Abstract
Diagnostic tests for the coronavirus infection 2019 (COVID-19) are critical for prompt diagnosis, treatment and isolation to break the cycle of transmission. A positive real-time reverse-transcriptase polymerase chain reaction (RT-PCR), in conjunction with clinical and epidemiologic data, is the current standard for diagnosis, but several challenges still exist. Serological assays help to understand epidemiology better and to evaluate vaccine responses but they are unreliable for diagnosis in the acute phase of illness or assuming protective immunity. Serology is gaining attention, mainly because of convalescent plasma gaining importance as treatment for clinically worsening COVID-19 patients. We provide a narrative review of peer-reviewed research studies on RT-PCR, serology and antigen immune-assays for COVID-19, briefly describe their lab methods and discuss their limitations for clinical practice.
Introduction
There has been a lack of guidance for practicing physicians in interpreting published evidence and limitations of emerging lab tests for coronavirus disease 2019 (COVID-19). Current diagnosis requires a combination of symptoms/signs of respiratory infection and a positive reverse-transcriptase polymerase chain reaction (RT-PCR) for severe acute respiratory syndrome coronavirus-2(SARS-CoV-2).1–4 The Chinese Centers for Disease Control and Prevention (China CDC) first submitted the SARS-CoV-2 genome to Global Initiative on Sharing All Influenza Data. The CDC in United States (U.S.) then obtained emergency use authorization (EUA) from the Food and Drug Administration(FDA) for the first RT-PCR.5,6 Tests were limited to the CDC, but later extended to other agencies and Clinical Laboratory Improvement Amendments (CLIA) certified labs for rapid diagnosis to interrupt the transmission cycle via quick isolation.7,8 With increasing EUAs, careful considerations are necessary to interpret test results.9
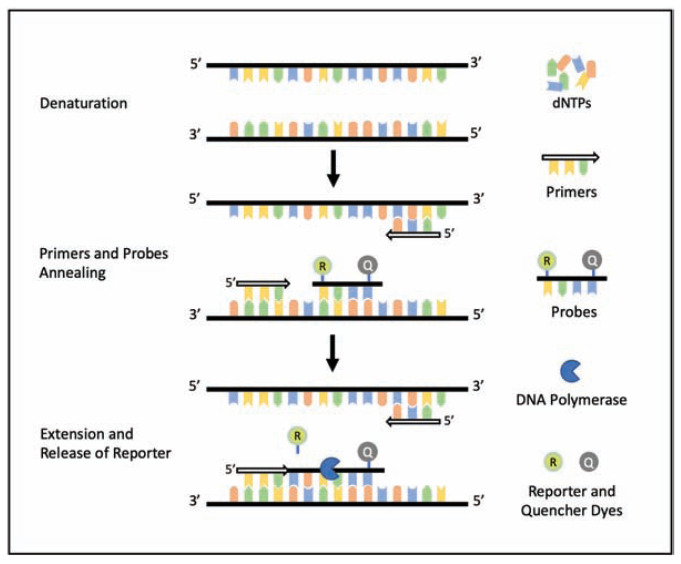
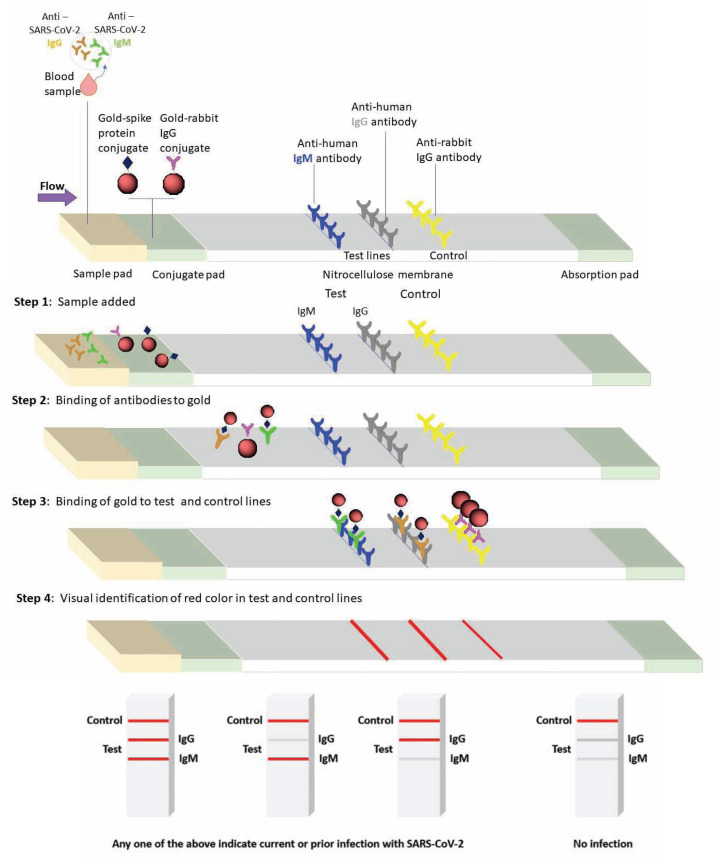
RT-PCR detects viral ribonucleic acid (RNA) using flourogenic primers/probes and thermal cycling steps (Figure 1). It is sensitive, safe and cost-effective than viral culture methods, but not without limitations.8,10,11 Serology has also gained attention based on certain advantages i.e. estimation of sero-prevalence, asymptomatic infections, contact tracing and vaccine effectiveness. In SARS, serology was reliable for diagnosis based on a negative test during the acute illness followed by a positive serology in convalescent samples, or a fourfold rise from acute to convalescent titers, tested in parallel.12–17 Lateral flow immunoassay (LFIA), chemi-luminescence immunoassay or the traditional enzyme linked immunosorbent assay (ELISA) are common methods, that rely on IgG/IgM’s affinity to recombinant spike (S) and nucleocapsid (N) proteins for SARS-CoV-2, and are point of care (POC) tests with a quick turnaround time (Figure 2), but certain disadvantages also exist.16
Figure 1.
