Abstract
Mammalian spermatogenesis is a complex developmental program that transforms mitotic testicular germ cells (spermatogonia) into mature male gametes (sperm) for production of offspring. For decades, it has been known that this several-weeks-long process involves a series of highly ordered and morphologically recognizable cellular changes as spermatogonia proliferate, spermatocytes undertake meiosis, and spermatids develop condensed nuclei, acrosomes, and flagella. Yet, much of the underlying molecular logic driving these processes has remained opaque because conventional characterization strategies often aggregated groups of cells to meet technical requirements or due to limited capability for cell selection. Recently, a cornucopia of single-cell transcriptome studies has begun to lift the veil on the full compendium of gene expression phenotypes and changes underlying spermatogenic development. These datasets have revealed the previously obscured molecular heterogeneity among and between varied spermatogenic cell types and are reinvigorating investigation of testicular biology. This review describes the extent of available single-cell RNA-seq profiles of spermatogenic and testicular somatic cells, how those data were produced and evaluated, their present value for advancing knowledge of spermatogenesis, and their potential future utility at both the benchtop and bedside.
Keywords: single-cell, transcriptome, stem cells, heterogeneity, testis
This review details the host of new and revolutionary single-cell RNA-seq results from mouse and human spermatogenic cells that are already informing basic biological concepts of testicular function with high translational significance for male infertility and contraception.
Heterogeneity in spermatogenesis
Gamete production is the limiting factor for transmission of genomic information to the next generation through fertilization. While the mammalian ovary produces a limited number of oocytes over the female reproductive lifespan [1–5], the testis sustains a high level of sperm output until death [6–8]. Life-long spermatogenesis is enabled by a highly productive, adult stem cell-based developmental system in which spermatogenic cell development is executed under the careful watch of the testicular soma. Spermatogonia are mitotic spermatogenic cells that can be broadly classified into undifferentiated and differentiating subsets based on nuclear histomorphological appearance [8–11] or expression of markers (e.g. KIT, ZBTB16) [12–19]. Spermatogonial stem cells (SSCs) are the most primitive spermatogonia and are a subset of undifferentiated spermatogonia capable of perpetual self-renewal [20] (Figure 1A). SSCs can also initiate spermatogenic differentiation by producing progenitor spermatogonia that are functionally distinct from SSCs because they have limited self-renewal capacity and are committed to continue differentiation [20]. Subsequently, differentiating spermatogonia undergo a species-specific number of amplifying mitotic divisions before producing spermatocytes that will undergo two meiotic divisions to produce haploid spermatids that undertake the multi-step spermiogenesis process to produce flagellated sperm [21]. Production of sperm from SSCs is completed over the course of roughly five (mice) to eight (human) weeks [22].
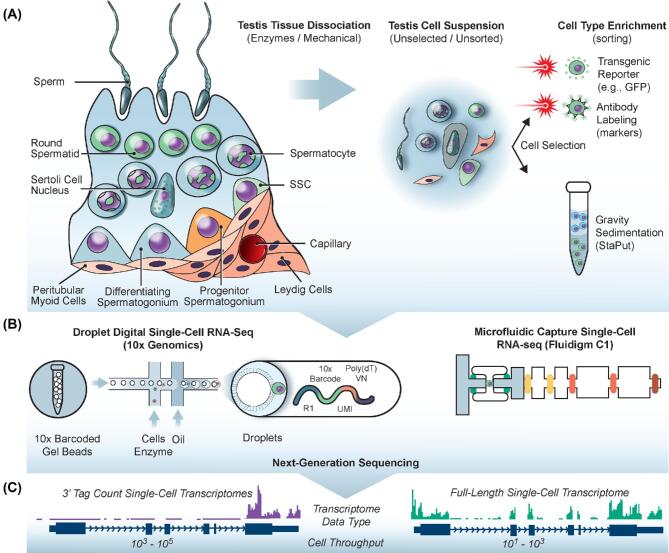
Figure 1.
