Abstract
Phylogenomic analyses have helped resolve many recalcitrant relationships in the angiosperm tree of life, yet phylogenetic resolution of the backbone of the Leguminosae, one of the largest and most economically and ecologically important families, remains poor due to generally limited molecular data and incomplete taxon sampling of previous studies. Here, we resolve many of the Leguminosae’s thorniest nodes through comprehensive analysis of plastome-scale data using multiple modified coding and noncoding data sets of 187 species representing almost all major clades of the family. Additionally, we thoroughly characterize conflicting phylogenomic signal across the plastome in light of the family’s complex history of plastome evolution. Most analyses produced largely congruent topologies with strong statistical support and provided strong support for resolution of some long-controversial deep relationships among the early diverging lineages of the subfamilies Caesalpinioideae and Papilionoideae. The robust phylogenetic backbone reconstructed in this study establishes a framework for future studies on legume classification, evolution, and diversification. However, conflicting phylogenetic signal was detected and quantified at several key nodes that prevent the confident resolution of these nodes using plastome data alone. [Leguminosae; maximum likelihood; phylogenetic conflict; plastome; recalcitrant relationships; stochasticity; systematic error.]
The legume family contains over 19,500 species in ca. 765 genera, 36 tribes, and 6 currently recognized subfamilies worldwide, making it the third largest angiosperm family in terms of species diversity [Lewis et al. 2005; LPWG (Legume Phylogeny Working Group) 2013, 2017]. Ranging in size from tiny annual herbs to giant long-lived trees, Leguminosae are often ecologically dominant across the tropical and temperate biomes (Lewis et al. 2005; LPWG 2017). Many legume species are economically important, providing highly nutritious plant proteins for both humans and livestock (Duranti 2006; Voisin et al. 2014). Additionally, ca. 88% of legume species have the ability to establish associations with nitrogen-fixing bacteria via root nodules and hence are important for sustainable agriculture and ecosystem function (Graham and Vance 2003; LPWG 2013; Sprent et al. 2017). Previous deep-level phylogenetic studies mainly on a few of plastid loci (e.g., Wojciechowski et al. 2004; Lavin et al. 2005; McMahon and Sanderson 2006; Bruneau et al. 2008; LPWG 2013) have greatly clarified phylogenetic relationships of legumes. However, relationships among subfamilies and some major clades at the tribal level, particularly within Caesalpinioideae and Papilionoideae, have been difficult to resolve despite over two decades of research (LPWG 2013), with different plastid loci sometimes yielding incongruent, albeit weakly supported, topologies (Wojciechowski et al. 2004; Cardoso et al. 2012; LPWG 2013; LPWG 2017).
Phylogenomic approaches have been applied to tackle difficult relationships in diverse groups of organisms (e.g., Rokas et al. 2003; Jian et al. 2008; Jarvis et al. 2014). In plants, an increasing number of studies have found conflicting phylogenetic signal among nuclear loci (e.g., Wickett et al. 2014; Smith et al. 2015; Parks et al. 2018; Walker et al. 2018a), with this conflict attributed mainly to biological factors such as hybridization, incomplete lineage sorting, hidden paralogy, and horizontal gene transfer (Galtier 2008). However, increasing attention is being paid to the role of other factors—such as uninformative genes and stochasticity, outlier genes, and systematic error—in generating conflict in phylogenomic analyses (e.g., Brown and Thomson 2017; Shen et al. 2017; Walker et al. 2018b). In legumes, a recent phylogenomic study of nuclear transcriptomic data and plastid genomes provided new insights into subfamilial relationships and early legume diversification, highlighting in particular the prevalence of uninformative loci across both the nuclear and plastid genomes and conflict at the family’s deepest nodes (Koenen et al. 2020). While nuclear conflict was thoroughly investigated by Koenen et al. (2020), conflict within the plastome was not fully explored. Plastome-scale data sets have been widely regarded as useful for resolving enigmatic and recalcitrant relationships (e.g., Xi et al. 2012; Goremykin et al. 2015; Zhang et al. 2017), in part because the plastome has long been considered to comprise a single evolutionary unit (Birky 1995; Vogl et al. 2003), meaning that genes can be concatenated in order to amplify phylogenetic signal. However, other recent studies have documented considerable conflict within the plastome (e.g., Gonçalves et al. 2019; Walker et al. 2019), suggesting that the operational assumption that the plastome represents a single evolutionary unit should be, if not abandoned, at least more thoroughly examined.
There are multiple factors that could potentially produce conflict in plastid phylogenies. Stochastic inferences from genes with low information content (due to short gene lengths or few variable sites) seem to be primary among them, but strongly supported conflicting genes/signals have been observed (Walker et al. 2019), warranting attention on other potential (biological) sources including selection and the possibility of ‘chimeric’ plastomes, that is, those harboring genes with distinct evolutionary histories. The potential for biparental inheritance has been documented in many angiosperm species (e.g., Corriveau and Coleman 1988; Zhang et al. 2003). Additionally, heteroplasmy (the presence of distinct plastomes within a single organism) has been directly documented in diverse plant species (reviewed by Ramsey and Mandel 2019). Heteroplasmy might in rare cases result in heteroplasmic recombination (Sullivan et al. 2017; Sancho et al. 2018), thus creating chimeric plastomes with potentially conflicting evolutionary histories. Sharing of genes among the plastid, nuclear, and mitochondrial genomes constitutes another (seemingly rare) source of gene conflict in plastid phylogenomics (e.g., Straub et al. 2013; Smith 2014). Among non-parasitic angiosperms, the legume family has one of the most complex histories of plastome evolution (e.g., Palmer and Thompson 1982; Palmer et al. 1987; Jansen et al. 2008; Lei et al. 2016), including major clades diagnosed by losses or expansions of the Inverted Repeat (IR) region, as well as an array of gene losses and inversions across the family’s phylogeny (Wang et al. 2018). In light of this complex history, conflicting phylogenetic signals in legume plastomes deserve close attention.
Using an extensive sampling of newly generated plastomes, our study aims to resolve many of the most problematic nodes of Leguminosae phylogeny while exploring the distribution of phylogenetic signal and conflict across plastome-inferred phylogenies in the context of the family’s complex history of plastome evolution. We estimated legume relationships using plastomes of 187 species from 35 tribes and all subfamilies, representing almost all major lineages within the family (LPWG 2017). We applied multiple strategies to minimize systematic errors, including removal of ambiguously aligned regions, saturated loci, and loci with low average bootstrap support, as well as recently developed methods for characterizing genomic conflict and evaluating phylogenetic signal within genomic data sets. Leguminosae represent an excellent system to explore the extent and impact of conflict on plastid phylogenomics, a topic that is only now being rigorously examined (e.g., Walker et al. 2019). We outline the significance of our results for understanding legume evolution and for guiding future phylogenomic studies employing the plastome.