Steps of a single thermal cycle of RT-PCR
To detect the presence of RNA, samples are combined with the primer/probe sets, reverse-transcriptase (RT), DNA polymerase, buffer, and deoxy-nucleoside triphosphates (dNTPs). This mixture then undergoes an initial annealing phase where the first strand of complimentary DNA (cDNA) is formed at the target sequence. Thermal cycling starts with denaturation to separate the strands, followed by annealing of specific primers and probes to each strand. Probes containing reporter (R) and quencher (Q) dyes are designed to anneal to parts of the target genetic sequence between the primers. In the next step, polymerase extends the primer (extension phase) towards the probe and releases R generating a visible fluorescence. This thermal cycle repeats to amplify the genetic material with every cycle. A cycle threshold defined as the number of cycles to attain a certain fluorescence, is often used as a determination of positivity. it is important that both the primers and probes should be designed to be both specific and sensitive by identifying highly conserved regions unique to the COVID-19.
Abbreviations: RT-PCR – reverse transcriptase polymerase chain reaction, dsDNA – double stranded deoxyribonucleic acid, PCR – polymerase chain reaction
Figure 2.
A schematic illustration of lateral flow immune-assay (LFIA) principle for antibody (IgG and IgM) testing for COVID-19. The LFIA uses a nitrocellulose membrane embedded with anti-human IgG, anti-human IgM and anti-rabbit IgG in designated spots for test and control, respectively. The sample for LFIA will use a finger-stick, single-use patient blood, which is drop-casted in the sample pad containing recombinant peptides of SARS CoV-2 conjugated with gold nanoparticle (AuNP; “red color indicator”) and rabbit IgG gold conjugates as a control. If patient’s blood has anti – SARS CoV-2 IgM or IgG antibodies, it will bind to the viral peptide-gold conjugate in sample pad, and capillary action drives the antigen-antibody complex and the control to the test and control spots respectively. A red color formation in the test spot is an indication of the presence of anti-SARS CoV-2 antibody (IgG or IgM - positive).
To assist the practicing physician in understanding COVID-19 lab tests in relevance to their schema, limitations, and statistical considerations for clinical practice, we performed a narrative review of research assays in COVID-19. Because other reviews have discussed the commercial assays with EUAs (available at link: https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergencyuse-authorizations,) we did not include them in our review.18 The Infectious Diseases Society of America (IDSA) has also issued a guidance document on RTPCR recently.19
Methods
In order to avoid pre-prints (non-peer reviewed), we searched PubMed only using the following search terms with no restriction on start time, till April 15, 2020. Search terms were “PCR” and either “COVID-19” or “SARS CoV-2,” and combinations of “serology” and “COVID-19,” “IgM”and”COVID-19,” and “IgG”and”COVID-19.” Individual studies were manually screened and included only human original research studies that reported test characteristics, methodology, performance, sample type and statistical outcomes. We individually reviewed every study separately for RT-PCR and serology and the information was tabulated.
Results
Fourteen studies reported RT-PCR research assays and seven studies reported serological assays for COVID-19. There were currently no original research studies on antigen immunoassays for COVID-19.
RT-PCR
Table 1 summarizes the peer-reviewed publications on RT-CPR, listed per publication date, and enumerates necessary test details that impact interpretation.
Table 1.
Studies on RT-PCR research assays
| Date, author, sample size | Time of collection from onset/hospitalization | Genes, Samples, Statistical Parameters/performance | Limitations |
|---|---|---|---|
| 01/23/20, Corman et al, 310 samples20 | N/A | Genes: E and RdRp, Specificity 100%. | Primers for RNA identical to 2019- nCoV target sequence & lack of testing with SARS-CoV-2. |
| 02/12/20 To et al, 12 patients31 | Median 2 days from hospitalization (range 0–7 days). | Sensitivity: 11/12 (91.7%). Specificity: 33/33 (100%) |
Easier sampling. Limitations include limited sample size. |
| 02/19/20, Fang et al, 51 patients34 | 3 ± 3 days for both chest CT and RT-PCR. | Sensitivities: Chest CT (50/51, 98%) Initial RT-PCR (36/51, 71%). |
Study limited by lack of information on laboratory procedure and positive controls |
| 02/26/20, Ai et al, 1014 patients33 | Median interval between chest CT and initial RT-PCR was 1 day (range 0–7 days). | 21/601 RT-PCR positive & not chest CT. 308/413 had positive chest CT & negative RT-PCR. 59% positive rate of RT-PCR. Chest CT sensitivity = 97%, specificity = 25%, accuracy = 68%, PPV = 65%, NPV = 83%. | |
| 03/04/20, Chan et al, 273 specimens25 | 0–18 days - respiratory specimens, 4–15 days for other specimens. | Sensitivity: 85% with samples from respiratory tract, 11.1% from non-respiratory tract. | Primers had 7–9 nucleotide differences with SARS-CoV |
| 03/05/20, Konrad et al, 669 respiratory samples21 | Not reported. | SARS-CoV E gene assay was more sensitive than the two RdRp gene assays; however, the E gene assay showed unspecified signals at late cycles. | Lack of information on procedure and positive controls. |
| 03/05/20, Pfefferle et al, 88 samples22 | N/A | Specificity: 88/88 (100%) Sensitivity: LOD of 275.72 copies/process at 95% detection probability |
Unable to validate the performance of the assay using clinical SARS-CoV-2 positive samples |
| 03/07/20, Liu et al, 4880 patients30 | Not reported. | Sensitivities and specificities not reported.. | Validity of test results was not assessed. |
