In all domains of life, RNA turnover is important for gene regulation and quality control. The process of RNA metabolism is regulated by many RNA-processing enzymes and assistant proteins, where these proteins usually exist as complexes. However, there is little known about the RNA metabolism, as well as about the RNA degradation complex. In the present study, we described an RNA degradosome-like complex in cyanobacteria and revealed an assembly mechanism different from that of E. coli. Moreover, CrhB could help RNase E in Anabaena sp. strain PCC 7120 degrade double-stranded RNA with a 5′ tail. In addition, CrhB and AnaRne have similar cytoplasm localizations, in contrast to the membrane localization in E. coli.
KEYWORDS: Anabaena sp. strain PCC 7120, CrhB, degradosome, enolase, RNase E
ABSTRACT
At present, little is known about the RNA metabolism driven by the RNA degradosome in cyanobacteria. RNA helicase and enolase are the common components of the RNA degradosome in many bacteria. Here, we provide evidence that both enolase and the DEAD-box RNA helicase CrhB can interact with RNase E in Anabaena (Nostoc) sp. strain PCC 7120 (referred to here as PCC 7120). Furthermore, we found that the C-terminal domains of CrhB and AnaEno (enolase of PCC 7120) are required for the interaction, respectively. Moreover, their recognition motifs for AnaRne (RNase E of PCC 7120) turned out to be located in the N-terminal catalytic domain, which is obviously different from those identified previously in Proteobacteria. We also demonstrated in enzyme activity assays that CrhB can induce AnaRne to degrade double-stranded RNA with a 5′ tail. Furthermore, we investigated the localization of CrhB and AnaRne by green fluorescent protein (GFP) translation fusion in situ and found that they both localized in the center of the PCC 7120 cytoplasm. This localization pattern is also different from the membrane binding of RNase E and RhlB in Escherichia coli. Together with the previous identification of polynucleotide phosphorylase (PNPase) in PCC 7120, our results show that there is an RNA degradosome-like complex with a different assembly mechanism in cyanobacteria.
IMPORTANCE In all domains of life, RNA turnover is important for gene regulation and quality control. The process of RNA metabolism is regulated by many RNA-processing enzymes and assistant proteins, where these proteins usually exist as complexes. However, there is little known about the RNA metabolism, as well as about the RNA degradation complex. In the present study, we described an RNA degradosome-like complex in cyanobacteria and revealed an assembly mechanism different from that of E. coli. Moreover, CrhB could help RNase E in Anabaena sp. strain PCC 7120 degrade double-stranded RNA with a 5′ tail. In addition, CrhB and AnaRne have similar cytoplasm localizations, in contrast to the membrane localization in E. coli.
INTRODUCTION
Cyanobacteria are Gram-negative bacteria characterized by oxygen-evolving photosynthesis and represent a morphologically diverse phylum, which is believed to have contributed to the initial oxygenation of the Earth’s atmosphere (1). This transformation eventually led to the emergence of aerobic respiration (2). As the earliest and most important producers of oxygen, cyanobacteria undoubtedly play an important part in the whole biosphere (3). However, with a changing environment, cyanobacteria face the same challenges as other organisms do. Tight regulation of gene expression is necessary for organisms to adapt to various growth and stress conditions, and this dynamic process is regulated at various stages while genetic information is transmitted from DNA to RNA to proteins (4). This process requires regulation of the steady-state level of transcripts that is controlled by both RNA synthesis and degradation.
All bacterial species have a variety of genes encoding enzymes involved in RNA metabolism. Mostly, these components are functionally, and often physically, linked (5). Among these enzymes, endoribonuclease RNase E is the most important. In addition to its highly conserved N-terminal nuclease activity, RNase E contains a variable C-terminal scaffolding domain that coordinates the formation of a multienzyme complex named the RNA degradosome (6). The typical Escherichia coli RNA degradosome is composed of RNase E, DEAD-box RNA helicase RhlB, phosphorolytic exoribonuclease polynucleotide phosphorylase (PNPase), and the glycolytic enzyme enolase (5, 7–9). Moreover, it has been reported that the degradosome associates with some minor components (such as RraA, RraB, and Hfq) and translating ribosomes (10–12).
The DEAD-box RNA helicase is a common component of the RNA degradosome (5). RNase E and PNPase are single-strand-specific enzymes, whereas RNA helicase can facilitate degradation of double-stranded structures via its ability to unwind secondary structures (13, 14). Two RNA helicase genes, alr1223 (crhB) and alr4718 (crhC), have been found in Anabaena (Nostoc) sp. strain PCC 7120 (referred to here as PCC 7120), and both their products are members of the DEAD-box family (15). All the DEAD-box RNA helicases have two conserved RecA-like domains, which contain 12 motifs involved in the binding of ATP (Q, I, II, and VI), in interaction with RNA (Ia, Ib, Ic, IV, IVa, and V), and in intramolecular interactions (III and Va) (4, 16). In PCC 7120, CrhC is an ATP-independent RNA binding protein with RNA-dependent ATPase activity and unwinding activity in the 5′→3′ direction (17) and is specifically expressed at low temperatures (18, 19). In contrast, CrhB is expressed under many environmental conditions and is most closely related to the single DEAD-box RNA helicase CrhR in Synechocystis sp. strain PCC 6803 (15, 18). Moreover, CrhR can combine with the components of both a polysome and an RNA degradosome (20). In PCC 7120, in addition to CrhB, there is another common component, AnaEno (enolase of PCC 7120), which is encoded by all3538. Enolase is a key enzyme of glycolysis. In E. coli, prevention of the assembly of enolase and the C terminus of RNase E affects the metabolism of many mRNAs related to energy-producing metabolic pathways (21). Furthermore, a recent study showed that enolase participates in RNase E/degradosome-based regulation of bacterial morphology under oxygen-limited conditions (22).
In this study, we expanded the characterization of the RNA degradation complex in PCC 7120, which is usually employed as a model strain for studying nitrogen fixation and heterocyst differentiation. Copurification analyses showed that both CrhB and AnaEno can form complexes with AnaRne (RNase E of PCC 7120) in vivo. Then, by far-Western blotting assays and bacterial two-hybrid and pulldown experiments performed in vitro, we revealed that the C-terminal domains of CrhB and AnaEno and the N-terminal domain of AnaRne take part in these interactions. Moreover, the functional significance of CrhB was demonstrated by degradation assays and the results indicated that CrhB can cause AnaRne to degrade a structured RNA with a 5′ tail. Furthermore, by means of experiments employing green fluorescence protein (GFP) fusion proteins whose expression was driven by the native promoters, we demonstrated that both CrhB and AnaRne colocalize in the cytoplasm. Combined with our previous finding that PNPase can form a complex with AnaRne (23), our data suggest that there is a multicomponent complex in PCC 7120 that is functionally similar to the RNA degradosome in E. coli. To our knowledge, this finding will further expand the literature on the convergence of (and differences in) the evolutionary characteristics of the RNA degradosome.
RESULTS
Identification of the interaction of AnaEno and CrhB with AnaRne in vivo.
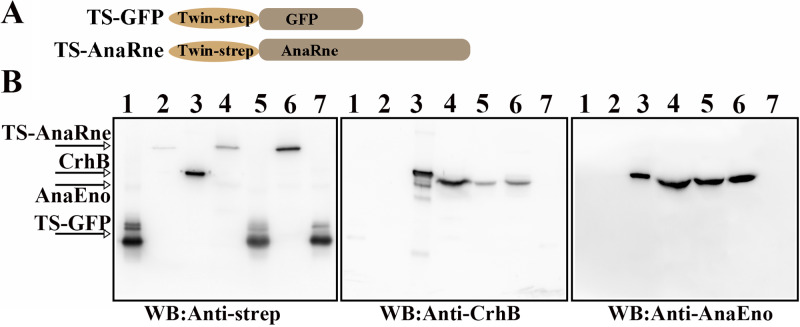
RNase E and PNPase can form a complex in PCC 7120, and this interaction is a common mechanism in cyanobacteria (23). Here, a copurification experiment was carried out to confirm that in addition, AnaEno and CrhB individually form a complex each with AnaRne. To facilitate this experiment, we constructed strains oeRne and oeGFP to overexpress AnaRne and GFP, respectively, with a C-terminal TwinStrep tag (named TS-AnaRne and TS-GFP, respectively; Fig. 1A). As shown in Fig. 1B, the corresponding signal bands of TS-AnaRne and TS-GFP were detected with a specific polyclonal antibody against the Strep tag (anti-Strep tag [anti-strep] antibody) in both the total-protein samples and the eluted samples (left, lanes 4 to 7). Moreover, there were bands of CrhB in the total-protein samples of both strain oeRne and strain oeGFP after probing with a specific polyclonal antibody against CrhB (anti-CrhB antibody; middle, lanes 4 and 5), whereas this band appeared only in the eluted sample of strain oeRne (middle, lane 6 versus the oeGFP control in lane 7), indicating that CrhB can interact with AnaRne and then bind to Strep-Tactin Sepharose. The slight difference in size between SCrhB (CrhB with a C-terminal Strep tag [a polypeptide sequence containing amino acids WSHPQFEK]) and CrhB may have been due to the Strep tag. The detection of AnaEno was similar to that of CrhB. AnaEno could be detected in the elution fraction of strain oeRne by Western blotting with a specific polyclonal antibody against AnaEno (anti-AnaEno antibody), which was not the case for strain oeGFP (right, lane 6 versus lane 7). These results indicated that the proteins that had copurified with AnaRne were CrhB and AnaEno.
