Cerato-platanins (CPs) are surface-active small proteins abundantly secreted by filamentous fungi. Consequently, immune systems of plants and other organisms recognize CPs and activate defense mechanisms. Some CPs are toxic to plants and act as virulence factors in plant-pathogenic fungi. Our analysis, however, demonstrates that the interactions with plants do not explain the origin and evolution of CPs in the fungal kingdom. We revealed a long evolutionary history of CPs with multiple cases of gene duplication and events of interfungal lateral gene transfers. In the mycoparasitic Trichoderma spp., CPs evolve under stabilizing natural selection and hamper the colonization of roots. We propose that the ability to modify the hydrophobicity of the fungal hyphosphere is a key to unlock the evolutionary and functional paradox of these proteins.
KEYWORDS: evolution, fungal-bacterial interactions, fungal-fungal interactions, gene duplication, lateral gene transfer, natural selection, plant immune response, protein secretion, rhizosphere colonization, small secreted cysteine-rich proteins, SSCPs
ABSTRACT
Cerato-platanins (CPs) form a family of fungal small secreted cysteine-rich proteins (SSCPs) and are of particular interest not only because of their surface activity but also their abundant secretion by fungi. We performed an evolutionary analysis of 283 CPs from 157 fungal genomes with the focus on the environmental opportunistic plant-beneficial and mycoparasitic fungus Trichoderma. Our results revealed a long evolutionary history of CPs in Dikarya fungi that have undergone several events of lateral gene transfer and gene duplication. Three genes were maintained in the core genome of Trichoderma, while some species have up to four CP-encoding genes. All Trichoderma CPs evolve under stabilizing natural selection pressure. The functional genomic analysis of CPs in Trichoderma guizhouense and Trichoderma harzianum revealed that only epl1 is active at all stages of development but that it plays a minor role in interactions with other fungi and bacteria. The deletion of this gene results in increased colonization of tomato roots by Trichoderma spp. Similarly, biochemical tests of EPL1 heterologously produced by Pichia pastoris support the claims described above. Based on the results obtained, we conclude that the function of CPs is probably linked to their surfactant properties and the ability to modify the hyphosphere of submerged mycelia and, thus, facilitate the nutritional versatility of fungi. The effector-like functions do not sufficiently describe the diversity and evolution of these proteins in fungi, as they are also maintained, duplicated, or laterally transferred in the genomes of nonherbivore fungi.
IMPORTANCE Cerato-platanins (CPs) are surface-active small proteins abundantly secreted by filamentous fungi. Consequently, immune systems of plants and other organisms recognize CPs and activate defense mechanisms. Some CPs are toxic to plants and act as virulence factors in plant-pathogenic fungi. Our analysis, however, demonstrates that the interactions with plants do not explain the origin and evolution of CPs in the fungal kingdom. We revealed a long evolutionary history of CPs with multiple cases of gene duplication and events of interfungal lateral gene transfers. In the mycoparasitic Trichoderma spp., CPs evolve under stabilizing natural selection and hamper the colonization of roots. We propose that the ability to modify the hydrophobicity of the fungal hyphosphere is a key to unlock the evolutionary and functional paradox of these proteins.
INTRODUCTION
Microbial interactions are ubiquitous and versatile, ranging from mutualism to parasitism and competition. Fungi have numerous mechanisms to communicate with other organisms, including plants, animals, and other microorganisms (1). Interestingly, most fungal relations are competitive or combative. For example, fungal-fungal wars are usually associated with mycoparasitism and the secretion of antibiotics, peptides, and cell wall-degrading enzymes (1–3). Secreted proteins play central roles in the interactions of fungi, acting as signals, toxins, and effectors (4). For example, small secreted cysteine-rich proteins (SSCPs) are common in fungal secretomes (5). Some SSCPs have been characterized as effectors that inhibit plant immune defense, and some are avirulent and able to elicit immune responses without causing cell death. Others, such as the surface-active hydrophobins (HFBs) and the relatively new protein family of cerato-platanins (CPs), still need broader investigations (Microascales) (6).
CPs form a family of fungal SSCPs whose founding member was named after Ceratocystis platani (Microascales) (6, 7). CPs have been reported to be universally present in Dikarya fungi (6, 8) and are abundantly secreted by fungi like Botrytis cinerea (Helotiales) and Trichoderma spp. (Hypocreales) (9, 10). They are known to cause local and systemic defense responses in plants; this property attracts most of the research attention (8, 11, 12). Plant-pathogenic fungi secrete CPs in the host cell, where these proteins act as virulence factors and effectors that suppress the plant’s basal defense (13–15). The function of CPs in nonphytopathogenic fungi is less understood. In Neurospora crassa (Sordariales), the CP-encoding Snodprot1 is a reported clock-controlled gene (16), while in Leptosphaeria maculans (Leptosphaeriales), it is regulated by light (17). In a few cases, these proteins are reported as involved in development, such as fruiting body formation in Agaricomycotina (6), oligosaccharide recognition (18, 19), spore formation, and hyphal growth (11). Biochemically, CPs are characterized by the presence of four position-conserved cysteine (Cys) residues that form two disulfide bridges, resulting in a 3-D structure with a double ψ-β-barrel fold similar to that of expansins (18). This structure allows CPs to self-assemble at interfaces and to alter surface hydrophobicity like HFBs, which have eight position-conserved Cys residues (8, 20).
The first review of the diversity of CPs in fungal genomes revealed that they are exclusively present in two groups of filamentous Dikarya (such as molds and mushrooms) and are absent in all lineages containing yeast and/or dimorphic fungi. It was concluded that CPs are expanded in Agaricomycotina (Basidiomycota), where up to a dozen individual genes can be present in a genome (Ganoderma species or Postia placenta [Polyporales]), while Pezizomycotina (Ascomycota) genomes usually contain either only one or a few of these genes (6). This analysis suggested that CPs could be present in the genome of the common ancestor of Dikarya fungi, but the genes were lost in all fungi that form yeasts in Ascomycota and in nonagaricoid Basidiomycota (6).
Interestingly, CPs are reported to be among the most abundantly secreted proteins not only in numerous phytopathogenic fungi (7, 9, 15) but also in mycoparasitic and environmentally opportunistic species of Trichoderma (Hypocreales), such as Trichoderma atroviride (21), Trichoderma virens (10), and T. harzianum (22). Among the three reported genes of these Trichoderma spp., one, epl1 (=sm1), was expressed under most of the conditions tested, while the other two, epl2 (=sm2) and epl3, were usually transcribed at a very low level or were not detectable (8, 12). These Trichoderma species are capable of establishing in the rhizosphere, where they beneficially influence plant growth and immunity and also antagonize other fungi, including plant pathogens (reviewed in reference 23). Although CPs of Trichoderma are not phytotoxic, they also trigger immune defense in plants and are thus termed eliciting plant response-like proteins (EPLs) (8), which contradicts the described role in virulence of their homologs in plant-pathogenic fungi like Botrytis cinerea (9), Sclerotinia sclerotiorum (both Helotiales) (15), and Magnaporthe grisea (Magnaporthales) (24).
Trichoderma is a large genus of primary mycoparasitic fungi (23) that are capable of interacting with plants, animals, and bacteria (reviewed in references 23 and 25). Despite the genus being relatively well studied, little is known about how the CP-encoding genes evolve and their roles in the whole spectrum of Trichoderma interactions. Here, we focused on the diversity and evolutionary analysis of CPs in 37 Trichoderma genomes and performed a functional genetic investigation of the EPL1 proteins in two Trichoderma species, T. guizhouense (TgEPL1) and T. harzianum (ThEPL1), that are plant growth promoting (26, 27), mycoparasitic (3, 28), and cellulolytic (29).
RESULTS
Evolutionary analysis of CPs in filamentous fungi reveals a history of lateral gene transfers and gene duplications.
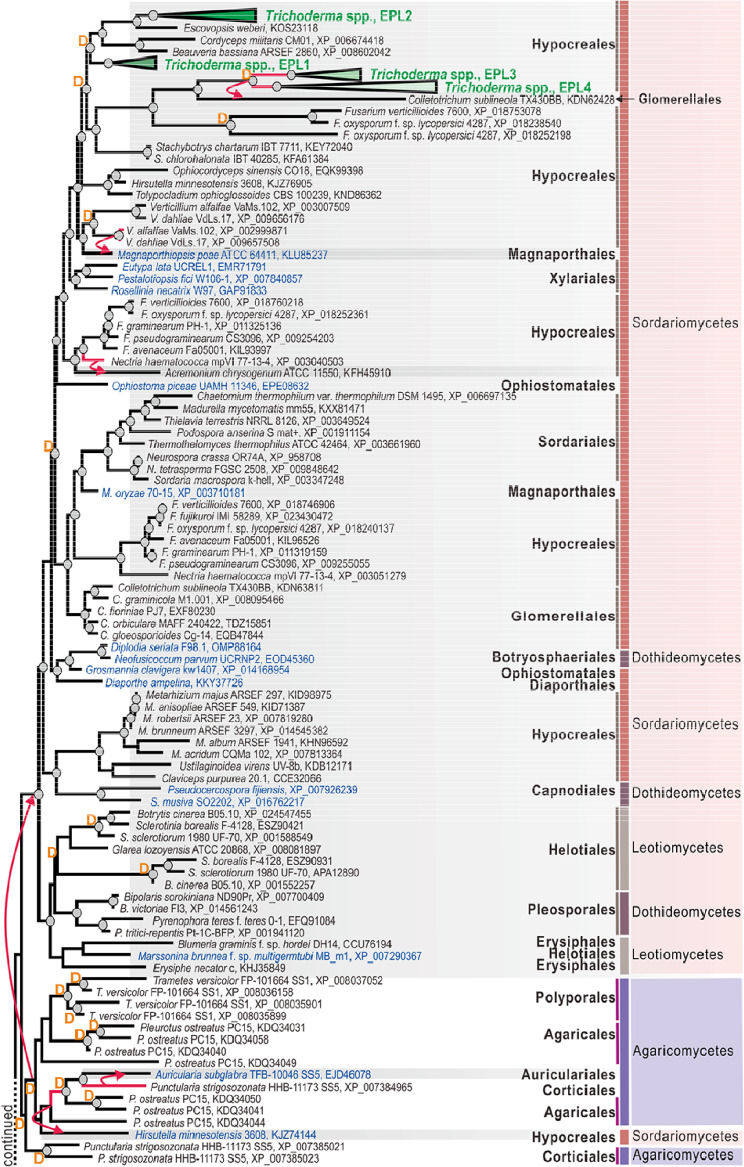
We first performed a broad-scale genome mining for CP-encoding genes in genomes (Table S1 in the supplemental material) of Trichoderma spp. and other fungi. We screened a total of 157 fungal genomes, including 150 from Ascomycota and 7 from Basidiomycota, deposited in the National Center for Biotechnology Information (NCBI) and the Joint Genome Institute (JGI) databases. Using three CPs from T. atroviride strain IMI 206040 (12) as the starting point, a total of 283 CP protein sequences were retrieved (Fig. 1). The maximum-likelihood (ML) phylogeny in Fig. 1 shows multiple cases of taxonomically incongruent phylogenetic positions when CP proteins from unrelated fungi form statistically supported clades. Interestingly, topological incongruence was observed on different taxonomic levels. For example, within Ascomycota, CPs from Capnodiales (Dothideomycetes) and Hypocreales (Sordariomycetes) occupied the same clade, supported by significant bootstrap values (Fig. 1). More surprisingly, several clades were formed by proteins from several orders of Agaricomycetes, while one such mixed clade had a hypocrealean (Ascomycota) protein as the next neighbor (Fig. 1).
