaccharides and isomalto-oligosaccharides in HLA-B27 transgenic rats. PLoS One 9:e111717. doi: 10.1371/journal.pone.0111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos A, San Mauro M, Díaz DM. 2006. Prebiotics and their long-term influence on the microbial populations of the mouse bowel. Food Microbiol 23:498–503. doi: 10.1016/j.fm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Dahiya JP, Hoehler D, Van Kessel AG, Drew MD. 2007. Effect of different dietary methionine sources on intestinal microbial populations in broiler chickens. Poult Sci 86:2358–2366. doi: 10.3382/ps.2007-00133. [DOI] [PubMed] [Google Scholar]
- 21.Baker DH. 1992. Applications of chemically defined diets to the solution of nutrition problems. Amino Acids 2:1–12. doi: 10.1007/BF00806073. [DOI] [PubMed] [Google Scholar]
- 22.Bermingham EN, Maclean P, Thomas DG, Cave NJ, Young W. 2017. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 5:e3019. doi: 10.7717/peerj.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Wang X, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C. 2018. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, An JU, Kim W, Lee S, Cho S. 2017. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog 9:68. doi: 10.1186/s13099-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffery ND, Barker AK, Alcott CJ, Levine JM, Meren I, Wengert J, Jergens AE, Suchodolski JS. 2017. The association of specific constituents of the fecal microbiota with immune-mediated brain disease in dogs. PLoS One 12:e0170589. doi: 10.1371/journal.pone.0170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belzer C, Gerber GK, Roeselers G, Delaney M, DuBois A, Liu Q, Belavusava V, Yeliseyev V, Houseman A, Onderdonk A, Cavanaugh C, Bry L. 2014. Dynamics of the microbiota in response to host infection. PLoS One 9:e95534. doi: 10.1371/journal.pone.0095534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaut M, Clavel T. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137:751S–755S. doi: 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- 28.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feehily C, Karatzas KA. 2013. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol 114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x. [DOI] [PubMed] [Google Scholar]
- 30.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. 2019. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 4:396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezaiki L, Lamberet G, Derre A, Gruss A, Gaudu P. 2008. Lactococcus lactis produces short-chain quinones that cross-feed group B Streptococcus to activate respiration growth. Mol Microbiol 67:947–957. doi: 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 32.Fenn K, Strandwitz P, Stewart EJ, Dimise E, Rubin S, Gurubacharya S, Clardy J, Lewis K. 2017. Quinones are growth factors for the human gut microbiota. Microbiome 5:161. doi: 10.1186/s40168-017-0380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568:505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franza T, Delavenne E, Derre-Bobillot A, Juillard V, Boulay M, Demey E, Vinh J, Lamberet G, Gaudu P. 2016. A partial metabolic pathway enables group b streptococcus to overcome quinone deficiency in a host bacterial community. Mol Microbiol 102:81–91. doi: 10.1111/mmi.13447. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. 2007. Biofilms in the normal human large bowel: fact rather than fiction. Gut 56:1481–1482. [PMC free article] [PubMed] [Google Scholar]
- 38.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. 2019. Role of the microbiome in human development. Gut 68:1108–1114. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. 2017. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol 17:508–517. doi: 10.1038/nri.2017.58. [DOI] [PubMed] [Google Scholar]
- 40.Salminen S, Gibson GR, McCartney AL, Isolauri E. 2004. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53:1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hytönen MK, Lohi H. 2016. Canine models of human rare disorders. Rare Dis 4:e1241362. doi: 10.1080/21675511.2016.1241362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 43.German AJ, Holden SL, Moxham GL, Holmes KL, Hackett RM, Rawlings JM. 2006. A simple, reliable tool for owners to assess the body condition of their dog or cat. J Nutr 136:2031S–2033S. doi: 10.1093/jn/136.7.2031S. [DOI] [PubMed] [Google Scholar]
- 44.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmieder R, Lim YW, Rohwer F, Edwards R. 2010. TagCleaner: identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics 11:341. doi: 10.1186/1471-2105-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 48.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 53.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. 2018. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH; the UniProt Consortium. 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, Paley S, Subhraveti P, Karp PD. 2018. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res 46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomics and 16S sequencing data were submitted to ENA under primary accession number PRJEB34360.