Materials and Methods
Plastome Sampling, Sequencing, Assembly, and Annotation
We sequenced plastomes for 151 species and downloaded those of 36 additional species from NCBI (https://www.ncbi.nlm.nih.gov); collectively these species represent 35 of the 36 tribes (Lewis et al. 2005) and major lineages of all six newly defined subfamilies (LPWG 2017) of Leguminosae, as well as eight outgroup taxa (Supplementary Table S1 available on Dryad at https://doi.org/10.5061/dryad.1vhhmgqpb). Illumina sequencing of long-range PCR products or genomic DNA was undertaken. Plastomes were de novo assembled using SPAdes or GetOrganelle (Camacho et al. 2009; Bankevich et al. 2012; Langmead and Salzberg 2012; Wick et al. 2015; Jin et al. 2019) for total DNA reads or using CLC Genomics Workbench (CLC Bio) for long-range PCR reads. Details of plastome assembly and annotation are available in the Supplementary Materials and Methods available on Dryad.
Sequence Alignment and Cleanup, Data Set Generation, and Phylogenetic Analysis
We developed new custom python scripts (https://github.com/Kinggerm/PersonalUtilities/) to automatically extract all annotated regions from plastomes and to rapidly concatenate the alignments of separate loci. Each locus was individually aligned using MAFFT (Katoh and Standley 2013). After excluding loci of low quality or with fewer than four species, we obtained three basic data sets: the PC (coding regions), PN (noncoding regions), and PCN (the concatenated PC and PN) data sets. Three strategies were then applied to reduce systematic error from the three basic data sets: pruning the ambiguously aligned regions, excluding loci with high levels of substitutional saturation (Supplementary Table S2, Fig. S1 available on Dryad), and excluding loci with low average ultrafast bootstrap (UFBoot) support (i.e., <70% and 80%). These strategies resulted in an additional 23 modified data sets; thus, including the original three data sets, 26 data sets were used in subsequent analyses.
We generated phylogenetic trees for each of the 26 concatenated data matrices (Supplementary Table S3 available on Dryad) as well as for individual genes and spacers using IQ-TREE (Nguyen et al. 2015; Chernomor et al. 2016; Hoang et al. 2018). Following these analyses, four data sets (PN-GB-strict, PCN-GB-strict, PN-slope2, and PCN-GB-slope2; GB stands for the program Gblocks, which was used to remove ambiguously aligned regions; see Supplementary Methods available on Dryad for more detailed description of these data sets) were excluded from subsequent analyses. PN-GB-strict was excluded due to its support for an outlier topology. As a result, we excluded PCN-GB-strict, as it includes the PN-GB-strict data set. PN-slope2 was excluded due to insufficient taxon sampling. Similarly, PCN-GB-slope2 was excluded due to its inclusion of the problematic PN-slope2. Thus, moving forward, 22 data sets were subjected to further analysis.
Quantification of Phylogenetic Signal for Alternative Tree Topologies
Following the methods of Smith et al. (2011), Shen et al. (2017) and Walker et al. (2018b), we evaluated phylogenetic signal within three sets of conflicting topologies. For the first set of conflicting topologies, concerning the root of Leguminosae, we compared signal for three alternative resolutions (i.e., the percentage of loci supporting each topology) across each of the 22 generated data sets. For these three topologies, we also calculated the gene-wise log-likelihood support (GLS), the site-wise log-likelihood scores (SLS), the summed difference in SLS (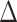 SLS) and in GLS (
SLS) and in GLS (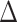 GLS) among the alternative hypotheses in each conflicting topology for each of the 22 data sets (Supplementary Fig. S2 available on Dryad). To reduce the conflict at the root of Leguminosae, we then removed and binned the loci supporting alternative topologies in the three main data sets (PC, PN, and PCN), and identified and removed outlier loci in these data sets assuming that for each data set the average
GLS) among the alternative hypotheses in each conflicting topology for each of the 22 data sets (Supplementary Fig. S2 available on Dryad). To reduce the conflict at the root of Leguminosae, we then removed and binned the loci supporting alternative topologies in the three main data sets (PC, PN, and PCN), and identified and removed outlier loci in these data sets assuming that for each data set the average 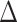 SLS of a locus follows a Gaussian-like distribution. (Supplementary Fig. S3 available on Dryad). This resulted in six additional reduced data sets (bringing the total number of data sets to 28). We reconstructed the phylogenetic trees for these six data sets and recalculated phylogenetic signal to compare the effect of the abovementioned two removals. We also applied these phylogenetic signal analyses for two alternative positions of Griffonia (Supplementary Fig. S4a available on Dryad) and two alternative positions of Pterogyne (Supplementary Fig. S4b available on Dryad ) in the PC, PN, and PCN data sets (see Supplementary Methods available on Dryad for more details).
SLS of a locus follows a Gaussian-like distribution. (Supplementary Fig. S3 available on Dryad). This resulted in six additional reduced data sets (bringing the total number of data sets to 28). We reconstructed the phylogenetic trees for these six data sets and recalculated phylogenetic signal to compare the effect of the abovementioned two removals. We also applied these phylogenetic signal analyses for two alternative positions of Griffonia (Supplementary Fig. S4a available on Dryad) and two alternative positions of Pterogyne (Supplementary Fig. S4b available on Dryad ) in the PC, PN, and PCN data sets (see Supplementary Methods available on Dryad for more details).
Test of Topological Concordance
We estimated topological concordance among phylogenetic trees by all-to-all Robinson–Foulds distance using IQ-TREE and Principal Coordinates Analysis (PCoA) clustering in R (R Development Core Team 2015), and Robinson–Foulds symmetric differences and the UPGMA clustering method using TreeSpace (Jombart et al. 2017).
We quantified conflict and concordance among the 28 data set trees using the bipartition method of PhyParts (Smith et al. 2015, Supplementary Fig. S5 available on Dryad), using an iterative approach to identify the topology most concordant with all data sets (see Supplementary Methods available on Dryad for more details). We also assessed conflicts among gene trees by mapping 226 rooted gene trees constructed by RAxML (Stamatakis 2014) against the PCN (the tree with the highest concordance with the other data set trees, Supplementary Fig. S6 available on Dryad). Finally, because recent studies have recommended the use of coalescent methods for analyzing plastid loci (e.g., Gonçalves et al. 2019), we used ASTRAL (Zhang et al. 2018) to infer species tree using the 226 locus trees from RAxML. We ran two default analyses in which i) all bipartitions were included and ii) bipartitions with <10% bootstrap support were collapsed prior to the analyses, as recommended in Zhang et al. (2017). Additional details of the methods are available in the Supplementary Materials and Methods available on Dryad.
