Microbial communities impact life on Earth in profound ways, including driving global nutrient cycles and influencing human health and disease. These community functions depend on the interactions that resident microbes have with the environment and each other. Thus, identifying genes that influence these interactions will aid the management of natural communities and the use of microbial consortia as biotechnology. Here, we identified genes that influenced Escherichia coli fitness during cooperative growth with a mutualistic partner, Rhodopseudomonas palustris. Although this mutualism centers on the bidirectional exchange of essential carbon and nitrogen, E. coli fitness was positively and negatively affected by genes involved in diverse cellular processes. Furthermore, we discovered an unexpected purine cross-feeding interaction. These results contribute knowledge on the genetic foundation of a microbial cross-feeding interaction and highlight that unanticipated interactions can occur even within engineered microbial communities.
KEYWORDS: cross-feeding, Escherichia coli, Rhodopseudomonas, coculture, fermentation, microbial ecology, nitrogen metabolism, purines, synthetic ecology, transposon sequencing
ABSTRACT
Microbial interactions abound in natural ecosystems and shape community structure and function. Substantial attention has been given to cataloging mechanisms by which microbes interact, but there is a limited understanding of the genetic landscapes that promote or hinder microbial interactions. We previously developed a mutualistic coculture pairing Escherichia coli and Rhodopseudomonas palustris, wherein E. coli provides carbon to R. palustris in the form of glucose fermentation products and R. palustris fixes N2 gas and provides nitrogen to E. coli in the form of NH4+. The stable coexistence and reproducible trends exhibited by this coculture make it ideal for interrogating the genetic underpinnings of a cross-feeding mutualism. Here, we used random barcode transposon sequencing (RB-TnSeq) to conduct a genome-wide search for E. coli genes that influence fitness during cooperative growth with R. palustris. RB-TnSeq revealed hundreds of genes that increased or decreased E. coli fitness in a mutualism-dependent manner. Some identified genes were involved in nitrogen sensing and assimilation, as expected given the coculture design. The other identified genes were involved in diverse cellular processes, including energy production and cell wall and membrane biogenesis. In addition, we discovered unexpected purine cross-feeding from R. palustris to E. coli, with coculture rescuing growth of an E. coli purine auxotroph. Our data provide insight into the genes and gene networks that can influence a cross-feeding mutualism and underscore that microbial interactions are not necessarily predictable a priori.
IMPORTANCE Microbial communities impact life on Earth in profound ways, including driving global nutrient cycles and influencing human health and disease. These community functions depend on the interactions that resident microbes have with the environment and each other. Thus, identifying genes that influence these interactions will aid the management of natural communities and the use of microbial consortia as biotechnology. Here, we identified genes that influenced Escherichia coli fitness during cooperative growth with a mutualistic partner, Rhodopseudomonas palustris. Although this mutualism centers on the bidirectional exchange of essential carbon and nitrogen, E. coli fitness was positively and negatively affected by genes involved in diverse cellular processes. Furthermore, we discovered an unexpected purine cross-feeding interaction. These results contribute knowledge on the genetic foundation of a microbial cross-feeding interaction and highlight that unanticipated interactions can occur even within engineered microbial communities.
INTRODUCTION
Within every ecosystem, microbial cells interact with both the environment and each other, and these interactions shape ecosystem functions (1). Microbial interactions are diverse and include nutrient competition and cross-feeding (2), cell-cell signaling (3), adhesion (4), and chemical warfare (5). Unfortunately, the identification of interaction mechanisms has far outpaced our understanding of the genetic and physiological bases of microbial interactions.
Much knowledge on microbial interactions has come from synthetic communities (i.e., cocultures) that offer simplicity and tractability over natural communities (1, 6). Our lab previously developed an anaerobic coculture pairing fermentative Escherichia coli with the N2-fixing, photoheterotroph Rhodopseudomonas palustris (7). In this coculture, the sole carbon source is glucose, which R. palustris cannot utilize, and the sole nitrogen source is N2 gas, which E. coli cannot utilize. Consequently, each species depends on the other for an essential nutrient: R. palustris gets carbon in the form of E. coli fermentation products, and E. coli gets nitrogen in the form of NH4+ that is excreted by an engineered R. palustris strain (Nx) (Fig. 1A). The stable coexistence and reproducible trends (7–9) make this coculture ideal for investigating the genetic and physiological characteristics that influence a cross-feeding mutualism.
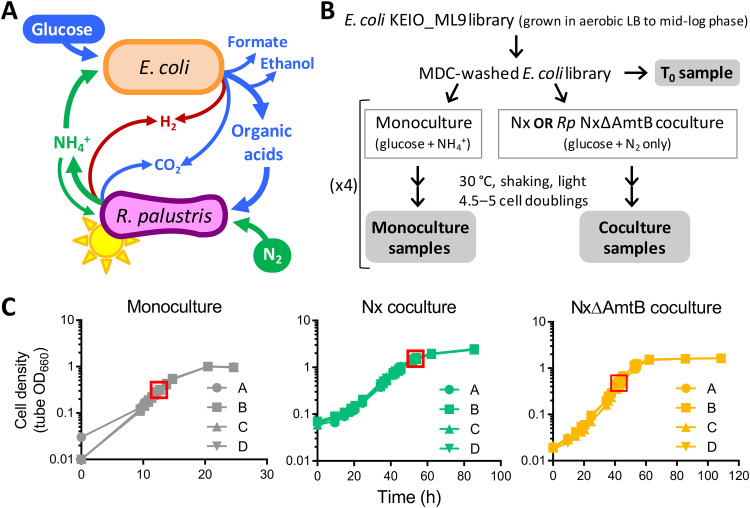
FIG 1.
Overview of RB-TnSeq screen for E. coli genes that influence fitness during mutualistic growth with R. palustris. (A) Mutualistic growth between E. coli and R. palustris requires carbon transfer, in the form of glucose fermentation products, from E. coli to R. palustris and nitrogen transfer, in the form of NH4+, from R. palustris to E. coli. (B) Experimental design. There was one T0 sample and four biological replicates for all mono- and cocultures. Gray bubbles indicate samples that were used for BarSeq. (C) Quadruplicate growth curves of KEIO_ML9 monocultures or cocultures with R. palustris Nx or R. palustris NxΔAmtB. Red boxes indicate sampling points. Because Nx cocultures were harvested at densities above the linear range of the spectrophotometer, growth beyond a tube OD (i.e., measured in the culture tube) of 1.0 was simultaneously monitored in cuvettes using diluted samples to ensure cultures were growing exponentially at harvest.
Recently, our lab used transcriptome sequencing (RNA-Seq) and proteomics to interrogate how cross-feeding impacts the physiology of each species (10). This study revealed that the E. coli NtrC-mediated nitrogen starvation response (NSR) is crucial for fitness in coculture (10). However, RNA-Seq does not necessarily identify genes that confer fitness under the tested conditions. Indeed, several highly upregulated genes did not contribute to E. coli fitness in coculture (10). One approach for directly identifying fitness determinants is transposon sequencing (Tn-Seq), which screens for the fitness of millions of individual transposon mutants simultaneously using next-generation sequencing (11). Random barcode Tn-Seq (RB-TnSeq) uses random DNA barcodes to uniquely identify each Tn insertion site, thereby reducing the sample preparation needed for fitness analysis (12). RB-TnSeq has been successfully used to identify genetic fitness determinants in numerous bacteria under diverse conditions (13–16), including in multispecies communities (17).
Here, we used RB-TnSeq to identify E. coli genes that affected fitness during cooperative growth with R. palustris. We identified numerous genes that had negligible impact on E. coli fitness in monoculture but promoted or reduced E. coli fitness in coculture. These mutualism-dependent fitness determinants were involved in diverse physiological processes ranging from amino acid metabolism to signal transduction to membrane biogenesis. Unexpectedly, we also identified E. coli genes that were important for monoculture fitness but dispensable in coculture, including genes for de novo purine synthesis. These results revealed an unprompted cross-feeding interaction that was not predictable a priori. Overall, our study provides insight into E. coli-R. palustris interactions and lays the groundwork for understanding the genetic underpinnings of a cross-feeding mutualism. In addition, our results highlight that microbes can interact in unanticipated and potentially covert manners, even in engineered communities.
RESULTS AND DISCUSSION
Experimental setup of RB-TnSeq screen for E. coli fitness determinants in coculture.
Previously, Wetmore et al. (12) generated and characterized an E. coli RB-TnSeq library in the K-12 parent strain of the KEIO deletion collection, BW25113 (18). This library, named KEIO_ML9, comprises 152,018 uniquely bar-coded mutants covering 3,728 of the 4,146 protein-coding genes; essential genes, along with genes that are too short or repetitive to uniquely map insertion sties and nonessential genes with low barcode abundance at the start of the experiment, are absent from the library (12). To identify E. coli genes contributing to fitness in coculture with R. palustris, we grew the KEIO_ML9 library under the following three conditions (Fig. 1B): (i) “Nx coculture,” that is, coculture with R. palustris Nx (CGA4005), a strain that excretes NH4+ at an arbitrarily defined level of 1× (7); (ii) “NxΔAmtB coculture,” that is, coculture with R. palustris NxΔAmtB (CGA4021), a strain that excretes 3× NH4+ (7); and (iii) “monoculture,” that is, monoculture using the same anaerobic medium used for cocultures but supplemented with 15 mM NH4Cl to enable E. coli growth in the absence of R. palustris. Monoculture was used to identify genes that affected E. coli fitness due to the anaerobic, minimal-medium environment. We examined E. coli fitness in both Nx and NxΔAmtB cocultures because E. coli growth is differentially constrained in these two cocultures. Specifically, the 1× NH4+ cross-feeding level in Nx coculture constrains E. coli growth such that it is coupled to that of R. palustris, with both species exhibiting a doubling time of approximately 10 h (7); in NxΔAmtB cocultures, the 3× level of NH4+ cross-feeding allows E. coli to grow faster than R. palustris, although E. coli still experiences a degree of nitrogen limitation and exhibits slower growth than in monocultures with excess NH4+ (7). We hypothesized that genes influencing the E. coli-R. palustris interaction would exhibit fitness effects of different magnitudes in these two cocultures, with effects being more pronounced in Nx cocultures.