| 03/11/20, Wang et al, 1070 specimens32 | Pharyngeal swabs on 1–3 days after hospitalization. Variable time for other specimens. | Sensitivities: BAL (14/15; 93%), sputum (75/104; 72%), nasal (5/8; 63%), bronchoscope brush biopsy (6/13; 46%), pharyngeal (126/398; 32%), feces (44/153; 29%), blood (3/307; 1%), urine (0/72, 0%). | Insufficient data to analyze correlation between symptoms or disease course and test results. Small sample size for some specimen types. |
| 03/25/20, Long et al, 204 patients35 | Study group = 2.6 ± 1.7 days, control group = 3.2 ± 1.6 days. | Sensitivities: Chest CT (35/36, 97.2%), initial RT-PCR (30/36, 83.3%). | RT-PCR only performed in patients with fever due to limited test kit supplies. Small sample size. |
| 03/26/20, Li et al. 610 patients36 | Initial RT-PCR done prior to admission. Repeated 1–2 days if negative or weakly positive. | Not reported | Study limited by lack of detailed patient information or RT-PCR protocol. |
| 03/30/20, CDC, 2071 respiratory specimens28 | Not reported. | Specificity: 60/60 for both assays. 49/2071 respiratory specimens tested positive. |
Study limited by positive controls. |
| 04/08/20, Nalla et al. 300 specimens27 | Not reported. | 100% specific. Sensitivity: CDC N2 and Corman E-gene: 10/10 and able to reliably detect samples with only 6 genomic equivalents of viral RNA. | Limited sample size |
| 04/09/20, Xiao et al. 70 patients37 | Symptoms to nucleic acid conversion, median (IQR): Consecutive false-negative patients = 36 (28–40), all other patients = 21 (18–26). | 15/70 (21.4%) positive on RT-PCR after two consecutive negative results. | Study does not mention details about the commercial detection kit. |
Abbreviations: RT- PCR – reverse transcriptase polymerase chain reaction, RNA – ribonucleic acid, CDC – Centers for Diseases Control and Prevention, LOD – Limit of detection, CT-computed tomography, COVID – coronavirus disease, N-nucleocapsid, RdRp – RNA dependent RNA polymerase, E- envelope, BAL – broncho-alveolar lavage, SARS-CoV-2 - severe acute respiratory syndrome conronavirus-2, N/A – Not available.
Most Sensitive and Specific Primers and Probes
The first robust RT-PCR for COVID-19 was developed in January 2020 in Germany by Corman et al.20 Their primers and probes targeted the highly specific RNA-dependent RNA polymerase (RdRp) and envelope (E) genes of SARS CoV-2 and did not cross-react with other respiratory viruses or bacteria. The E-gene was first performed, followed by the RdRp-gene assay that differentiated SARS CoV-2 from SARS CoV. Follow-up studies further optimized the process to minimize handson time.21,22 Chu et al. developed a sequential quantitative real-time RT-PCR that identified two different regions – open reading frame 1b (ORF1b) and nucleocapsid (N) of the SARS CoV-2 genomes, but as these are highly conserved sequences among Sarbecovirus, specificity was compromised by crossreactivity23. SARS CoV-2’s open reading frame 1ab (ORF1ab) and N genes were also targeted by Wang et al.24 Another approach taken by Chan et al. utilized primers and probes designed for RdRp/helicase (Hel), S and N genes.25,26 These novel real-time RT-PCR assays, which contained 7–9 nucleotide differences from human SARS-CoV, were found to be more sensitive than the RdRpgene assay, while also not cross-reacting with other human-pathogenic coronaviruses and respiratory pathogens. More recently, Nalla et al. compared the primers and probes developed by Corman et al. with the N1, N2, and N3 sets developed by the CDC.27,28 While all sets continued to be highly specific, the CDC N2 and Corman et al.’s E-gene sets were the most sensitive. In addition to the academic development of primer/probe sets for diagnosis of COVID-19, several EUA-granted commercial products have been developed.29 As of March 15, 2020, the CDC’s protocol for EUA assays contain N1 and N2 only excluding the N3 detection.
Performance of Different Specimen Types
As more primers and probe sets became available, a variety of specimens were tested. Among all specimens, lower respiratory tract specimens continue to have the highest sensitivity, followed by upper respiratory tract (naso- or oropharynx). 30 One study found that sets designed by Chan et al. were able to detect SARS CoV-2 in the saliva of 11/12 patients.31 Other researchers have also tested sputum, blood, and feces using the ORF1ab and N gene primers/probes.32 Despite several case reports of SARS CoV-2 detection in feces and blood, these specimens have lowest number of viral particles.
Chest Imaging and RT PCR
CT of chest has been reported to have a similar sensitivity to PCR and reported as a screening tool when RT-PCR is falsely negative.33–35 A few early studies, where a high number of negative RT-PCR tests on those who satisfied clinical criteria for COVID-19 pneumonia and abnormal CT chest, projected the idea that radiological findings may be used as a surrogate. In one retrospective study, only 40%(241/610) with abnormal CT findings tested positive by RT-PCR, but 18 had a positive RTPCR after two consecutive negative results.36 Xiao et al. also found 15/70 patients testing positive after two previous consecutive negative results.37 Ai et al. showed that from 1014 patients undergoing both chest CT and RT-PCR with throat swab sample, 21 who had positive RT-PCR results had a normal initial chest CT, but 308 with negative RT-PCR had chest CT images suggestive of COVID-19.33
Serology
Table 2 summarizes peer-reviewed research publications on serology research assays for diagnosis of COVID-19. One of the first large studies used lateral flow immunoassay (LFIA) to qualitatively detect IgM and IgG antibodies to the S protein of SARS-CoV-2 with a high sensitivity and specificity at various points of patient stays.38 All of the finger-stick blood and venous blood serum and plasma, were sensitive to detect IgG and IgM. Reported turnaround time of 15 minutes combined with simple design enables POC testing. Currently, several commercial agencies are developing accurate and sensitive LFIAs for point of care diagnostics of SARS-
Table 2.