Results and Discussion
New Insights into Deep Phylogenetic Relationships of Leguminosae
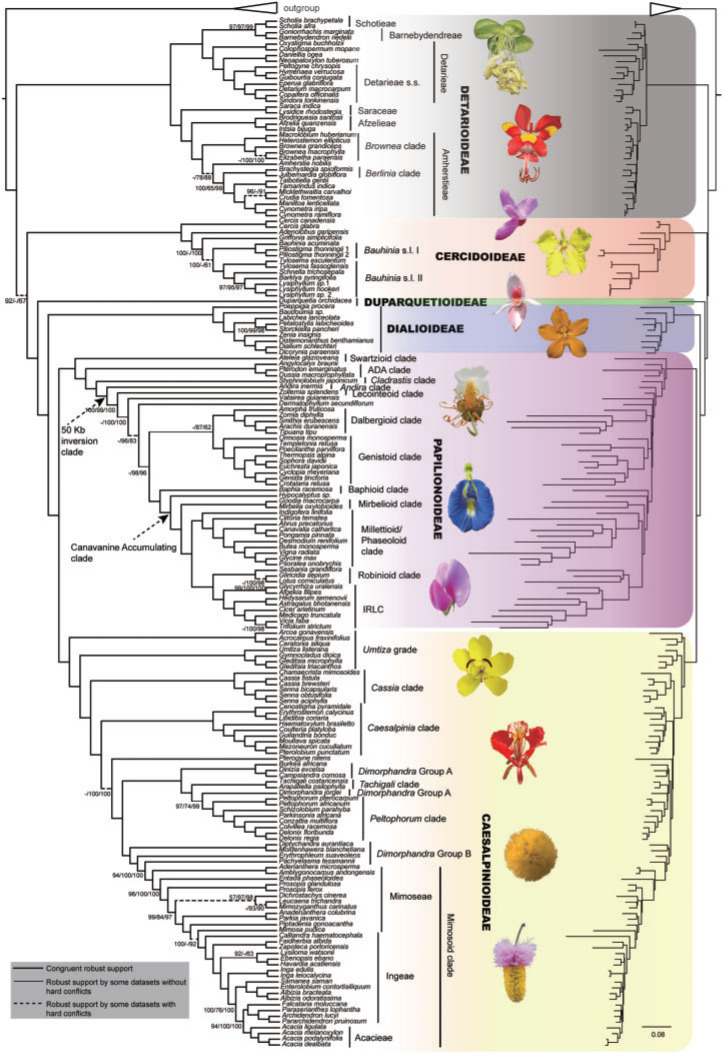
Using an increased sampling of species and methods for dissecting signal and conflict among loci, our plastid phylogenomic study has resolved with strong support many recalcitrant deep relationships within Leguminosae (detailed statistics of the assembled plastome sequences are provided in Supplementary Table S1 available on Dryad, and the characteristics of all 32 modified data sets, including the four excluded data sets, are provided in Table 1; additional details about the 81 coding loci, the 145 noncoding loci, and the 32 data sets are found in Dryad https://doi.org/10.5061/dryad.1vhhmgqpb; all phylogenetic trees are found in Supplementary file S1 available on Dryad). With the exclusion of data sets that produced an outlier tree topology (Supplementary Figs. S7 and S8 available on Dryad), contained insufficient parsimony-informative sites, and/or had limited taxon sampling (Table 1), the remaining 28 data sets produced largely congruent topologies with respect to major legume relationships regardless of the properties (coding or noncoding) of the data set and the various strategies for removing sites, loci, or outlier loci. The PCN tree from the iterative topological concordance analyses was the most concordant summary of the 28 data sets analyzed, and thus was used as our main reference or summary tree (Fig. 1; Supplementary Figs. S5 and S9 and Table S4 available on Dryad). However, conflicting topologies were detected at several nodes among different data sets (see below) despite the multiple strategies we used to reduce systematic error. Hence, while these strategies are useful for dissecting phylogenetic signal, they may not always lead to a full resolution of difficult relationships such as those encountered in some parts of the Leguminosae phylogeny.
Table 1.
Characteristics of all analyzed plastome data sets for reconstructing the deep evolutionary history of the Leguminosae
| No. of | No. of | No. of parsimony- | |
|---|---|---|---|
| Data sets | loci | sites (bp) | informative sites (bp) |
| PC | 81 | 89,989 | 28,494 |
| PC-GB-relaxed | 81 | 73,850 | 26,178 |
| PC-GB-default | 81 | 68,840 | 24,726 |
| PC-GB-strict | 81 | 57,490 | 18,593 |
| PC-slope1 | 76 | 84,615 | 27,071 |
| PC-slope2 | 69 | 67,297 | 19,947 |
| PC-BS70 | 31 | 66,448 | 22,698 |
| PC-BS80 | 17 | 54,116 | 18,799 |
| PC-10-removed | 71 | 83,742 | 26,241 |
| PC-outlier-removed | 80 | 89,872 | 28,259 |
| PN | 145 | 214,876 | 66,139 |
| PN-GB-relaxed | 145 | 59,621 | 24,088 |
| PN-GB-default | 145 | 27,013 | 11,713 |
| PN-GB-strict | 145 | 11,979 | 3,094 |
PN-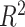
|
98 | 100,491 | 28,866 |
| PN-slope1 | 90 | 81,129 | 22,622 |
| PN-slope2 | 33 | 13,827 (187 spp.) | 2,658 |
| PN-BS70 | 88 | 182,998 | 58,797 |
| PN-BS80 | 58 | 150,901 | 49,506 |
| PN-26-removed | 119 | 189,669 | 58,702 |
| PN-outlier-removed | 140 | 212,961 | 65,284 |
| PCN | 226 | 304,865 | 94,633 |
| PCN-GB-relaxed | 226 | 133,471 | 50,266 |
| PCN-GB-default | 226 | 95,853 | 36,439 |
| PCN-GB-strict | 226 | 69,469 | 21,687 |
PCN-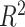
|
179 | 190,480 | 57,360 |
| PCN-slope1 | 166 | 165,744 | 49,693 |
| PCN-slope2 | 102 | 81,126 | 22,605 |
| PCN-BS70 | 119 | 249,446 | 81,674 |
| PCN-BS80 | 75 | 205,017 | 68,395 |
| PCN-36-removed | 190 | 273,411 | 84,943 |
| PCN-outlier-removed | 224 | 304,652 | 94,150 |
Notes: All data sets are explained in Materials and Methods section.
Figure 1.