FIG 1.
Evolution of cerato-platanin (CP) proteins in Hypocreales and the representative Dikarya fungi. CPs obtained via statistically confirmed lateral gene transfer (LGT) are annotated with a red arrow from the donor fungi (red branches) to the putative receiver (shaded in gray). The blue font highlights OTUs that occupy a position on the tree that is incongruent with fungal phylogeny (http://tolweb.org/fungi). D marks the gene duplication events revealed by NOTUNG analysis (see Materials and Methods for details). The corresponding taxonomic information (including the order, class, and phylum) for each fungus is given on the right. The clades containing CPs from 37 Trichoderma species genomes were collapsed and marked in green font. The details are provided in Fig. 2. The maximum-likelihood (ML) phylogram of CPs was constructed using IQ-TREE 1.6.12 (bootstrap replicate n = 1,000). Circles at the nodes indicate IQ-TREE ultrafast bootstrap values of >75. Protein accession numbers are provided for every OTU except those of Trichoderma species (see Fig. 2).
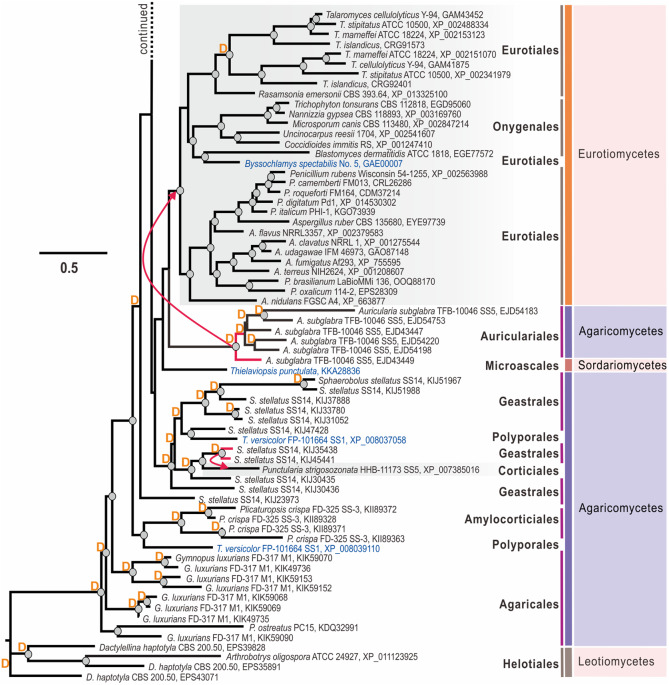
To test whether the CPs underwent lateral gene transfer (LGT) events during their evolution (30–32), we reconciled the protein trees to the multilocus species phylogeny (Fig. S1) in NOTUNG (33) and T-Rex (34), as was performed for plant cell wall-degrading enzymes of Trichoderma (31). The results showed numerous statistically confirmed LGT events that putatively occurred at the early stages of Dikarya evolution. Thus, our analysis revealed that the diversity of CPs in Eurotiomycetes (Eurotiales and Onigeneales) possibly originated after the LGT event, from the hypothetical taxonomic unit (HTU) ancestral to the modern Auriculariales (Agaricomycetes, Basidiomycota). Interestingly, a CP protein from the microascalean Thielaviopsis punctulata (Sordariomycetes, Ascomycota) appeared as a sister branch of the Eurotiales-Auriculariales clade, but the scenario of a respective LGT event did not get statistical support (Fig. 1). Furthermore, the diversity of CPs in the three other classes of Ascomycota (Sordariomycetes, Leotiomycetes, and Dothideomycetes) originated after another putative LGT event from Basidiomycota fungi (Fig. 1). In this case, the most likely donor taxon was the ancestor of Polyporales and Agaricales mushrooms. In addition to these two taxonomically broad and ancient transfers, the strict sense NOTUNG and T-Rex analyses revealed several more recent events that took place between the phyla (from agaricalean Pleurotus spp. to hypocrealean Hirsutella spp.) or within each phylum. Thus, in Basidiomycota, Sphaerobolus spp. donated a CP-encoding gene to Punctularia strigosozonata (Corticiales), while another CP-encoding gene of P. strigosozonata was transferred to Auricularia subglabra (Auriculariales) (Fig. 1). Similarly, several statistically confirmed transfers were also recorded in Ascomycota. For example, a CP-encoding gene of Verticillium (Hypocreales) was putatively transferred to Magnaporthiopsis poae (Magnaporthales) and, in another case, from Nectria haematococca to Acremonium chrysogenum (both Hypocreales). The HTU ancestral to epl3 from Trichoderma spp. was transferred to Colletotrichum sublineola (Glomerellales). Several other cases of taxonomically incongruent phylogeny of CPs could be explained by LGT, but this scenario was not supported due to the strict criteria applied in NOTUNG and insufficient genome sampling. In addition to LGT, our analysis revealed an exceptionally high number of gene duplication (GD) events that happened most frequently in Basidiomycota CP-encoding genes. Thus, up to nine cases of GD were recorded in Sphaerobolus stellatus and five in Auricularia subglabra. GDs also occurred frequently in the evolution of Ascomycota CPs (Fig. 1).
In summary, this analysis reveals that CPs likely originated in Basidiomycota, but the maintenance of these genes in the core genome of Dikarya filamentous fungi can be explained by several ancient events of lateral gene transfer from Basidiomycota to Ascomycota. The subsequent diversification is best described by birth-and-death evolution (35, 36), as genes frequently duplicated but many copies were also subsequently lost.
All four Trichoderma CPs evolve under purifying selection pressure.
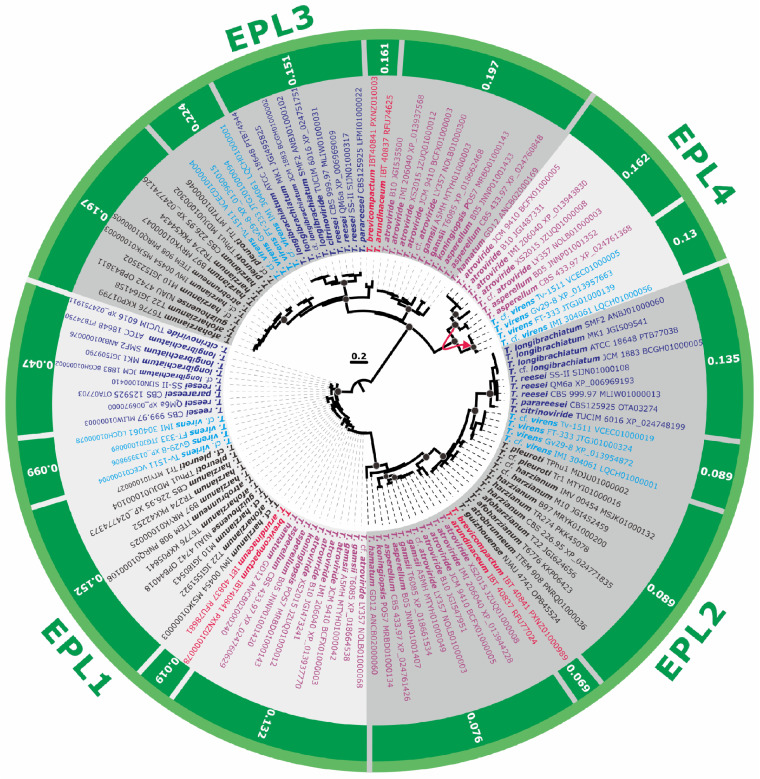
CPs of 37 Trichoderma genomes formed the four distinct clades that also originated through several GD events (Fig. 1 and 2). Thus, the clade containing the paralogous proteins EPL1 (GenBank accession number XP_013937770) and EPL2 (GenBank accession number XP_013944228) of Trichoderma and several proteins from other hypocrealean fungi (Escovopsis weberi, Cordyceps militaris, and Beauveria bassiana) was paralogous to a clade containing the other two paralogous Trichoderma CPs, EPL3 (GenBank accession number XP_013937568) and EPL4 (nom. nov.; GenBank accession number XP_013943830), which only occur in a few Trichoderma species (see below), and proteins from Fusarium spp. (also paralogous) and Stachybotrys spp. (Fig. 1). Remarkably, EPL1, EPL2, and EPL3 were present in all 37 Trichoderma genomes. The NOTUNG analysis revealed that EPL4 originated from the GD event in the ancestor of the extant section Trichoderma. As the gene is absent from the genomes of Trichoderma gamsii (which is closely related to T. atroviride) and Trichoderma hamatum, which is monophyletic with the latter two (37), it was probably then lost in several strains of this section. Interestingly, a statistically significant LGT event from the ancestral to this section HTU to the distantly related T. virens was detected (Fig. 2).
FIG 2.
Maximum-likelihood phylogram (proteins) and natural selection pressure analyses (genes) of CPs in 37 genomes of Trichoderma spp. The phylogram was constructed using IQ-TREE 1.6.12 (bootstrap replicate n = 1,000). Circles at the nodes indicate IQTree ultrafast bootstrap support values of >75. The outer numbers represent the ratios of nonsynonymous/synonymous substitution rates (ω = dN/dS) for the natural selection pressure for all branches tested as estimated using EasyCodeML. No other types of natural selection pressure were found for the tested genes from the Trichoderma genomes included (a ratio of 0 < ω < 1 indicates purifying natural selection). The red arrow shows the case of LGT (see Fig. 1 for details). Protein accession numbers are provided for every OTU. Colored fonts highlight the same infrageneric groups of Trichoderma (37, 57).