Application of single cell RNA-seq to the study of spermatogenesis. (A) The complex testicular tissue is dissociated enzymatically and with mechanical force into individual cells. Many scRNA-seq studies explored the transcriptomes of these cell unselected/unsorted cell suspensions, while others used various strategies reliant on transgenic reporters, antibody labeling or cell density to enrich for particular cell types. (B) Unsorted or selected cell suspensions were then used for single-cell RNA-seq. Two popular platforms are the 10x Genomics Chromium (left) and Fluigidm C1 (right). In the 10x Genomics droplet-digital RNAseq approach, individual cells are capsulated in aqueous droplets together with microbeads that deliver barcoded primers for reverse transcription. The Fluidigm C1 physically captures individual cells on microfluidic chips and automatically generates full-length cDNA. (C) After next-generation sequencing of single-cell RNA-seq libraries, the 10x Genomics and Fluidigm C1 produce different data types depicted by the “tracks” of colored sequence located above the genome annotation. 10x Genomics libraries utilize 3′ end-counting chemistry, which maximizes cell throughput and better controls for PCR duplication bias through the use of unique molecular identifiers (UMIs). Fluidigm C1 libraries are full-length transcriptomes, meaning that mRNA variants (e.g., spliceoforms, alternative TSS usage) can be recognized, but with the drawback of reduced cell throughput and greater expense. This figure is available in color at Biology of Reproduction online.
This simplistic characterization of the spermatogenic process, though, ignores decades of morphological studies which have described in intricate detail the identity, numbers, and kinetics of multiple spermatogenic cell types and subtypes (Figure 1A) [10]. Consider that in mice, undifferentiated spermatogonia (including functionally defined SSCs and progenitor spermatogonia) exist in multiple clonal generations of undifferentiated spermatogonia, Asingle (1 cell), Apaired (2 cell clones), and Aaligned (4–16 cell clones), which give rise to multiple sequential generations of differentiating spermatogonia that are themselves morphologically distinguishable (Types A1, A2, A3, A4, intermediate, and B spermatogonia) [23–34]. At the time of entry into prophase I of meiosis, Type B spermatogonia will transition into preleptotene spermatocytes that subsequently follow the meiotic program characterized by the well-defined primary spermatocyte (leptonema, zygonema, pachynema, diplonema) and secondary spermatocyte phases [35]. Finally, spermiogenesis is separable into two broad phases encompassing round spermatid and elongating/condensing spermatids and occurs in 16 distinct steps over more than two weeks [36–38]. During spermiogenesis, these steps coincide with genome repackaging in which the vast majority of histones are sequentially replaced by transition proteins and then protamines, formation of an acrosome and assembly of the flagellum [39]. While spermatogenesis in higher primates utilizes different terminology to describe spermatogenic cell types and occurs with more limited clonal amplification, the process is considered to be highly conserved [22].
Across an entire adult testis in steady-state, spermatogenesis is both highly ordered and asynchronous, which allows for continual sperm production [15, 40, 41]. At any given position along the length of the seminiferous tubules of the testis, spermatogenic development occurs in repeating fashion, termed the cycle of the seminiferous epithelium, which is characterized by a recurrent set of defined cellular associations between different spermatogenic cell types [10, 42]. Each set of associations between different types of spermatogonia, spermatocytes and spermatids is considered to be a “stage” of the cycle of the seminiferous epithelium—there are 12 stages in mice [35] and rhesus monkeys [43], but only 6 stages in humans [44–46], and at any given time, stages appear to proceed in a wave-like fashion along the length of the seminiferous tubules [36–38, 47]. In mice, a pulse of retinoic acid (RA) production at the mid-point of the seminiferous epithelial cycle (stages VII-VIII) drives spermatogonial differentiation and coincides with meiotic entry and spermatid release (spermiation) [48, 49]. At any given position along the length of mouse seminiferous tubules, the differentiation inducing RA pulse occurs every 8.6 days [14–16, 50]. Thus, spermatogenic development is highly heterogeneous in time and space.