Cladogram (left) and phylogram (right) of the maximum-likelihood tree of Leguminosae derived from the plastid phylogenomic analysis of a concatenated data set including 81 coding and 145 noncoding loci (PCN data set). Relationships inconsistent with the other inferred trees (Supplementary Table S4 and File S1 available on Dryad) are indicated. Nodal support values for the PC/PN/PCN data sets (see text for data set composition) are from IQ-TREE ultrafast bootstrapping analyses. Only support values <100% UFBoot are shown. Hyphens (-) identify splits not supported by the PC or PN data sets. Thick solid lines indicate internodes that were congruently and robustly supported by different data sets. Thin solid lines indicate internodes that were robustly supported by partial data sets without significant conflicts in other data sets. Dashed lines indicate internodes that were robustly supported by partial data sets but had alternative topologies in other data sets. The tree shown is the same as the PCN.tre in Supplementary File S1 available on Dryad, with the outgroup taxa removed. Images of representative species from clades across the family from top to down are: Colophospermum mopane (Benth.) Leonard (from https://www.dreamstime.com), Amherstia nobilis Wall. (photo courtesy: Dr. K. Karthigeyan, https://doi.org/10.13140/rg.2.1.3932.3287), Cercis siliquastrum L. (photographer: Phil Bendle, http://ketenewplymouth.peoplesnetworknz.info), Tylosema fassoglensis (Schweinf.) Torre & Hillc. (https://upload.wikimedia.org), Duparquetia orchidacea Baill. (photographer: M. de la Estrella), Petalostylis labicheoides R. Br. (http://www.bkaussi.de), Bobgunnia madagascariensis (Desv.) J.H.Kirkbr. & Wiersema (photographer: M. Séleck, http://copperflora.org), Clitoria ternatea L. (photographer: F. Guadagni, http://effegua.myphotos.cc), Lathyrus latifolius L. (photographer: B. Tanneberger, https://www.flickr.com), Senna pendula H.S. Irwin & Barneby (photographer: L. P. Queiroz), Delonix regia (Hook.) Raf. (http://www.peakpx.com), Vachellia farnesiana (L.) Wight & Arn. (photographer: T. M. Perez, https://twitter.com), Dichrostachys cinerea (L.) Wright & Arn. (https://jooinn.com).
Notwithstanding the monotypic Duparquetioideae, the other five subfamilies were recovered as monophyletic with strong support (UFBoot = 100%) in all analyses (Fig. 1 and Supplementary Fig. S5 and Table S4 available on Dryad). A relationship of (Duparquetioideae, (Dialioideae, (Caesalpinioideae, Papilionoideae))), abbreviated as DDCP, was strongly supported in all analyses, as recently reported by (LPWG 2017) based on matK and 81 plastid coding genes. This relationship was also recovered in the analyses of Koenen et al. (2020), except that Duparquetioideae was not sampled in their nuclear data set. However, the relationships among DDCP, Cercidoideae, and Detarioideae remained unresolved in our analyses, with all three possible relationships supported by different data sets in our analyses. The topology of (Cercidoideae, (Detarioideae, DDCP)) was strongly supported in the PC-GB-default data set (UFBoot 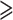 94%) and the PCN-GB-default data set (UFBoot
94%) and the PCN-GB-default data set (UFBoot 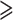 93%), consistent with previous studies based on a few plastid loci (e.g., Doyle et al. 2000; Bruneau et al. 2001; Kajita et al. 2001) as well as the plastome analyses of Koenen et al. (2020). The topology of (Detarioideae, (Cercidoideae, DDCP)) was supported by the multiple PC and PCN data sets (see Supplementary Methods available on Dryad); this relationship was also weakly supported in the study of Bruneau et al. (2008). The topology of ((Cercidoideae, Detarioideae), DDCP) was strongly supported by most PN-derived data sets with UFBoot
93%), consistent with previous studies based on a few plastid loci (e.g., Doyle et al. 2000; Bruneau et al. 2001; Kajita et al. 2001) as well as the plastome analyses of Koenen et al. (2020). The topology of (Detarioideae, (Cercidoideae, DDCP)) was supported by the multiple PC and PCN data sets (see Supplementary Methods available on Dryad); this relationship was also weakly supported in the study of Bruneau et al. (2008). The topology of ((Cercidoideae, Detarioideae), DDCP) was strongly supported by most PN-derived data sets with UFBoot 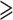 90% (Supplementary Table S4 available on Dryad); the same topology was reconstructed based on 101 single-copy nuclear genes (Bootstrap Support = 61%; Cannon et al. 2015), while the nuclear analyses of Koenen et al. (2020) recovered Cercidoideae + Detarioideae as sister to DDCP. The ASTRAL analyses were largely consistent with results from the concatenation analyses. Like the concatenation analyses, the ASTRAL results showed poor resolution at the root node, with Cercidoideae + Detarioideae sister to the rest of the family (with low support) when all bipartitions were included (Supplementary Fig. S10 available on Dryad), and with Detarioideae sister to the rest of the family when branches with <10% bootstrap support were collapsed (Supplementary Fig. S11 available on Dryad). The difficulty in confidently resolving these deepest relationships of Leguminosae has been attributed to rapid diversification of these lineages (Lavin et al. 2005; Koenen et al. 2020) and ancient polyploidization (Cannon et al. 2015).
90% (Supplementary Table S4 available on Dryad); the same topology was reconstructed based on 101 single-copy nuclear genes (Bootstrap Support = 61%; Cannon et al. 2015), while the nuclear analyses of Koenen et al. (2020) recovered Cercidoideae + Detarioideae as sister to DDCP. The ASTRAL analyses were largely consistent with results from the concatenation analyses. Like the concatenation analyses, the ASTRAL results showed poor resolution at the root node, with Cercidoideae + Detarioideae sister to the rest of the family (with low support) when all bipartitions were included (Supplementary Fig. S10 available on Dryad), and with Detarioideae sister to the rest of the family when branches with <10% bootstrap support were collapsed (Supplementary Fig. S11 available on Dryad). The difficulty in confidently resolving these deepest relationships of Leguminosae has been attributed to rapid diversification of these lineages (Lavin et al. 2005; Koenen et al. 2020) and ancient polyploidization (Cannon et al. 2015).
Given the inability of both nuclear (Koenen et al. 2020) and plastid (this study) genomic data sets to fully resolve the legume root, it seems possible that this represents a hard polytomy, with a more-or-less simultaneous origin of major legume lineages. Future studies might explore the implications of a hard polytomy for understanding early morphological and genomic diversification in this important family.
In contrast to these problematic deep relationships, our analyses significantly clarified relationships within the Leguminosae subfamilies (Fig. 1 and Supplementary Fig. S5 and Table S4 available on Dryad). Within Caesalpinioideae, the two clades of the Umtiza grade [((Arcoa, (Acrocarpus, Ceratonia)) and (Umtiza, (Gleditsia, Gymnocladus))] were subsequent sisters to remaining members of the subfamily in all data sets except the PN-GB-default data set. A robustly supported Cassia clade was resolved as sister to the remaining Caesalpiniodieae, which is divided into two clades (see Supplementary Results available on Dryad). Within Papilionoideae, our study strongly supported the Swartzioid clade, the ADA clade (comprising the tribes Amburaneae, Dipterygeae, and Angylocalyceae; Cardoso et al., 2012, 2013), and the Cladrastis clade as successive sisters to the 50-kb inversion clade (Fig. 1), whereas previous studies recovered, with weak support, an ADA and Swartzioid clade as the first diverging lineage (Wojciechowski et al. 2004) or the ADA clade and the Swartzioid clade as successive sisters to remaining papilionoids (Cardoso et al. 2012, 2013). Within subfamily Detarioideae (e.g., Bruneau et al. 2001, 2008; de la Estrella et al. 2017, 2018), we recovered the six tribes recognized by de la Estrella et al. (2018) with strong support and we were able to resolve the previously problematic relationships amongst these tribes (see Supplementary Results available on Dryad). Within subfamily Cercidoideae, Cercis and Adenolobus were robustly supported as successively sister to the remaining lineages, which is consistent with the results from Bruneau et al. (2008) and Sinou et al. (2009); Sinou et al. (2020), and Bauhinia 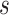 .