FIG 1.
Analyses of the interaction of CrhB and AnaEno with AnaRne in PCC 7120 through copurification. (A) Schematic presentation of TS-GFP and TS-AnaRne in PCC 7120. TS-GFP was expressed as a negative control. (B) Western blotting (WB) detected the proteins interacting with AnaRne in the elution fraction with anti-strep (left), anti-CrhB (middle), and anti-AnaEno antibodies (right). Cell lysates of oeRne or oeGFP were applied to the Strep-Tactin column. TS-AnaRne served as bait, while TS-GFP as a negative control. Lanes 1, Strep-tagged GFP (SGFP); lanes 2, Strep-tagged AnaRne (SAnaRne); lanes 3, Strep-tagged CrhB (SCrhB; left and middle) or Strep-tagged AnaEno (SAnaEno [AnaEno with a C-terminal Strep tag]; right); lanes 4, total protein of strain oeRne; lanes 5, total protein of strain oeGFP; lanes 6, an elution sample from oeRne; lanes 7, an elution sample from oeGFP. Purified proteins SGFP, SAnaRne, SCrhB, and SAnaEno served as controls to evaluate the size of the corresponding proteins. Results are representative of three independent experiments.
The C-terminal domain of CrhB directly interacts with AnaRne.
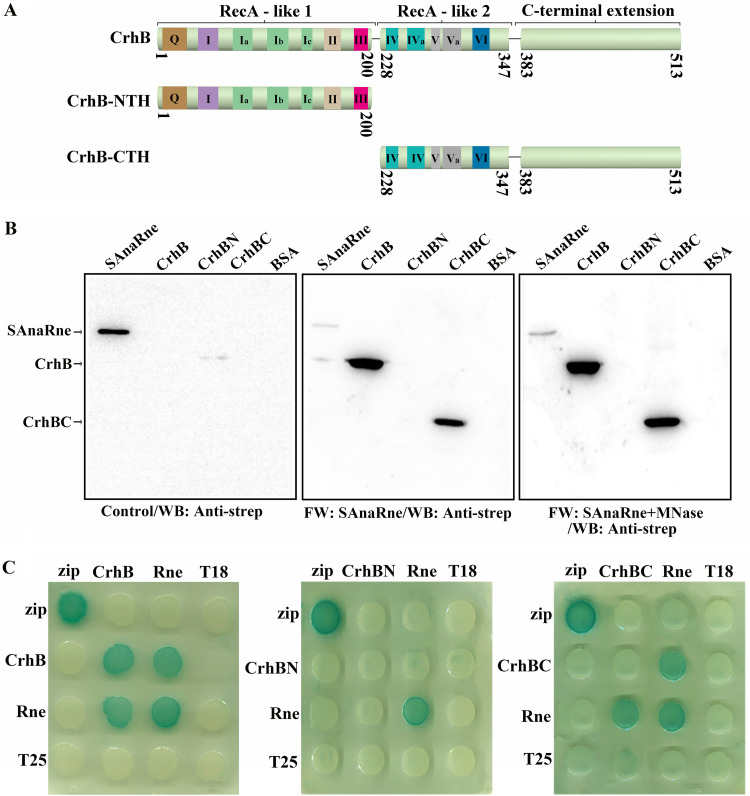
Following the identification of the interaction between CrhB and AnaRne in vivo, we set out to analyze this association in vitro. A previous study revealed that E. coli RNase E (EcRne) can bind to RNA and interact with some proteins in an RNA-mediated manner (23, 24). For this reason, it was necessary to check whether there is a direct interaction between AnaRne and CrhB. Thus, a far-Western blotting assay was performed to characterize this interaction by means of recombinantly expressed and purified AnaRne and CrhB. In this assay, we employed micrococcal nuclease (MNase) to avoid the possible interference from RNA. As presented in Fig. 2B, whether it was treated with MNase (right panel) or not (middle panel), the signal of the interaction between AnaRne and CrhB was evident, suggesting that the interaction was direct rather than being mediated by RNA. Furthermore, the region of CrhB required for this binding was pinpointed. The CrhB was first divided into two halves according to the conserved domains of the DEAD-box RNA helicases: the N-terminal half (residues 1 to 200, CrhB-NTH) and the C-terminal half (residues 228 to 513, CrhB-CTH) (Fig. 2A). After that, their ability to interact with AnaRne was studied by far-Western blotting. There was an obvious signal in the CrhB-CTH group, but there was none in the CrhB-NTH group (Fig. 2B). From these data, we concluded that CrhB-CTH participated in the direct interaction with AnaRne. Furthermore, we complemented our analysis by two-hybrid experiments in the E. coli system and found that both CrhB and CrhB-CTH can bind to AnaRne, whereas CrhB-NTH cannot (Fig. 2C). These results indicated that CrhB-CTH is necessary for this interaction. In addition, this assay uncovered a self-interaction of CrhB and AnaRne in the E. coli system.
FIG 2.
CrhB-CTH can interact with AnaRne directly. (A) Schematic of the domain composition of CrhB according to the alignment. The numbers indicate the amino acid positions. (B) Far-Western blotting assay was conducted to evaluate the interaction between recombinant AnaRne and CrhB (or the truncated variants). The PVDF membrane represented on the left was incubated with skim milk as a control, the one in the middle was incubated with SAnaRne, and the one on the right was incubated with SAnaRne and MNase. SAnaRne and BSA served as a positive control and a negative control, respectively. Other proteins were purified with the C-terminal His tag. “FW” represents far-Western blotting. The final results were detected with the anti-strep antibody. The amount of protein used here was 1 μg, with the exception that 20 ng was loaded for SAnaRne. (C) Bacterial two-hybrid analysis of the interactions of CrhB (or its truncated variants) and AnaRne. “Rne” denotes AnaRne, “CrhBN” represents CrhB-NTH, and “CrhBC” stands for CrhB-CTH. The horizontal rows represent the E. coli BTH101 cells carrying the recombinant “T18” protein fusion plasmids, whereas the vertical columns represent the cells carrying the recombinant “T25” protein fusion plasmids. These results were based on three independent experiments.
Interaction of CrhB with the catalytic domain of AnaRne.
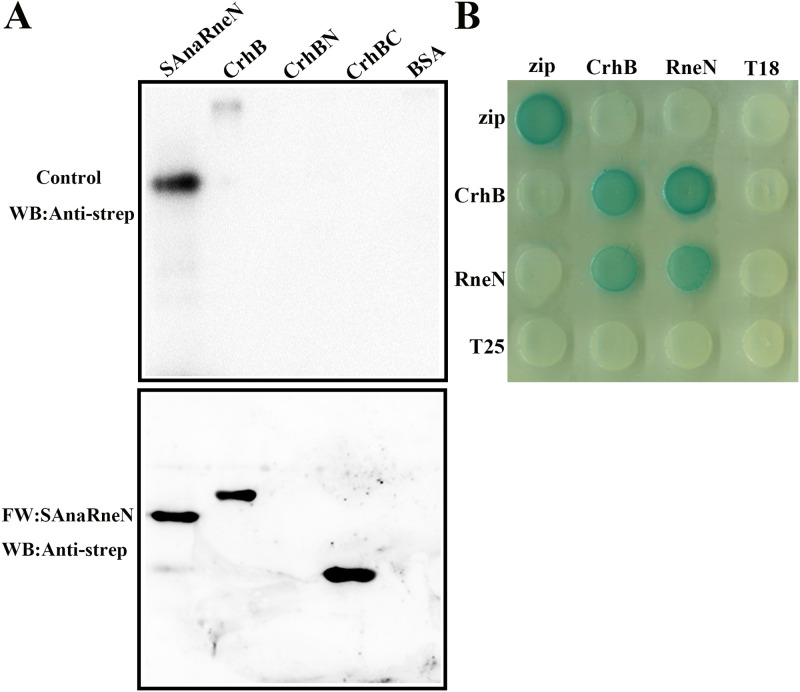
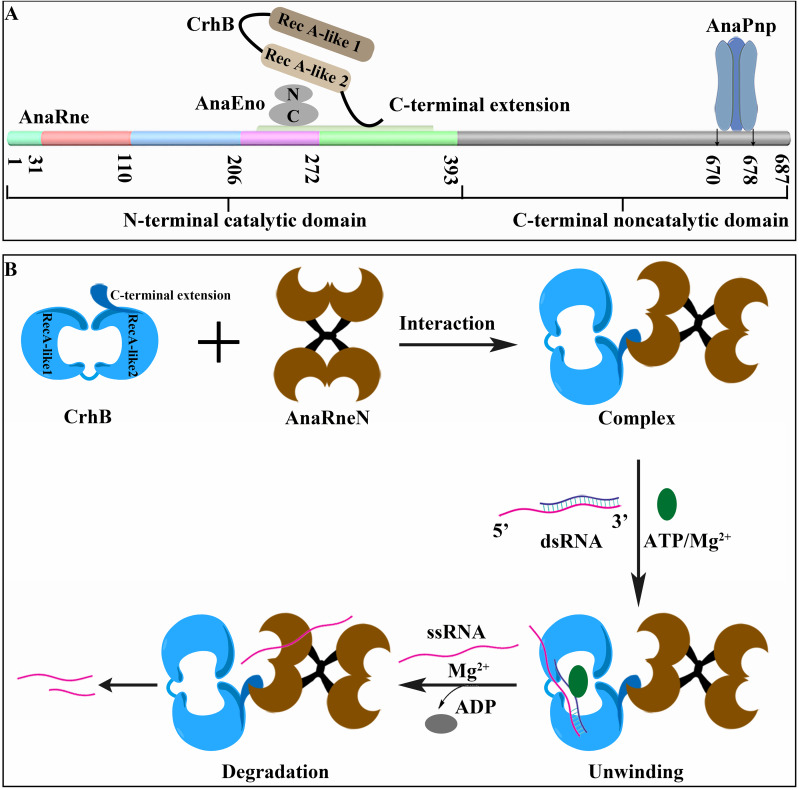
Just as in the case of EcRne, AnaRne consists of an N-terminal catalytic domain (residues 1 to 396 of AnaRne, i.e., AnaRneN) and a C-terminal noncatalytic domain (residues 401 to 687 of AnaRne, i.e., AnaRneC). And AnaPnp (PNPase in PCC 7120) can also interact with AnaRneC (23). In addition, residues 719 to 731 of the EcRne noncatalytic C terminus are responsible for the interaction with RhlB (25). Thus, to verify whether AnaRneC also forms the scaffolding for the RNA degradosome assembly, we purified recombinant proteins Strep-tagged AnaRneN (SAnaRneN [AnaRneN with a C-terminal Strep tag]) and Strep-tagged AnaRneC (SAnaRneC [AnaRneC with a C-terminal Strep tag]) from E. coli cells and determined the CrhB-binding domain of AnaRne by a far-Western blotting assay performed in vitro. As illustrated in Fig. 3, there were interaction signals for AnaRneN (Fig. 3A), but there were none for AnaRneC (see Fig. S1A in the supplemental material). This phenomenon meant that the CrhB-binding domain of AnaRne was located in the N-terminal catalytic domain.