A common feature of all the CPs is the characteristic pattern of four cysteine residues, while most other residues were highly polymorphic. The low conservation in amino acid sequences of CPs raises the question of how the evolutionary mechanism drove the rapid divergency of CP-encoding genes, although gene groups from different species were statistically supported in terminal branches within Trichoderma. The natural selection pressure analysis (Fig. 2) carried out using the EasyCodeML program (38) for each Trichoderma section/clade gave a ratio of 0 < ω < 1 for all of the lineages tested indicated that CPs in Trichoderma evolve during a purifying (stabilizing) selection pressure. Therefore, together with the observation of GD, this suggests that the Trichoderma CPs also evolve by a birth-and-death mechanism in which new genes are created by repeated GDs and result in the maintenance of some copies for a considerable evolutionary time in the genome while other copies are rapidly lost or converted to pseudogenes (35).
EPL1 is massively secreted by Trichoderma spp., and epl1 is the predominant gene expressed during development.
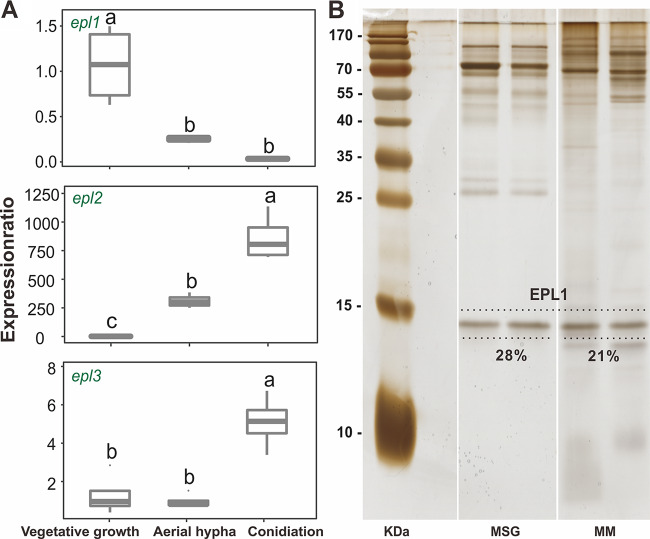
Two species of Trichoderma, T. guizhouense (strain NJAU 4742) and T. harzianum (strain CBS 226.95, ex-type), from the Harzianum clade, were adopted in the functional investigation of CPs. T. guizhouense NJAU 4742, as an aeroaquatic strain, showed better growth in and on liquid media than T. harzianum CBS 226.96, which hardly grew under aquatic conditions and had a significantly shorter life span. Thus, for conciseness, results for T. guizhouense are presented below, while the outcome of the parallel experiments with T. harzianum are provided in the supplemental material (see below). For conclusions, both species are considered. Thus, in order to test the secretion of CPs in T. guizhouense, the fungus was grown in a 30% Murashige and Skoog basal salt mixture (Sigma-Aldrich, USA) supplemented with 1% glucose (MSG) and in minimal medium supplemented with 4% glycerol (MM). The collected culture filtrates were then analyzed by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS-PAGE). The results shown in Fig. 3 confirm the presence of a small secreted protein with a size of ca. 15 kDa in both media. The protein identity given by the matrix-assisted laser desorption ionization-tandem time of flight (MALDI-TOF/TOF) mass spectrometry (MS) analysis confirmed the same results for these protein bands (Fig. S2). A tandem mass spectrometry (MS-MS) ion search based on peptide mass fingerprinting of the most prominent tryptic peptides offered the only hit to the EPL1 protein of T. guizhouense NJAU 4742 when aligned within its genome (NCBI accession number GCA_002022785.1). The semiquantitative analysis of the SDS-PAGE results revealed that EPL1 accounted for 28% and 21%, respectively, of the whole proteome of T. guizhouense NJAU 4742 when grown in MSG and MM media, respectively. The data obtained from T. harzianum showed a similar pattern (File S1 in the supplemental material). Thus, EPL1 was detected as at least one of the major secreted proteins in T. guizhouense and T. harzianum under the conditions tested.
FIG 3.
Transcriptional and proteomic determination of CPs in T. guizhouense NJAU 4742. (A) Expression dynamics of epls at the three developmental stages of the fungus: submerged vegetative growth, aerial hypha formation, and conidiation. Boxes without a same letter indicate a significant difference at P < 0.05 (Tukey multiple-comparison test). (B) Silver-stained sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS-PAGE) gels with culture filtrates of T. guizhouense NJAU 4742 collected from the 30% Murashige and Skoog basal salt mixture (Sigma-Aldrich, USA) supplemented with 1% glucose (MSG) or the minimal medium supplemented with 4% glycerol (MM).
The development of Trichoderma includes interchanges between penetration into the substrate for nutrition and growing out of it for reproduction. Therefore, in this study, the asexual life cycle of Trichoderma was divided into three stages, including one nutritional stage (submerged vegetative growth) and two stages related to reproduction: aerial hypha formation and conidiation. The reverse transcription-quantitative PCR (RT-qPCR) results revealed that the transcription level of epl1 was highest in the vegetative stage and decreased significantly (P < 0.05) later. In contrast, transcription of epl2 increased dramatically (>700-fold) during development and epl3 was expressed only minimally at the conidiation stage (Fig. 3). More importantly, it also showed that epl1 was the predominant gene expressed during all three developmental stages of fungal growth tested, while epl2 and epl3 were only detectable at the later time points (Fig. 3). Hence, epl1 was selected as the target gene for the functional genetic investigation.
Construction of epl1 deletion and overexpression Trichoderma mutants.
A fragment containing the two homologous arms and a selectable marker, the hygromycin B gene (hph), was constructed to replace the entire open reading frame (ORF) of EPL1 as illustrated in File S2. A total of 47 stable transformants of T. guizhouense and 35 of T. harzianum were screened for the designed gene disruption, and four of them were positive for both species. All vectors and PCR products were confirmed by sequencing. An expression vector harboring a copy of the epl1 gene with its terminator under the control of the constitutive promoter Pcdna1 from T. reesei was constructed (pPcdna1::epl1::Tepl1) to overexpress epl1 in T. guizhouense and T. harzianum. Among 15 putative transformants of T. guizhouense (and ten of T. harzianum), three (for both species) were found to have the expected overexpression cassette. Two strains of each genotype, namely, Δepl1-3 and Δepl1-4 among the deletion strains and OEepl1-6 and OEepl1-8 among the overexpression strains for T. guizhouense, were randomly selected for further verification by SDS-PAGE analysis, as shown in File S2, and the following experiments. Strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this study
| TUCIM IDa | Strain | Comment | Reference or source |
|---|---|---|---|
| 4742 | T. guizhouense NJAU 4742 | Wild type | 28 |
| 6353 | Δepl1-3 | epl1 deletion | This study |
| 6354 | Δepl1-4 | epl1 deletion | This study |
| 6611 | OEepl1-6 | epl1 overexpression | This study |
| 6612 | OEepl1-8 | epl1 overexpression | This study |
| 916 | T. harzianum CBS 226.95 | Wild type | 57 |
| 6344 | Δepl1-1 | epl1 deletion | This study |
| 6345 | Δepl1-2 | epl1 deletion | This study |
| 6607 | OEepl1-1 | epl1 overexpression | This study |
| 6608 | OEepl1-2 | epl1 overexpression | This study |
| 4812 | Fusarium oxysporum FOC4 | Wild type | 28 |
| 5319 | F. fujikuroi | Wild type | 28 |
| 3753 | Rhizoctonia solani | Wild type | 28 |
| 4679 | Botrytis cinerea | Wild type | 28 |
| - | Sclerotinia sclerotiorum | Wild type | 28 |
| 4076 | Athelia rolfsii | Wild type | 28 |
| 3737 | Alternaria alternata | Wild type | 28 |
| - | Escherichia coli DH5α | Commercial strain | Takara |
| - | Bacillus amyloliquefaciens 9 | Wild type | 58 |
| - | Ralstonia solanacearum RS1115 | Wild type | 59 |
| 6622 | Pichia pastoris | EPL1 producer | This study |
TUCIM (TU Wien Collection of Industrial Microorganisms, Vienna University of Technology, Vienna, Austria) identification number. -, no ID provided.
EPL1 plays only a minor role in Trichoderma interactions with other fungi and bacteria.
To determine whether EPL1, as one of the major secreted proteins of Trichoderma spp., participates in interactions with other organisms, we used RT-qPCR to analyze the transcription of epl1 in response to the presence of another fungal colony (T. guizhouense NJAU 4742 itself, Fusarium oxysporum f. sp. cubense 4 [FOC4], or Rhizoctonia solani TUCIM 3753 [Cantharellales, Basidiomycota]), a bacterial colony (Escherichia coli DH5α, Ralstonia solanacearum RS1115, or Bacillus amyloliquefaciens 9), and a tomato plant (Solanum lycopersicum cv. HEZUO903). The results shown in Table 2 demonstrate that the expression of epl1 was slightly but statistically significantly induced by the presence of R. solanacearum and tomato seedlings and was reduced in the interactions with F. oxysporum and E. coli (P < 0.05).
TABLE 2.
Expression pattern of epl1 in T. guizhouense NJAU 4742 during interaction with fungi, bacteria, or tomato seedlings
| Partner | Expression ina
: |
|
|---|---|---|
| Solo culture | Interacting culture | |
| Solanum lycopersicum cv. HEZUO903 (tomato) | 1.05 ± 0.37 | 2.85 ± 0.89* |
| Ralstonia solanacearum RS1115 | 1.04 ± 0.14 | 3.97 ± 1.35* |
| Bacillus amyloliquefaciens 9 | 1.11 ± 0.35 | 0.96 ± 0.67 |
| Escherichia coli DH5α | 1.00 ± 0.17 | 0.69 ± 0.07* |
| Trichoderma guizhouense NJAU 4742 | 1.01 ± 0.08 | 0.81 ± 0.15 |
| Fusarium oxysporum FOC4 | 1.10 ± 0.23 | 0.31 ± 0.08* |
| Rhizoctonia solani | 1.04 ± 0.15 | 1.34 ± 0.20 |
The solo-cultured sample was used as the control. Gene expression was measured by RT-qPCR. tef1 gene was used as the internal control. Expression ratio of the target gene is the fold change relative to the value for the control sample, calculated using the 2−ΔΔCT method (n≥ 4). *, a significant difference was found between the paired samples at P < 0.05, calculated based on t test.
Therefore, a broader in vitro dual confrontation test of the Trichoderma mutants against seven fungi (including FOC4, Fusarium fujikuroi, R. solani, B. cinerea, S. sclerotiorum, Athelia rolfsii [Atheliales], and Alternaria alternata [Pleosporales]) was applied as shown in Fig. S3. However, no morphological difference was noted due to the deletion or overexpression of epl1, except that both of the epl1 deletion and overexpression mutants of T. harzianum showed reduced antagonism against A. rolfsii compared to that of the wild-type strain (File S1). Additionally, no effect of epl1 deletion or overexpression on fungal-bacterial (including E. coli, R. solanacearum, and B. amyloliquefaciens) interaction was found between strains (Fig. S4). The above-described results indicate that EPL1 played a minor role in biotic interactions for Trichoderma spp.