.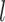 . was resolved into two strongly supported clades (Fig. 1; see Supplementary Results available on Dryad). The placement of Griffonia was unresolved in past analyses, and in our analyses, it was strongly supported as either sister to the two Bauhinia
. was resolved into two strongly supported clades (Fig. 1; see Supplementary Results available on Dryad). The placement of Griffonia was unresolved in past analyses, and in our analyses, it was strongly supported as either sister to the two Bauhinia 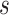 .
.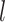 . clades (by all PC data sets and some PCN data sets) or as sister to Bauhinia
. clades (by all PC data sets and some PCN data sets) or as sister to Bauhinia 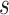 .
.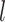 . I (i.e., the Phanera clade of Sinou et al. 2020) (by all PN data sets and some PCN data sets; Fig. 1 and Supplementary Table S4 available on Dryad).
. I (i.e., the Phanera clade of Sinou et al. 2020) (by all PN data sets and some PCN data sets; Fig. 1 and Supplementary Table S4 available on Dryad).
Conflicting Phylogenetic Signals in the Plastome
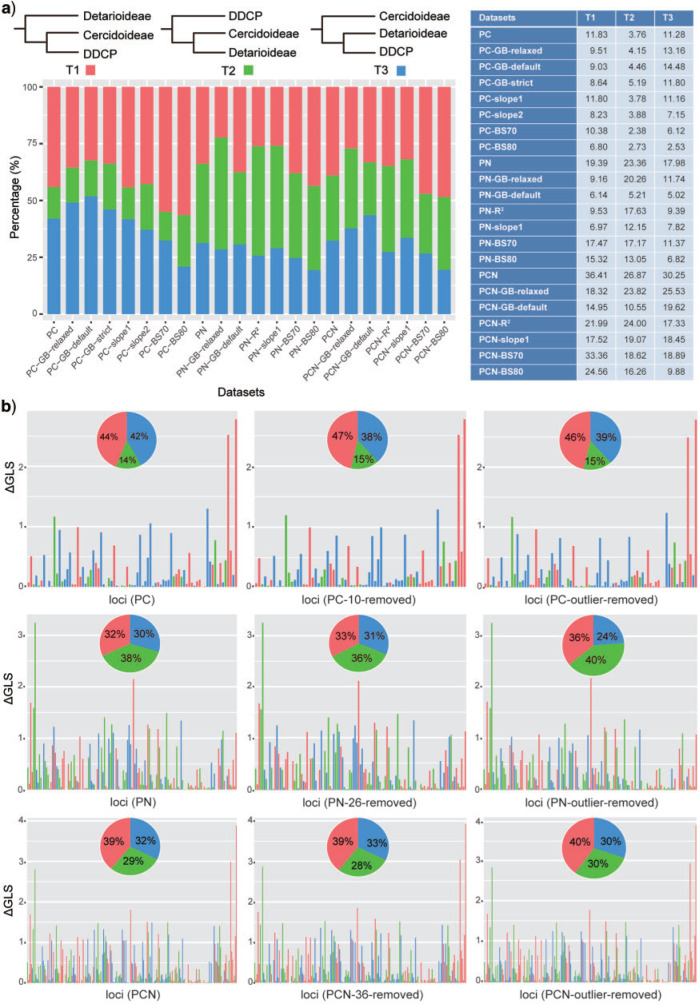
Although our plastid analyses largely resolved recalcitrant relationships across Leguminosae phylogeny, we identified multiple instances of strongly supported conflict among plastid loci and among sequence types (coding vs. non-coding) at several long-controversial nodes in the family (e.g., the root of legumes and the positions of the genera Griffonia and Pterogyne). Strategies to reduce systematic error (including the removal of outlier genes, saturated nucleotide positions, and poorly supported genes; see Supplementary Materials available on Dryad for more details) were effective for resolving many previously contentious relationships, but not for the root of legumes and the positions of the genera Griffonia and Pterogyne, for example, where conflict/concordance analysis of the gene trees (Supplementary Fig. S6 available on Dryad) revealed considerable strongly supported gene tree conflict. Concerning the root of legumes, subsets of genes supported three main alternative resolutions (Fig. 2, Supplementary Table S5 available on Dryad). The alternative positions of Griffonia and Pterogyne seemed to be largely driven by distinct phylogenetic signal in the coding versus non-coding regions of the plastome (Supplementary Fig. S4 available on Dryad). It is possible that placements of these genera in PN data sets are driven by sequence saturation, as we inferred many of the PN regions to exhibit significant signatures of saturation (in contrast to the PC regions; Supplementary Tables S2 and S6 available on Dryad). Both of these genera are relatively phylogenetically isolated (i.e., on long branches) and thus would be susceptible to misplacement with extensive homoplasy (due to long-branch attraction).
Figure 2.
The distribution of phylogenetic signal for three alternative topological hypotheses at the root of Leguminosae. a) (upper left) The three alternative topological hypotheses; (bottom left), 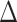 GLS proportion of loci supporting each of three alternative hypotheses across 22 data sets; (right) the summed
GLS proportion of loci supporting each of three alternative hypotheses across 22 data sets; (right) the summed 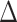 GLS values for each data set; b) the distribution of the
GLS values for each data set; b) the distribution of the 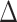 GLS values for three basic data sets (the PC, PN, and PCN data sets) and the derived data sets (details in Supplementary Methods available on Dryad) including (1) inconsistent loci removed and (2) outlier loci removed. The pies indicate the
GLS values for three basic data sets (the PC, PN, and PCN data sets) and the derived data sets (details in Supplementary Methods available on Dryad) including (1) inconsistent loci removed and (2) outlier loci removed. The pies indicate the 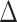 GLS proportion supported by each alternative topology, and collectively show the signal distribution before and after removal of the inconsistent and outlier loci.
GLS proportion supported by each alternative topology, and collectively show the signal distribution before and after removal of the inconsistent and outlier loci.