Studies on SARS CoV-2 Serology Research Assays
| Date/author/study type | Method, sample type/number | Time from onset to test | Statistical parameters | Limitations |
|---|---|---|---|---|
| 02/17/20 Zhang et al. Case series45 | ELISA kits developed using SARSr-CoV Rp3 NP as the antigen. | Day 10 of treatment | 1st day of sampling: IgM and IgG titers were relatively low or undetectable 5th day of sampling: increase in both IgM and IgG seen in nearly all patients |
No data after day 5. No data listed for specificity of ELISA kits. |
| 02/27/20 Li et al. Diagnostic Accuracy Cross-sectional study38 | LFIA - IgM and IgG antibodies to recombinant antigen that is the receptor binding domain of SARS-CoV-2 Spike Protein. | Day 8 to day 33 after onset |
Sensitivity: IgG + IgM: 88.66%; IgG: 70.53%; IgM: 82.62% Specificity: IgG + IgM: 116/128 (90.63%); IgG: 126/128 (91.41%); IgM: 117/128 (91.41%) |
This test cannot confirm virus presence or provide quantitative information. Day of sample collection was inconsistent from patient to patient |
| 03/19/20 Haveri et al. Case series43 | Serum on days 4, 9, 10, and 20 from onset of symptoms. IgM and IgG antibodies against SARS-CoV-2 were analyzed using IFA | Day 3 to day 23 after onset | Day 4: antibodies were undetectable. Day 9: IgG titer: 80; IgM titer: 80. Day 20: IgG: 1,280; IgM titer: 320 | This study was able to track IgM and IgG temporally up to day 23 d.p.o. Sample collection was inconsistent from patient to patient. No specificity data noted. |
| 03/21/20 Guo et al. Diagnostic Accuracy Comparative Study42 | 208 samples from 140 cases cases. Indirect ELISA for IgM, IgA, and IgG. 135 samples from adults with respiratory tract infections & 150 healthy adults were controls | 1–7 days post-symptom onset (41 samples), 8–14 days (84 samples), 14+ days (83 samples) |
IgM: Sensitivity: 90.4%; Specificity: 100% IgG: Sensitivity: 77.9%; Specificity: 100% IgA: Sensitivity: 93.3%; Specificity: 100% |
Cross-sectional sample of specimens was used to determine the kinetics of antibodies; however, each patient has different kinetics for the development of antibodies. |
| 03/28/20 Zhao et al. Diagnostic Accuracy Comparative Study40 | 173 patients, serial samples to a total of 535 samples. ELISA IgM, and IgG. for receptor binding domain of the Spike protein, | 1–39 days after onset. Groups : 1–7 days (94 patients), 8–14 (135 patients), 15–39 (90 patients) |
Ab: Sensitivity:93.1% Specificity: 99.1% IgM: Sensitivity: 82.7%; Specificity: 98.6% IgG: Sensitivity: 64.7%; Specificity: 99% |
Sensitivities varied depending on which day after onset samples were collected. Patients were selected based on positive RT-PCR, potentially missing atypical patients with lower respiratory viral load |
| 03/30/20 Cassaniti et al. Diagnostic Accuracy Cross-sectional study41 | 110 subjects, VivaDiag COVID-19 IgM/IgG Rapid LFIA. | Positive control collected at median 7 days (IQR 4–11). |
COVID-19:
Sensitivity of combined IgM and IgG: 63.3% were positive; 16.7% were weakly positive; 16.7% were negative, 1/30 was positive for IgM, negative for IgG 50 patients evaluated in ED Sensitivity: 18.4%; Specificity: 91.7%. |
Advantage of study is that patients enrolled from the ED provided a relatively consistent time frame of symptom onset. Limitation of the study is that there is no temporal data to follow. |
| 03/30/20 Liu et al. Diagnostic Accuracy Case-Control study44 | 214 patients, 100 healthy controls. The recombinant nucleocapsid (rN) protein-based ELISA IgM or IgG. | Samples divided into 1–5 (22), 6–10 (38), 11–15 (54), 16–20 (55), 21–30 (32), 31–35 (6), and >35 (7) days post disease onset |
rN-based ELISA Sensitivity: 80.4%; Specificity: 100% rS-based ELISA Sensitivity:82.2%; Specificity: 100% |
Temporal data available, showing that IgM and/or IgG of SARS-CoV-2 might be positive at 11–15 d.p.o. Decreased positive rate of IgM was observed at > 35 d.p.o. A limitation is that samples were not collected from these patients after discharge. |
Abbreviations: ELISA – enzyme-linked immunosorbent assay, IFA – immunofluorescence assay, d.p.o – days post-disease onset, LFIA – lateral flow immunoassay, IgM-Immunoglobulin M, IgG-immunoglobulin G, IgA-immunoglobulin A, RT-PCR – reverse Transcriptase polymerase chain reaction, SARS-CoV-2 -severe acute respiratory syndrome coronavirus-2.