Removal of epl1 from Trichoderma spp. was associated with a reduced JA-mediated defense response in a tomato plant.
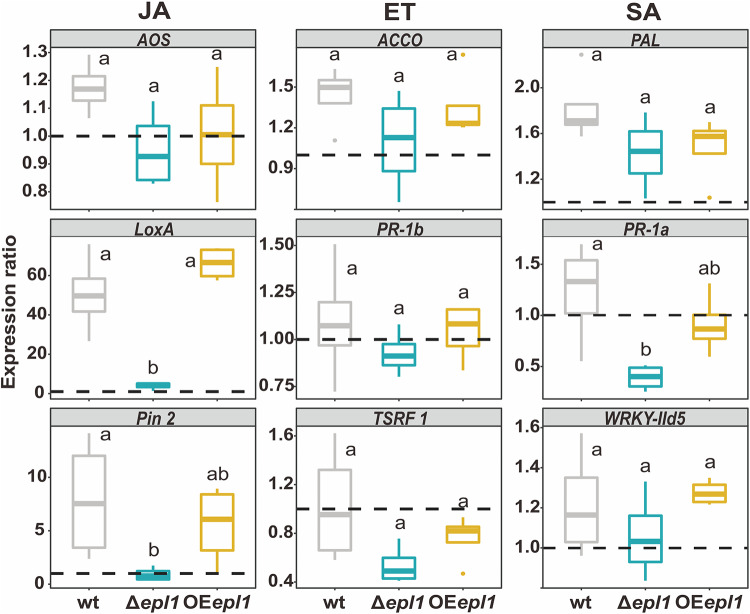
To investigate the effect of epl1’s presence in Trichoderma spp. on triggering plant immune responses, the expression of nine defense-related genes in tomato seedlings was analyzed 48 h after inoculation with epl1 mutants and the wild-type (wt) strain. Seedlings without Trichoderma inoculation were used as controls. As shown by the results in Fig. 4, seedlings inoculated with the wt and OEepl1 strains upregulated the expression of LOX A and PIN2 genes (up to 66- and 8-fold, respectively, compared to the control), which are involved in the jasmonic acid (JA) signaling pathway, while Δepl1 strains did not show a significant increase in mRNA copies of these genes. Inoculation of T. guizhouense in tomato seedlings generated only a weak wave (0.5- to 1.5-fold) of expression of ethylene (ET) signaling pathway-related genes (ACCO, PR-1b, and TSRF1), and no significant difference was noticed between the mutants and the wt. Additionally, compared to its expression in the wt strain, the expression of one salicylic acid (SA) defense gene, PR-1a, was significantly lower in the seedlings inoculated with the Δepl1 strains. T. harzianum mutants gave similar results (File S1). Therefore, EPL1 triggered the plant immune response caused by Trichoderma inoculation, through the JA-mediated pathway.
FIG 4.
Immune response of tomato seedlings to T. guizhouense colonization. JA, jasmonic acid-mediated signaling pathway; ET, ethylene-mediated signaling pathway; and SA, salicylic acid-mediated signaling pathway. Expression ratio of the immune defense genes is the fold change, calculated using the 2−ΔΔCT method (n = 12), relative to their expression in the control sample that was not colonized by T. guizhouense NJAU 4742. PGK gene was used as the internal housekeeping gene. The dashed lines represent the expression rates of the corresponding genes in the control samples grown without T. guizhouense. Boxes without the same letter indicate a significant difference at P < 0.05 (Tukey multiple-comparison test).
Removal of epl1 from Trichoderma spp. improves root colonization.
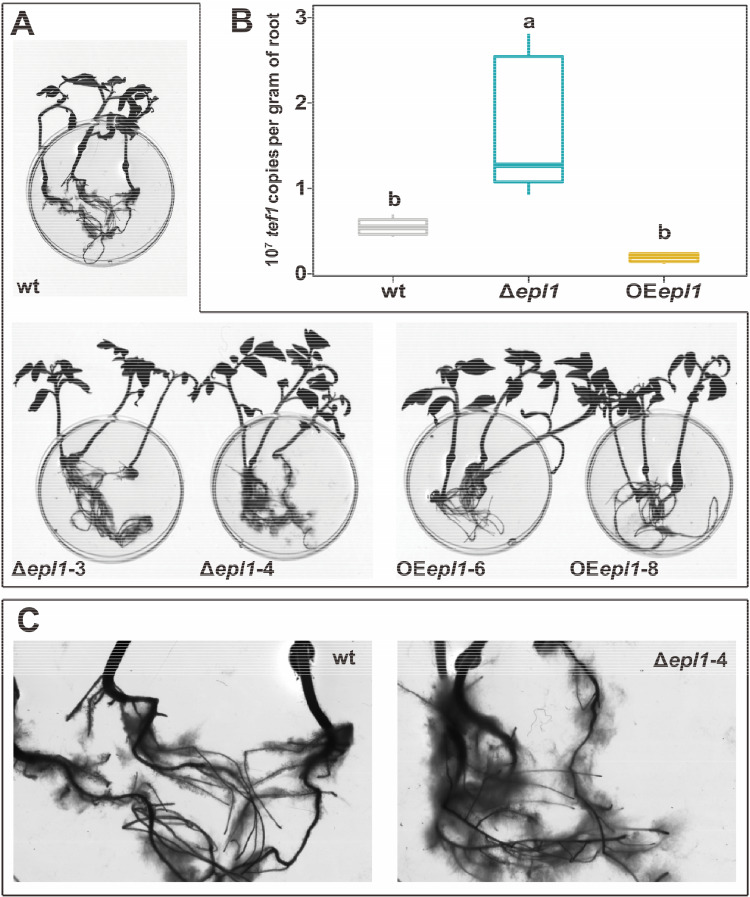
As the plant’s immune response directly mediates the colonization process of fungi in roots (13), tomato roots grown in the hydroponic system were collected for colonization estimation after incubation with Trichoderma spores for 48 h. The copy number of the Trichoderma tef1 gene on roots was determined using qPCR and was used to calculate the index of colonization for each strain. The results shown in Fig. 5 demonstrate that deletion of epl1 significantly (P < 0.05) increased its colonization amount (to ca. 3-fold) compared to that of the wt strain, while overexpression of epl1 had no significant effect on colonization (P > 0.05). It interestingly suggests that the presence of TgEPL1 had a negative effect on the colonization of tomato roots by T. guizhouense. A similar effect was found for ThEPL1 in T. harzianum (shown in File S1).
FIG 5.
Quantitative determination of T. guizhouense colonization on tomato roots. (A) Scanned images of tomato seedlings colonized by T. guizhouense. Diameter of petri plate is 6 cm. (B) A box plot showing the quantification of T. guizhouense tef1 gene copies per gram of root. The values were obtained by RT-qPCR with RNA isolated from the roots (n ≥ 4). Boxes without a same letter indicate a significant difference at P < 0.05 (Tukey multiple-comparison test). (C) A magnified field of the roots colonized by the wild-type strain of T. guizhouense NJAU 4742 (left) and its epl1 deletion mutant (right).
Recombinant TgEPL1 reduces surface hydrophobicity of materials.
In order to test the properties of EPL1, the recombinant pPICZαA::epl1 vector was transformed into the Pichia pastoris KM71H strain, and 51 zeocin resistance-positive transformants were obtained and checked for the right construction using PCR and sequencing. One of the positive transformants was further confirmed for the production of recombinant TgEPL1 proteins. As shown in the SDS-PAGE and Western blotting results (File S2), an extracellular band greater than 15 kDa was found in the P. pastoris fermentation filtrate.
The ability to modify the surface hydrophobicity of EPL1 as a surface-active protein was illustrated by the results of the water contact angle (WCA) measurement with the recombinant TgEPL1. The data shown in Table 3 demonstrate the potential of recombinant TgEPL1 to change the surface hydrophobicity of materials. Specifically, a coating of 10 μM recombinant TgEPL1 on (hydrophobic) poly(ethylene terephthalate) (PET) significantly reduced the surface hydrophobicity, by up to 34%, compared to that of the control, but the effect on glass (hydrophilic) was not significant (P > 0.05). This result assumes the role of EPL1 in modulating the surface hydrophobicity of the host/environment.
TABLE 3.
Surface-modulating property of TgEPL1 protein recombinantly produced in Pichia pastoris
| Surfacea | Mean WCA (°) ± SD forb
: |
|
|---|---|---|
| Control | PpEPL1 | |
| Glass | 31.96 ± 3.16 | 35.54 ± 2.78 |
| PET | 87.06 ± 1.62 | 29.64 ± 2.52* |
PET, poly(ethylene terephthalate).
Surface hydrophobicity was monitored by the water contact angle measurement (WCA). *, a significant difference was found between the values for the EPL1-coated sample and the control at P < 0.05, calculated based on t test.
Recombinant TgEPL1 in vitro prevents root colonization of T. guizhouense and triggers plant immunity.
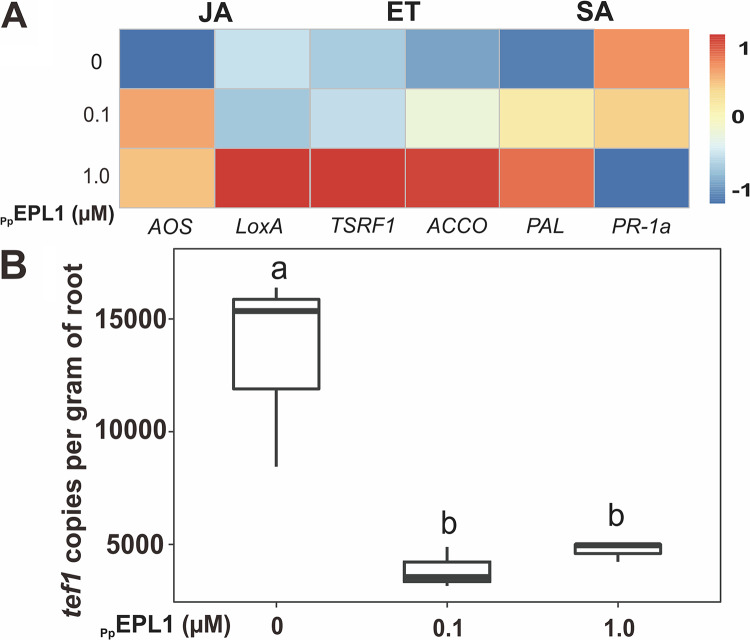
Due to multiple molecular mechanisms, such as bioactive secondary metabolites, enzymes, and peptides, possibly employed by Trichoderma in interacting with plants (25), in vitro tests with the purified protein are necessary to identify the function of a specific gene/protein. Thus, the recombinant TgEPL1 from the P. pastoris fermentation filtrate was purified as described previously (39). Recombinant TgEPL1 (Fig. 6) significantly (P < 0.05) triggered the SA- and JA-mediated defense in tomato seedlings, shown by the upregulation of the related genes PR-1a, PAL, LOX A, and AOS, whereas no elicitation effect of the recombinant TgEPL1 protein on ET-mediated defense was noted. Correspondingly, the addition of the recombinant TgEPL1 protein significantly (P < 0.05) prevented the colonization of T. guizhouense on roots (to ca. 30%) compared to its colonization without the addition of recombinant TgEPL1. Hence, the in vitro test of the purified recombinant TgEPL1 protein supported the hypothesis that EPL1 causes an immune response in plants, which in turn prevents further root colonization by the fungus.