To enable accurate and informative comparisons between conditions, we took care in how we inoculated, incubated, and harvested the cultures. First, all cultures were inoculated using minimal-medium-washed E. coli cells from a single KEIO_ML9 culture grown to mid-log phase in aerobic Luria-Bertani broth (LB) with kanamycin (Fig. 1B). Aerobic LB served as a “nonselective” condition to preserve library diversity (12). However, it was imperative to wash the cells using minimal medium to remove LB components that could disrupt the coculture mutualism (e.g., amino acids) and the kanamycin to which R. palustris was not resistant. Second, all cultures were inoculated using the same volume of washed KEIO_ML9 library, for an E. coli starting optical density at 660 nm (OD660) of ∼0.01. Third, R. palustris was inoculated at strain-specific cell densities, such that the initial E. coli/R. palustris ratio approximated the final species ratio known to occur in each coculture type (1:10 and 1:1 for Nx and NxΔAmtB cocultures, respectively) (7). Fourth, all cultures were incubated under identical conditions (i.e., lying flat at 30°C with horizontal shaking and constant illumination) and were harvested in exponential phase after 4.5 to 5 cell doublings (Fig. 1C; after approximately 12, 54, and 42 h for monocultures, Nx cocultures, and NxΔAmtB cocultures, respectively). We assessed fitness after 4.5 to 5 cell doublings to enable discrimination between subtle and strong phenotypes (19). Finally, mutant fitness was assessed using a 4-fold biological replication for each culture type. All RB-TnSeq experiments in this study met the quality-control standards previously described (see Table S1 in the supplemental material) (12).
Identification of E. coli genes affecting fitness in mono- or coculture.
The KEIO_ML9 library has a median of 16 unique barcodes (i.e., strains) per gene (12). A fitness score for each E. coli strain was calculated as the log2 difference in barcode abundance between the washed KEIO_ML9 inoculum (T0) and the population after 4.5 to 5 generations in the mono- or coculture condition. The fitness scores for all strains with insertional disruptions of a given gene were then used collectively to calculate a fitness score for that gene. Genes with no impact on fitness in a given condition had fitness scores close to zero. Negative fitness scores reflected genes that were beneficial to fitness (i.e., gene disruption resulted in a fitness disadvantage), and positive fitness scores indicated genes that were detrimental to fitness (i.e., gene disruption resulted in a fitness advantage). A moderated t statistic (t-score) was used to estimate how reliably a fitness value differed from zero (12). For each gene, we averaged the fitness values and t-scores across culture replicates and defined a strong fitness effect as a |mean fitness| of >1 and a |mean t-score| of >3.
Of the 3,728 protein-coding genes covered in the KEIO_ML9 library, we could calculate fitness scores for 3,564 genes (95.6% of covered genes) (Table S2). Most genes had a negligible impact on fitness (Fig. S1). A total of 306 unique genes strongly affected fitness either negatively or positively in one or multiple culture types (Table S3). The largest number of fitness-affecting genes was identified in cocultures with R. palustris Nx, with 167 genes having negative fitness scores and 105 having positive fitness scores (Fig. 2A). Fewer genes affected E. coli fitness in coculture with the 3×-NH4+-excreting R. palustris NxΔAmtB, with 122 gene insertions negatively affecting fitness and two insertions positively affecting fitness. Only 117 genes were identified to affect E. coli monoculture fitness, all of which had negative scores (Fig. 2A). Principal-component analysis showed clear clustering according to culture type, with the first two principal components explaining approximately 40% of the variability (Fig. 2B), and the E. coli fitness profiles were more similar between the two types of cocultures than between the monoculture and either coculture (Fig. 2C; Fig. S2). In addition, fitness effects were generally stronger in Nx coculture than in NxΔAmtB coculture (Fig. 2C), as we originally hypothesized given that NH4+ cross-feeding levels are more favorable for E. coli growth in NxΔAmtB coculture than in Nx coculture.
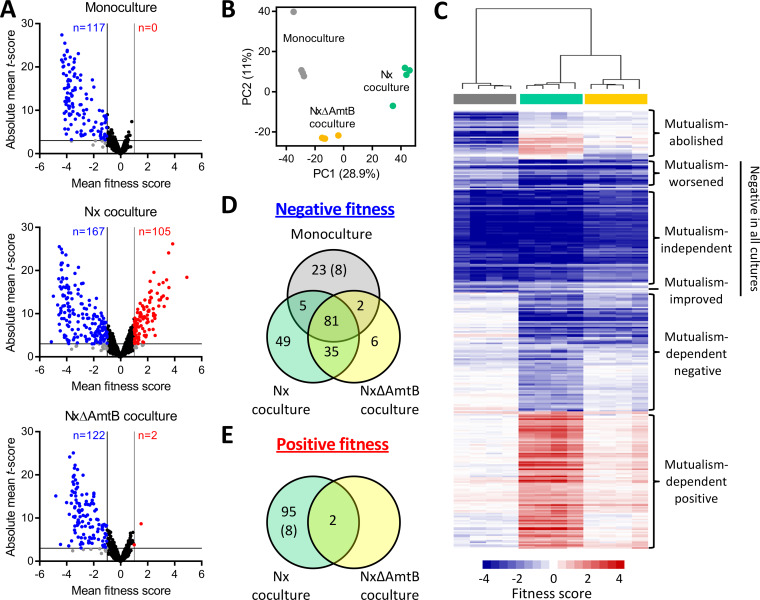
FIG 2.
Summary of RB-TnSeq results identifying E. coli fitness determinants in monoculture or coculture with R. palustris. (A) Volcano plots showing mean fitness scores and absolute mean t-scores for 3,564 E. coli gene mutants grown in monoculture (top), Nx coculture (middle), or NxΔAmtB coculture (bottom). Each circle represents a single gene, with fitness scores and t-scores averaged across culture replicates (n = 4). Vertical lines indicate a fitness score threshold of |fitness|>1. Horizontal lines indicate a t-score threshold of |t-score| > 3. Genes with strong negative or positive fitness scores are indicated in blue or red, respectively. Black circles indicate genes that did not meet the fitness score threshold; gray circles indicate genes that met the fitness score threshold but not the t-score threshold. (B) Two-component principal component (PC) analysis using fitness scores for 3,564 E. coli genes for individual monocultures (gray), Nx cocultures (teal), or NxΔAmtB cocultures (yellow). Percentages indicate variance captured by each of the first two PCs. (C) Heatmap showing fitness scores and designated effect categories for the 306 genes having strong fitness effects in at least one culture type. Genes and samples were clustered hierarchically (average linkage, uncentered correlation). Columns are fitness scores from independent replicates, with culture type indicated by the colored bars below the cluster tree (gray, monoculture; teal, Nx coculture; yellow, NxΔAmtB coculture). (D and E) Venn diagrams of genes having strong negative (D) or positive (E) fitness scores in the indicated culture types. Eight genes had negative fitness scores in monoculture but positive fitness scores in Nx coculture (indicated by parentheses).
Of the 306 genes identified as having strong fitness effects, 81 genes had negative fitness scores in all three cultures (Fig. 2D). Based on the magnitudes of effect in the different cultures, these 81 genes fell into three subgroups (Fig. 2C). Genes that had similar fitness scores in all three cultures (e.g., thrA and pfkA) were considered to have environment-dependent but “mutualism-independent” fitness effects. Genes with fitness scores that were more severe in one or both cocultures than in monoculture (e.g., lrp and clpP) were deemed to have “mutualism-worsened” fitness scores. Conversely, genes with negative fitness scores that were less severe in one or both cocultures than in monoculture (e.g., ilvY and hisI) were considered to have “mutualism-improved” negative fitness scores (Fig. 2C). For simplicity, these 81 genes were classified collectively as “negative in all cultures” for later analyses (Table S3).
Separately, 35 genes had negative fitness scores in both cocultures but had negligible fitness effects in monoculture (Fig. 2D). An additional 49 genes had negative fitness scores in Nx cocultures only, and an additional six genes had negative fitness scores in NxΔAmtB cocultures only (Fig. 2D). Together, we classified these 90 genes as having “mutualism-dependent negative” fitness scores (Fig. 2C; Table S3). Notably, 31 genes had negative fitness scores in monoculture but either no fitness effect (23 genes) or a positive fitness score (8 genes) in coculture (Fig. 2D and E). These genes were classified as exhibiting “mutualism-abolished fitness defects” (Fig. 2C; Table S3). An additional 97 genes had no fitness effect in monoculture but had positive fitness scores in one or both cocultures (Fig. 2E) and were thus classified as having “mutualism-dependent positive” fitness scores (Fig. 2C; Table S3). Finally, there were seven genes that had negative fitness scores in monoculture and one type of coculture (Fig. 2C; Table S3); given their scarcity and their ambiguous fitness effects, we did not designate a label for these seven genes.
As initial validation of our approach, we examined the fitness scores for genes that were previously tested in coculture. Six genes that have no impact on E. coli fitness in coculture (ddpA, ddpX, rutA, argT, patA, and potF) (10) were also shown by RB-TnSeq to have negligible effects in coculture (Table S2). Moreover, amtB and ntrC, which are crucial for E. coli fitness in coculture (9, 10), had negligible impact on monoculture fitness but had two of the top three mutualism-dependent negative fitness scores (Table 1). Thus, RB-TnSeq could accurately distinguish between genes that did and did not affect E. coli fitness in coculture.
TABLE 1.