The topology for the legume root predominantly favored in our analyses (i.e., that shown in Fig. 1) differs from the plastid results of Koenen et al. (2020), who recovered Cercidoideae as sister to the rest of the family with moderate support. However, their plastid analyses were based entirely on amino acid analyses of the coding regions. Walker et al. (2019) found that, even across the phylogenetic breadth of angiosperms, the coding regions of the plastome did not show significant signs of saturation, and consequently nucleotides proved much more informative, a result supported by our analyses (Supplementary Methods, Table S2 available on Dryad). Only a handful of genes showed signals of saturation, and excluding them did not significantly impact topological inferences, perhaps explaining previous suggestions that plastid genes were largely uninformative (e.g., Koenen et al. 2020). Of course, we also observed the majority of the plastid genes to have low information content, but nevertheless we were able to identify both coding and non-coding loci with strong signal for many nodes of the legume phylogeny. Our analyses show that, in addition to many uninformative regions, the plastome shows complex, and often conflicting, patterns of strong phylogenetic signal.
The sources of conflict in plastome phylogenies remain understudied and poorly understood. While most plastid regions examined appear largely uninformative (at least for many nodes, consistent with Walker et al. (2019)), we nevertheless recovered strongly supported conflict at 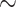 32% of nodes and strongly supported gene tree concordance at many others (Supplementary Fig. S6 available on Dryad). Stochasticity (stemming from rapid radiations and limited phylogenetic signal/information) and systematic error likely explain much of the observed conflict, and our efforts to reduce systematic error did indeed alleviate some of the observed conflict (Supplementary Fig. S6 available on Dryad). Nevertheless, other biological sources, such as heteroplasmic recombination, deserve consideration in light of the remaining strongly supported conflict. Potential for heteroplasmy (based on pollen screenings) was documented in 19/61 legume species examined (Corriveau and Coleman 1988; Zhang et al. 2003), and heteroplasmy has been directly documented in four legume genera: Astragalus (Lei et al. 2016), Cicer (Kumari et al. 2011), Medicago (Johnson and Palmer 1989; Lee et al. 1988), and Lens (Rajora and Mahon 1995). Plastid recombination is generally regarded as rare (Birky 1995), but several recent studies have highlighted potential cases of heteroplasmic recombination (Sullivan et al. 2017; Sancho et al. 2018), and this phenomenon has been documented in the laboratory (Medgyesy et al. 1985). We hesitate to attribute any of our observed conflict to such causes, as explicit documentation of heteroplasmic recombination is a challenging task, beyond the scope of this study. Nevertheless, it is possible that the observed conflicts relate to the complex history of plastome structural evolution in legumes (e.g., Palmer and Thompson 1982; Palmer et al. 1987; Jansen et al. 2008; Lei et al. 2016; Wang et al. 2018), a topic that clearly deserves further attention in future studies. The results presented here, characterizing conflict across Leguminosae phylogeny, provide a critical roadmap for future investigations of plastome conflict and evolution across the family.
32% of nodes and strongly supported gene tree concordance at many others (Supplementary Fig. S6 available on Dryad). Stochasticity (stemming from rapid radiations and limited phylogenetic signal/information) and systematic error likely explain much of the observed conflict, and our efforts to reduce systematic error did indeed alleviate some of the observed conflict (Supplementary Fig. S6 available on Dryad). Nevertheless, other biological sources, such as heteroplasmic recombination, deserve consideration in light of the remaining strongly supported conflict. Potential for heteroplasmy (based on pollen screenings) was documented in 19/61 legume species examined (Corriveau and Coleman 1988; Zhang et al. 2003), and heteroplasmy has been directly documented in four legume genera: Astragalus (Lei et al. 2016), Cicer (Kumari et al. 2011), Medicago (Johnson and Palmer 1989; Lee et al. 1988), and Lens (Rajora and Mahon 1995). Plastid recombination is generally regarded as rare (Birky 1995), but several recent studies have highlighted potential cases of heteroplasmic recombination (Sullivan et al. 2017; Sancho et al. 2018), and this phenomenon has been documented in the laboratory (Medgyesy et al. 1985). We hesitate to attribute any of our observed conflict to such causes, as explicit documentation of heteroplasmic recombination is a challenging task, beyond the scope of this study. Nevertheless, it is possible that the observed conflicts relate to the complex history of plastome structural evolution in legumes (e.g., Palmer and Thompson 1982; Palmer et al. 1987; Jansen et al. 2008; Lei et al. 2016; Wang et al. 2018), a topic that clearly deserves further attention in future studies. The results presented here, characterizing conflict across Leguminosae phylogeny, provide a critical roadmap for future investigations of plastome conflict and evolution across the family.
Acknowledgments
We thank Prof. M. van der Bank (The African Centre for DNA Barcoding, South Africa), Mr. S.M. Omosowon (Imperial College London, United Kingdom), Mr. N. Zamora (Museo Nacional de Costa Rica, Costa Rica), Dr. J. Cai, Mr. C. Liu, Mr. Y.-J. Guo, Mr. J.-D Ya and Dr. T. Zhang (Kunming Institute of Botany, Chinese Academy of Sciences, China), Royal Botanic Garden Edinburgh (United Kingdom), Royal Botanic Gardens Kew (United Kingdom), Institute of Botany (Chinese Academy of Sciences, China), Muséum national d’Histoire naturelle (France), Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, China for assistance in sample acquisitions. We are grateful to Dr. X.-X. Shen from Zhejiang University, China and Prof. T.H. Struck from University of Oslo, Norway for helpful discussions of our results. We sincerely thank Prof. S.A. Smith at the University of Michigan, USA and anonymous reviewers for helping to review the manuscript. All experiments for this study were facilitated by the Germplasm Bank of Wild Species at the Kunming Institute of Botany, Chinese Academy of Sciences. The use of DNA from Brazilian species is authorized by SISGEN  R79ED6F.
R79ED6F.
Supplementary Material
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.1vhhmgqpb.
Funding
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences [XDB31010000], the National Natural Science Foundation of China [key international (regional) cooperative research project No. 31720103903], the Large-scale Scientific Facilities of the Chinese Academy of Sciences [No. 2017-LSF-GBOWS-02], an open research project for “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, CNPq Research Productivity Fellowships [306736/2015-2 and 303585/2016-1], Prêmio CAPES de Teses [23038.009148/2013-19], FAPESB [APP0037/2016], and the Natural Sciences and Engineering Research Council (NSERC) of Canada to A.B..