FIG 3.
Identification of the CrhB interaction domain in AnaRne. (A) Far-Western blotting assays designed to determine the regions of AnaRne that interacted with CrhB. One of the PVDF membranes was incubated with SAnaRneN (bottom), and the other was incubated with skim milk as a control (top). SAnaRneN served as a positive control, while BSA served as a negative control. The other proteins were purified with the C-terminal His tag. The final results were also detected with the anti-strep antibody. The amount of proteins loaded was 1 μg, except for the positive controls (20 ng). (B) Bacterial two-hybrid analysis of the interaction between AnaRneN and CrhB. “RneN” represents AnaRneN. The horizontal rows represent the E. coli BTH101 cells carrying the recombinant “T18” protein fusion plasmids, whereas the vertical columns represent the cells carrying the recombinant “T25” protein fusion plasmids. These results were based on three independent experiments.
In agreement with the results described above, the interaction between AnaRneN and CrhB was demonstrated in two-hybrid experiments again (Fig. 3B). In contrast, there was no signal for AnaRneC (Fig. S1B). This finding also showed that CrhB is most likely to interact with the N-terminal regions of AnaRne.
The C-terminal extension of CrhB interacts with AnaRneN.
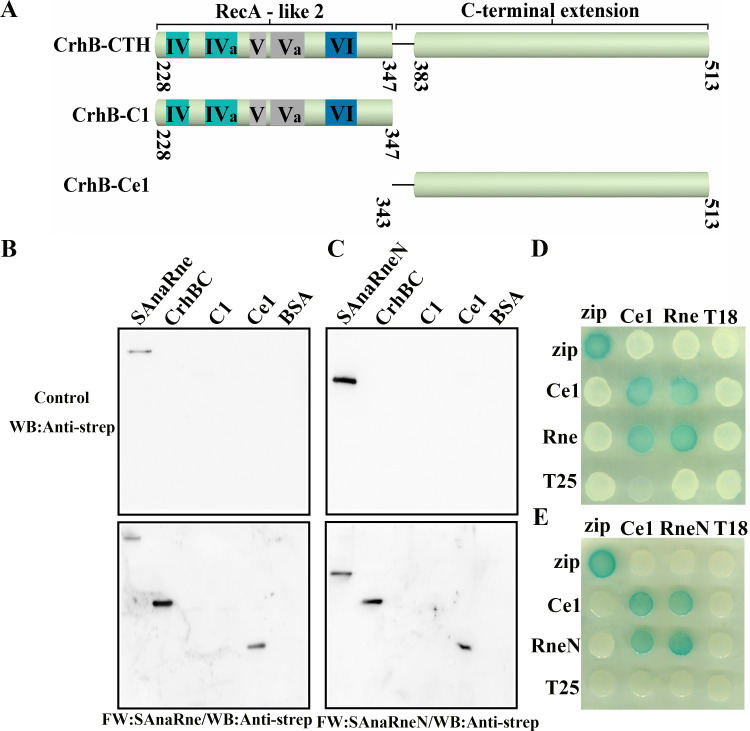
Like other DEAD-box proteins, CrhB-CTH contains one RecA-like fold (CrhB-C1) and a C-terminal extension (CrhB-Ce1). Generally, the C-terminal extension is responsible for the specific interactions with RNA or regulatory proteins (20, 26, 27). In addition, our study indicated that CrhB-CTH can associate with AnaRne. Therefore, we then tested whether the C-terminal extension of CrhB is required for its interaction with AnaRne. We constructed a series of truncations of CrhB-CTH (Fig. 4A), and their interaction with AnaRne was tested by far-Western blotting assays and two-hybrid experiments. As presented in Fig. 4B, an obvious signal for CrhB-Ce1 binding to AnaRne was detected, whereas there was none for CrhB-C1. Furthermore, we detected an interaction signal for AnaRneN and CrhB-Ce1 (Fig. 4C). Next, two-hybrid experiments were conducted to verify these interactions and the results showed that CrhB-Ce1 was able to interact with both AnaRne and AnaRneN (Fig. 4D and E). This finding is consistent with the data from the far-Western blotting assays, indicating that the C-terminal extension of CrhB can drive the interaction with AnaRne.
FIG 4.
The C-terminal extension of CrhB mediates the association with AnaRneN. (A) Schematic of CrhB-CTH and its truncated derivatives according to the alignment. The numbers indicate the amino acid positions. (B and C) Far-Western blotting assay evaluating the role of the deletion variants of CrhB-CTH in the interaction of CrhB-CTH and AnaRne. The PVDF membranes represented at the bottom of panels B and C were incubated with SAnaRne (B) and SAnaRneN (C), respectively, and the other two represented at the top were incubated with skim milk as controls. “C1” stands for CrhB-C1, and “Ce1” means CrhB-Ce1. SAnaRne and SAnaRneN were chosen as positive controls, while BSA was a negative control. The other proteins were purified with the C-terminal His tag. The anti-strep antibody was used to detect the interaction. The amount of protein was 1 μg, except for the positive controls (20 ng). (D and E) Bacterial two-hybrid assay to verify the interaction between CrhB-Ce1 and AnaRne. The rows represent the E. coli BTH101 cells carrying the recombinant “T18” protein fusion plasmids, while the columns represent the cells carrying the recombinant “T25” protein fusion plasmids. These results were based on three independent experiments.
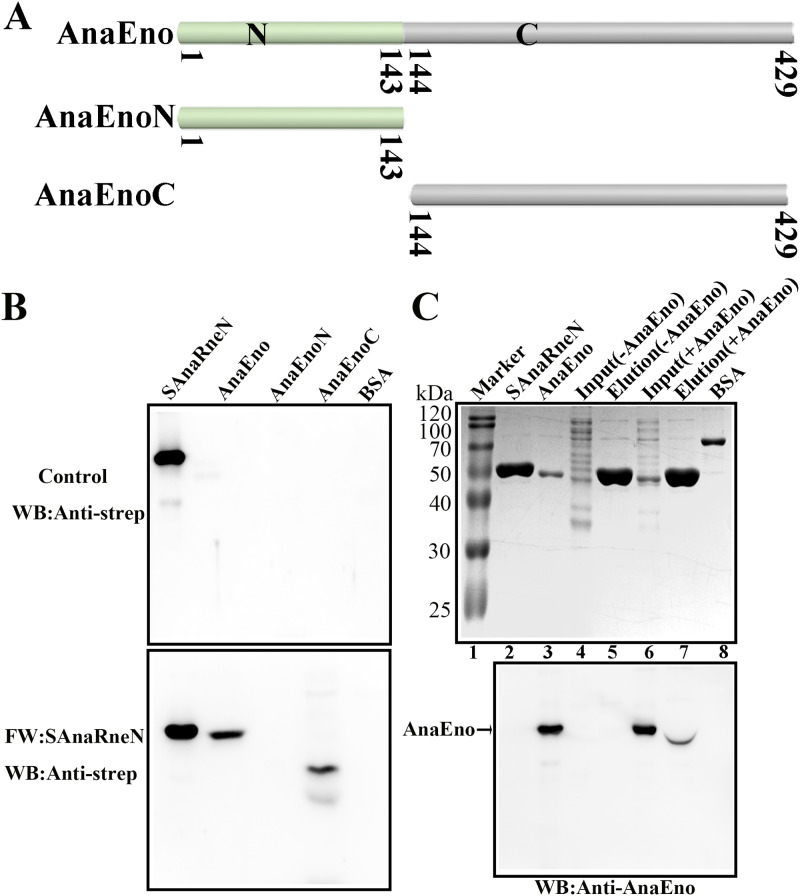
The C-terminal domain of AnaEno is associated with the catalytic domain of AnaRne.