Top genes with mutualism-dependent negative or positive fitness scoresa
| Geneb | Gene product/general rolec | Monoculture |
Nx coculture |
NxΔAmtB coculture |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fitness score |
|Mean t-score| | Fitness score |
|Mean t-score| | Fitness score |
|Mean t-score| | |||||
| Mean | SD | Mean | SD | Mean | SD | |||||
| Negative fitness effects | ||||||||||
| ntrC* | Nitrogen transcriptional regulator | –0.54 | 0.14 | 3.26 | –4.44 | 0.42 | 10.20 | –3.81 | 0.45 | 10.87 |
| gltB* | Glutamate synthase subunit | –0.04 | 0.07 | 0.37 | –4.32 | 0.06 | 24.14 | –3.51 | 0.09 | 25.09 |
| amtB* | Ammonium transporter | –0.02 | 0.10 | 0.18 | –4.26 | 0.21 | 14.10 | –3.32 | 0.27 | 15.11 |
| gltD* | Glutamate synthase subunit | 0.07 | 0.08 | 0.55 | –4.07 | 0.19 | 16.03 | –3.30 | 0.14 | 16.73 |
| clpX | ATP-dependent protease subunit | –0.64 | 0.15 | 2.88 | –3.86 | 0.69 | 7.04 | –3.21 | 0.30 | 8.29 |
| glnK* | Nitrogen assimilation regulator | –0.13 | 0.08 | 0.51 | –3.82 | 0.35 | 3.74 | –2.90 | 0.47 | 4.35 |
| ptsP | PEP-protein phosphotransferase | 0.59 | 0.03 | 5.00 | –3.61 | 0.25 | 13.46 | –2.60 | 0.28 | 13.16 |
| yeeX | Unknown | 0.08 | 0.04 | 0.33 | –3.44 | 0.46 | 4.44 | –2.64 | 0.35 | 5.37 |
| npr | Phosphorelay protein | 0.54 | 0.06 | 3.26 | –3.37 | 0.66 | 5.29 | –3.07 | 0.70 | 8.88 |
| hldE | Fused kinase/adenyltransferase | –0.71 | 0.14 | 2.61 | –3.19 | 0.73 | 4.59 | –2.15 | 0.50 | 5.27 |
| cra | Catabolite repressor/activator | –0.64 | 0.08 | 3.51 | –3.16 | 0.43 | 8.09 | –1.71 | 0.52 | 6.87 |
| asnB | Asparagine synthetase | 0.07 | 0.07 | 0.51 | –3.14 | 0.20 | 15.58 | –2.08 | 0.17 | 13.42 |
| mdh | Malate dehydrogenase | 0.15 | 0.09 | 1.13 | –2.92 | 0.47 | 12.30 | –2.29 | 0.39 | 13.01 |
| waaF | Lipopolysaccharide biosynthesis | –0.49 | 0.25 | 1.71 | –2.92 | 0.37 | 5.00 | –2.30 | 0.33 | 5.50 |
| pcnB | Poly(A) polymerase | –0.47 | 0.17 | 2.67 | –2.83 | 0.39 | 9.93 | –1.07 | 0.32 | 6.09 |
| yejM | Inner membrane protein | –0.09 | 0.12 | 0.59 | –2.70 | 0.22 | 11.25 | –2.45 | 0.18 | 10.71 |
| bssS | Biofilm regulator | –0.81 | 0.04 | 3.82 | –2.68 | 0.51 | 7.30 | –0.85 | 0.23 | 3.68 |
| waaC | Lipopolysaccharide biosynthesis | –0.96 | 0.22 | 3.43 | –2.63 | 0.33 | 4.74 | –2.22 | 0.21 | 5.35 |
| tatB | Protein translocase | –0.33 | 0.16 | 1.39 | –2.54 | 0.12 | 6.82 | –2.02 | 0.41 | 6.01 |
| ompC | Outer membrane porin | –0.27 | 0.09 | 1.84 | –2.54 | 0.14 | 11.07 | –1.92 | 0.22 | 11.65 |
| yieP | Transcriptional regulator | –0.11 | 0.12 | 0.69 | –2.52 | 0.24 | 9.79 | –1.36 | 0.45 | 7.92 |
| pal | Outer membrane lipoprotein | –0.97 | 0.60 | 2.22 | –2.51 | 0.80 | 3.61 | –2.81 | 0.36 | 4.05 |
| thiL | Thiamine synthesis | –0.78 | 0.20 | 4.83 | –2.45 | 0.27 | 8.88 | –1.38 | 0.41 | 7.41 |
| fabF | Fatty acid synthesis | –0.04 | 0.12 | 0.27 | –2.35 | 0.25 | 8.96 | –1.15 | 0.42 | 6.40 |
| ilvI | Acetolactate synthase | –0.67 | 0.10 | 5.06 | –2.33 | 0.61 | 13.07 | –0.77 | 0.13 | 5.49 |
| clpA | ATP-dependent protease subunit | –0.34 | 0.10 | 2.46 | –2.30 | 0.19 | 11.99 | –1.32 | 0.33 | 8.38 |
| Positive fitness effects | ||||||||||
| sspA | Stringent starvation protein | –0.06 | 0.17 | 0.25 | 4.91 | 0.64 | 18.43 | 1.01 | 0.54 | 3.79 |
| glnB* | Nitrogen regulatory protein | –0.62 | 0.23 | 2.99 | 3.85 | 0.65 | 26.17 | 0.31 | 0.45 | 1.91 |
| sucA | αKG dehydrogenase E1 | –0.64 | 0.16 | 2.26 | 3.58 | 0.63 | 16.06 | 0.24 | 0.40 | 0.96 |
| gshA | Glutathione biosynthesis | 0.52 | 0.21 | 3.45 | 3.56 | 0.59 | 24.08 | 0.62 | 0.39 | 4.10 |
| rsmH | 16S rRNA methyltransferase | –0.24 | 0.22 | 0.96 | 3.44 | 0.18 | 13.51 | 0.47 | 0.11 | 1.95 |
| hpt | Purine salvage | –0.35 | 0.12 | 1.76 | 3.41 | 0.38 | 17.82 | 0.56 | 0.28 | 2.68 |
| srmB | ATP-dependent RNA helicase | –0.42 | 0.41 | 1.16 | 3.23 | 0.67 | 11.68 | 0.40 | 0.36 | 1.26 |
| rsmA | 16S rRNA dimethyltransferase | –0.06 | 0.35 | 0.23 | 3.17 | 0.30 | 14.11 | 0.94 | 0.24 | 4.10 |
| rpoS | RNA polymerase sigma factor S | 0.08 | 0.13 | 0.41 | 3.14 | 0.58 | 17.20 | 1.51 | 0.25 | 8.67 |
| cyaA | Adenylate cyclase | 0.11 | 0.08 | 0.64 | 3.04 | 0.64 | 13.82 | 0.61 | 0.28 | 3.52 |
| lepA | Translation elongation factor | –0.01 | 0.15 | 0.04 | 2.94 | 0.32 | 13.20 | 0.40 | 0.38 | 1.62 |
| purR | Transcriptional repressor | –0.01 | 0.16 | 0.06 | 2.84 | 0.26 | 16.79 | 0.81 | 0.25 | 4.50 |
| nfuA | Iron-sulfur cluster carrier protein | –0.02 | 0.06 | 0.09 | 2.84 | 0.34 | 13.17 | 0.48 | 0.32 | 2.51 |
| upp | Pyrimidine salvage | –0.36 | 0.24 | 1.16 | 2.83 | 0.51 | 11.78 | -0.11 | 0.22 | 0.34 |
| tyrR | Transcriptional regulator | –0.11 | 0.08 | 0.63 | 2.72 | 0.36 | 14.75 | 0.40 | 0.41 | 2.32 |
| fadR | Transcriptional regulator | –0.02 | 0.05 | 0.07 | 2.56 | 0.36 | 4.97 | 0.76 | 0.22 | 4.15 |
| ybfE | Unknown | –0.06 | 0.23 | 0.28 | 2.55 | 0.24 | 9.64 | 0.17 | 0.37 | 0.50 |
| cpxA | Sensor histidine kinase | 0.48 | 0.04 | 3.80 | 2.53 | 0.50 | 8.89 | 0.67 | 0.46 | 4.09 |
| bipA | Ribosome assembly factor | –0.30 | 0.06 | 2.45 | 2.47 | 0.30 | 19.58 | 0.03 | 0.21 | 0.22 |
| nuoI | NADH dehydrogenase subunit | 0.12 | 0.37 | 0.44 | 2.44 | 0.46 | 9.52 | 0.43 | 0.35 | 1.53 |
| maoP | Ori macrodomain organization | –0.66 | 0.16 | 3.46 | 2.43 | 0.09 | 10.05 | 0.78 | 0.18 | 4.11 |
Genes had negative or positive fitness scores, respectively, in one or both cocultures but negligible fitness effects in monoculture (based on thresholds of |mean fitness| > 1 and |mean t-score| > 3). The top genes in each category (negative or positive) were identified as sorted by mean fitness score in Nx coculture.
Genes indicated by an asterisk (*) are participants in the NSR.
More detailed descriptions are provided in Table S3.
Functional classification of E. coli genes affecting fitness in mono- or coculture.
To investigate the cellular activities that influenced E. coli fitness in mono- and coculture, we functionally classified the genes with strong fitness effects using clusters of orthologous groups (COGs) (20). Overall, the 306 genes represented diverse functions, although almost half (48.7%) were involved in metabolism (Fig. 3A). A total of 239 genes pertained to 16 functional categories, with “amino acid metabolism and transport” predominating, and 67 genes had multiple or poorly defined functions (Fig. 3B). Six COG categories were not represented in our screen: “RNA processing and modification,” “chromatin structure and dynamics,” “cell motility,” “nuclear structure,” “defense mechanisms,” and “cytoskeleton.”