COV-2. Sona-Nanotech in collaboration with GE Healthcare Life sciences, utilizes S1 domain of the virus for detection and uses gold nanoparticles as the label. Another company, Pharmact, developed a 20-minute assay for detecting N, S1, and S2 domains of the protein.39 This test also detects IgM and IgG antibodies present in the patient’s blood. The company has validated the test kit by confirming the infection in patients by PCR analysis. The kit showed a higher sensitivity for IgM after four days and the IgG after 11 days after the onset of symptoms. Traditional laboratory-based ELISAs have also been used to quantify total antibodies, IgM, and IgG.40–46 A strong IgG and IgM response in blood compartment was reported by Amanat et al.47 Zhao et al. found that the seroconversion rate for total antibodies was 93.1% and high titers of Ab were found to be associated with more severe disease. However, 6.9% of patients remained seronegative, possibly due to lack of blood samples from lost follow-up.40
Timing of Serology Testing
Median seroconversion time for total antibody, IgM, and IgG were 11, 12, and 14 days respectively, observed in >98.8% of COVID-19 patients with a rapid increase after six days from symptom onset. The sensitivity for IgM and IgG increased significantly after two weeks of symptoms.40 Guo et al. also found similar results in respect to seroconversion rate with high sensitivity and specificity.42 Lou et al. observed that those with prolonged incubation period, seroconverted in 21 days versus shorter incubation periods, who seroconverted in 13 days.46 Zhao et al. reported a median seroconversion time of 11–14 days, depending on the assay used.40 Okba et al. also found in their limited longitudinal serum samples, that IgG seroconversion was reliably detected at two weeks from onset.48 Lastly, To et al. compared serial viral loads with antibody levels against the SARS-CoV-2 internal nucleoprotein (NP) and surface spike protein receptor binding domain (RBD) using enzyme immunoassay.49 They found that IgG levels correlated with virus neutralization titer and that for the patients with samples available 14 days or longer after symptom onset, both IgG and IgM were highly sensitive for both anti-NP and anti-RBD.
Discussion
Clinical and epidemiological factors are primary drivers of pretest probability, crucial for interpreting any test result. Variations in available resources [E.g. materials needed by lab and clinical personal collecting samples] also affect testing decisions and interpretations. Understanding test characteristics is vital for interpreting positive or negative results.50
Statistical Considerations for Interpreting Diagnostic Test Results
Sensitivity, a test characteristic, is the proportion of people with disease who will have a positive result, whereas positive predictive value provides the odds of actually having the disease (accuracy) from a positive result. Table 3 lists the general effect of sensitivity and specificity of any dichotomous (positive or negative) diagnostic test on outcomes, and is worthy of review.51 The effect of disease prevalence over positive predictive value without any change in sensitivity and specificity of the diagnostic test is illustrated Table 4. A useful and detailed review of how to interpret a diagnostic result in clinical decision making is available from Armstrong et al.52 Cheng et al. provide a very useful heat map in their review and classify test choices based on four uses: screening, diagnosis of symptomatic cases, diagnosis of asymptomatic shedding and epidemiologic surveillance.53
Table 3.
Potential Outcomes of Screening Tests with Varying Levels of Sensitivity and Specificity [Reprinted with permission from © 2020 BioMed Central Ltd. Part of Springer nature. 2012, BMC Research Notes; Vol 5, Iss 1; TGK Bentley et al. Implications of the impact of prevalence on test thresholds and outcomes: lessons from tuberculosis, p563]
| Test characteristic | High sensitivity | High specificity |
|---|---|---|
| Impact on results | ↑ True positives | ↑ True negatives |
| Testing goal | Identify people with disease | Identify people without disease |
| Treatment goal | Treat disease Prevent future illness and, in the case of infection, possible disease spread |
Avoid unnecessary treatment |
| Potential harms of opposite test characteristic | Low sensitivity: | Low specificity: |
| ↑ False negatives ↑ Potential future illness and suffering ↑ Potential future spread of disease (in the case of infection) |
↑ False positives ↑ Bodily harms, toxicity, and financial costs of unnecessary treatment ↑ Social stigmatization ↓ confidence in screening program |
Table 4.
Effect of Prevalence on the Positive Value of Diagnostic Test
| Disease (gold standard) | ||||||
|---|---|---|---|---|---|---|
| Dichotomous Diagnostic Test | Present | Absent | Total | |||
| + | a (True +) | b (False +) | a+b | PPV = a/(a+b) | ||
| − | c (False −) | d (True −) | c+d | NPV = d/(c+d) | ||
| Total | a+c | b+d | ||||
| Sensitivity = a/(a+c) | Specificity = d/(b+d) | |||||
| Disease Prevalence = 1%* | Test | + | 950 | 990 | 1940 | PPV = 49%* |
| − | 50 | 98010 | 98060 | NPV = 99.9% | ||
| Total | 1000 | 99000 | 100,000 | |||
| Sensitivity = 95% * | Specificity = 99%* | |||||
| Disease Prevalence = 5%* | Test | + | 4750 | 950 | 5700 | PPV = 83.3%* |
| − | 250 | 94050 | 94300 | NPV = 99.7% | ||
| Total | 5000 | 95000 | 100,000 | |||
| Sensitivity = 95%* | Specificity = 99%* | |||||
Abbreviations: PPV – positive predictive value, NPV – negative predictive value.
Without any changes in sensitivity and specificity of the diagnostic test, the positive predictive value increased from 49% to 83.3% with an increase in prevalence of disease from 1% to 5%.