FIG 6.
The impact of recombinant TgEPL1 on the tomato immune system and on root colonization by T. guizhouense. (A) A heat map of relative expression of genes related to immune defense in tomato seedlings treated with different concentrations of the protein. The recombinant TgEPL1 protein was applied at concentrations of 0.1 μM and 1 μM. (B) Quantitative determination of T. guizhouense NJAU 4742 hyphae developing on tomato roots with or without TgEPL1 application. Box plots represent the determination of tef1 gene copies per gram of root, obtained by RT-qPCR (n ≥ 4). Boxes without a same letter indicate a statistically significant difference at P < 0.05 (Tukey multiple-comparison test).
DISCUSSION
The evolutionary and functional genetic analysis of Trichoderma CPs further contributed to the so-called “CP paradox” in fungi (11): together with the previously published studies (6, 8, 18), our results (Fig. 1) showed that none of the so-far-revealed studies of CPs in different fungi (either interactions or development and regulation) sufficiently explained the common features of CPs, such as massive secretion and long evolutionary history in the core genome of most filamentous fungi. For example, the well-documented role of CPs in plant pathogenicity (9, 15, 24) will not be a good predictor for the diversity and evolution of CPs in genomes of fungi that have no known interactions with living plants but are either carnivores (e.g., Beauveria spp. and Metarhizium spp.), fungivores (e.g., Escovopsis spp.), or strict saprotrophs (e.g., Auricularia spp. and Pleurotus spp.). In Trichoderma spp., at least three CP proteins (EPL1, EPL2, and EPL3) were detected in such species as Trichoderma pleuroti, which causes the green mold disease of Pleurotus spp. (40), or Trichoderma reesei, which is rarely isolated from soil (41) and thus has no known root endophytic potential. Although further investigations might reveal herbivorous properties of these Trichoderma spp., the expansion and multiple gene duplications of CPs in the genomes of wood rot fungi (e.g., Auricularia spp., Trametes spp. and Pleurotus spp.) and specialized fungal pathogens like E. weberi, saprotrophic Sphaerobolus spp., and Aspergillus spp., contradicts phytotoxicity as the primary function of CPs while supporting the conclusion of plant virulence factor being a secondary role of CPs in fungi. Because at least one or several CPs are massively secreted under a variety of conditions in many fungi (9, 10, 42), including this study, plants may evolve a mechanism to respond to the presence of fungi by detecting these proteins. It is worth mentioning that plants have evolved several layers of defense strategies to suppress colonization by other organisms, especially fungi (43). For example, Arabidopsis thaliana uses the plant defensin gene (PDF) family, including PDF1, PDF1.2, PDF1.2c, and PDF1.3, to inhibit the growth of a broad range of fungi (44). In another case, plants prevent the penetration of Trichoderma asperellum by depositing dense materials and synthesizing phenolic compounds to restrict fungal dispersion to the epidermis and outer cortex (45). This explains why the CPs from nonphytopathogenic fungi also trigger immune defense responses in plants, as at the early contact stage, plants response to any partner regardless of whether it is a plant-beneficial or pathogenic one. The obtained role in pathogenicity probably resulted in the evolutionary fixation of respective properties and the emergence of the effector or effector-like functions. We hypothesize that in phytopathogenic fungi, the role in virulence developed as a functional exaptation other than the primary role, which remains putative (see below). The results obtained in this study (Fig. 4 and 6; File S1 in the supplemental material) further add to this paradox: EPL1 of the rhizosphere-competent and plant-beneficial Trichoderma spp. prevents the root colonization of the fungus but triggers the immune response of plants.
We also revealed the persistent stabilizing (purifying) natural selection pressure operating on all CP-encoding genes in Trichoderma (Fig. 2), which highlights their functional significance. Moreover, most CPs evolved over numerous events of gene duplication that frequently resulted in the maintenance of several copies in the genome. The comparison of CP diversity in Trichoderma spp. and the phylogenomic chronogram of Hypocreales (37) allows for the conclusion that all paralogous Trichoderma CPs emerged within the last 200 million years but before the more recent evolutionary history of the extant species (last 50 million years) (Fig. 1). However, the recent phylogenetic analysis of plant cell wall-degrading enzymes in Trichoderma revealed that the cellulolytic ability of these fungi is a relatively recent evolutionary achievement that emerged, along with the formation of the ancestors of the genus and clades (20 to 70 million years ago), through multiple cases of lateral gene transfer of the respective genes from plant-pathogenic fungi (31). Moreover, previous ecological surveys also allowed the conclusion that Trichoderma’s endophytic abilities are present in the most evolutionarily advanced species, namely, the opportunistic ones (23, 37), meaning that these fungivorous fungi probably evolved toward interacting with plants. The long evolutionary history of CPs in Pezizomycotina genomes, including the Trichoderma genome, suggests that they also perform another function that is not linked to the herbivorous nature of some species.
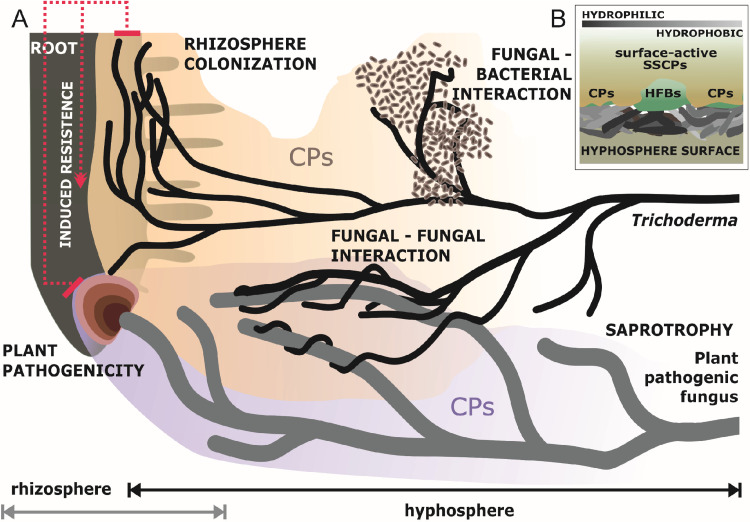
The results of the present work allow the proposal that the ability of CPs to alter surface hydrophobicity best explains their massive and almost unconditional (or constitutive) secretion by fungi in submerged cultures (also seen in references 43 and 46) and their long evolutionary history. Our data allow speculation that CPs contribute to the ability of fungi to modify the substrates where they feed by making surfaces more hydrophilic and thus accessible for enzymes and, subsequently, for the acquisition of nutrients (Fig. 7). This function is probably complementary to those of the other surface-active proteins, namely, HFBs, which are amphiphilic and mainly associated with the hydrophobicity of aerial fungal structures required for reproduction, such as aerial hyphae (47), fruiting bodies (48), and conidial spores. Therefore, it is also possible that other SSCPs play a similar and/or complementary role in the hyphosphere, making the substrate more suitable for absorptive nutrition. The activation of CPs in response to the presence of other organisms (mainly plants but also bacteria and other fungi, as revealed by the results in Table 2 of this study) may correspond to the requirement to outbalance surfactants released by the partner organisms. The recombinant TgEPL1 protein, purified from P. pastoris cultures, retained the surface and biological activity of the original protein (Fig. 5). This result indicates a role of EPL1 in modulating the surface hydrophobicity of the host/environment, which may favor attachment of the fungus that, for example, subsequently helps or prevents an interaction. Due to the presence of both hydrophobic and hydrophilic patches, HFBs are amphipathic and, thus, are able to modulate the hydrophobicity of hydrophobic and hydrophilic surfaces, while EPLs were only known to decrease hydrophobicity and possibly can be described as “hydrophilins” (8). Thus, EPLs are possibly more related to the trophic growth stage of the fungus that requires more hydrophilic surfaces. Thus, further investigation may focus on the role of CPs in the hyphosphere of submerged mycelium and their roles in the nutritional versatility of some opportunistic or specialized fungi.
FIG 7.
The role of CPs in fungal interactions. (A) Shown is a schematic diagram of the involvement of cerato-platanin proteins (CPs) in interactions between Trichoderma hyphae and other organisms (including bacteria, fungi, and plants) (A). Trichoderma modulates the hyphosphere via the massive secretion of surface-active proteins like CPs and HFBs to modify the surface hydrophobicity of the host/substrate, which may favor the attachment and nutrition of the fungus (B).
Moreover, it is interesting to notice that the evolutionary history of CP genes in fungi contains a high frequency of gene duplication. However, the paralogous copies were lost in many species. In this case, it suggests that the CPs evolve by a death-and-birth mechanism (35, 36). This model of evolution assumes new genes appear by gene duplication and some of the duplicated genes are maintained in the genome for a long term, while others could possibly be lost or become nonfunctional (20, 49). Coincidentally, little phenotypic change can be noticed when epl2/sm2 is removed from T. atroviride and T. virens, respectively, except an influence on the fungal-plant interaction, as for epl1 (12). Therefore, different CPs may have overlapping redundant functions for the fungus and behave similarly. CPs from different Trichoderma species clustered into the same phylogenetic clade were found to be statistically similar. Such a long-term conservation of amino acid sequences of the CP subgroups in Trichoderma spp. can be supported by the strong purifying selection, which allows for little mutation, leading the Trichoderma CPs to be recognized by the plant immune system. Hence, it will be meaningful to know the reason why CPs are duplicated in some phytopathogenic and/or some strictly saprotrophic fungi and how CPs evolve in these genomes.
MATERIALS AND METHODS
Microbial strains and plant materials.