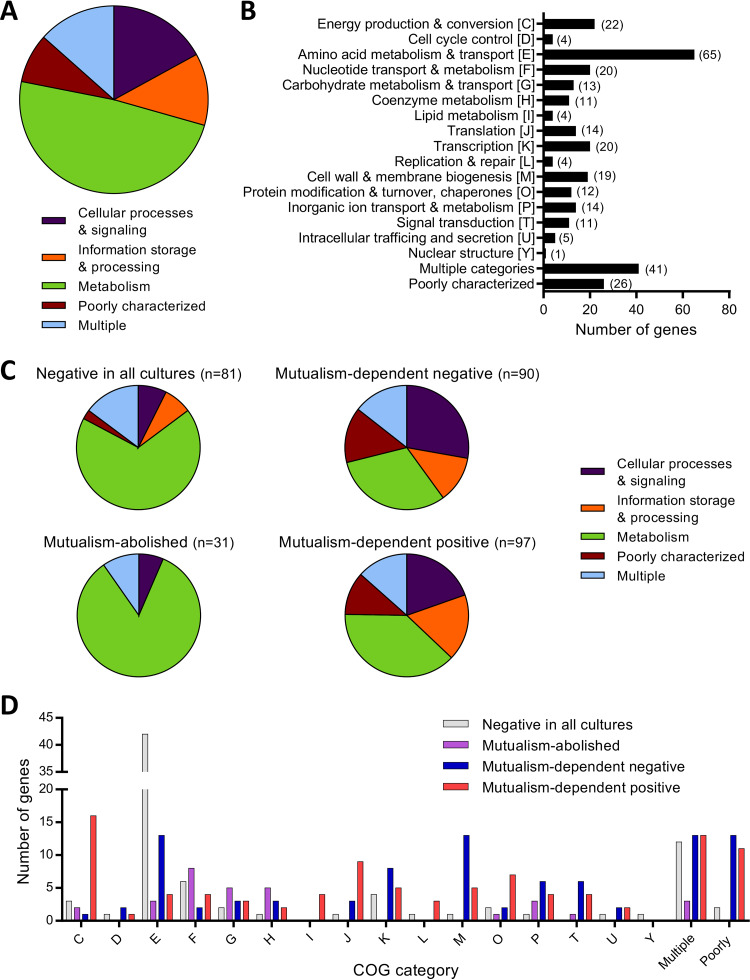
FIG 3.
Functional summary of genes exhibiting strong fitness effects in one or more culture type. Functional categories, based on COGs, for (A and B) all 306 genes with a |mean fitness score| of >1 and a |mean t-score| of >3 and (C and D) gene subsets exhibiting the designated fitness effects. Genes listed as “poorly characterized” include genes with general functional predictions only (COG category R), genes with unknown function (COG category S), and genes with no COG classification. Genes listed as “multiple” have more than one COG category assignment. Gene names, descriptions, fitness scores, t-scores, and COG classifications are provided in Table S3. COG categories are the same in panels B and D. (B) Numbers in parentheses denote the number of genes in each category. The pie charts in panels A and C represent the same genes in panels B and D, respectively, but with COG categories grouped into generalized cell functions, as follows: cellular processing and signaling, COG categories DMNOTUVYZ; information storage and processing, COG categories ABJKL; and metabolism, COG categories CEFGHIPQ.
When we examined functional classes according to the fitness effect categories described above, there were clear differences in predominant functions among the different groups. Of the 81 genes with negative fitness scores in all cultures, 55 (67.9%) were related to metabolism (Fig. 3C), including 42 of the 65 “amino acid metabolism and transport” genes in the full 306-gene set (Fig. 3B and D). This was unsurprising considering that many amino acid synthesis genes are expected to be essential in the minimal medium we used, which lacks amino acids. Metabolism-related genes accounted for 31.1 and 38.1% of the mutualism-dependent negative and positive groups, respectively (Fig. 3C), but these similar proportions were due to distinct gene classes. Specifically, in the mutualism-dependent negative group, 19 (67.8%) of the 28 metabolism-related genes were involved in “amino acid metabolism and transport” (n = 13) and “inorganic ion transport and metabolism” (n = 6) (Fig. 3D). In contrast, the metabolism-related genes in the mutualism-dependent positive group were predominantly linked to “energy production and conversion” (16 of 37 genes, 43.2%), with the remaining genes being fairly evenly spread among other metabolic functions (Fig. 3D).
The prevalence of genes related to other general functions was also distinct between the mutualism-dependent negative and positive groups (Fig. 3C). The mutualism-dependent negative group had the largest proportion of genes related to “cellular processes and signaling” among the groups (27.8%) (Fig. 3C), and these were primarily involved in “cell wall and membrane biogenesis” (n = 13), “transcription” (n = 8), and “signal transduction” (n = 6) (Fig. 3D). This group also had the largest proportion of “poorly characterized” genes among the groups (14.4%) (Fig. 3C). Meanwhile, the mutualism-dependent positive group had a smaller proportion of genes involved in “cellular processes and signaling” (19.6%) and had the largest proportion of genes related to “information storage and processing” (17.5%) among the groups (Fig. 3C).
Finally, the mutualism-abolished fitness defect group consisted almost entirely (83.9%) of metabolism-related genes (Fig. 3C). The largest proportion of these genes was involved in “nucleotide metabolism and transport” (25.8%), followed by “coenzyme metabolism” and “carbohydrate metabolism” (16.1% each) (Fig. 3D); “nucleotide metabolism and transport” and “coenzyme metabolism” were poorly represented (1.2 to 7.4%) in the other groups. Overall, these data indicate that genes involved in diverse cellular functions impact E. coli fitness in coculture with R. palustris.
Only a portion of the nitrogen starvation response benefits E. coli fitness in coculture.
As mentioned above, the transcriptional regulator NtrC and the NH4+ transporter AmtB were previously shown to be decisive E. coli fitness determinants in coculture (9, 10), and our TnSeq data supported these results (Table 1). NtrC and AmtB are central players in the NSR (21). Other NSR-related genes also exhibited mutualism-dependent fitness effects in our screen (Fig. 4A). In fact, NSR-related genes comprised five of the top seven mutualism-dependent fitness scores (Table 1). These data further authenticated the importance of the E. coli NSR for mutualistic coexistence with R. palustris. However, not all NSR genes contributed to E. coli fitness in coculture. For example, the nitrogen assimilation control (nac) gene had limited impact on fitness in any culture type (mean fitness: 0.20 for monoculture, −0.58 for Nx coculture, and −0.48 for NxΔAmtB coculture) (Fig. 4A). Nac is the second main transcriptional regulator of the NSR, behind NtrC, and of the 23 operons within the NtrC regulon, which includes nac, nine are regulated indirectly via Nac (22). E. coli nac expression is upregulated >40-fold in coculture compared to monoculture (10). However, only four Nac-regulated genes are differentially expressed in coculture versus monoculture (10), and none of these met our thresholds for strongly impacting fitness in coculture (Table S4). Thus, although the low NH4+ cross-feeding level in coculture activates the NSR and the resulting NtrC-mediated induction of some NSR genes is crucial for E. coli fitness, the Nac-mediated arm of the E. coli NSR appears to be dispensable for mutualistic coexistence with R. palustris.
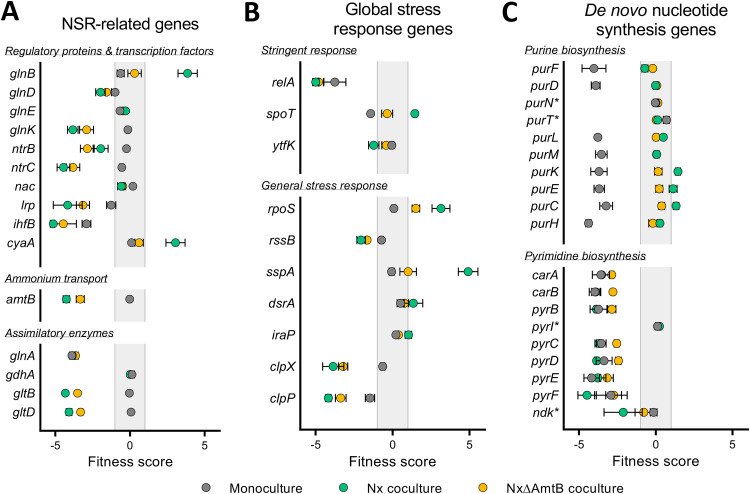
FIG 4.
Fitness scores for select E. coli genes. (A to C) Mean fitness scores for select genes involved in the NSR (A), global stress responses (B), and de novo nucleotide biosynthesis (C) for E. coli grown in monoculture (gray), Nx coculture (teal), or NxΔAmtB coculture (yellow). Error bars indicate standard deviations (n = 4). In panel C, asterisks (*) denote genes known to have redundant functions or known to be dispensable for de novo synthesis.
Notably, whereas disruption of many NSR genes negatively affected fitness in coculture, glnB had a strong positive fitness score in Nx coculture (Table 1 and Fig. 4A). In response to nitrogen availability, GlnB (also known as PII) regulates glutamine synthetase (GS) adenylation and NtrC phosphorylation (23). In the absence of GlnB, the cell cannot sense when there is ample nitrogen and the NSR is activated via NtrC even under high-nitrogen conditions (21). We posit that the glnB mutant exhibits better fitness upon introduction into coculture due to constitutively active NtrC. Specifically, when the KEIO_ML9 library was initially grown in LB, NtrC (and the NSR) would be inactive in most cells due to abundant nitrogen. However, in the glnB mutant, constitutively active NtrC would be activating transcription of the NSR genes, including amtB (23). Consequently, unlike the rest of the population, the glnB mutant would already have high levels of AmtB upon inoculation in coculture and therefore be primed for mutualistic growth. Although possible, we reason it unlikely that the glnB fitness effect is mediated via GS adenylation because disruption of glnE, encoding the adenylyltransferase responsible for GS adenylation, had a negligible fitness effect in all culture types (Fig. 4A). Future studies will be needed to verify how GlnB affects fitness in coculture.