References
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A.. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C.W. 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. USA 92:11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.M., Thomson R.C.. 2017. Bayes factors unmask highly variable information content, bias, and extreme influence in phylogenomic analyses. Syst. Biol. 4:517–530. [DOI] [PubMed] [Google Scholar]
- Bruneau A., Forest F., Herendeen P.S., Klitgaard B.B., Lewis G.P.. 2001. Phylogenetic relationships in the Caesalpinioideae (Leguminosae) as inferred from chloroplast trnL intron sequences. Syst. Bot. 26:487–514. [Google Scholar]
- Bruneau A., Mercure M., Lewis G.P., Herendeen P.S.. 2008. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany 86:697–718. [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S.B., McKain M.R., Harkess A., Nelson M.N., Dash S., Deyholos M.K., Peng Y., Joyce B. Stewart C.N. Jr, Rolf M., Kutchan T., Tan X., Chen C., Zhang Y., Carpenter E., Wong G.K., Doyle J.J., Leebens-Mack J.. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol. Biol. Evol. 32:193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso D., de Queiroz L.P., Pennington R.T., Lima H.C., Fonty E., Wojciechowski M.F., Lavin M.. 2012. Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. Am. J. Bot. 99:1991–2013. [DOI] [PubMed] [Google Scholar]
- Cardoso D., Pennington R.T., de Queiroz L.P., Boatwright J.S., Van Wyk B.E., Wojciechowski M.F., Lavin M.. 2013. Reconstructing the deep-branching relationships of the papilionoid legumes. S. Afr. J. Bot. 89:58–75. [Google Scholar]
- Chernomor O., von Haeseler A., Minh B.Q.. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corriveau J. L., Coleman A.W.. 1988. A rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 75:1443–1458. [Google Scholar]
- Doyle J.J., Chappill J.A., Bailey C.D., Kajita T.. 2000. Towards a comprehensive phylogeny of legumes: evidence from rbcL sequences and non-molecular data In: Herendeen P.S., Bruneau A., editors. Advances in legume systematics, Part 9. Richmond: Royal Botanic Gardens, Kew; p. 1–20. [Google Scholar]
- Duranti M. 2006. Grain legume proteins and nutraceutical properties. Fitoterapia 77:67–82. [DOI] [PubMed] [Google Scholar]
- de la Estrella M., Forest F., Wieringa J.J., Fougere-Danezan M., Bruneau A.. 2017. Insights on the evolutionary origin of Detarioideae, a clade of ecologically dominant tropical African trees. New Phytol. 214:1722–1735. [DOI] [PubMed] [Google Scholar]
- de la Estrella M., Forest F., Klitgård B., Lewis G.P., Mackinder B.A., de Queiroz L.P., Wieringa J.J., Bruneau A.. 2018. A new phylogeny-based tribal classification of subfamily Detarioideae, an early branching clade of florally diverse tropical arborescent legumes. Sci. Rep. 8:6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Daubin V.. 2008. Dealing with incongruence in phylogenomic analyses. Philos. Trans. R Soc. Lond. B Biol. Sci. 363:4023–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves D.J.P., Simpson B.B., Ortiz E.M., Shimizu G.H., Jansen R.K.. 2019. Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 138:219–232. [DOI] [PubMed] [Google Scholar]
- Goremykin V.V., Nikiforova S.V., Cavalieri D., Pindo M., Lockhart P.. 2015. The root of flowering plants and total evidence. Syst. Biol. 64:879–891. [DOI] [PubMed] [Google Scholar]
- Graham P.H., Vance C.P.. 2003. Legumes: importance and constraints to greater use. Plant Physiol. 131:872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.K., Wojciechowski M.F., Sanniyasi E., Lee S.-B., Daniell H.. 2008. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48:1204–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T., Suh A., Weber C.C., da Fonseca R.R., Li J., Zhang F., Li H., Zhou L., Narula N., Liu L., Ganapathy G., Boussau B., Bayzid M.S., Zavidovych V., Subramanian S., Gabaldon T., Capella-Gutierrez S., Huerta-Cepas J., Rekepalli B., Munch K., Schierup M., Lindow B., Warren W.C., Ray D., Green R.E., Bruford M.W., Zhan X., Dixon A., Li S., Li N., Huang Y., Derryberry E.P., Bertelsen M.F., Sheldon F.H., Brumfield R.T., Mello C.V., Lovell P.V., Wirthlin M., Schneider M.P.C., Prosdocimi F., Samaniego J.A., Velazquez A.M.V., Alfaro-Nunez A., Campos P.F., Petersen B., Sicheritz-Ponten T., Pas A., Bailey T., Scofield P., Bunce M., Lambert D.M., Zhou Q., Perelman P., Driskell A.C., Shapiro B., Xiong Z., Zeng Y., Liu S., Li Z., Liu B., Wu K., Xiao J., Xiong Y., Zheng Q., Zhang Y., Yang H., Wang J., Smeds L., Rheindt F.E., Braun M., Fjeldsa J., Orlando L., Barker F.K., Jonsson K.A., Johnson W., Koepfli K.P., O’Brien S., Haussler D., Ryder O.A., Rahbek C., Willerslev E., Graves G.R., Glenn T.C., McCormack J., Burt D., Ellegren H., Alstrom P., Edwards S.V., Stamatakis A., Mindell D.P., Cracraft J., Braun E.L., Warnow T., Jun W., Gilbert M.T.P., Zhang G.. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian S., Soltis P.S., Gitzendanner M.A., Moore M.J., Li R., Hendry T.A., Qiu Y.-L., Dhingra A., Bell C.D., Soltis D.E.. 2008. Resolving an ancient, rapid radiation in Saxifragales. Syst. Biol. 57:38–57. [DOI] [PubMed] [Google Scholar]
- Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C.D., Yi T.-S., Li D.-Z. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv 256479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Kendall M., Almagro-Garcia J., Colijn C.. 2017. Treespace: statistical exploration of landscapes of phylogenetic trees. Mol. Ecol. Resour. 17:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.B., Palmer J.D.. 1989. Heteroplasmy of chloroplast DNA in Medicago. Plant Mol. Biol. 12:3–11. [DOI] [PubMed] [Google Scholar]
- Kajita T., Ohashi H., Tateishi Y., Bailey C.D., Doyle J.J.. 2001. rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst. Bot. 26:515–536. [Google Scholar]
- Katoh K., Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen E.J.M., Ojeda D.I., Steeves R., Migliore J., Bakker F.T., Wieringa J.J., Kidner C., Hardy O.J., Pennington T.R., Bruneau A., Hughes C.E.. 2020. Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near simultaneous evolutionary origin of all six subfamilies. New Phytol. 225:1355–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M., Clarke H.J., des Francs-Small C.C., Small I., Khan T.N., Siddique K.H.. 2011. Albinism does not correlate with biparental inheritance of plastid DNA in interspecific hybrids in Cicer species. Plant Sci. 180:628–633. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F.. 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54:575–594. [DOI] [PubMed] [Google Scholar]
- Lee D.J., Blake T.K., Smith S.E.. 1988. Biparental inheritance of chloroplast DNA and the existence of heteroplasmic cells in alfalfa. Theor. Appl. Genet. 76: 545–549. [DOI] [PubMed] [Google Scholar]