T. guizhouense strain NJAU 4742 (28, 31) and T. harzianum CBS 226.95 (type strain of T. harzianum) were used as the wild types throughout this study. Seven other fungi from the TU Collection of Industrial Microorganisms (TUCIM) at TU Wien, Vienna, Austria, namely, Fusarium oxysporum FOC4 (TUCIM 4812), F. fujikuroi (TUCIM 5319), Rhizoctonia solani (TUCIM 3753), Botrytis cinerea (TUCIM 4679), Sclerotinia sclerotiorum, Athelia rolfsii (formerly Sclerotium rolfsii; TUCIM 4076), and Alternaria alternata (TUCIM 3737), were used to perform the fungal-fungal interaction assay with Trichoderma (28). Three bacterial strains from three different genera, Escherichia coli DH5α, Bacillus amyloliquefaciens 9, and Ralstonia solanacearum RS1115, were used to perform the fungal-bacterial interaction assay with T. guizhouense. All strains (listed in Table 1) were, if not otherwise stated, maintained on potato dextrose agar (PDA; BD Difco, USA) at 25°C. Tomato seeds (Solanum lycopersicum cv. HEZUO903) were purchased from Jiangsu Academy of Agricultural Sciences, Nanjing, China.
Cultivation of Trichoderma spp.
Trichoderma spores were collected from 7-day-old PDA cultures and filtrated. For fermentation in shake flasks, 100 μl of Trichoderma spores (106 spores per ml) was cultivated in 100 ml of 30% Murashige and Skoog basal salt mixture (Sigma-Aldrich, USA) supplemented with 1% glucose (MSG) and in minimal medium supplemented with 4% glycerol (MM) (see recipes in reference 50), respectively, at 25°C for 48 h. Culture filtrates were collected for SDS-PAGE.
To study the expression pattern of epls during fungal development, static cultivation was applied. Amounts of 10 μl of the spores described above were inoculated into 10 ml of MSG. Fungal biomass was collected at the stages of (submerged) vegetative growth, aerial hypha formation, and conidiation for RNA extraction.
Generation of epl1 deletion and overexpression mutants of Trichoderma spp.
For constructing the epl1 gene replacement cassette, a 1.2-kb upstream flanking fragment (5′ homologous arm) and a 1.2-kb downstream flanking fragment (3′ homologous arm) of the epl1 gene were amplified from Trichoderma genomic DNA by PCR using the primer pairs e1-upF/e1-upR and e1-dnF/e1-dnR, respectively (all primers used are listed in Table 4). The hygromycin B expression cassette (hph, 2.3 kb) was PCR amplified from the pPcdna1 plasmid using the primer pair e1hph-F/e1hph-R and was inserted between the two flanking arms described above via overlapping PCR with the primers e1-upF/e1-dnR. The amplified fragment (4.7 kb) was then purified and used for the standard polyethylene glycol (PEG)-mediated protoplast transformation for Trichoderma (3). For overexpressing epl1 in Trichoderma spp. under the constitutive promoter Pcdna1, the primer pair OEe1-F/OEe1-R was used to amplify a 1.6-kb fragment, which contains the ORF of epl1 and a 1.2-kb terminator region, from the genomic DNA of the wt strain. The PCR product was purified and fused into the ClaI-digested pPcdna1 plasmid with the In-Fusion HD cloning kit (TaKaRa, Japan), resulting in plasmid pPcdna1::epl1::Tepl1. Protoplast transformation was performed using the PscI-linearized plasmid. Stable transformants were verified by sequencing and maintained on PDA medium containing 200 μg ml−1 of hygromycin B (Thermo Fisher Scientific, USA).
TABLE 4.
Primers used in this study
| Primer | Sequence (5′–3′) | Comment |
|---|---|---|
| e1-upF | CACACTGCGTCATCAATACG | Deletion of epl1 in T. guizhouense NJAU 4742 |
| e1-upR | CATATTGATGTAAGGTAGCTCTCGGATCCCTTGACTATTGTGTAGAGTG | |
| e1-dnF | TATTCCATCTAAGCCATAGTACCCTCGAGATTGCATTGTGGTATATGGC | |
| e1-dnR | GGAGATATAATCTGGCAATG | |
| e1hph-F | GGATCCGAGAGCTACCTTACAT | |
| e1hph-R | CTCGAGGGTACTATGGCTTAGAT | |
| The1-upF | TCGGCACTGCTTCGCACTAA | Deletion of epl1 in T. harzianum CBS 226.95 |
| The1-upR | CATATTGATGTAAGGTAGCTCTCGGATCCATATCAACGAAAGTCGAGGTGAGT | |
| The1-dnF | TATTCCATCTAAGCCATAGTACCCTCGAGATTGTGGTATATGGCGGGATT | |
| The1-dnR | CGATGTAAGTCATCACCGTTCTACT | |
| The1hph-F | Refer to e1hph-F | |
| The1hph-R | Refer to e1hph-R | |
| e1-F2 | GAAAGCAAGCCTACCAAGCTACCT | Verification of epl1 deletion in T. guizhouense NJAU 4742 |
| e1-R2 | AGTACAACCTAACAGCTGAGCACG | |
| e1-F3 | AGATTCCTCGCTTCCCATACAT | |
| e1-R3 | TAGATGGTGTGGCCGCTGTA | |
| e1-F4 | AGAAGGGCGTCGAGCATTGT | |
| e1-R4 | AAGGAAGGCTTGAGGTACTTGG | |
| The1-F2 | GTATATGCTGGTACACGCCGTC | Verification of epl1 deletion in T. harzianum CBS 226.95 |
| The1-R2 | Refer to e1-R2 | |
| The1-F3 | TTCACTGCTGCCGTTTCTGC | |
| The1-R3 | Refer to e1-R3 | |
| The1-F4 | Refer to e1-F4 | |
| The1-R4 | CATGTGTAATGACCTGTGGCTAC | |
| OEe1-F | AACAACTTCTCTCATCGATATGCAATTGTCCAGCCTCTTC | Construction of OEepl1 vector for T. guizhouense NJAU 4742 |
| OEe1-R | CCTGCAGGTCGACATCGATTGGCAGCGGAGAGGGTTAT | |
| ThOEe1-F | AACAACTTCTCTCATCGATATGCAATTGTCCAGCCTCTTC | Construction of OEepl1 vector for T. harzianum CBS 226.95 |
| ThOEe1-R | CCTGCAGGTCGACATCGATCGATGTAAGTCATCACCGTTCTACT | |
| OEe1-F2 | ATGCAAGGTCGATTCCAATCAT | Verification of OEepl1 in Trichoderma spp. |
| OEe1-R2 | Refer to e1-R3 | |
| qtef1-F | TACAAGATCGGTGGTATTGGAACA | Quantification of tef1 in T. guizhouense NJAU 4742 and T. harzianum CBS 226.95 |
| qtef1-R | AGCTGCTCGTGGTGCATCTC | |
| qepl1-F | Refer to e1-F3 | Quantification of epl1 in T. guizhouense NJAU 4742 |
| qepl1-R | Refer to e1-R3 | |
| qepl2-F | GCTATGATGACCCTTCTCGTTCT | Quantification of epl2 in T. guizhouense NJAU 4742 |
| qepl2-R | TATAACAGGCTCCGCATTGTG | |
| qepl3-F | AAGATGCCGTTGGCTTCATT | Quantification of epl3 in T. guizhouense NJAU 4742 |
| qepl3-R | CGATGGCAAGGAGTGATATAGTTT | |
| Thqepl1-F | TCAACGTCGTCTCCTGCTCC | Quantification of epl1 in T. harzianum CBS 226.95 |
| Thqepl1-R | Refer to e1-R3 | |
| Thqepl2-F | TTACTGCTGCTATCCTCTCTGTGG | Quantification of epl2 in T. harzianum CBS 226.95 |
| Thqepl2-R | TGGTGATGAGGCCATTGGG | |
| Thqepl3-F | AGCAGTTGTTGTCCCTGTTACC | Quantification of epl3 in T. harzianum CBS 226.95 |
| Thqepl3-R | ATGAACCCGATATTCTTTCTCCA | |
| Ppepl1-F | GAAGAAGGGGTATCTCTCGAGAAAAGAGATACCGTCTCGTATGATACCGG | Construction of Ppepl1 vector for Pichia pastoris KM71H |
| Ppepl1-R | GAGTTTTTGTTCTAGAATAAGACCGCAGTTCTTGACAGC | |
| PpAOX1-F2 | GACTGGTTCCAATTGACAAGC | Verification of epl1-expressing mutants for Pichia |
| PpAOX1-R2 | GCAAATGGCATTCTGACATCC | |
Interaction assays between Trichoderma spp. and other organisms.
To test the effect of plants on epl expression, the wild-type strain of Trichoderma was cocultured with three 15-day-old tomato seedlings on an MSG plate, as described in our previous work (50), using the solo-cultured Trichoderma as the control.
The fungal-fungal interaction was assessed by dual confrontation assays. Agar plugs of fresh Trichoderma culture and the partner fungus, which were pregrown on PDA at 25°C for 72 h, were placed on opposite poles of a PDA plate. Images of each plate were recorded by a Canon EOS 70D camera after incubation at 25°C in darkness for 14 days. Fungal biomass was collected from the interacting and solo sides after connection for 24 h (as shown in Fig. S3 in the supplemental material).
Three bacterial strains were selected to investigate the fungal-bacterial interaction. An amount of 100 μl of each bacterial suspension (ca. 103 cell per ml) was spread on a PDA plate, and two fresh culture plugs from one Trichoderma strain were inoculated onto the same plate. The cocultured plates were incubated at 25°C in darkness for 3 days, and images were recorded. Fungal biomass was collected for RNA extraction from the interacting (after connection for 24 h) and the solo-cultured samples (Fig. S4).
RNA extraction and RT-qPCR assay.
Total RNAs were extracted using the RNeasy plant minikit (Qiagen, Germany) according to the manufacturer’s instructions. cDNAs were synthesized using the RevertAid first-strand cDNA kit (Thermo Scientific, USA) with the oligo(dT)18 primer. RT-qPCR was performed to determine the expression of epl genes, with the reaction mixture comprised of 10 μl of iQ SYBR green PCR supermix (Bio-Rad, USA), 0.5 μl of each primer (10 μM), 0.5 μl of cDNA (ca. 100 ng μl−1), and water to 25 μl. The thermal program was set up in a qTOWER real-time PCR system (Jena Analytics, Germany) as follows: one cycle of 6 min at 95°C, followed by 40 cycles of denaturation for 30 s at 95°C and annealing for 60 s at 60°C, with a melting curve from 60°C to 95°C.
Root colonization and plant immunity response assays.