Metabolic pathway choice influences E. coli fitness in coculture.
The second and fourth top mutualism-dependent negative fitness scores went to gltB and gltD (Table 1). These genes are NSR related, but their effect was likely assimilatory rather than regulatory. gltB and gltD encode the two subunits of glutamate synthetase (GOGAT), which is one of two E. coli glutamate synthesis enzymes. GOGAT transfers an amino group from glutamine to α-ketoglutarate (αKG), resulting in two molecules of glutamate (24). In the other pathway, glutamate is synthesized directly from αKG and NH4+ by glutamate dehydrogenase (GDH), encoded by gdhA. The use of GOGAT or GDH depends on NH4+ concentrations, with GOGAT being important under low-NH4+ conditions due to a much higher affinity for NH4+ (24). The gdhA gene exhibited negligible fitness effects under each condition (Fig. 4A), presumably because GOGAT can compensate for the absence of GDH under both low- and high-NH4+ conditions (21). In contrast, gltB and gltD had negative fitness scores in both cocultures (Fig. 4A), with the score magnitudes inversely correlating with NH4+ cross-feeding levels. These data suggest that the low NH4+ levels in coculture compel E. coli to use GOGAT for mutualistic growth.
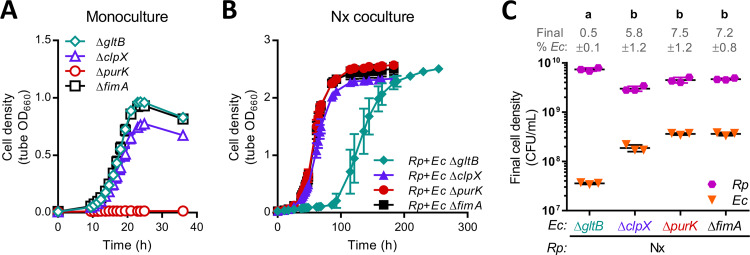
To directly assess the effect of GOGAT on growth in coculture, we grew the KEIO ΔgltB mutant in both monoculture and Nx coculture. We used the KEIO ΔfimA mutant as a control strain, as fimA had a negligible impact on fitness (|mean fitness <0.1) in all culture types (Table S2) and carries the same kanamycin resistance cassette as the other mutant strains. As expected, the ΔfimA mutant exhibited robust growth in both monoculture and Nx coculture (Fig. 5A), with growth trends comparable to those observed with strain MG1655 (7). In agreement with the RB-TnSeq data, the ΔgltB mutant grew well in monoculture, matching the ΔfimA mutant growth trends (Fig. 5A). In contrast, ΔgltB Nx cocultures exhibited an extended lag period of ∼85 h versus ∼26 h in ΔfimA Nx cocultures (Fig. 5B). The gltB RB-TnSeq fitness score near −5 in Nx cocultures, which indicates negligible growth, came from samples harvested at ∼54 h, which would be within this extended lag period. From these results, we conclude that GOGAT is important for E. coli fitness in coculture.
FIG 5.
Growth of select KEIO mutants in monoculture and Nx coculture. Growth of individual KEIO mutant monocultures (A) or cocultures pairing individual KEIO mutants with R. palustris Nx (B). The KEIO ΔfimA mutant was used as a control because it carries the same kanamycin resistance cassette as the other mutants and fimA had negligible fitness effects by RB-TnSeq (Table S2). The data are presented as means ± the standard deviations (n = 3). Some error bars are too small to visualize. OD measurements were taken within culture tubes (tube OD). (C) Final cell densities of individual E. coli KEIO mutants and R. palustris Nx after growth in coculture. CFU were measured at the final time points in the respective growth curves in panel B. Different letters indicate statistical differences between groups (P < 0.05, one-way analysis of variance with Tukey’s posttest, n = 3). Error bars indicate the standard deviations.
Nonetheless, we were intrigued by the eventual growth of ΔgltB Nx cocultures. The abrupt growth after an extended lag phase (Fig. 5B) suggested that the absence of GOGAT was overcome in some manner. When we examined final cell densities, we found that the proportion of the ΔgltB mutant within Nx cocultures was >10-fold less than that of the ΔfimA mutant, resulting from both lower E. coli cell densities and higher R. palustris cell densities (Fig. 5C). Thus, one explanation is that the skewed species ratio in ΔgltB Nx coculture provided higher NH4+ levels per E. coli cell, thereby allowing GDH-mediated glutamate synthesis and coculture growth; the extended lag could represent the period during which the coculture was reaching this alternate species ratio. It is also possible that the ΔgltB mutant acquired suppressor mutations that conferred elevated GDH activity, which can rescue GOGAT mutant growth under low NH4+ conditions (25). Overall, enzyme choice for glutamate synthesis clearly affects E. coli fitness during mutualistic growth, as GDH does not compensate for the absence of GOGAT in coculture.
Multiple global regulatory systems influence E. coli fitness in coculture.
The NSR is the primary cellular response to nitrogen limitation in E. coli. However, the E. coli NSR is coupled to the stringent response under low-nitrogen conditions because NtrC directly activates transcription of relA (26). E. coli RelA is the primary enzyme for the synthesis of the alarmone (p)ppGpp; the remaining (p)ppGpp is synthesized by the (p)ppGpp synthase/hydrolase SpoT (27). Accumulation of (p)ppGpp coordinates the redistribution of resources to enable continued growth and survival amid nutrient stress (27). Both relA and spoT were identified by RB-TnSeq as strongly influencing E. coli fitness in one or multiple culture types (Fig. 4B; Table S1). relA had negative fitness scores in all cultures, but the effects were stronger in cocultures than in monoculture. In contrast, spoT had a negative fitness score in monoculture but a positive fitness score in Nx coculture and a negligible effect in NxΔAmtB coculture. SpoT is essential in relA+ E. coli because uncontrolled, high levels of (p)ppGpp inhibit growth (28). However, viable spoT insertion mutants have been identified by both RB-TnSeq (15) and transposon-directed insertion site sequencing (29). It is possible that the RB-TnSeq spoT strains produce truncated SpoT proteins with some degree of function or harbor compensatory secondary mutations that enable growth (30, 31), and thus we mention the spoT data for transparency only. Given that (i) E. coli (p)ppGpp levels increase in response to nitrogen limitation (32), (ii) RelA mutants are severely impaired for the stringent response (33), and (iii) disruption of relA was associated with a negative fitness effect that was exacerbated in coculture (Fig. 4B), we infer that the E. coli stringent response is activated in coculture and contributes to fitness during mutualistic growth.
Since (p)ppGpp affects the expression of ∼500 E. coli genes (33), we can currently only speculate as to which stringent response targets might affect fitness in coculture. Several (p)ppGpp-regulated genes differentially affected E. coli fitness in coculture versus monoculture, including upp, nuoA to nuoN, cyoDE, sucACD, ptsG, and plsB (Table S3). Most intriguing to us, three interlinked global regulators that are influenced by (p)ppGpp were also hit in our screen: leucine responsive protein (Lrp), integration host factor (IHF), and the alternative sigma factor RpoS (Fig. 4A and B). Lrp affects transcription of ∼236 E. coli genes and coordinates multiple regulatory networks to adjust cellular metabolism in response to environmental perturbations (34). Lrp is also considered part of the nitrogen assimilation network because its regulon includes gltBD (21). The second regulator, IHF, has many functions including serving as an important NSR transcription factor (21) and regulating anaerobic fermentative metabolism in E. coli (35). The expression of both Lrp and IHF are upregulated by the stringent response (33, 36), and disruption of lrp or ihfB resulted in mutualism-worsened fitness (Fig. 4A). Lrp and IHF might be beneficial in coculture by activating enzymes like GOGAT, but their effects could also be unrelated to the NSR given the diverse genes in their regulons. The third global regulator, RpoS (σ38), mediates the general stress response and directly or indirectly regulates ∼10% of all E. coli genes (37). The general stress response is stimulated by nitrogen starvation (38), although not via NtrC (39), and like Lrp and IHF, RpoS levels increase with (p)ppGpp levels (40). Unlike disruption of lrp and ihfB, however, disruption of rpoS resulted in enhanced fitness in both cocultures (Fig. 4B). Thus, the general stress response appears to be detrimental to E. coli fitness in coculture.
Lending support to this notion, several regulators of RpoS exhibited negative or positive mutualism-dependent fitness effects in correlation with the gene’s expected effect on RpoS levels (Fig. 4B). For example, the protease ClpXP, in conjunction with the adapter protein RssB, degrades RpoS (37). Disruption of any of these genes results in RpoS accumulation (41, 42), and all three had negative fitness scores in coculture (Fig. 4B). Because clpX had the fifth most negative mutualism-dependent fitness score (Table 1), we examined KEIO ΔclpX mutant growth in both monoculture and Nx coculture. Whereas RB-TnSeq indicated clpX to have a minor fitness effect in monoculture but a major effect in coculture (Fig. 4B), the KEIO ΔclpX mutant exhibited comparable minor growth defects in both culture types, as evidenced by lower final cell densities, lower growth yields, and slightly lower growth rates compared to control ΔfimA cultures (Fig. 5A and B; Fig. S3). We hypothesize that the exacerbated negative fitness effect of clpX in RB-TnSeq cocultures is due to competition against the other E. coli mutants in the population. In other words, some E. coli mutants, such as the ΔclpX mutant, might exhibit better growth when partnered as a clonal population with R. palustris compared to being part of a pool of potential mutant competitors. Conversely, some E. coli mutants might benefit from the activities of other members the mutant pool but would grow poorly as a clonal population.