Similarly, to identify the interaction domains of AnaEno and AnaRne in vitro, an additional far-Western blotting assay was performed. In this assay, we employed recombinantly expressed and purified AnaEno and its truncated versions. First, we performed a Simple Modular Architecture Research Tool (SMART) analysis and subdivided AnaEno into AnaEnoN and AnaEnoC according to the sequence conservation results (Fig. 5A). As depicted in Fig. 5B, when incubated with SAnaRneN, both AnaEno and AnaEnoC engaged in reciprocal interactions with AnaRneN, but AnaEnoN did not. In contrast, during incubation with SAnaRneC, the interaction signal was not detectable (Fig. S2), implying that SAnaRneC might not have participated in this interaction; again in contrast, residues 833 to 850 in the C-terminal half of EcRne are responsible for the interaction with enolase (25). Furthermore, we complemented our analysis with a pulldown experiment using SAnaRneN as the bait. Given that AnaRneN and AnaEno have similar molecular weights (∼50 kDa), it was difficult to distinguish AnaEno by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Fig. 5C, top). Thus, a Western blotting assay was carried out with anti-AnaEno antibody to detect AnaEno. As expected, after incubation of SAnaRneN-coupled resin with the cell lysate containing AnaEno, there was a clear-cut signal at the same position, corresponding to 50 kDa (Fig. 5C, bottom, lane 7), suggesting that AnaEno can bind to SAnaRneN, whereas this was not the case in the negative control (Fig. 5C, bottom, lane 5). Thus, we concluded that the C-terminal domain of AnaEno was able to interact with the catalytic domain of AnaRne.
FIG 5.
The C-terminal domain of AnaEno can interact with AnaRneN. (A) Schematic of AnaEno and its truncated derivatives based on the SMART analysis. SMART analysis is available as a Web resource (http://smart.embl.de) designed to identify and annotate protein domains and to analyze protein domain architectures (51). The numbers indicate the amino acid positions. (B) Far-Western blotting assay investigating the interaction between AnaEno and AnaRneN. One of the PVDF membranes was incubated with SAnaRneN (bottom). The other (top) was incubated with skim milk as a control. The final results were detected with the anti-strep antibody. SAnaRneN purified from E. coli was employed as a positive control, while BSA was employed as a negative control. Other proteins were purified with the C-terminal His tag. The amount of protein was 1 μg, with the exception that it was 20 ng for the positive control. (C) The pulldown assay detecting the interaction of AnaRneN and AnaEno. Cell lysates expressing AnaEno or not were applied to Strep-Tactin Sepharose with bound SAnaRneN and eluted with d-biotin. Eluted samples were loaded onto a 10% gel for SDS-PAGE (top) and were next subjected to WB with the anti-AnaEno antibody for detection (bottom). Marker lane (lane 1), Blue Plus II protein markers (TransGen) (14 to 120 kDa); lane 2, SAnaRneN (purified from E. coli) (positive control); lane 3, AnaEno (purified from E. coli) (positive control); lane 4 [Input (-AnaEno)], a cell lysate without AnaEno input; lane 5, [Elution (-AnaEno)], an elution sample after incubation with SAnaRneN and cell lysate as described for lane 4; lane 6 [Input (+AnaEno)], cell lysate expressing AnaEno input; lane 7 [Elution (+AnaEno)], an elution sample following incubation with SAnaRneN and cell lysate as described for lane 6; lane 8 (BSA), negative control. Results presented here are representative of three independent experiments.
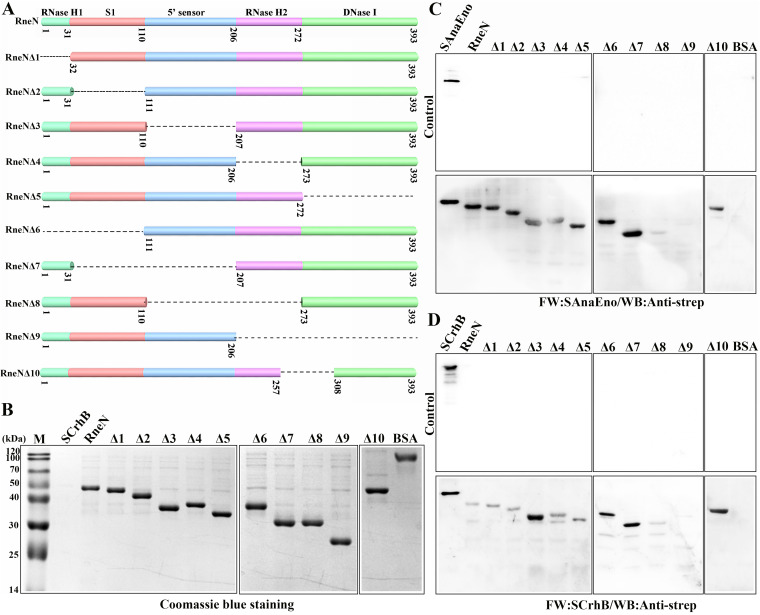
AnaEno and CrhB interact with the N-terminal RNase H2 and DNase I regions of AnaRne.
The data presented so far indicated that AnaEno and CrhB both mainly combined with the N-terminal catalytic domain of AnaRne. It is known that the conserved N-terminal domain of RNase E is mostly responsible for the catalytic activity, and few studies have focused on its interaction with other components. Therefore, to identify the specific motifs in AnaRneN that associate with AnaEno and CrhB, we first performed a sequence alignment of RNase E homologs from 16 representative cyanobacterial strains (23) corresponding to EcRne (Fig. S3). The N-terminal catalytic domain of AnaRne was divided into five subdomains in accordance with established folds (Fig. 6A). Then, we constructed a series of deletion mutants of AnaRneN (Fig. 6A) and detected these proteins purified from E. coli by SDS-PAGE (Fig. 6B). Finally, their interactions with AnaEno and CrhB were investigated by far-Western blotting. Of note, interaction signals were detectable in any single-subdomain deletion mutants (Fig. 6C and D, Δ1 to Δ5), but there was no signal when the RNase H2 and DNase I subdomains were deleted together (Fig. 6C and D, Δ9).
FIG 6.
A far-Western blotting assay designed to identify the specific motif for AnaRneN that can interact with AnaEno and CrhB. (A) Schematic representation of AnaRneN and the deletion mutants. Structural subdomains of AnaRneN are shown at the top. The numbers beneath indicate the amino acid positions. The missing parts are represented by dashed lines. (B) SDS-PAGE analysis of AnaRneN and its truncated variants. The proteins were all purified from E. coli. The gel was stained with Coomassie blue. (C and D) Far-Western blotting assay analyzing the interaction between AnaEno (C) or CrhB (D) and the different truncations of AnaRneN. The PVDF membranes represented at the bottom of panels C and D were incubated with SAnaEno (C) or SCrhB (D), whereas the others represented at the top of the panels were incubated with skim milk as controls. The interaction signals were detected with the anti-strep antibody. The samples used here are indicated above the lanes. SAnaEno, SCrhB, and BSA were used as controls. The other proteins were purified via the use of a C-terminal His tag. The amount of protein loaded in each lane was 1 μg, with the exception that it was 20 ng for SAnaEno and SCrhB. Data corresponding to protein levels and interactions are representative of results from three independent experiments.
According to the results described above, we carefully analyzed the alignment of RNase E proteins from all sequenced cyanobacterial strains and noted that there is a conserved subregion across RNase H2 and DNase I subdomains. Next, we constructed a deletion mutant lacking this subregion (Fig. 6C and D, Δ10). Unexpectedly, there were still signal bands in the far-Western blotting experiment (Fig. 6C and D, Δ10). Given that the N-terminal catalytic domain of RNase E folds into a compact structure, the AnaEno- and CrhB-binding sites might therefore correspond to a surface of AnaRne that contains residues from different regions of the primary amino acid sequence. Accordingly, on the basis of these results, we are sure only that the RNase H2 and DNase I subdomains of AnaRneN are likely responsible for these interactions.