Twenty-one-day-old tomato seedlings were cultivated in a hydroponic system containing 40 ml of 25% MS basal salt mix (pH 6.5), and 0.4 ml of spores (106 ml−1) from each Trichoderma strain was inoculated into the hydroponic system. After incubation at 25°C under cycled-light conditions (light/dark, 12 h/12 h) for 48 h, quantification of Trichoderma fungi colonized on tomato roots was performed by RT-qPCR with total RNAs isolated from the interacting organisms. A 137-bp fragment of the tef1 exon was cloned into a pMD 19-T vector (TaKaRa, Japan) and then used as a DNA standard. Templates for the standard curve were made using 10-fold serial dilutions of the recombinant plasmid harboring one copy of the tef1 gene fragment. The amount of colonized Trichoderma was then calculated based on the standard curve and described as the copy number of tef1 per gram of root.
The expression of genes corresponding to different plant defense pathways was examined for the tomato roots colonized by mutants, using the noncolonized ones as a control. The qPCR was set up as described above.
Production of the recombinant TgEPL1 in Pichia pastoris.
The EasySelect Pichia expression kit (Invitrogen, USA) was used to express the TgEPL1 protein in P. pastoris strain KM71H yeast according to the manufacturer’s instructions. The epl1 coding region from the end of the predicted signal peptide to the stop codon was amplified by PCR (CloneAmp HiFi PCR premix; Clontech, USA) from the cDNA of the wt T. guizhouense with the primer pair PPe1-F/PPe1-R. The signal peptide was excluded using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) (51). The cloned fragment was inserted into the plasmid of pPICZαA between the XhoI and XbaI sites using the In-Fusion HD cloning kit (Clontech, USA). The recombinant protein coded by this construction harbors the yeast α-factor signal sequence at the N terminus and a 6×His epitope at the C terminus. The resulting plasmid, pPICZαA::epl1, was linearized with Sac I and then transformed into Pichia cells by pulsed cell transformation, and one of the Western blot-positive transformants was chosen to obtain the recombinant TgEPL1 protein (see Pichia fermentation in reference 28). The protein was purified via affinity chromatography using His-Talon cobalt-loaded resins (Clontech, USA) and redissolved in high-performance liquid chromatography-grade (HPLC) water.
Protein biochemical and biophysical assays.
The protein concentration of the fungal culture filtrates was quantified with a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, USA), using bovine serum album (BSA) as the standard, according to the manufacturer’s instructions. Protein samples were loaded into a 15% polyacrylamide gel and run with a constant voltage (200 V) for 1 h, followed by silver staining using the SilverQuest silver staining kit (Life Technologies, Germany). Qualitative identification of the protein on SDS-PAGE gel was performed by using the quantification tool of the software Image Lab 3.0 with default parameters. The protein identification of the four targeted protein bands (indicated by dotted lines in Fig. 3) was performed by Luming Biotechnology, Inc. (Shanghai, China), using MALDI-TOF/TOF mass spectrometry. Immunological visualization of the recombinant TgEPL1 was carried out by using a mouse anti‐His tag-horseradish peroxidase (HRP) antibody (Genescript, USA) following the protocol supplied with the One‐Hour Western standard kit (GenScript, China) for Western blotting.
The water contact angle (WCA) measurement for surface hydrophobicity determination was performed by using a Krüss EasyDrop DSA20E (Krüss GmbH, Germany). Purified proteins and surface materials (glass and PET) were prepared as described previously (52). PpEPL1 protein coating was applied at a concentration of 10 μM.
In vitro bioactivity test of recombinant TgEPL1.
The colonization test was further performed for the wt strain with the addition of two concentrations of pure Pichia-produced EPL1 (recombinant TgEPL1). As described above, tomato seedlings were cultivated in a hydroponic system with Trichoderma spores, and recombinant TgEPL1 was applied at concentrations of 0.1 μM and 1 μM, using water as the control. The colonization rate and elicitation of plant defense response were then compared by RT-qPCR between the EPL1-treated roots and the nontreated ones.
Genome mining and phylogenetic analysis.
A total of 157 fungal genomes, including 150 from Ascomycota and 7 from Basidiomycota, deposited in the National Center for Biotechnology Information (NCBI) and the Joint Genome Institute (JGI) databases, was used as the sequence resource. The protein sequences of the three reported CPs (GenBank accession numbers XP_013937770, XP_013944228, and XP_013937568) from T. atroviride IMI 206040 (12, 42) were used in the BLAST query of each fungal genome. Hits with an E value of <10−4 were retrieved. Sequences were aligned by using MUSCLE integrated in AliView 1.23 (53, 54), and the CP‐specific cysteine sequence pattern of ‐C‐C X X C‐C‐ (X represents any possible amino acid) was manually verified during alignment. N and C termini were trimmed manually when necessary. Maximum-likelihood (ML) phylogenetic analysis was constructed by using IQ-TREE 1.6.12 (no. of ultrafast bootstraps, 1,000) (55) with 262 sites of the CP sequences. The amino acid substitution model was selected with ModelFinder, which is integrated into the IQ-TREE program, according to the Bayesian information criterion (BIC).
Natural selection pressure assay and tests for horizontal gene transfer.
To identify specific branches evolving under natural positive selection pressure, EasyCodeML version 1.21 (38) was used to calculate the ratio of nonsynonymous/synonymous substitution rates (ω = dN/dS). Inference of lateral gene transfer, gene loss (GL), and gene duplication (GD) was performed by using NOTUNG (33) as described in our previous research (31), with modifications. Briefly, an edge weight threshold of 0.9 was applied, with assigned costs of HGT, GD, and GL at rates of 9, 3, and 1, respectively. The predicted HGT events were also compared to those revealed using T-Rex (34).
Statistical analysis.
The relative expression levels (fold changes) of genes of interest were calculated according to the cycle threshold (2−ΔΔCT) method, using tef1 as the housekeeping gene (56). Data regarding the gene expression and the absolute gene copy number were obtained, if not otherwise stated, from at least three biological replicates. The line chart, box plot, and heat map representations of data were obtained by using R (version 3.2.2). The results represent the mean values and standard deviations and were statistically analyzed by analysis of variance (ANOVA) and Tukey multiple-comparison test or t test with a P value of <0.05 using STATISTICA 6 (StatSoft, USA).
Data availability.
The protein sequences of EPL1 from T. guizhouense NJAU 4742 and T. harzianum CBS 226.95 are available in NCBI GenBank under accession numbers OPB44018 and XP_024774373, respectively. The genomes of Trichoderma spp. used in this paper for in silico work were retrieved from the NCBI or JGI database with the accession numbers listed in Table S1. Accession numbers of CPs that were used for LGT analysis are presented in Fig. 1 and 2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gunseli Bayram Akcapinar (Sabanci University, Istanbul, Turkey) for cooperation on the production of recombinant proteins in Pichia pastoris. We thank the JGI Community Sequencing Program, proposal 1966-CSP2016, and Matteo Lorito, Sheridan Woo, Maria R. Ercolano, and Francesco Vinale (University of Naples Federico II, Italy) for permission to explore the diversity of epl genes in the genomes of T22 and M10 strains of Trichoderma. Equally, we appreciate the permission of Peter Urban (University of Pecs, Hungary) and Laszlo Kredics (University of Szeged, Hungary) for their agreement to use sequences of epl genes present in genomes of green mold Trichoderma spp. deposited in GenBank.
This work was supported by grants from the National Natural Science Foundation of China (grant number KJQN201920), Ministry of Science and Technology of Jiangsu Province (grant number BK20180533), and China Postdoctoral Science Foundation (grant number 2018M630567), all to F.C. The work in Vienna (Austria) was supported by the Austrian Science Fund (FWF) via grant number P25613-B20 and the Vienna Science and Technology Fund (WWTF) via grant number LS13-048, both to I.S.D.