Regardless of potential intraspecific competition effects on clpX mutant growth trends, the general stress response could hamper E. coli fitness in coculture by several mechanisms. First, RpoS could directly control expression of target genes that influence E. coli fitness. Alternatively, because sigma factors compete for a limited amount of RNA polymerase (43), induction of the general stress response might limit gene expression driven by other sigma factors important in coculture, such as RpoN. RpoN drives expression of many NSR genes, including ntrC (21), and has also been implicated in the control of membrane and cell wall biogenesis (44), a function that was well represented by genes exhibiting mutualism-dependent fitness effects (Fig. 3D). Notably, E. coli sigma factor competition is influenced by ppGpp (45), Lrp (46), and IHF (47), as well as two other genes hit in our screen, namely, ptsN (48) (Table 2), encoding part of the regulatory PTSNtr system (49), and cyaA (47) (Table 1), encoding the cAMP-synthesizing adenylate cyclase. Thus, sigma factor competition resulting from the activation of multiple stress responses might curtail E. coli fitness in coculture.
TABLE 2.
Top genes exhibiting mutualism-abolished fitness defectsa
| Gene | Gene product/general roleb | Monoculture |
Nx coculture |
NxΔAmtB coculture |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fitness score |
|Mean t-score| | Fitness score |
|Mean t-score| | Fitness score |
|Mean t-score| | |||||
| Mean | SD | Mean | SD | Mean | SD | |||||
| purH | Purine biosynthesis | –4.38 | 0.17 | 21.62 | 0.27 | 0.15 | 2.14 | –0.18 | 0.27 | 1.53 |
| sucD | Succinyl-CoA synthetase subunit | –4.14 | 0.76 | 5.69 | –0.93 | 0.26 | 2.45 | –0.44 | 0.13 | 1.24 |
| purF | Purine biosynthesis | –4.02 | 0.78 | 11.85 | –0.71 | 0.19 | 3.35 | –0.20 | 0.11 | 1.18 |
| purD | Purine biosynthesis | –3.91 | 0.28 | 16.73 | –0.02 | 0.13 | 0.06 | 0.08 | 0.13 | 0.55 |
| sucC | Succinyl-CoA synthetase subunit | –3.81 | 0.11 | 10.47 | –0.89 | 0.43 | 4.51 | –0.14 | 0.15 | 0.79 |
| purL | Purine biosynthesis | –3.77 | 0.06 | 18.86 | 0.50 | 0.09 | 3.70 | 0.03 | 0.13 | 0.21 |
| purK | Purine biosynthesis | –3.70 | 0.53 | 13.84 | 1.45 | 0.17 | 9.73 | 0.17 | 0.24 | 1.13 |
| purE | Purine biosynthesis | –3.68 | 0.34 | 6.70 | 1.13 | 0.26 | 5.00 | 0.23 | 0.22 | 0.96 |
| purM | Purine biosynthesis | –3.54 | 0.38 | 9.72 | 0.06 | 0.16 | 0.31 | 0.05 | 0.07 | 0.21 |
| purC | Purine biosynthesis | –3.23 | 0.42 | 4.95 | 1.32 | 0.18 | 4.24 | 0.39 | 0.19 | 1.44 |
| pdxJ | Vitamin B6 biosynthesis | –3.06 | 0.44 | 8.89 | –0.91 | 0.26 | 2.83 | –0.52 | 0.11 | 2.19 |
| pdxA | Vitamin B6 biosynthesis | –2.95 | 0.09 | 15.11 | –0.94 | 0.40 | 5.73 | –0.18 | 0.15 | 1.36 |
| eda | KHG/KDPG aldolase | –2.78 | 0.33 | 4.25 | 1.32 | 0.35 | 3.88 | –0.51 | 0.38 | 1.22 |
| gcvR | Transcriptional regulator | –2.76 | 0.35 | 8.30 | –0.63 | 0.16 | 2.92 | –0.53 | 0.22 | 2.63 |
| ptsG | Glucose transport | –2.55 | 0.26 | 13.72 | 1.00 | 0.23 | 7.38 | 0.11 | 0.08 | 0.78 |
| pdxB | Vitamin B6 biosynthesis | –2.51 | 0.29 | 12.70 | –0.71 | 0.42 | 4.03 | –0.09 | 0.03 | 0.63 |
| argD | Arginine/ornithine biosynthesis | –1.98 | 0.11 | 15.35 | –0.50 | 0.04 | 4.07 | –0.25 | 0.05 | 2.14 |
| ptsN | Phosphotransferase system enzyme | –1.84 | 0.12 | 7.28 | 0.91 | 0.32 | 4.59 | –0.74 | 0.18 | 3.33 |
| pgl | 6-Phosphogluconolactonase | –1.49 | 0.61 | 4.46 | 1.38 | 0.21 | 5.37 | 0.04 | 0.26 | 0.17 |
| spoT | (p)ppGpp synthase/hydrolase | –1.43 | 0.07 | 6.83 | 1.45 | 0.12 | 8.32 | –0.35 | 0.36 | 1.75 |
| nadA | NAD biosynthesis | –1.39 | 0.24 | 6.90 | –0.07 | 0.17 | 0.41 | –0.03 | 0.07 | 0.15 |
Genes had negative fitness scores in monoculture but negligible effects or positive fitness scores in one or both cocultures (based on our thresholds of |mean fitness| > 1 and |mean t-score| > 3). The top genes were identified as sorted by mean fitness score in monoculture.
More detailed descriptions are provided in Table S3.
E. coli mutants defective for de novo purine biosynthesis are rescued in coculture with R. palustris.
Among the hits from our screen, we were most intrigued by the genes exhibiting mutualism-abolished fitness defects (Table 2) because, unlike some other coculture systems (50–52), our coculture does not enforce coexistence through an engineered auxotrophy. One mechanism by which coculturing could restore E. coli mutant fitness is by slowing the growth of all E. coli strains. In our coculture, the E. coli growth rate is dictated by the NH4+ cross-feeding level (7). Thus, E. coli mutants with growth rate defects in monoculture could have higher fitness scores in coculture simply due the nitrogen-limiting conditions that slow the growth of all E. coli mutants in the competition. Indeed, one of the genes exhibiting mutualism-abolished fitness defects that could be explained by slower growth of competing strains was ptsG (Table 2), which encodes the permease of the glucose phosphotransferase transport system. Disruption of ptsG in E. coli results in poor glucose uptake and slower growth on glucose minimal medium (53). This mutation would have little consequence in coculture as long as the decreased growth rate of the ptsG mutant was equivalent or faster than the growth rate dictated by the NH4+ level. Notably, the positive fitness score of ptsG in Nx coculture (Table 2) indicates that the mutant grew better than the average E. coli strain in the population, perhaps due to directing carbon more efficiently to biomass (53). An alternative mechanism by which coculturing could restore fitness of an E. coli mutant is by decreasing the importance of a given metabolic pathway. For example, nitrogen starvation results in decreased glucose uptake rates in E. coli (54), potentially further reducing the importance of ptsG in coculture.
However, most of the top genes exhibiting mutualism-abolished fitness defects were not readily explained by slowed growth of all E. coli strains, as the genes were involved in synthesizing essential cellular building blocks, such as purines (Table 2). Of the 14 E. coli genes involved in AMP and GMP synthesis, purB, guaA, and guaB were not in the library, and purA could not be analyzed for fitness due to insufficient abundance in the initial (T0) library sample. Eight of the ten remaining genes had severe negative fitness scores in monoculture but had negligible effects or positive fitness scores in coculture (Fig. 4C). The two genes that did not exhibit fitness defects in monoculture, purN and purT, are redundant in function (55) and thus would not have a phenotype when mutated individually. Importantly, de novo purine synthesis mutants are purine auxotrophs (56). Thus, the success of the pur mutants specifically in coculture suggested that coculturing alleviated purine auxotrophy. Coculturing did not eliminate the fitness defects of de novo pyrimidine synthesis mutants (Fig. 4C), indicating that coculture-mediated rescue of nucleotide auxotrophy was specific to purines.
To verify that coculture with R. palustris could rescue E. coli purine auxotroph growth, we tested growth of the KEIO ΔpurK mutant in monoculture and Nx coculture. As expected, the ΔpurK mutant did not grow in monoculture (Fig. 5A), as no purines were provided. However, in agreement with the RB-TnSeq data, ΔpurK Nx cocultures exhibited comparable growth trends to those of control ΔfimA Nx cocultures (Fig. 5B and C; Fig. S3). These data demonstrate that coculture with R. palustris can restore E. coli purine auxotroph growth to wild-type levels. In support of this conclusion, E. coli pur gene transcription is downregulated in coculture with R. palustris (10). Although this downregulation was originally ascribed to slower E. coli growth (10), de novo purine synthesis is repressed by the presence of purines in the medium (57), as could occur if provided by R. palustris. Importantly, although R. palustris can fully satisfy the purine requirement of E. coli, the level of purine cross-feeding is not enough to satisfy the entire E. coli nitrogen requirement; our past work with E. coli NH4+ transporter mutants conclusively implicates NH4+ cross-feeding in this role (9).
Conclusions.
Using RB-TnSeq, we identified E. coli genes that were beneficial or detrimental to E. coli fitness in coculture with R. palustris. This rich data set provided both general gene fitness trends and some specific molecular insights into the mutualistic relationship, such as the importance or lack thereof of various regulatory and metabolic pathways and the existence of unexpected purine cross-feeding. Similar to findings from TnSeq experiments on cocultures mimicking coinfections (58–60), we observed that diverse cellular functions impact E. coli fitness in coculture with R. palustris. These observations underscore that the genetic and physiological architectures underlying microbial interactions, cooperative or otherwise, are complex and often difficult to predict, even in engineered systems. However, architectures common to microbial interactions may become apparent as additional genome-scale fitness studies of microbial consortia are performed. For example, genes involved in cell wall and membrane biogenesis impacted E. coli fitness in coculture with R. palustris, and membrane transport was also important for E. coli growth in a cheese rind community (17), suggesting that membrane-related functions may be of broad importance for E. coli growth in the presence of additional species. Our work further demonstrates that experimentation on microbial consortia will invariably unmask hidden, but nonetheless important, aspects of microbial physiology.