A wealth of advancements in our collective understanding of the fundamental biological mechanisms responsible for the ongoing spermatogenesis have emerged in the molecular biology era. Gene expression patterns among spermatogenic cell types have been reported numerous times and have generally relied upon analyses of bulk RNA from two sources: (1) whole testes of mice during the first wave of spermatogenesis and (2) enriched, but mixed aggregates of particular spermatogenic cell types [32, 37, 51]. For instance, it has been very popular to generate enriched populations of adult pachytene spermatocytes and round spermatids from suspensions of adult testes based on cell density using StaPut gravity sedimentation [52–54] (Figure 1A). However, this and similarly “crude” methods such as cell sorting (FACS) for DNA ploidy, transgenic reporters, or cell surface antibody labeling do not produce purified cell populations and instead group multiple cell types together (e.g. the seven steps of round spermatids) (Figure 1A). Likewise, during the first weeks after birth in the mouse testis, succeeding spermatogenic cell types emerge sequentially during what is called the “first wave” of spermatogenesis [55], providing a window into the molecular changes that accompany emergence of each new cell type. But, first-wave spermatogenic cells may also exhibit unique features compared with their counterparts from steady-state adult spermatogenesis [31, 56]. Most importantly, though, these approaches fail to reveal the variation within and between cell types, and may completely ignore the phenotypes of rare cell populations, such as SSCs or transitional cell types later in the lineage, because such cells are masked by ensemble averaging across the aggregate population(s). Therefore, there is a need to overcome the limitations of conventional gene expression approaches, to effectively probe the gene expression signatures associated with each spermatogenic step and distinguish unique patterns within heterogeneous cell populations that may reflect distinct subsets.
Distinguishing spermatogenic cell phenotypes with single-cell resolution
Until relatively recently, measuring gene expression at the mRNA level across the entire transcriptome (e.g. gene expression microarray, RNA-seq) required input of purified RNA from hundreds-to-millions of cells. Given the low resolution of most spermatogenic cell selection methods, effectively discerning the gene expression patterns of every unique spermatogenic cell type was impossible because of ensemble averaging. Transcriptome profiling at the single-cell level (i.e. single-cell RNA-seq or scRNA-seq) can overcome this limitation by comprehensively measuring mRNA levels within all spermatogenic cells to detect the variation across the lineage and heterogeneity among cells at any given step or phase [57].
Single-cell RNA-seq methods
A variety of scRNA-Seq methodologies have been reported [58], including two which have become more accessible to the research community following successful commercialization: (1) microfluidic capture SMART-Seq using the Fluidigm C1 platform [59] and (2) droplet-digital RNA-seq using the 10x Genomics platform [60] (Figure 1B and C). In general, the SMART-Seq chemistry affords greater sensitivity and permits analysis of the full length transcriptome (e.g. detection of splice variants), but at much lower cell throughput (10s-100s) and with greater normalization difficulty and considerable expense (>$50/cell) (Figure 1C) [61]. The greater single-cell transcriptome depth achieved using the Fluidigm C1-SMART-Seq approach can facilitate enhanced detection of low-abundance transcripts and lower false-negative detection (dropout) rate, but with the tradeoff of profiling fewer cells and often requiring cell type enrichment (e.g. FACS), thereby introducing selection bias or potentially missing rare cell populations (Figure 1A). Reciprocally, droplet-digital RNA-seq methods employ 3′ end-counting chemistries that are multiple orders of magnitude cheaper (<$1/cell), afford easy bioinformatics normalization of PCR duplication, and much greater cell throughput (1000s–10 000s) (Figure 1C) [60]. This facilitates profiling of cells without pre-selection and detection of rare populations, but with reduced transcriptome depth in any given cell which leads to increase probability of gene dropout. In actuality, these alternate methods are complementary and each can be chosen to match experimental objectives (Figure 1). While the commercialized scRNA-seq methods require expensive instrumentation and disposables/reagents, it is possible to use home-grown methodologies such as Drop-Seq [62], Microwell-Seq [63], mechanical cell picking/single-cell sorting with Smart-Seq2 [64, 65] or single-cell combinatorial indexing RNA-seq (sci-RNA-seq) [66] to reduce the cost [61, 67]. For instance, Microwell-Seq utilizes 3′ end-counting chemistry and was used recently to report the mouse single cell atlas in which cells from nearly every tissue in the mouse body were characterized [63]. Relatively cheaply, this one study investigated more than 400 000 individual primary mouse cells, including 19 659 cells from mouse testes (although no effort was made to even distinguish between germ cells and somatic cells, let alone among spermatogenic cell types) [63].