RT-PCR
While RT-PCR is the current gold standard for the laboratory diagnosis of COVID-19, potential pre-analytical (ineffective symptom screening, sampling errors, prolonged transportation time, sample contamination, etc.) and analytical errors (insufficient sample, non-validated tests, instrument malfunction, testing out of diagnostic window, viral RNA recombination, etc.) that compromise the results should be kept in mind.53, 54 While a false positive result may result in wasteful use of health care and public health resources, a false negative could lead to failure of isolation and foster community spread.54 If SARS CoV-2 RNA quantity is below the threshold of RT-PCR analytical sensitivity (very early symptomatic or post-symptomatic/recovery), then results will be reported as negative, emphasizing the importance of appropriate sample collection technique and transport. The CDC has published clear guidelines for appropriate collection.55 In contrast to SARS CoV infection (SARS 2002–2003) where RT-PCR sensitivity was low during the early symptomatic period, early shedding (pre- or early symptomatic phase of illness) of SARS CoV-2 virions is remarkable in COVID-19, accounting for its rapid spread in pandemic proportions.56,57 Like other viral illnesses, the shedding of infectious SARS CoV-2 virions is also suspected to drop over time from onset of illness, but data is insufficient at this time.49 Wolfel et al. evaluated SARS CoV-2 replication by assessing sub-genomic RNA (a marker of replication) from serial throat, sputum, blood, and fecal samples in nine mildly symptomatic COVID-19 patients.56 They noticed early viral replication in throat and sputum samples that lasted for almost two weeks beyond the symptomatic period. Hence, RTPCR from naso- or oro-pharyngeal samples will continue to remain as the reference standard for diagnosis during the acute phase of COVID-19.
CT chest should not be used as a standalone test for diagnosis of COVID-19, as it lacks specificity given that the typical changes (such as ground glass opacities) can also be found in a variety of other lung diseases/infections.33 A CT chest should be considered for suspected pulmonary embolism, pneumothorax or another thoracic pathology, that will require it for further management.58,59 CT chest in COVID-19, delays care for other non-COVID patients in dire need of it, as the disinfection process in most radiology units takes 1–2 hours.59 Studies comparing CT chest with RT-PCR should be interpreted with caution due to differences in the RT-PCR research assays across the globe and the potential pre-analytical and analytical errors discussed above. If RT-PCR is negative and clinical suspicion for COVID-19 remains high, a repeat RT-PCR from an upper airway sample or from lower respiratory tract in intubated patients (higher positivity rates) with in-line suction may be considered.33 The CDC’s test-based strategy to release from isolation, was based on early studies that reported negative RT-PCRs in recovering patients. As a result of which, we now know that prolonged viral shedding is not uncommon, ranging from two to six weeks from onset.60,61 Whether such persistent positivity by RT-PCR for prolonged periods imply continued infectious viral shedding remains to be determined.56,57,62
Serology
Considering the different antibodies that recovering patients may develop, it is important that serologic assays target those specific to the SARS CoV-2 structural proteins that differentiate it from the commonly circulating coronaviruses or SARS CoV. Despite the promising sensitivity and specificity for antibodies to N and S proteins of SARS CoV-2, human studies show wide variability (Table 2). With most studies suggesting seroconversion between days 11–14 after symptom onset, serology should not be considered for diagnosis during the acute phase of COVID-19.44 Serology helps to determine an existing immune response in the host i.e. a surrogate of an immune response, but is not a concrete proof of protective immunity.48,63 Humoral response is B cell mediated, which is only one arm of our protective immunity and may not last longer depending on the robustness of initial immune response.64,65 In a phase 1 SARS CoV vaccine trial, T cell mediated responses (virus-specific memory cells) persisted for several years, while neutralizing antibodies only lasted for 1–2 years.65,66 The most recent primer on serological assay from the IDSA, cautions about the high false positivity rates of serology tests when pre-test probability or prevalence of disease is <5%.67
In the absence of vaccines or proven effective treatment, the use of convalescent plasma (CP) from patients who recovered form COVID-19 is being explored as a treatment option.68–70 CP has been used in the past for other infections, including the SARS and MERS epidemics.71,72 In March 2020, the FDA supported the use of convalescent plasma to treat patients with severe or life-threatening COVID-19, under expanded access protocol.73 Under Mayo Clinic’s lead, as of April 26, 2020, 4,524 patients have been enrolled and 1,726 patients had received plasma across the U.S. However, the biggest challenge has been ensuring the availability of convalescent plasma.74 Donors must have had a prior documented COVID-19 infection. COVID-19 antibody testing can help widen the pool of plasma donors by determining a serologic evidence of prior infection.75 The effectiveness of CP therapy depends on the neutralizing antibody titer exhibiting virus-killing function in donor plasma.76 A defined titer of neutralizing antibodies has not been established and not all COVID-19 patients mount robust antibody response. Neutralization assays are not widely available and require biosafety level 3 facilities with skilled experts. Currently, the American Red Cross has a defined COVID-19 neutralizing antibody titer (e.g. > 1:80); however, it is not a needed criteria to refer a donor or transfuse plasma, because specimens are stored for testing later. ELISA and IFA, which are relatively easy and safe, have predicted neutralization activity, in MERS-CoV.77 In one study, ELISA IgG predicted neutralization activity with >95% specificity at a cutoff optical density ratio of 1.6, suggesting it as a substitute in resource-limited situations.78 With increasing COVID-19 prevalence, there is need for reliable serology assays to identify vaccine responses in trials and recovered patients who have detectable antibodies for therapeutic purposes i.e. donation of convalescent plasma as a potential treatment or for identification of past infection18.