MATERIALS AND METHODS
Strains and media.
All R. palustris strains were derived from the wild-type strain CGA009 (61). R. palustris Nx (CGA4005) harbors (i) a nifA* allele, resulting in constitutive nitrogenase expression and NH4+ excretion, (ii) the ΔhupS mutation, which prevents H2 oxidation, and (iii) the ΔuppE mutation, which prevents cell-cell aggregation and biofilm formation (7). R. palustris strain NxΔAmtB (CGA4021) harbors the same three mutations as strain Nx and additional deletions of amtB1 and amtB2 (7). All E. coli strains were derived from the E. coli K-12 strain BW25113, the parent strain of the KEIO single gene deletion collection (18). The RB-TnSeq E. coli library, KEIO_ML9, was made and characterized previously (12). The single-gene deletion KEIO mutants used in this study were JW4277 (ΔfimA::Km), JW3179 (ΔgltB::Km), JW0428 (ΔclpX::Km), and JW0511 (ΔpurK::Km) (18). KEIO mutant genotypes were confirmed by PCR.
For routine cultivation, R. palustris colonies were isolated on defined mineral (PM) (62) agar supplemented with 10 mM succinate and 0.1% (wt/vol) yeast extract. Plates were incubated anaerobically in a jar with a BD GasPack sachet at 30°C. E. coli KEIO mutant colonies were isolated on Luria-Bertani agar containing 30 μg/ml kanamycin and incubated aerobically at 30°C in the dark. All anaerobic plates and monocultures, and all cocultures were illuminated using a 60-W soft white halogen bulb (750 lumens). All anaerobic liquid mono- and cocultures were laid flat and shaken at 150 rpm. Other aspects of the growth conditions for the KEIO_ML9 library are described below.
Anaerobic monocultures and cocultures were cultivated in 10 ml defined coculture medium (MDC) (7) in 27-ml anaerobic test tubes flushed with 100% N2 and sealed with rubber stoppers and aluminum crimps prior to autoclaving. After autoclaving, MDC was supplemented with 25 mM glucose and cation solution (1% [vol/vol]; 100 mM MgSO4 and 10 mM CaCl2). In addition, monocultures received 15 mM NH4Cl, whereas cocultures received 15 mM NaCl.
Analytical procedures.
Culture growth was assessed by determining the OD660 using a Genesys 20 visible spectrophotometer (Thermo-Fisher). OD measurements for all anaerobic monocultures and cocultures were taken in culture tubes without sampling. Growth rates were calculated using OD values between 0.1 and 1.0, where cell density and OD660 are linearly correlated. Cell densities of aerobic E. coli monocultures were measured in cuvettes. All final culture densities were also measured in cuvettes, and samples were diluted as necessary to achieve values <1 OD660. High-performance liquid chromatography (Shimadzu) was used to quantify glucose, as described previously (63).
RB-TnSeq culture preparation, incubation, and harvesting.
A 2-ml aliquot of the KEIO_ML9 library, stored in 25% (vol/vol) glycerol at –80°C, was thawed on ice and used to inoculate 48 ml of LB with 30 μg/ml kanamycin in a 250-ml aerobic flask. The culture was incubated at 30°C with shaking at 150 rpm until mid-exponential phase (OD660 = 0.44). Cells from 35 ml of culture were pelleted (4,000 rpm, 5 min, room temperature), washed four times with 33 ml of MDC, and resuspended in MDC to an OD660 of 1.0.
Single colonies of R. palustris Nx or NxΔAmtB were inoculated to carbon-limited MDC supplemented with 3 mM acetate. Once the OD of these starter cultures plateaued, the cultures were diluted or concentrated as necessary in MDC to achieve desired cell densities (see below).
The following protocol was used to inoculate three types of cultures, each in quadruplicate: (i) KEIO_ML9 monoculture; (ii) Nx coculture, pairing KEIO_ML9 with R. palustris Nx; and (iii) NxΔAmtB coculture, pairing KEIO_ML9 with R. palustris NxΔAmtB. Each monoculture and coculture was inoculated with 100 μl of washed KEIO_ML9 library. Separately, 1 ml of washed KEIO_ML9 library was centrifuged, and the cell pellet (input sample, T0) was stored at –80°C for barcode sequencing (BarSeq) analysis. For R. palustris, cells from duplicate Nx and NxΔAmtB starter cultures were resuspended to an OD660 of 0.6; then, the two strains were treated differently to account for the strain-specific R. palustris/E. coli ratios observed previously (7). Specifically, for R. palustris NxΔAmtB, 100 μl of the resuspended cells was used directly to inoculate each NxΔAmtB coculture. For R. palustris Nx, 1-ml aliquots of the resuspended cells were concentrated 10-fold into 100 μl of MDC and then inoculated to start each Nx coculture. Each R. palustris starter culture replicate was used to inoculate two corresponding cocultures, for a total of four cocultures of each type.
Each culture was allowed to double 4.5 to 5 times (Fig. 1C). The samples were then harvested (1 ml for Nx cocultures; 2 ml for monocultures and NxΔAmtB cocultures) and centrifuged, and the cell pellets were stored at –80°C for BarSeq analysis.
BarSeq sample preparation and sequencing.
Total genomic DNA (gDNA) was isolated from the T0, monoculture, and coculture cell pellets by using a Wizard genomic DNA purification kit (Promega) or a Bactozol kit (Molecular Research Center). To offset expected differences in E. coli abundance between monocultures, Nx cocultures, and NxΔAmtB cocultures (7), different amounts of total purified gDNA were used as the template in 50-μl BarSeq PCRs, as follows: 100 ng of gDNA from T0 and monocultures, 300 ng of gDNA from Nx cocultures, and 200 ng of gDNA from NxΔAmtB cocultures. BarSeq PCRs were performed using the previously described BarSeq98 protocol and primers, which add a unique experimental index to each sample to enable multiplex sequencing (12).
PCR product analysis, pooling, and Illumina sequencing were performed at the Indiana University Center for Genomics and Bioinformatics. The quality and concentration of the amplified PCR products were determined using an Agilent 2200 TapeStation. Equimolar concentrations of PCR product from all samples were pooled, and the pooled PCR products were purified using Agencourt AMpure beads (Beckman Coulter) and then sequenced using a single Illumina NextSeq 75 cycle high-output run.
Fitness data analysis, COG assignment, and statistical analyses.
Gene fitness values were determined from the BarSeq data using the described pipeline (12). Gene fitness scores and t-scores were averaged across quadruplicate replicates for each culture type, and genes with |mean fitness score| of >1 and a |mean t-score| of >3 in at least one culture type were considered to have strong fitness effects. Although polar effects are a potential confounding issue in transposon studies, we believe these effects to be minimal in the present study because of the following: (i) the transposon does not contain a transcriptional terminator; (ii) fitness scores were calculated only using mutants harboring insertions within the central 80% (10 to 90%) of a gene; and (iii) polar effects stemming from transposon insertions within operons were previously shown to have a negligible impact on ML9 fitness data (12). Indeed, in the present study there were instances where transposon insertions in different genes within a known operon resulted in markedly different fitness scores, including nagBACD, tatAB (tatCD was not represented in the library), and rfaQGPSBIJYZK (Table S2).
COG assignments for the 306 genes exhibiting strong fitness effects were curated manually by sequentially converting gene locus tags to K numbers and then to COG numbers using the files at https://www.genome.jp/dbget-bin/get_linkdb?-t+genes+gn:T00007 and https://www.genome.jp/kegg/files/ko2cog.xl. Since this method did not identify COG numbers for all genes, some COG numbers were assigned directly from the locus tags using the JGI Gene Search database (https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=GeneSearch&page=searchForm). The COG numbers were then used to assign COG categories based on the files available at two sites (ftp://ftp.ncbi.nih.gov/pub/COG/COG2014/static/lists/listEsccol.html and ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/COG0303/cogs.csv). Finally, COG categories were tallied using Microsoft Excel.
Principal-component analysis was performed in ClustVis (64) using all calculated fitness scores (3,564 genes). Results were plotted using GraphPad Prism 6.0. Hierarchical clustering (average linkage, uncentered coordination) was performed in Cluster 3.0 (65), and the resulting data were visualized using Java TreeView (66).
Mutant fitness validation using KEIO mutants.
Overnight cultures of KEIO mutants received glycerol to final concentration of 25% (vol/vol), and 250-μl aliquots were stored at –80°C to mimic the conditions used for RB-TnSeq experiments. Individual aliquots were thawed on ice and used to inoculate triplicate 6-ml aerobic starter cultures. At mid-exponential phase (OD660 = 0.40 to 0.55), cultures were pelleted (4,000 rpm, 5 min, room temperature), washed four times with 5 ml of MDC, and resuspended in MDC to an OD660 of 0.5. Triplicate R. palustris Nx starter cultures were prepared under carbon-limiting conditions as described above. The Nx cultures were resuspended to an OD660 of 0.3, and 1-ml cell aliquots were pelleted (4,000 rpm, 5 min, room temperature) for use below.
KEIO mutant monocultures were inoculated using 100 μl of washed cells. For cocultures, one cell pellet of each Nx replicate was resuspended in 100 μl of a washed KEIO mutant cell suspension and each resulting 100-μl pairing was used to inoculate one of three Nx coculture replicates. Cell densities (CFU/ml culture) were determined by selective plating using PM agar with 10 mM succinate but without (NH4)2SO4 or yeast extract for R. palustris and Luria-Bertani agar for E. coli. Statistical analyses were performed in GraphPad Prism 6.0.
Data availability.