- Lei W., Ni D., Wang Y., Shao J., Wang X., Yang D., Wang J., Chen H., Liu C.. 2016. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceous. Sci. Rep. 6:21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.P., Schrire B., Mackinder B., Lock M. 2005. Legumes of the World. Richmond: Royal Botanic Gardens, Kew. [Google Scholar]
- LPWG (Legume Phylogeny Working Group). 2013. Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62:217–248. [Google Scholar]
- LPWG (Legume Phylogeny Working Group). 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66:44–77. [Google Scholar]
- McMahon M.M., Sanderson M.J.. 2006. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst. Biol. 55:818–836. [DOI] [PubMed] [Google Scholar]
- Medgyesy I., Fejes E., Maliga P.. 1985. Interspecific chloroplast DNA recombination in a Nicotiana somatic hybrid. Proc. Natl. Acad. Sci. USA 82:6960–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D., Thompson W.F.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29:537–550. [DOI] [PubMed] [Google Scholar]
- Palmer J.D., Osorio B., Aldrich J., Thompson W.F.. 1987. Chloroplast DNA evolution among legumes: loss of an inverted repeat occurred prior to other sequence rearrangements. Curr. Genet. 11:275–286. [Google Scholar]
- Parks M.B., Wickett N.J., Alverson A.J.. 2018. Signal, uncertainty, and conflict in phylogenomic data for a diverse lineage of microbial Eukaryotes (Diatoms, Bacillariophyta). Mol. Biol. Evol. 35:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org. [Google Scholar]
- Rajora O.P., Mahon J.D.. 1995. Paternal plastid DNA can be inherited in lentil. Theor. Appl. Genet. 90:607–610. [DOI] [PubMed] [Google Scholar]
- Ramsey A.J., Mandel J.R.. 2019. When one genome is not enough: organellar heteroplasmy in plants. Ann. Plant Rev. 2:1–40. [Google Scholar]
- Rokas A., Williams B.L., King N., Carroll S.B.. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804. [DOI] [PubMed] [Google Scholar]
- Sancho R., Cantalapiedra C.P., López-Alvarez D., Gordon S.P., Vogel J.P., Catalán P., Contreras-Moreira B.. 2018. Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 218:1631–1644. [DOI] [PubMed] [Google Scholar]
- Shen X.-X., Hittinger C.T., Rokas A.. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1:126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinou C., Forest F., Lewis G.P., Bruneau A.. 2009. The genus Bauhinia s.l. (Leguminosae): a phylogeny based on the plastid trnL-trnF region. Botany 87:947–960. [Google Scholar]
- Sinou C., Cardinal-McTeague W., Bruneau A.. 2020. Testing generic limits in Cercidoideae (Leguminosae): insights from plastid and duplicated nuclear gene sequences. Taxon. doi: 10.1002/(ISSN)1996-8175. [DOI] [Google Scholar]
- Smith D.R. 2014. Mitochondrion-to-plastid DNA transfer: it happens. New Phytol. 202:736–738. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Wilson N.G., Goetz F.E., Feehery C., Andrade S.C., Rouse G.W., Giribet G., Dunn C.W.. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480: 364–367. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Moore M.J., Brown J.W., Yang Y.. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol. 15:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J.I., Ardley J., James E.K.. 2017. Biogeography of nodulating legumes and their nitrogen-fixing symbionts. New Phytol. 215:40–56. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S.C.K., Cronn R.C., Edwards C., Fishbein M., Liston A.. 2013. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol. 5:1872–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A.R., Schiffthaler B., Thompson S.L., Street N.R., Wang X.R.. 2017. Interspecific plastome recombination reflects ancient reticulate evolution in Picea (Pinaceae). Mol. Biol. Evol. 34:1689–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl C., Badger J., Kearney P., Li M., Clegg M., Jiang T.. 2003. Probabilistic analysis indicates discordant gene trees in chloroplast evolution. J. Mol. Evol. 56:330–340. [DOI] [PubMed] [Google Scholar]
- Voisin A.S., Gueguen J., Huyghe C., Jeuffroy M.H., Magrini M.B., Meynard J.M., Mougel C., Pellerin S., Pelzer E.. 2014. Legumes for feed, food, biomaterials and bioenergy in Europe: a review. Agron. Sustain. Dev. 34:361–380. [Google Scholar]
- Walker J.F., Yang Y., Feng T., Timoneda A., Mikenas J., Hutchison V., Edwards C., Wang N., Ahluwalia S., Olivieri J., Walker-Hale N., Majure L.C., Puente R., Kadereit G., Lauterbach M., Eggli U., Flores-Olvera H., Ochoterena H., Brockington S.F., Moore M.J., Smith S.A.. 2018a. From cacti to carnivores: improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. Am. J. Bot. 105:446–462. [DOI] [PubMed] [Google Scholar]
- Walker J.F., Brown J.W., Smith S.A.. 2018b. Analyzing contentious relationships and outlier genes in phylogenomics. Syst. Biol. 67:916–924. [DOI] [PubMed] [Google Scholar]
- Walker J.F., Walker-Hale N., Vargas O.M., Larson D.A., Stull W.G.. 2019. Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7:e7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-H., Wicke S., Wang H., Jin J.-J., Chen S.-Y., Zhang S.-D., Li D.-Z., Yi T.-S.. 2018. Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci. 9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Schultz M.B., Zobel J., Holt K.E.. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett N.J., Mirarab S., Nguyen N., Warnow T., Carpenter E., Matasci N., Ayyampalayam S., Barker M.S., Burleigh J.G., Gitzendanner M.A., Ruhfel B.R., Wafula E., Der J.P., Graham S.W., Mathews S., Melkonian M., Soltis D.E., Soltis P.S., Miles N.W., Rothfels C.J., Pokorny L., Shaw A.J., DeGironimo L., Stevenson D.W., Surek B., Villarreal J.C., Roure B., Philippe H., dePamphilis C.W., Chen T., Deyholos M.K., Baucom R.S., Kutchan T.M., Augustin M.M., Wang J., Zhang Y., Tian Z., Yan Z., Wu X., Sun X., Wong G.K., Leebens-Mack J.. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 111:E4859-E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M.F., Lavin M., Sanderson M.J.. 2004. A phylogeny of legumes (Leguminosae) based on analyses of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91:1846–1862. [DOI] [PubMed] [Google Scholar]
- Xi Z., Ruhfel B.R., Schaefer H., Amorim A.M., Sugumaran M., Wurdack K.J., Endress P.K., Matthews M.L., Stevens P.F., Mathews S.. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. USA 109:17519–17524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu Y., Sodmergen. 2003. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 44:941–951. [DOI] [PubMed] [Google Scholar]
- Zhang C., Sayyari E., Mirarab S.. 2017. ASTRAL-III: Increased scalability and impacts of contracting low support branches In: Meidanis J., Nakhleh L., editors. Comparative genomics. RECOMB-CG 2017. Cham: Springer (Lecture notes in computer science; vol. 10562). [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S.. 2018. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.-D., Jin J.-J., Chen S.-Y., Chase M.W., Soltis D.E., Li H.-T., Yang J.-B., Li D.-Z., Yi T.-S.. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214:1355–1367. [DOI] [PubMed] [Google Scholar]