Antigen Immuno-Assays
On May 9, 2020 the U.S.FDA granted the first EUA for COVID-19 antigen test (Quidel Corporation) for use at the POC, which is only 80% sensitive compared to RT-PCR and 100% specific for detection of N protein of SARS CoV-2 from naso-pharyngeal and nasal swabs.29 Antigen immuno-assays offer the potential for rapid, inexpensive, POC diagnosis of COVID-19.53 Antigen tests are cheaper and simpler than RT-PCR and allow for early diagnosis. Antigen detection assays are available for adenovirus, respiratory syncytial virus (RSV), and influenza.79 Well studied antigen assays for influenza and RSV have a high specificity.80,81 They have also been developed for SARS-CoV and MERS-CoV.82,83 The principal limitation is low sensitivity compared to RT-PCR.80,81 Immuno-assays previously developed for the SARS-CoV and MERS-CoV coronaviruses may guide the development of similar tests for the novel SARS-CoV-2. Lau et al. utilized ELISA to detect SARS-CoV N protein using hyper-immune polyclonal antibodies. This assay demonstrated high specificity (≥96%) for nasopharyngeal, urine, and fecal specimens collected from patients without SARS, with respective sensitivities of 52%, 5%, and 55% from SARS-positive patients.83 Chen and colleagues developed a capture ELISA using monoclonal antibodies specific to the MERS-CoV nucleo-capsid protein. This antigen detection assay demonstrated high specificity and sensitivity; yet, the study was limited by using only simulated MERS-CoV-positive specimens for determining sensitivity.82 It is possible that antigen tests for SARS-CoV-2 may also be limited by poor sensitivity, particularly given the variability of viral shedding seen in COVID-19 patients.53 Numerous other immuneassay test kits for COVID-19 diagnosis are under current development.84
Conclusions and Future Perspectives
A combination of clinical, epidemiologic and laboratory considerations are essential for accurate diagnosis of emerging infections such as COVID-19. Community prevalence or disease activity plays an important role is choosing the laboratory test of choice and in interpretation of results. During the pandemic spread, RT-PCR being the most sensitive and specific test, it is the current laboratory standard for diagnosis during the acute phase of COVID-19. It is now widely available in most health care settings, but it cannot differentiate between infectious and noninfectious viral shedding during recovery. With increasing community spread, reliable serology testing is necessary for both epidemiologic and therapeutic reasons. Viral genomic recombination or acquired mutations, leading to phenotypic changes, could result in failure of these detection assays and pose threat to reliable interpretation of results.85 Longitudinal studies are required to define a reliable test strategy.64 Antigen based immuno-assays may have a role in the future for resource limited settings, but there are none available at this time.
Footnotes
Daniel Shyu, James Dorroh, and Caleb Holtmeyer, are in the Department of Medicine. Detlef Ritter, MD, is in the Department of Pathology and Anatomical Sciences. Anandhi Upendran, PhD, is in the Department of Medical Pharmacology and Physiology and at the University of Missouri Institute of Clinical And Translational Science (MU-iCATS). Raghuraman Kannan, PhD, is in both the Departments of Radiology and Bioengineering. Dima Dandachi, MD, and Christian Rojas-Moreno, MD, are in the Department of Medicine – Division of Infectious Diseases. Stevan P. Whitt, MD, and Hariharan Regunath, MD, MSMA member since 2019 and Missouri Medicine Editorial Board Member for Infectious Disease, (above), are in the Divisions of Infectious Diseases, Pulmonary, Critical Care and Environmental Medicine. All are at the University of Missouri - Columbia, Columbia, Missouri.
Disclosure
None reported.
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. The New England journal of medicine. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan W, Zhao X, Ma X, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019−2020. 2020;2(4):61–62. [PMC free article] [PubMed] [Google Scholar]
- 6.Patel A, Jernigan DB nCo VCDCRT. Initial Public Health Response and Interim Clinical Guidance for the 2019 Novel Coronavirus Outbreak - United States, December 31, 2019-February 4, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(5):140–146. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emergency Use Authorizations. 2020. [Accessed 04/04/2020]. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations. 2020.
- 8.Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better Tests, Better Care: Improved Diagnostics for Infectious Diseases. Clinical Infectious Diseases. 2013;57(suppl_3):S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petherick A. Developing antibody tests for SARS-CoV-2. The Lancet. 2020;395(10230):1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deepak S, Kottapalli K, Rakwal R, et al. Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr Genomics. 2007;8(4):234–251. doi: 10.2174/138920207781386960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valones MA, Guimaraes RL, Brandao LA, de Souza PR, de Albuquerque Tavares Carvalho A, Crovela S. Principles and applications of polymerase chain reaction in medical diagnostic fields: a review. Braz J Microbiol. 2009;40(1):1–11. doi: 10.1590/S1517-83822009000100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio. 2020;11(2) doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.President Donald J. Trump Is Ensuring States Have The Testing Capacity Needed To Safely Open Up America Again. 2020. [Accessed 04/28/2020]. https://www.whitehouse.gov/briefings-statements/president-donald-j-trump-ensuring-states-testing-capacity-needed-safely-open-america/ 2020.
- 14.Coronavirus (COVID-19) Update: Serological Test Validation and Education Efforts. 2020. [Accessed 04/28/2020]. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-serological-test-validation-and-education-efforts. 2020.
- 15.Organization WH. WHO guidelines for the global surveillance of severe acute respiratory syndrome (SARS): updated recommendations, October 2004. Geneva: World Health Organization; 2004. [Google Scholar]
- 16.Meyer B, Drosten C, Müller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang J, Wang MX, Ang IYH, et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J Clin Med. 2020;9(3) doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus-2: A Narrative Review. Ann Intern Med. 2020 doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson KE, Azar MM, Banerjee R, et al. Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA’s Diagnostics Committee. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konrad R, Eberle U, Dangel A, et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfefferle S, Reucher S, Norz D, Lutgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu DKW, Pan Y, Cheng SMS, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JF, Yip CC, To KK, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated. J Clin Microbiol. 2020 doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nalla AK, Casto AM, Huang MW, et al. Comparative Performance of SARS-CoV-2 Detection Assays using Seven Different Primer/Probe Sets and One Assay Kit. J Clin Microbiol. 2020 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention Division of Viral Diseases. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Revision: 03. FDA; 2020. [Google Scholar]
- 29.U.S. Food and Drug Administration. Emergency Use Authorizations. 2020. [Accessed 04/14/2020]. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd.