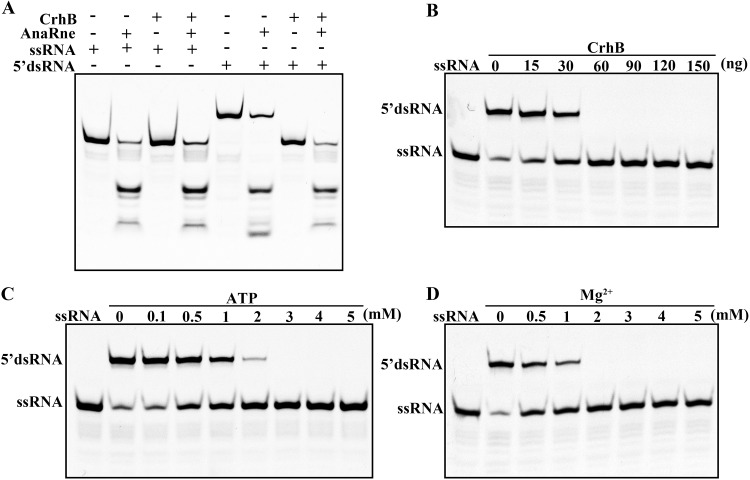
CrhB helps AnaRne to degrade the 5′ tailed duplex by its unwinding activity.
RhlB of E. coli is an ATP-dependent RNA helicase, which unfolds structured RNA to promote the degradation of the substrate by partner ribonucleases. In accordance with the interaction of CrhB and AnaRne, the functional consequences of this interaction were then examined in vitro. We first used the 22-mer RNA (single-stranded RNA [ssRNA]) and the artificial double-stranded RNA (dsRNA) substrates containing the 5′ 9-nucleotide overhang (5′ dsRNA) as substrates in AnaRne activity assays in the presence or absence of CrhB. As depicted in Fig. 7A, AnaRne almost completely degraded ssRNA, which was not the case for the 5′ dsRNA in the absence of CrhB. Nevertheless, when CrhB was added into the same reaction system, the degradation of 5′ dsRNA by AnaRne was able to reach a level comparable to that of ssRNA (Fig. 7A). The partial degradation of the dsRNA by AnaRne may have been due to the incomplete annealing of a single chain. Then, the unwinding-activity assay of recombinant purified CrhB confirmed that CrhB could unwind 5′ dsRNA (Fig. 7B). In addition, this reaction was ATP and Mg2+ dependent (Fig. 7C and D). These data suggested that the unwinding ability of CrhB can promote the degradation of dsRNA by AnaRne rapidly.
FIG 7.
Influence of CrhB on the catalytic activity of AnaRne in vitro. (A) Enzyme activity assays of AnaRne in the presence or absence of CrhB. +, presence of protein(s) or RNA; −, absence of protein(s) or RNA. (B) Unwinding activity of CrhB on 5′ dsRNA. CrhB concentrations were adjusted as indicated. (C) The RNA unwinding activity of CrhB depends on ATP. ATP concentrations in the reaction are shown above the lanes. (D) The RNA unwinding activity of CrhB is Mg2+ dependent. The Mg2+ concentrations are shown above the lanes. The RNA sequences are described in Materials and Methods. Data correspond to results from three independent experiments.
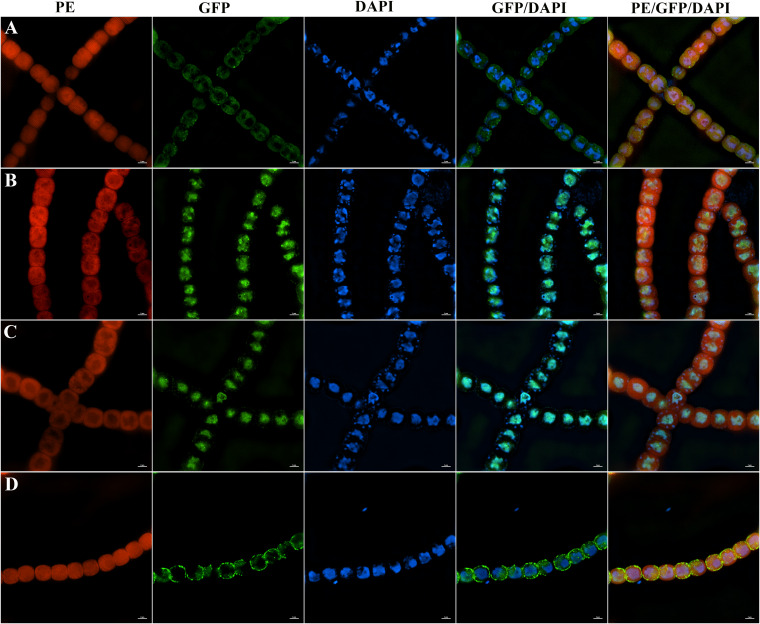
CrhB and AnaRne both localize in the center of the cytoplasm.
RNase E is associated with the membrane in E. coli, and this property is common among RNase E/G homologs in many other bacteria. Additionally, the RNA degradosome component RhlB can also associate with a membrane depending on a direct protein-protein interaction with RNase E (28). For this reason, the functional AnaRne-GFP (AnaRne fused to GFP) and CrhB-GFP (CrhB fused to GFP) fusions synthesized from their native promoters in situ were constructed to determine whether the same property exists in PCC 7120. The fusion of GFP in situ was unable to interfere with the functionalities of these target proteins, as the growth phenotype of the translation fusion strains was similar to that of the wild type (WT) (Fig. 8). The septum site-determining protein MinD combined with the cytoplasmic membrane (29) served as a control for membrane localization (Fig. 8D). As illustrated in Fig. 8, CrhB-GFP and AnaRne-GFP fusion proteins manifested a distribution relatively highly concentrated in the center of the cytoplasm (Fig. 8B and C). This localization pattern was different from that in E. coli. After that, we performed DAPI (4′,6-diamidino-2-phenylindole) staining to examine the nucleoid DNA and further elucidated whether CrhB and AnaRne colocalize with nucleoid DNA. As shown in the merged images in Fig. 8, the cellular localizations of CrhB and AnaRne were mostly but not completely consistent with those of DNA. However, there were many obvious fluorescence spots mainly distributed around the nucleoid, implying that the randomly distributed clusters of CrhB and AnaRne were in near proximity to DNA (Fig. 8B and C). In addition, the GFP alone was distributed throughout the cell, which was different from the results seen with CrhB and AnaRne (Fig. S4). These results indicated that CrhB and AnaRne mainly localized in the center of the cytoplasm. The association between DNA and the RNA-processing enzymes therefore suggests that mRNA decay in PCC 7120 may be arranged spatially according to chromosomal organization.
FIG 8.
The localization of CrhB and AnaRne in PCC 7120. (A) Localization of DNA (DAPI) in WT PCC 7120. (B) Covisualization of the localization of CrhB-GFP and DNA (DAPI) in strain CrhB-GFP. (C) Covisualization of the localization of AnaRne-GFP and DNA (DAPI) in strain AnaRne-GFP. (D) Covisualization of the localization of MinD-GFP and DNA (DAPI) in strain MinD-GFP. Cells were stained with DAPI (blue). Each photograph was obtained by an N-SIM microscopy system (Nikon) and shows three different channels for phycoerythrin (PE) fluorescence (red), GFP fluorescence (green), and DAPI (blue). From left to right: PE, GFP, DAPI, merge of GFP and DAPI channels, and merge of all three channels. The strain cells were analyzed using an N-SIM microscopy system and wavelengths of 405 nm (for DAPI), 488 nm (for GFP), and 561 nm (for PE). Scale bars correspond to 2 μm.
DISCUSSION
Following up on previous work, which revealed that AnaPnp (23) can form a complex with AnaRne in PCC 7120, we demonstrated in this study that both AnaEno and CrhB can also interact with AnaRne in vivo according to a copurification assay. These interactions were next confirmed by far-Western blotting assay and by bacterial two-hybrid and pulldown assays in vitro. Moreover, CrhB could help AnaRne degrade double-stranded RNA with a 5′ tail (Fig. 9B). Of note, as in E. coli, there are at least four classic components (RNase E, DEAD-box RNA helicase CrhB, enolase, and PNPase) of the RNA degradosome in PCC 7120 that have been identified so far. In addition, AnaRne acts as a platform that coordinates the interaction. Consequently, we concluded that there is an RNA degradosome-like complex in PCC 7120.
FIG 9.
Schematic representation of the RNA degradosome-like complex in strain PCC 7120 (A) and the enzymatic mechanism of AnaRne and CrhB interaction (B). The N-terminal domain of AnaRne is shown as described above, with the binding sites for CrhB and AnaEno indicated. The disordered C-terminal domain is gray, with interaction sites for AnaPnp indicated.