F.C. and I.S.D. conceived of and designed the study. R.G., F.C., M.D., S.J., Z.Z., K.C., and I.S.D. carried out the experiments. F.C. and R.G. carried out the data analysis and prepared the supplemental material. R.G. F.C., and I.S.D. prepared the figures. F.C. and I.S.D. wrote and revised the manuscript with comments from Q.S. All authors read and approved the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, Hacquard S, Hervé V, Labbé J, Lastovetsky OA, Mieszkin S, Millet LJ, Vajna B, Junier P, Bonfante P, Krom BP, Olsson S, van Elsas JD, Wick LY. 2018. Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev 42:335–352. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 2.Hiscox J, O’Leary J, Boddy L. 2018. Fungus wars: basidiomycete battles in wood decay. Stud Mycol 89:117–124. doi: 10.1016/j.simyco.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Miao Y, Rahimi MJ, Zhu H, Steindorff A, Schiessler S, Cai F, Pang G, Chenthamara K, Xu Y, Kubicek CP, Shen Q, Druzhinina IS. 2019. Guttation capsules containing hydrogen peroxide: an evolutionarily conserved NADPH oxidase gains a role in wars between related fungi: the role of hydrogen peroxide in fungal wars. Environ Microbiol 21:2644–2658. doi: 10.1111/1462-2920.14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortíz-Castro R, Contreras-Cornejo HA, Macías-Rodríguez L, López-Bucio J. 2009. The role of microbial signals in plant growth and development. Plant Signal Behav 4:701–712. doi: 10.4161/psb.4.8.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krijger J-J, Thon MR, Deising HB, Wirsel SG. 2014. Compositions of fungal secretomes indicate a greater impact of phylogenetic history than lifestyle adaptation. BMC Genomics 15:722. doi: 10.1186/1471-2164-15-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Kovalchuk A, Keriö S, Asiegbu FO. 2013. Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 105:1479–1488. doi: 10.3852/13-115. [DOI] [PubMed] [Google Scholar]
- 7.Pazzagli L, Cappugi G, Manao G, Camici G, Santini A, Scala A. 1999. Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J Biol Chem 274:24959–24964. doi: 10.1074/jbc.274.35.24959. [DOI] [PubMed] [Google Scholar]
- 8.Gaderer R, Bonazza K, Seidl-Seiboth V. 2014. Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol 98:4795–4803. doi: 10.1007/s00253-014-5690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frías M, González C, Brito N. 2011. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol 192:483–495. doi: 10.1111/j.1469-8137.2011.03802.x. [DOI] [PubMed] [Google Scholar]
- 10.Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM. 2006. Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact 19:838–853. doi: 10.1094/MPMI-19-0838. [DOI] [PubMed] [Google Scholar]
- 11.Baccelli I. 2015. Cerato-platanin family proteins: one function for multiple biological roles? Front Plant Sci 5:769. doi: 10.3389/fpls.2014.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaderer R, Lamdan NL, Frischmann A, Sulyok M, Krska R, Horwitz BA, Seidl-Seiboth V. 2015. Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol 15:2. doi: 10.1186/s12866-014-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bent AF, Mackey D. 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 14.Barsottini MRDO, de Oliveira JF, Adamoski D, Teixeira P, do Prado PFV, Tiezzi HO, Sforça ML, Cassago A, Portugal RV, de Oliveira PSL, de M Zeri AC, Dias SMG, Pereira GAG, Ambrosio A. 2013. Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol Plant Microbe Interact 26:1281–1293. doi: 10.1094/MPMI-05-13-0148-R. [DOI] [PubMed] [Google Scholar]
- 15.Yang G, Tang L, Gong Y, Xie J, Fu Y, Jiang D, Li G, Collinge DB, Chen W, Cheng J. 2018. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol 217:739–755. doi: 10.1111/nph.14842. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Nowrousian M, Kupfer D, Colot HV, Berrocal-Tito G, Lai H, Bell-Pedersen D, Roe BA, Loros JJ, Dunlap JC. 2001. Analysis of expressed sequence tags from two starvation, time-of-day-specific libraries of Neurospora crassa reveals novel clock-controlled genes. Genetics 157:1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson LM, Idnurm A, Howlett BJ. 2002. Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus. Mol Plant Pathol 3:487–493. doi: 10.1046/j.1364-3703.2002.00144.x. [DOI] [PubMed] [Google Scholar]
- 18.de Oliveira AL, Gallo M, Pazzagli L, Benedetti CE, Cappugi G, Scala A, Pantera B, Spisni A, Pertinhez TA, Cicero DO. 2011. The structure of the elicitor cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J Biol Chem 286:17560–17568. doi: 10.1074/jbc.M111.223644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frischmann A, Neudl S, Gaderer R, Bonazza K, Zach S, Gruber S, Spadiut O, Friedbacher G, Grothe H, Seidl-Seiboth V. 2013. Self-assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride. J Biol Chem 288:4278–4287. doi: 10.1074/jbc.M112.427633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubicek CP, Baker SE, Gamauf C, Kenerley CM, Druzhinina IS. 2008. Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol Biol 8:4. doi: 10.1186/1471-2148-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salas-Marina MA, Isordia-Jasso MI, Islas-Osuna MA, Delgado-Sánchez P, Jiménez-Bremont JF, Rodríguez-Kessler M, Rosales-Saavedra MT, Herrera-Estrella A, Casas-Flores S. 2015. The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front Plant Sci 6:77. doi: 10.3389/fpls.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes EV, Ulhoa CJ, Cardoza RE, Silva RN, Gutiérrez S. 2017. Involvement of Trichoderma harzianum Epl-1 protein in the regulation of Botrytis virulence- and tomato defense-related genes. Front Plant Sci 8:880. doi: 10.3389/fpls.2017.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. 2011. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol 9:749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 24.Jeong JS, Mitchell TK, Dean RA. 2007. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol Lett 273:157–165. doi: 10.1111/j.1574-6968.2007.00796.x. [DOI] [PubMed] [Google Scholar]
- 25.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 26.Cai F, Yu G, Wang P, Wei Z, Fu L, Shen Q, Chen W. 2013. Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum. Plant Physiol Biochem 73:106–113. doi: 10.1016/j.plaphy.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Li R-X, Cai F, Pang G, Shen Q-R, Li R, Chen W. 2015. Solubilisation of phosphate and micronutrients by Trichoderma harzianum and its relationship with the promotion of tomato plant growth. PLoS One 10:e0130081. doi: 10.1371/journal.pone.0130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Bayram Akcapinar G, Atanasova L, Rahimi MJ, Przylucka A, Yang D, Kubicek CP, Zhang R, Shen Q, Druzhinina IS. 2016. The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self-defence: NMP1 of the fungicidal mould Trichoderma guizhouense. Environ Microbiol 18:580–597. doi: 10.1111/1462-2920.12966. [DOI] [PubMed] [Google Scholar]
- 29.Grujić M, Dojnov B, Potočnik I, Atanasova L, Duduk B, Srebotnik E, Druzhinina IS, Kubicek CP, Vujčić Z. 2019. Superior cellulolytic activity of Trichoderma guizhouense on raw wheat straw. World J Microbiol Biotechnol 35:194. doi: 10.1007/s11274-019-2774-y. [DOI] [PubMed] [Google Scholar]
- 30.Schönknecht G, Weber APM, Lercher MJ. 2014. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution: insights and perspectives. BioEssays 36:9–20. doi: 10.1002/bies.201300095. [DOI] [PubMed] [Google Scholar]
- 31.Druzhinina IS, Chenthamara K, Zhang J, Atanasova L, Yang D, Miao Y, Rahimi MJ, Grujic M, Cai F, Pourmehdi S, Salim KA, Pretzer C, Kopchinskiy AG, Henrissat B, Kuo A, Hundley H, Wang M, Aerts A, Salamov A, Lipzen A, LaButti K, Barry K, Grigoriev IV, Shen Q, Kubicek CP. 2018. Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLoS Genet 14:e1007322. doi: 10.1371/journal.pgen.1007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barreiro C, Gutiérrez S, Olivera ER. 2019. Fungal horizontal gene transfer: a history beyond the phylogenetic kingdoms, p 315–336. In Villa TG, Viñas M (ed), Horizontal gene transfer. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 33.Chen K, Durand D, Farach-Colton M. 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- 34.Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res 40:W573–W579. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nei M, Rogozin IB, Piontkivska H. 2000. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci U S A 97:10866–10871. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouché S, Plissonneau C, Croll D. 2018. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr Opin Microbiol 46:34–42. doi: 10.1016/j.mib.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Kubicek CP, Steindorff AS, Chenthamara K, Manganiello G, Henrissat B, Zhang J, Cai F, Kopchinskiy AG, Kubicek EM, Kuo A, Baroncelli R, Sarrocco S, Noronha EF, Vannacci G, Shen Q, Grigoriev IV, Druzhinina IS. 2019. Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20:485. doi: 10.1186/s12864-019-5680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao F, Chen C, Arab DA, Du Z, He Y, Ho S. 2019. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol 9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Przylucka A, Akcapinar GB, Bonazza K, Mello-de-Sousa TM, Mach-Aigner AR, Lobanov V, Grothe H, Kubicek CP, Reimhult E, Druzhinina IS. 2017. Comparative physiochemical analysis of hydrophobins produced in Escherichia coli and Pichia pastoris. Colloids Surf B Biointerfaces 159:913–923. doi: 10.1016/j.colsurfb.2017.08.058. [DOI] [PubMed] [Google Scholar]
- 40.Marik T, Urbán P, Tyagi C, Szekeres A, Leitgeb B, Vágvölgyi M, Manczinger L, Druzhinina IS, Vágvölgyi C, Kredics L. 2017. Diversity profile and dynamics of peptaibols produced by green mould Trichoderma species in interactions with their hosts Agaricus bisporus and Pleurotus ostreatus. Chem Biodiversity 14:e1700033. doi: 10.1002/cbdv.201700033. [DOI] [PubMed] [Google Scholar]
- 41.Druzhinina IS, Kubicek CP. 2016. Familiar stranger: ecological genomics of the model saprotroph and industrial enzyme producer Trichoderma reesei breaks the stereotypes. Adv Appl Microbiol 95:69–147. doi: 10.1016/bs.aambs.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Seidl V, Marchetti M, Schandl R, Allmaier G, Kubicek CP. 2006. Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J 273:4346–4359. doi: 10.1111/j.1742-4658.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 43.Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 44.Thomma B, Cammue B, Thevissen K. 2002. Plant defensins. Planta 216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- 45.Yedidia I, Benhamou N, Kapulnik Y, Chet I. 2000. Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol Biochem 38:863–873. doi: 10.1016/S0981-9428(00)01198-0. [DOI] [Google Scholar]
- 46.Baroncelli R, Piaggeschi G, Fiorini L, Bertolini E, Zapparata A, Pè ME, Sarrocco S, Vannacci G. 2015. Draft whole-genome sequence of the biocontrol agent Trichoderma harzianum T6776. Genome Announc 3:e00647-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Askolin S, Penttilä M, Wösten HAB, Nakari-Setälä T. 2005. The Trichoderma reesei hydrophobin genes hfb1 and hfb2 have diverse functions in fungal development. FEMS Microbiol Lett 253:281–288. doi: 10.1016/j.femsle.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 48.Yamada M, Sakuraba S, Shibata K, Inatomi S, Okazaki M, Shimosaka M. 2005. Cloning and characterization of a gene coding for a hydrophobin, Fv-hyd1, specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes. Appl Microbiol Biotechnol 67:240–246. doi: 10.1007/s00253-004-1776-2. [DOI] [PubMed] [Google Scholar]
- 49.Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Przylucka A, Akcapinar GB, Chenthamara K, Cai F, Grujic M, Karpenko J, Livoi M, Shen Q, Kubicek CP, Druzhinina IS. 2017. HFB7—a novel orphan hydrophobin of the Harzianum and Virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genet Biol 102:63–76. doi: 10.1016/j.fgb.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 52.Espino-Rammer L, Ribitsch D, Przylucka A, Marold A, Greimel KJ, Herrero Acero E, Guebitz GM, Kubicek CP, Druzhinina IS. 2013. Two novel class II hydrophobins from Trichoderma spp. stimulate enzymatic hydrolysis of poly(ethylene terephthalate) when expressed as fusion proteins. Appl Environ Microbiol 79:4230–4238. doi: 10.1128/AEM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Chaverri P, Branco-Rocha F, Jaklitsch W, Gazis R, Degenkolb T, Samuels GJ. 2015. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107:558–590. doi: 10.3852/14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang N, Yang D, Wang D, Miao Y, Shao J, Zhou X, Xu Z, Li Q, Feng H, Li S, Shen Q, Zhang R. 2015. Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16:685. doi: 10.1186/s12864-015-1825-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Y, Hou Y, Huang D, Hao Z, Wang X, Wei Z, Jousset A, Tan S, Xu D, Shen Q, Xu Y, Friman V-P. 2017. Application of biochar reduces Ralstonia solanacearum infection via effects on pathogen chemotaxis, swarming motility, and root exudate adsorption. Plant Soil 415:269–281. doi: 10.1007/s11104-016-3159-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein sequences of EPL1 from T. guizhouense NJAU 4742 and T. harzianum CBS 226.95 are available in NCBI GenBank under accession numbers OPB44018 and XP_024774373, respectively. The genomes of Trichoderma spp. used in this paper for in silico work were retrieved from the NCBI or JGI database with the accession numbers listed in Table S1. Accession numbers of CPs that were used for LGT analysis are presented in Fig. 1 and 2.