- 30.Liu R, Han H, Liu F, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To KK, Tsang OT, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ai T, Yang Z, Hou H, et al. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang Y, Zhang H, Xie J, et al. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long C, Xu H, Shen Q, et al. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Yao L, Li J, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Yi Y, Luo X, et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020 doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020 doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11) doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Liu L, Kou G, et al. Evaluation of Nucleocapsid and Spike Protein-based ELISAs for detecting antibodies against SARS-CoV-2. Journal of Clinical Microbiology. 2020 doi: 10.1128/JCM.00461-20. JCM.00461-00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou B, Li T, Zheng S, et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv. 2020 doi: 10.1183/13993003.00763-2020. 2020.2003.2023.20041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020 doi: 10.1038/s41591-020-0913-5. 2020.2003.2017.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okba NMA, Muller MA, Li W, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit 23 March 2020: Value of Diagnostic Testing for SARS–CoV-2/COVID-19. mBio. 2020;11(2):e00722–00720. doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentley TGK, Catanzaro A, Ganiats TG. Implications of the impact of prevalence on test thresholds and outcomes: lessons from tuberculosis. BMC Res Notes. 2012;5:563–563. doi: 10.1186/1756-0500-5-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armstrong KA, Metlay JP. Annals Clinical Decision Making: Using a Diagnostic Test. Annals of Internal Medicine. 2020;172(9):604–609. doi: 10.7326/M19-1940. [DOI] [PubMed] [Google Scholar]
- 53.Tang YW, Schmitz JE, Persing DH, Stratton CW. The Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges. J Clin Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clinical chemistry and laboratory medicine. 2020 doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 55.Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19) 2020. [Accessed 04/26/2020]. https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html. 2020.
- 56.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 57.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nature Medicine. 2020 doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 58.Hope MD, Raptis CA, Shah A, Hammer MM, Henry TS. A role for CT in COVID-19? What data really tell us so far. Lancet (London, England) 2020;395(10231):1189–1190. doi: 10.1016/S0140-6736(20)30728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hope MD, Raptis CA, Henry TS. Chest Computed Tomography for Detection of Coronavirus Disease 2019 (COVID-19): Don’t Rush the Science. Annals of Internal Medicine. 2020 doi: 10.7326/M20-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou B, She J, Wang Y, Ma X. The duration of viral shedding of discharged patients with severe COVID-19. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atkinson B, Petersen E. SARS-CoV-2 shedding and infectivity. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Q, Zhu L, Ni Z, Meng H, You L. Duration of serum neutralizing antibodies for SARS-CoV-2: Lessons from SARS-CoV infection. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altmann DM, Douek DC, Boyton RJ. What policy makers need to know about COVID-19 protective immunity. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Channappanavar R, Zhao J, Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunologic Research. 2014;59(1):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin JE, Louder MK, Holman LA, et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26(50):6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.IDSA COVID-19 Antibody Testing Primer. 2020. [Accessed 04/20/2020]. https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf. 2020.
- 68.Roback JD, Guarner J. Convalescent Plasma to Treat COVID-19: Possibilities and Challenges. JAMA. 2020 doi: 10.1001/jama.2020.4940. [DOI] [PubMed] [Google Scholar]
- 69.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan K, Liu B, Li C, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A; 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mair-Jenkins J, Saavedra-Campos M, Baillie JK, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lai ST. Treatment of severe acute respiratory syndrome. Eur J Clin Microbiol Infect Dis. 2005;24(9):583–591. doi: 10.1007/s10096-005-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanne JH. Covid-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ. 2020;368:m1256. doi: 10.1136/bmj.m1256. [DOI] [PubMed] [Google Scholar]
- 74.Arabi YM, Hajeer AH, Luke T, et al. Feasibility of Using Convalescent Plasma Immunotherapy for MERS-CoV Infection, Saudi Arabia. Emerg Infect Dis. 2016;22(9):1554–1561. doi: 10.3201/eid2209.151164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franchini M, Marano G, Velati C, Pati I, Pupella S, Liumbruno GM. Operational protocol for donation of anti-COVID-19 convalescent plasma in Italy. Vox Sang. 2020 doi: 10.1111/vox.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko JH, Seok H, Cho SY, et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir Ther. 2018;23(7):617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- 78.Ko JH, Müller MA, Seok H, et al. Suggested new breakpoints of anti-MERS-CoV antibody ELISA titers: performance analysis of serologic tests. Eur J Clin Microbiol Infect Dis. 2017;36(11):2179–2186. doi: 10.1007/s10096-017-3043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang N, Wang L, Deng X, et al. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chartrand C, Tremblay N, Renaud C, Papenburg J. Diagnostic Accuracy of Rapid Antigen Detection Tests for Respiratory Syncytial Virus Infection: Systematic Review and Meta-analysis. J Clin Microbiol. 2015;53(12):3738–3749. doi: 10.1128/JCM.01816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Merckx J, Wali R, Schiller I, et al. Diagnostic Accuracy of Novel and Traditional Rapid Tests for Influenza Infection Compared With Reverse Transcriptase Polymerase Chain Reaction: A Systematic Review and Meta-analysis. Ann Intern Med. 2017;167(6):394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y, Chan KH, Kang Y, et al. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2015;4(4):e26. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lau SK, Woo PC, Wong BH, et al. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in sars patients by enzyme-linked immunosorbent assay. J Clin Microbiol. 2004;42(7):2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.SARS-CoV-2 diagnostic pipeline. 2020. [Accessed 4/19/20.]. https://www.finddx.org/covid-19/pipeline/?section=show-all#diag_tab.
- 85.Yao H, Lu X, Chen Q, et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. medRxiv. 2020 [Google Scholar]