Tn insertion site-barcode linkages were published previously (15) and are available at http://genomics.lbl.gov/supplemental/bigfit/. The raw reads used for BarSeq-based enumeration of barcodes from cultures in this study are available from the NCBI SRA under BioProject PRJNA613549.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. L. Foster for providing KEIO mutants, J. Liu at the IU Center for Genomics and Bioinformatics for assistance with Illumina sequencing, and A. M. Randich and members of the McKinlay laboratory for helpful discussions.
This study was supported by U.S. Army Research Office grant W911NF-14-1-0411.
We declare there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Widder S, Isaac Newton Institute Fellows, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, Kettle H, Flint HJ, Haas AF, Laroche B, Kreft J-U, Rainey PB, Freilich S, Schuster S, Milferstedt K, van der Meer JR, Grosskopf T, Huisman J, Free A, Picioreanu C, Quince C, Klapper I, Labarthe S, Smets BF, Wang H, Soyer OS. 2016. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J 10:2557–2568. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seth EC, Taga ME. 2014. Nutrient cross-feeding in the microbial world. Front Microbiol 5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR. 2018. Bacterial quorum sensing and microbial community interactions. mBio 9:e01749-18. doi: 10.1128/mBio.01749-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schluter J, Nadell CD, Bassler BL, Foster KR. 2015. Adhesion as a weapon in microbial competition. ISME J 9:139–149. doi: 10.1038/ismej.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelsic ED, Zhao J, Vetsigian K, Kishony R. 2015. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindemann SR, Bernstein HC, Song HS, Fredrickson JK, Fields MW, Shou W, Johnson DR, Beliaev AS. 2016. Engineering microbial consortia for controllable outputs. ISME J 10:2077–2084. doi: 10.1038/ismej.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaSarre B, McCully AL, Lennon JT, McKinlay JB. 2017. Microbial mutualism dynamics governed by dose-dependent toxicity of cross-fed nutrients. ISME J 11:337–348. doi: 10.1038/ismej.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCully AL, LaSarre B, McKinlay JB. 2017. Growth-independent cross-feeding modifies boundaries for coexistence in a bacterial mutualism. Environ Microbiol 19:3538–3550. doi: 10.1111/1462-2920.13847. [DOI] [PubMed] [Google Scholar]
- 9.McCully AL, LaSarre B, McKinlay JB. 2017. Recipient-biased competition for an intracellularly generated cross-fed nutrient is required for coexistence of microbial mutualists. mBio 8:e01620-17. doi: 10.1128/mBio.01620-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCully AL, Behringer MG, Gliessman JR, Pilipenko EV, Mazny JL, Lynch M, Drummond DA, McKinlay JB. 2018. An Escherichia coli nitrogen starvation response is important for mutualistic coexistence with Rhodopseudomonas palustris. Appl Environ Microbiol 84:e00404-18. doi: 10.1128/AEM.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helmann TC, Deutschbauer AM, Lindow SE. 2019. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc Natl Acad Sci U S A 116:18900–18910. doi: 10.1073/pnas.1908858116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentchel KL, Reyes Ruiz LM, Curtis PD, Fiebig A, Coleman ML, Crosson S. 2019. Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater. ISME J 13:523–536. doi: 10.1038/s41396-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 16.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A, Golden SS. 2015. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci U S A 112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin M, Pierce EC, Dutton RJ. 2018. Changes in the genetic requirements for microbial interactions with increasing community complexity. Elife 7:e37072. doi: 10.7554/eLife.37072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol Microbiol 86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 20.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Heeswijk WC, Westerhoff HV, Boogerd FC. 2013. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol Mol Biol Rev 77:628–695. doi: 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci U S A 97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninfa AJ, Jiang P. 2005. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol 8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Helling RB. 1998. Pathway choice in glutamate synthesis in Escherichia coli. J Bacteriol 180:4571–4575. doi: 10.1128/JB.180.17.4571-4575.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan D. 2007. Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci U S A 104:9475–9480. doi: 10.1073/pnas.0703360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. 2014. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun 5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. 1991. Overexpression of the relA gene in Escherichia coli. J Biol Chem 266:3760–3767. [PubMed] [Google Scholar]
- 29.Goodall ECA, Robinson A, Johnston IG, Jabbari S, Turner KA, Cunningham AF, Lund PA, Cole JA, Henderson IR. 2018. The essential genome of Escherichia coli K-12. mBio 9:e02096-17. doi: 10.1128/mBio.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montero M, Rahimpour M, Viale AM, Almagro G, Eydallin G, Sevilla Á, Cánovas M, Bernal C, Lozano AB, Muñoz FJ, Baroja-Fernández E, Bahaji A, Mori H, Codoñer FM, Pozueta-Romero J. 2014. Systematic production of inactivating and non-inactivating suppressor mutations at the relA locus that compensate the detrimental effects of complete spot loss and affect glycogen content in Escherichia coli. PLoS One 9:e106938. doi: 10.1371/journal.pone.0106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tedin K, Bremer H. 1992. Toxic effects of high levels of ppGpp in Escherichia coli are relieved by rpoB mutations. J Biol Chem 267:2337–2344. [PubMed] [Google Scholar]
- 32.Villadsen IS, Michelsen O. 1977. Regulation of PRPP and nucleoside tri and tetraphosphate pools in Escherichia coli under conditions of nitrogen starvation. J Bacteriol 130:136–143. doi: 10.1128/JB.130.1.136-143.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci U S A 105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kargeti M, Venkatesh KV. 2017. The effect of global transcriptional regulators on the anaerobic fermentative metabolism of Escherichia coli. Mol Biosyst 13:1388–1398. doi: 10.1039/c6mb00721j. [DOI] [PubMed] [Google Scholar]
- 36.Aviv M, Giladi H, Schreiber G, Oppenheim AB, Glaser G. 1994. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp, and by autoregulation. Mol Microbiol 14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 37.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyaneshwar P, Paliy O, McAuliffe J, Jones A, Jordan MI, Kustu S. 2005. Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc Natl Acad Sci U S A 102:3453–3458. doi: 10.1073/pnas.0500141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandel MJ, Silhavy TJ. 2005. Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J Bacteriol 187:434–442. doi: 10.1128/JB.187.2.434-442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. 2017. DksA and ppGpp regulate the sigma(S) stress response by activating promoters for the wmall RNA DsrA and the anti-adapter protein IraP. J Bacteriol 200:e00463-17. doi: 10.1128/JB.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pratt LA, Silhavy TJ. 1996. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci U S A 93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Gottesman S. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol 180:1154–1158. doi: 10.1128/JB.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Notley-McRobb L, King T, Ferenci T. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol 184:806–811. doi: 10.1128/jb.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jishage M, Kvint K, Shingler V, Nystrom T. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev 16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colland F, Barth M, Hengge-Aronis R, Kolb A. 2000. sigma factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and lrp transcription factors. EMBO J 19:3028–3037. doi: 10.1093/emboj/19.12.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CR, Cho SH, Kim HJ, Kim M, Peterkofsky A, Seok YJ. 2010. Potassium mediates Escherichia coli enzyme IIA(Ntr)-dependent regulation of sigma factor selectivity. Mol Microbiol 78:1468–1483. doi: 10.1111/j.1365-2958.2010.07419.x. [DOI] [PubMed] [Google Scholar]
- 49.Pfluger-Grau K, Gorke B. 2010. Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol 18:205–214. doi: 10.1016/j.tim.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Harcombe W. 2010. Novel cooperation experimentally evolved between species. Evolution 64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 51.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. 2014. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J 8:953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziesack M, Gibson T, Oliver JKW, Shumaker AM, Hsu BB, Riglar DT, Giessen TW, DiBenedetto NV, Bry L, Way JC, Silver PA, Gerber GK. 2019. Engineered interspecies amino acid cross-feeding increases population evenness in a synthetic bacterial consortium. mSystems 4:e00352-19. doi: 10.1128/mSystems.00352-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picon A, Teixeira de Mattos MJ, Postma PW. 2005. Reducing the glucose uptake rate in Escherichia coli affects growth rate but not protein production. Biotechnol Bioeng 90:191–200. doi: 10.1002/bit.20387. [DOI] [PubMed] [Google Scholar]
- 54.Chubukov V, Sauer U. 2014. Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol 80:2901–2909. doi: 10.1128/AEM.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marolewski A, Smith JM, Benkovic SJ. 1994. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from Escherichia coli. Biochemistry 33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- 56.Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol 188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koch AL, Putnam FW, Evans EA Jr. 1952. The purine metabolism of Escherichia coli. J Biol Chem 197:105–112. [PubMed] [Google Scholar]
- 58.Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, Wu W, Mobley HLT. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13:e1006434. doi: 10.1371/journal.ppat.1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibberson CB, Stacy A, Fleming D, Dees JL, Rumbaugh K, Gilmore MS, Whiteley M. 2017. Coinfecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat Microbiol 2:17079. doi: 10.1038/nmicrobiol.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. 2019. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc Natl Acad Sci U S A 116:19685–19694. doi: 10.1073/pnas.1907619116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 62.Kim M-K, Harwood CS. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiology Lett 83:199–203. doi: 10.1111/j.1574-6968.1991.tb04440.x-i1. [DOI] [Google Scholar]
- 63.McKinlay JB, Zeikus JG, Vieille C. 2005. Insights into Actinobacillus succinogenes fermentative metabolism in a chemically defined growth medium. Appl Environ Microbiol 71:6651–6656. doi: 10.1128/AEM.71.11.6651-6656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metsalu T, Vilo J. 2015. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 66.Saldanha AJ. 2004. Java Treeview: extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tn insertion site-barcode linkages were published previously (15) and are available at http://genomics.lbl.gov/supplemental/bigfit/. The raw reads used for BarSeq-based enumeration of barcodes from cultures in this study are available from the NCBI SRA under BioProject PRJNA613549.