It is important to recognize that individual single-cell transcriptome datasets may vary considerably based on a variety of technical parameters, including cell number, multiplet rate, sequencing depth, and replication, all of which drive important outcomes that should be considered when designing and evaluating such studies (Tables 1 and 2). The number of cells profiled is one basis of the statistical power to uncover differentially expressed genes, but is also critical for detection of rare cell types. The rarer the cell type of interest, the more cells need to be profiled, and vice versa. Experiments intended to compare relatively abundant cell populations may require relatively low cell throughput. Yet, consider that an experiment designed to detect SSCs, which have a reported abundance of 1 in 3000 cells from adult mouse testes [68], would need to profile 300 000 adult mouse testis cells to detect only 100 SSCs in the absence of cell enrichment/selection. Detecting rare cells is further complicated by the frequency of cell multiplets in the dataset. Multiplets are data points which, rather than arising from bona fide single-cells, actually arise from two or more cells, and the frequency of multiplets can be determined empirically and can be partially addressed bioinformatically [69]. Sequencing depth per cell is also an important parameter that influences gene/transcript detection and all downstream analyses. Depths may vary widely between studies, but are typically within the range of 2.5 × 104–2.5 × 105 reads per cell for droplet-digital scRNA-seq studies [62] to 2–5 × 106 reads per cell for SMART-Seq-based studies [70]. In either case, increasing degrees of sequencing depth leads to enhanced sensitivity for detecting genes and transcripts, though in a non-linear fashion [71], and the degree of sequencing saturation can be calculated empirically for a given dataset. Lastly, scRNA-seq studies may utilize widely different strategies of replication, including multiple independent preparations of cells (biological replicates) and independent assessments of the same cell preparations (technical replicates), which understandably, impact the robustness and reliability of the data. In general, it appears that the value of technical replication is less than biological replication, particularly for commercial scRNA-seq approaches, while greater biological replication is valuable in experiments comparing cells from genetically diverse vs. uniform sources [72]. As noted for other technical parameters, replication sufficiency can and should be determined empirically using statistical methods.
Table 1.
Mouse spermatogenesis single-cell RNA-seq datasets.
| Chen Y et al. (2018), Cell Res | Ernst C et al. (2019). Nat Commun | Green CD et al (2018), Dev Cell | Grive JK et al. (2019), PLoS Genet | Han X et al. (2018), Cell | Hermann BP et al. (2018), Cell Rep | Jung M et al. (2018), BioRxiv | La HM et al. (2018), Nat Commun | Liao J et al. (2019), Development | Makino Y et al. (2019). Sci Rep | Neirijunck et al. (2018), Endocrinology | Lukassen S et al. (2018), Sci Rep | Song HW et al. (2016). Cell Rep | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total cell number | 1204 | 53 510 (from 2 P5, 1 P10, 1 P15, 1 P20, 1 P25, 1 P30, 1 P35, 2 8–9 week mice) | 34 633 | 1200–2500 per sample (from 1 P6, 1 P14, 1 P18, 1 P25, 1 P30, 1 8week mice) | 19 659 (>50 mice) | 31 163 (from 21 adult mice, 30 P6 mice) | 62 600 (from 11 to 38 week WT, Mlh3 KO, Cul4a KO, Hormad1 KO, and Cnp mice) | Not mentioned | 71 | 175 (80 from P1.5, 48 from P3.5, and 47 from P5.5) | Interstitial steroidogenic progenitor cells (6362 cells), SCs (1932 cells), fetal Leydig cells (521 cells), primordial germ cells (483 cells), and endothelial cells (180 cells) from E16.5 | 2500 (1250 each from 2 mice) | 181 (71 from P3 WT, 53 from P7 WT, 57 from Rhox10 KO P3) |
| First-Wave | + | + | – | + | – | + | – | – | + | + | – | – | + |