It is worth noting that there are some differences in the assembly mechanisms of the RNA degradosome between PCC 7120 and E. coli. Some studies have shown that the RhlB-recognition motif in EcRne corresponds to residues 719 to 731 of the noncatalytic C-terminal domain (30), and residues 833 to 850 have been mapped to enolase (25), although similar segments could not be identified in AnaRne. In contrast, we found that the domains interacting with CrhB and AnaEno are located in the N-terminal catalytic domain of AnaRne. This interaction mode is somewhat similar to the degradosome of Caulobacter crescentus, in which the segment interacting with RhlB is located in the N terminus of RNase E (6). Regarding AnaEno and CrhB, as in most RNA degradosomes, the C-terminal domains are mainly involved in the interactions. Considering the close interaction between a DEAD-box helicase and the RNase E catalytic domain, it is believed that in this mode, helicase can first unwind the structured RNA substrates that need to be further processed or degraded and then pass them directly to the active site of RNase E (6). Besides, this mode could be attributed to the conformation of AnaRne, and the specific interaction sites and mechanism still need verification. On the basis of the results described above, we schematically summarized a primary model of the protein interaction partners in the PCC 7120 RNA degradosome-like complex in Fig. 9A. Obviously, as mentioned above, the different locations of the partners and the absence of a membrane attachment motif make this complex more valuable for research.
Many reports have revealed that RNase E is a membrane-binding protein and is located on the inner cytoplasmic membrane according to a membrane-targeting sequence in E. coli (28, 31, 32). Additionally, RhlB in the degradosome is also membrane associated in a manner depending on a direct protein-protein interaction with RNase E (28). This membrane binding could enhance the rate of RNA processing and decay through stabilized protein folding, thereby leading to stronger binding affinity of the degradosome for its substrates (32). As for cyanobacteria, CrhC in PCC 7120 shows a cytoplasmic membrane association, and CrhR in Synechocystis sp. strain PCC 6803 localizes to both the cytoplasmic and thylakoid membrane regions (20, 33). The findings of our study mean that CrhB and AnaRne mainly localize in PCC 7120 cytoplasm and in close proximity to nucleoid DNA. This result is similar to a previous finding in Caulobacter crescentus indicating that RNase E can associate with nucleoid DNA (34). Considering the evolutionary antiquity of cyanobacteria and the fact that the transcriptional and translational tasks in bacteria are not completed in different organelles as in eukaryotes, we speculated that this localization model may facilitate a rapid interaction between the proteins responsible for RNA processing and the nucleic acid in the vicinity. In addition, this mechanism may be especially important for promoting the formation of the degradosome complex.
The multienzyme assembly that regulates processing, quality control, and degradation of a variety of RNA transcripts is present in many bacteria and eukaryotes. For example, within gammaproteobacteria, there is a set of conserved short sequence motifs in the noncatalytic region of RNase E that can interact with RhlB, enolase, and PNPase, whereas the main executors in eukaryotes are exoribonucleases and RNA helicases (35, 36). Nevertheless, on the basis of the sequence differences in the C-terminal extension between AnaRne and EcRne, a recent study suggested that there is no degradosome in cyanobacteria (37). Our report provides evidence that there is an RNA degradosome-like complex in cyanobacteria. Taken together, our results should lay the foundation for further research on the RNA degradosome and RNA metabolism in cyanobacteria and shed light on the modular interactions that have been utilized by this multienzyme machine in the course of evolution.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. The strains of cyanobacterium Anabaena sp. strain PCC 7120 employed in this study included the wild type (WT); GFP translation fusion strains AnaRne-GFP, CrhB-GFP, and MinD-GFP; and overexpression strains oeRne and oeGFP. These microbes were grown in BG-11 medium (containing NaNO3, 1,500 mg/liter; K2HPO4·3H2O, 40 mg/liter; MgSO4·7H2O, 75 mg/liter; CaCl2·2H2O, 36 mg/liter; Na2CO3, 20 mg/liter; citric acid, 6 mg/liter; EDTA, 1 mg/liter; ferric ammonium citrate, 6 mg/liter; H3BO3, 2.86 mg/liter; MnCl2·4H2O, 1.81 mg/liter; ZnSO4·7H2O, 0.222 mg/liter; CuSO4·5H2O, 0.079 mg/liter; NaMoO4·5H2O, 0.39 mg/liter; Co(NO3)2·6H2O, 0.0494 mg/liter) with continuous shaking (150 rpm) at 30°C. Antibiotics were added at the following concentrations: neomycin, 150 mg/liter; streptomycin, 10 mg/liter; spectinomycin, 10 mg/liter.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1/λpir | RP4-mob+, λpir, host for R6K-based suicide plasmids | Invitrogen |
| E. coli BL21(DE3) | F− ompT hsdS (rB− mB−) gal dcm | Invitrogen |
| E. coli BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1 (Smr) hsdR2 mcrA1 mcrB1 | Invitrogen |
| Anabaena sp. strain PCC 7120 oeRne | Neor, overexpression of the protein of AnaRne in PCC 7120 | This study |
| Anabaena sp. strain oeGFP | Neor, overexpression of the protein of GFP in PCC 7120 | This study |
| Anabaena sp. strain AnaRne-GFP | Spr and Smr, AnaRne and GFP translation fusion strain | This study |
| Anabaena sp. strain CrhB-GFP | Spr and Smr, CrhB and GFP translation fusion strain | This study |
| Anabaena sp. strain MinD-GFP | Spr and Smr, MinD and GFP translation fusion strain | This study |
| Plasmids | ||
| pCT | Kmr, protein expression vector in PCC 7120 | Our laboratory |
| pCT-rne | pCT carrying rne, C-terminal TwinStrep tag | This study |
| pCT-gfp | pCT carrying gfp, C-terminal TwinStrep tag | This study |
| pRL271 | Cmr, mutant construction vector | Our laboratory |
| pRL271-rne-gfp | Cmr, Spr, and Smr, pRL271 carrying rne and gfp, C-terminal TwinStrep tag | This study |
| pRL271-crhB-gfp | Cmr, Spr, and Smr, pRL271 carrying crhB and gfp, C-terminal TwinStrep tag | This study |
| pRL271-minD-gfp | Cmr, Spr, and Smr, pRL271 carrying minD and gfp, C-terminal TwinStrep tag | This study |
| pHS-Tag | Kmr, protein expression vector derivative cured of pET28a | Our laboratory |
| pHS-rne | pHS-Tag carrying rne, N-terminal His tag, C-terminal Strep tag | This study |
| pHS-rneN | pHS-rne with deletion of residues 397–687 | This study |
| pHS-rneC | pHS-rne with deletion of residues 1–400 | This study |
| pHS-crhB | pHS tag carrying crhB, N-terminal His tag, C-terminal Strep tag | This study |
| pHS-enolase | pHS Tag carrying the enolase gene, N-terminal His tag, C-terminal Strep tag | This study |
| pET28a | Kmr, protein expression vector | Invitrogen |
| pET28a-crhB | pET28a carrying crhB, C-terminal His tag | This study |
| pET28a-crhBN | pET28a carrying crhBN (1–200), C-terminal His tag | This study |
| pET28a-crhBC | pET28a carrying crhBC (228–513), C-terminal His tag | This study |
| pET28a-crhBC1 | pET28a carrying crhBC1 (228–347), C-terminal His tag | This study |
| pET28a-crhBCe1 | pET28a carrying crhBCe1 (343–513), C-terminal His tag | This study |
| pET28a-enolase | pET28a carrying the enolase gene, C-terminal His tag | This study |
| pET28a-enoN | pET28a carrying enoN (1–143), C-terminal His tag | This study |
| pET28a-enoC | pET28a carrying enoC (144–429), C-terminal His tag | This study |
| pET28a-rneN | pET28a carrying rneN (1–396), C-terminal His tag | This laboratory |
| pET28a-rneNΔ1 | pET28a carrying rneN with deletion of residues 1–31 | This study |
| pET28a-rneNΔ2 | pET28a carrying rneN with deletion of residues 32–110 | This study |
| pET28a-rneNΔ3 | pET28a carrying rneN with deletion of residues 111–206 | This study |
| pET28a-rneNΔ4 | pET28a carrying rneN with deletion of residues 207–272 | This study |
| pET28a-rneNΔ5 | pET28a carrying rneN with deletion of residues 273–393 | This study |
| pET28a-rneNΔ6 | pET28a carrying rneN with deletion of residues 1–110 | This study |
| pET28a-rneNΔ7 | pET28a carrying rneN with deletion of residues 32–206 | This study |
| pET28a-rneNΔ8 | pET28a carrying rneN with deletion of residues 111–272 | This study |
| pET28a-rneNΔ9 | pET28a carrying rneN with deletion of residues 207–393 | This study |
| pET28a-rneNΔ10 | pET28a carrying rneN with deletion of residues 258–307 | This study |
| pUT18Ca | Carr, bacterial two-hybrid vector carrying complementary fragment T25 | Invitrogen |
| pKT25a | Kmr, bacterial two-hybrid vector carrying complementary fragment T18 | Invitrogen |
| pUT18Ca-zip | Carr, pUT18Ca carrying zip, as positive control | Invitrogen |
| pKT25a-zip | Kmr, pKT25a carrying zip, as positive control | Invitrogen |
| pUT18Ca-rne | Carr, pUT18Ca carrying rne | This laboratory |
| pKT25a-rne | Kmr, pKT25a carrying rne | This laboratory |
| pUT18Ca-crhB | Carr, pUT18Ca carrying crhB | This study |
| pKT25a-crhB | Kmr, pKT25a carrying crhB | This study |
The abbreviations Neor, Kmr, Spr, Smr, Cmr, and Carr refer to neomycin resistance, kanamycin resistance, spectinomycin resistance, streptomycin resistance, chloramphenicol resistance, and carbenicillin resistance, respectively.
E. coli strains were cultured in Lennox broth (LB) liquid medium or agar plates at 37°C. Antibiotics were applied at the following concentrations: carbenicillin, 100 mg/liter; chloramphenicol, 25 mg/liter; kanamycin, 50 mg/liter; streptomycin, 100 mg/liter; spectinomycin, 100 mg/liter. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added at 1 mM for expression induction.
Plasmid construction.
All plasmids used in this study are listed in Table 1, and the primers are described in Table 2. To express recombinant proteins AnaRne, AnaEno, and CrhB, the open reading frames (ORFs) of rne, the enolase gene, and crhB were amplified and cloned into expression vector pET28a (Invitrogen) or subjected to pHS tag digestion with NdeI and XhoI. The AnaEnoN and AnaEnoC truncated variants were generated by one-step PCR amplification, using pET28a-enolase as the template and the following primers: primer pair Pall3538F430his and Pall3538R1287his and primer pair Pall3538F1his and Pall3538R429his. A series of AnaRneN and CrhB truncated variants were also generated as described above, using pET28a-rneN and pET28a-crhB as the templates and the corresponding primers. The templates were then digested with DpnI.
TABLE 2.
Sequences of the oligonucleotides used in this study
For the overexpression of AnaRne in PCC 7120, we constructed the pCT-rne plasmid. First, we amplified the ORF of rne with primers Palr4331F5h and Palr4331R2061h matching the genome of PCC 7120. Second, we cloned the PCR products into shuttle vector pCT digested with XhoI and SmaI via the use of a ClonExpress II one-step cloning kit (Vazyme). This vector contains the copper-regulated petE promoter (38, 39) and a theophylline-controlled riboswitch (40, 41) to prevent leaky expression. As a control, pCT-gfp was constructed by amplification of the ORF of gfp from plasmid p8760 with primers Plgfp_F3 and Plgfp_R717 (42).
To construct plasmid pRL271-crhB-gfp expressing the fusion protein CrhB-GFP, we did the following. First, one crhB fragment was amplified from the PCC 7120 chromosome with primers Palr1223F648 and Palr1223R1539, and another was amplified with primers Palr1223F1549 and Palr1223R2470. Next, plasmid pSfgfp-Sp was digested with BamHI and SmaI to recover the gfp-carrying fragment. Finally, these fragments were cloned into linearized pRL271 with PstI, XhoI, and SpeI by the use of a ClonExpress II one-step cloning kit (Vazyme). Plasmids pRL271-rne-gfp and pRL271-minD-gfp were constructed by the same method.
Expression and purification of recombinant proteins.
E. coli BL21(DE3) was transformed with the protein expression plasmids. The resulting cultures were grown in LB medium to an optical density at 600 nm (OD600) of 0.5, and then the cultures were supplemented with 1 mM IPTG (induction for 4 h). The cells were harvested by centrifugation, resuspended in lysis buffer (300 mM KCl, 20 mM Tris-HCl [pH 8.0], 10% glycerol), and disrupted with a high-pressure homogenizer. The target proteins with the His tag in the lysate were purified on a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen), whereas the proteins with the Strep tag were purified on a Strep-Tactin column (IBA). Finally, each purified protein was dialyzed against protein storage buffer (20 mM Tris-HCl [pH 7.5], 300 mM KCl, 10% glycerol) and stored at −80°C. Protein concentrations were quantified by the Bradford assay with bovine serum albumin (BSA) as the standard, and protein purity was confirmed by SDS-PAGE.
Copurification of the protein associated with AnaRne from PCC 7120 cells.
Plasmids pCT-gfp and pCT-rne were transferred into WT Anabaena sp. strain PCC 7120 by triparental mating (43). The resulting strains, oeGFP and oeRne, were cultured in liquid BG-11 medium to an OD750 of 0.5 and induced with 1 μM CuSO4 and 2 mM theophylline for 12 h. The cells were harvested by centrifugation and resuspended in buffer W (150 mM NaCl, 100 mM Tris-HCl, pH 8.0) supplemented with 0.1% Triton X-100, 1× protease inhibitor cocktail (Sigma), and 5 mM EDTA. After that, the cells were disrupted with FastPrep-24 (MP), and the target protein was purified from the supernatant with Strep-Tactin Sepharose (IBA). The proteins in the elution fraction were analyzed by Western blotting with an anti-strep (GenScript), anti-CrhB, or anti-AnaEno antibody.
To obtain anti-CrhB and anti-AnaEno antibodies, we first purified recombinant antigens CrhB and AnaEno on a Ni-NTA affinity column and then injected them into rabbits for antibody production. The specificity of the anti-CrhB and anti-AnaEno antibodies was assessed by Western blotting of the total cellular protein of PCC 7120 (23).
Far-Western blotting assay.
Proteins with a His tag to be detected in this assay were separated by SDS-PAGE in 10% gels and electrotransferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were then treated as described elsewhere (23). For detection of the interaction, the membranes were blocked for 1 h with 5% skim milk–phosphate-buffered saline (PBS)-0.1% Tween 20 (PBS-T) at room temperature (PBS contains 0.01 mol/liter phosphate buffer, 0.137 mol/liter NaCl, and 0.003 mol/liter KCl [pH = 7.4]). After rinsing was performed (five times with PBS-T, 10 min each time), the membranes were incubated with 200 μg of incubation protein with Strep tag in a reaction mixture with PBS-T containing 1% skim milk. Next, the membranes were washed as described above and sequentially incubated with the anti-strep tag polyclonal antibody (GenScript) (1:1,000) for 1 h and with a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Sigma) (secondary antibody) (1:5,000). Finally, the signals were detected by means of a BioReady ECL kit (Bio-Rad).
The bacterial two-hybrid assay.
These assays were performed using the BACTH system strain, which lacks endogenous adenylate cyclase activity (44, 45). E. coli strain BTH101 was transformed with two recombinant protein fusion plasmids (T25 and T18) simultaneously, and the transformants were selected on LB agar containing carbenicillin and kanamycin. To test for the interaction of the two components, three individual colonies of each combination were grown overnight in LB with antibiotics. The overnight cultures (4 μl) were spotted onto LB agar containing carbenicillin, kanamycin, 0.5 mM IPTG, and 40 mg/liter 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The plates were incubated in the dark at 30°C for 24 h and then imaged. Experiments were conducted in triplicate (biological replicates).
Pulldown assay.
In this experiment, 500 μg of purified SAnaRneN was applied to 1 ml of Strep-Tactin Sepharose (IBA). The protein-coupled resin was incubated at 4°C for 2 h with a lysate of cells overexpressing AnaEno in E. coli. The Strep-Tactin column was loaded with proteins and washed with buffer W, and the bound proteins were eluted with buffer W supplemented with 50 mM d-biotin (Sigma). Finally, the result was obtained by SDS-PAGE and visualized by Western blotting with the anti-AnaEno antibody.
Preparation of RNA substrates.
Ribooligonucleotides used in this study were synthesized by GenScript. The 22-mer RNA oligonucleotide 5′-ACA GUA UUU GGU ACU GCG CUC U-3′ (single stranded) was labeled with 5′-FAM (6-carboxyfluorescein) and is part of the 5′-terminal sequences of RNAI, the antisense RNA of ColEI-type plasmids (46). This 22-mer oligonucleotide contains an RNase E cleavage site (47–50). To obtain the 5′ dsRNA, a short unlabeled RNA single-stranded oligonucleotide, 5′-AGA GCG CAG UAC C-3′, was hybridized to the 22-mer RNA strand. The duplex RNA substrate was created by combining the “long” RNA and “short” RNA (1:2 ratio). RNA annealing was performed in RNA annealing buffer (10 mM Tris-HCl [pH 8.0], 20 mM KCl) by heating to 100°C for 5 min followed by slow cooling to room temperature.
RNA helicase assays.
The RNA-unwinding activity of CrhB was assessed in 10-μl reaction mixtures containing 20 mM Tris-HCl (pH 8.5), 3 mM MgCl2, 1 mM dithiothreitol (DTT), 3 mM ATP, 50 nM dsRNA substrates, and 60 ng of CrhB, and the reaction was performed at 30°C for 30 min, unless stated otherwise in the corresponding figure legends. Reactions were quenched by the addition of 10 μl of 2× RNA loading dye (95% formamide, 0.02% SDS, 0.02% bromophenol blue, 0.01% xylene cyanol, 1 mM EDTA) (BioLabs). Reaction products were separated by the use of a 20% native polyacrylamide gel (19:1 bisacrylamide) in a mixture with 0.5× Tris-borate-EDTA (TBE) at 10 mA for 90 min and, finally, were detected by autoradiography via the use of a LAS-1000 Plus luminescent image analyzer (Fujifilm).
Enzyme activity assays.
RNA degradation assays were carried out in reaction buffer composed of 20 mM Tris-HCl (pH 8.0), 10 mM KCl, 3 mM MgCl2, 1 mM dithiothreitol, and 3 mM ATP at 30°C for 30 min with 100 nM protein, 50 nM RNA, and 10-μl final reaction volumes. The reaction was stopped by the addition of 10 μl of 2× RNA loading dye. The reaction products were separated on 20% native polyacrylamide gel (19:1 bisacrylamide) in 0.5× TBE. The resulting fragments of RNA were detected by autoradiography via the use of an LAS-1000 Plus luminescent image analyzer (Fujifilm).
Microscopic analysis and photography.
Microscopic analysis and photography were performed by means of a Nikon-structured illumination microscope (N-SIM) system (Nikon) or a fluorescence microscope (Nikon Eclipse 80i), and the details are provided in relevant figure legends. We stained the DNA with 1 mg/ml 4′,6-diamidino 2-phenylindole (DAPI; Sigma) for 20 min in the dark before examination.
Supplementary Material
ACKNOWLEDGMENTS
The research was financially supported by the National Natural Science Foundation of China (31570048) and National Key Research and Development Program of China (2018YFE0105600).
We declare that we have no conflict of interest.
W.C. conceived the study and edited the manuscript. H.Y., L.W., and W.C. designed the experiments. The experiments were performed by H.Y. and X.Q. The final results were analyzed by H.Y. The paper was written by H.Y. with input from the rest of us.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Schopf JW. 1975. Precambrian paleobiology: problems and perspectives. Annu Rev Earth Planet Sci 3:213–249. doi: 10.1146/annurev.ea.03.050175.001241. [DOI] [Google Scholar]
- 2.Herrero A, Stavans J, Flores E. 2016. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol Rev 40:831–854. doi: 10.1093/femsre/fuw029. [DOI] [PubMed] [Google Scholar]
- 3.Lyons TW, Reinhard CT, Planavsky NJ. 2014. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 4.Giraud C, Hausmann S, Lemeille S, Prados J, Redder P, Linder P. 2015. The C-terminal region of the RNA helicase CshA is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen Staphylococcus aureus. RNA Biol 12:658–674. doi: 10.1080/15476286.2015.1035505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Górna MW, Carpousis AJ, Luisi BF. 2012. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys 45:105–145. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 6.Voss JE, Luisi BF, Hardwick SW. 2014. Molecular recognition of RhlB and RNase D in the Caulobacter crescentus RNA degradosome. Nucleic Acids Res 42:13294–13305. doi: 10.1093/nar/gku1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 8.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpousis AJ. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol 61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 10.Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore CJ, Cohen SN, Georgiou G. 2006. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol 61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda Y, Yagi M, Morita T, Aiba H. 2011. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol 79:419–432. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsai Y-C, Du D, Domínguez-Malfavón L, Dimastrogiovanni D, Cross J, Callaghan AJ, García-Mena J, Luisi BF. 2012. Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res 40:10417–10431. doi: 10.1093/nar/gks739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 14.Khemici V, Poljak L, Toesca I, Carpousis AJ. 2005. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc Natl Acad Sci U S A 102:6913–6918. doi: 10.1073/pnas.0501129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamot D, Magee WC, Yu E, Owttrim GW. 1999. A cold shock-induced cyanobacterial RNA Helicase. J Bacteriol 181:1728–1732. doi: 10.1128/JB.181.6.1728-1732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redder P, Hausmann S, Khemici V, Yasrebi H, Linder P. 2015. Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol Rev 39:392–412. doi: 10.1093/femsre/fuv011. [DOI] [PubMed] [Google Scholar]
- 17.Yu E, Owttrim G. 2000. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res 28:3926–3934. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owttrim GW. 2013. RNA helicases. RNA Biol 10:96–110. doi: 10.4161/rna.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamot D, Owttrim G. 2000. Regulation of cold shock-induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 182:1251–1256. doi: 10.1128/jb.182.5.1251-1256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosana ARR, Whitford DS, Fahlman RP, Owttrim GW. 2016. Cyanobacterial RNA helicase, CrhR, localizes to the thylakoid membrane region and co-sediments with degradosome and polysome complexes in Synechocystis sp. PCC 6803. J Bacteriol 198:2089–2099. doi: 10.1128/JB.00267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein JA, Lin P-H, Cohen SN, Lin-Chao S. 2004. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci U S A 101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murashko ON, Lin-Chao S. 2017. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc Natl Acad Sci U S A 114:E8025–E8034. doi: 10.1073/pnas.1703731114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J-Y, Deng X-M, Li F-P, Wang L, Huang Q-Y, Zhang C-C, Chen W-L. 2014. RNase E forms a complex with polynucleotide phosphorylase in cyanobacteria via a cyanobacterial-specific nonapeptide in the noncatalytic region. RNA 20:568–579. doi: 10.1261/rna.043513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A 93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi B. 2018. Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes. Nucleic Acids Res 46:387–402. doi: 10.1093/nar/gkx1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Górna MW, Pietras Z, Tsai Y-C, Callaghan AJ, Hernández H, Robinson CV, Luisi BF. 2010. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA 16:553–562. doi: 10.1261/rna.1858010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietras Z, Hardwick SW, Swiezewski S, Luisi BF. 2013. Potential regulatory interactions of Escherichia coli RraA protein with DEAD-box helicases. J Biol Chem 288:31919–31929. doi: 10.1074/jbc.M113.502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl H, Turlan C, Khalid S, Bond PJ, Kebalo J-M, Peyron P, Poljak L, Bouvier M, Hamoen L, Luisi BF, Carpousis AJ. 2015. Membrane recognition and dynamics of the RNA degradosome. PLoS Genet 11:e1004961. doi: 10.1371/journal.pgen.1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pazos M, Casanova M, Palacios P, Margolin W, Natale P, Vicente M. 2014. FtsZ placement in nucleoid-free bacteria. PLoS One 9:e91984. doi: 10.1371/journal.pone.0091984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aït-Bara S, Carpousis AJ. 2010. Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria. J Bacteriol 192:5413–5423. doi: 10.1128/JB.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khemici V, Poljak L, Luisi BF, Carpousis AJ. 2008. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol 70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murashko ON, Kaberdin VR, Lin-Chao S. 2012. Membrane binding of Escherichia coli RNase E catalytic domain stabilizes protein structure and increases RNA substrate affinity. Proc Natl Acad Sci U S A 109:7019–7024. doi: 10.1073/pnas.1120181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Fahmawi B, Owttrim GW. 2003. Polar-biased localization of the cold stress-induced RNA helicase, CrhC, in the Cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol 50:1439–1448. doi: 10.1046/j.1365-2958.2003.03783.x. [DOI] [PubMed] [Google Scholar]
- 34.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Razew M, Warkocki Z, Taube M, Kolondra A, Czarnocki-Cieciura M, Nowak E, Labedzka-Dmoch K, Kawinska A, Piatkowski J, Golik P, Kozak M, Dziembowski A, Nowotny M. 2018. Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat Commun 9:97. doi: 10.1038/s41467-017-02570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron JC, Gordon GC, Pfleger BF. 2015. Genetic and genomic analysis of RNases in model cyanobacteria. Photosynth Res 126:171–183. doi: 10.1007/s11120-015-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghassemian M, Wong B, Ferreira F, Markley JL, Straus NA. 1994. Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120. Microbiology 140:1151–1159. doi: 10.1099/13500872-140-5-1151. [DOI] [PubMed] [Google Scholar]
- 39.Buikema WJ, Haselkorn R. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc Natl Acad Sci U S A 98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 41.Ma AT, Schmidt CM, Golden JW. 2014. Regulation of gene expression in diverse cyanobacterial species using Theophylline-responsive riboswitches. Appl Environ Microbiol 80:6704–6713. doi: 10.1128/AEM.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santini C-L, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu L-F. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem 276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 43.Ermakova M, Battchikova N, Richaud P, Leino H, Kosourov S, Isojarvi J, Peltier G, Flores E, Cournac L, Allahverdiyeva Y, Aro EM. 2014. Heterocyst-specific flavodiiron protein Flv3B enables oxic diazotrophic growth of the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc Natl Acad Sci U S A 111:11205–11210. doi: 10.1073/pnas.1407327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van den Bossche A, Hardwick SW, Ceyssens P-J, Hendrix H, Voet M, Dendooven T, Bandyra KJ, De Maeyer M, Aertsen A, Noben J-P, Luisi BF, Lavigne R. 2016. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. Elife 5:e16413. doi: 10.7554/eLife.16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin-Chao S, Cohen SN. 1991. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell 65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 47.Lin-Chao S, Wong TT, McDowall KJ, Cohen SN. 1994. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J Biol Chem 269:10797–10803. [PubMed] [Google Scholar]
- 48.McDowall KJ, Lin-Chao S, Cohen SN. 1994. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem 269:10790–107906. [PubMed] [Google Scholar]
- 49.McDowall KJ, Kaberdin VR, Wu S-W, Cohen SN, Lin-Chao S. 1995. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- 50.Liou G-G, Chang H-Y, Lin C-S, Lin-Chao S. 2002. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem 277:41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.