| Unselected steady-state spermtogenesis | – | + | + | + | – | + | + | – | – | – | – | + | – |
| Sorted Spermatogonia | + | – | + | + | – | + | + | + | – | – | – | – | – |
| Sorted Spermatocytes | + | – | + | + | – | + | + | – | – | – | – | – | – |
| Sorted Spermatids | + | – | + | + | – | + | + | – | – | – | – | – | – |
| Somatic cells | – | Sertoli cells, Leydig cells, Myoid cells, Endothelial cells, Marcrophages | Sertoli cells, Leydig cells, Myoid cells, Endothelial cells, Macrophage, Inntate Lymphosid | Sertoli cells, Endothelial, Hematopoietic cells, Smooth muscle | – | Sertoli cells, Peritubular cells (adult) | Sertoli cells, Leydig cells | – | – | Sertoli cells | Endothelial cells, fetal Leydig cells, Sertoli cells | Sertoli cells, Leydig cells | – |
| Validation methods | Knockout, ChIP-seq | RNA scope, ChIP-seq | IHC, smFISH | IHC | – | IHC, qRT-PCR | Knockout, IHC | Knockout, Transplantation, Bulk RNA-seq, IHC | IHC, Function (RA inhibitior) | Knockout, ChIP-qPCR, IHC, WB | Knockout in Sertoli cells | – | Knockout, IHC |
| scRNA-seq Chemistry/Method | 3′ sequencing and full length sequence (SMART-seq2) | 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing and barcoding (Oliginal Drop-seq) | 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing and barcoding (SMART-seq2 and Microwell-seq) | full length sequence (Fluidigm C1), 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing and barcoding (10x Genomics) | full length sequence (Fluidigm C1) | full length sequence (Fluidigm C1) | 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing and barcoding (10x Genomics) | full length sequence (Fluidigm C1) |
| Replication | Replication unclear | 1 replicate (P10, P15, P20, P25, P30, and P35) and 2 biological replicates (P5 and adult) | 6 x samples from individual adult mice. | 1 replicate | 2 replicates of adult testes | 5 x C1 P6 ID4-EGFP, 4 x C1 adult ID4-EGFP, 1 x 10x Genomics P6 ID4-EGFPbright, 1 x 10x Genomics P6 ID4-EGFPdim, 1 x 10x Genomics P6 unselected testis cells, 1 x 10x Genomics Adult ID4-EGFPbright, 1 x 10x Genomics Adult ID4-EGFPdim, 1 x 10x Genomics Adult unselected testis cells, 3 x 10x Genomics adult steady-state spermatogenesis (unselected), 1 x Adult StaPut Spermatocytes, 1 x Adult StaPut Round Spermatids | 11 x Unselected testis cells from adult mice, 1 x spermatogonia, 1 x primary spermatocytes, 1 x secondary spermatocytes, 1 x Spermatids | 2 x biolgical replicates, each from one mouse | 1 replicate | 1 replicate | 2 x E16.5 testes | 2 biological replicates | 2 replicates of ID4-positive testicular cells from Wildtype at P3 and P7, 3 replicates of Rhox10 knockout mice at P3 |
| GEO number | GSE107644 | E-MTAB-6946 | GSE112393 | GSE121904 | GSE108097 | GSE108970, GSE108974, GSE108977, GSE109049, GSE109033, and GSE109037 | GSE113293 | GSE112880 | GSE107711 | PRJNA524391 | GSE123119 | GSE104556 | GSE82174 |
Table 2.
Human spermatogenesis single-cell RNA-seq datasets.
| Guo J et al. (2017), Cell Stem Cell | Guo J et al. (2018), Cell Res | Hermann BP et al. (2018), Cell Rep | Neuhaus N et al. (2017) Mol Hum Reprod | Sohni A et al. (2019), Cell Rep | Wang M et al. (2018), Cell Stem Cell | |
|---|---|---|---|---|---|---|
| Total cell number | 92 (from 1 to 5 men) | ∼7792 (6492 from 2 of 17-years-old, 24-years-old, and 25-years-old and ∼1300 from 12 to 13 month) | 37 086 (from 15 men) | 105 (from 1 patients with spermatogonial arrest, from 7 normal) | 33 585 (18 723 from 2 adult men and 14 862 from 2 neonates) | 3028 (2854 from normal or OA and 174 from NOA) |
| Young spermatogonia | – | + | – | – | + | – |
| Unselected steady-state spermtogenesis | – | + | + | – | + | + |
| Sorted Spermatogonia | + | + | + | + | + | + |
| Sorted Spermatocytes | – | + | + | – | + | + |
| Sorted Spermatids | – | + | + | – | + | + |
| Sperm | – | + | – | – | – | – |
| Somatic cells | – | Macrophages, Sertoli cells, Leydig cells, Myoid cells, Endothelial cells | Sertoli cells, Leydig cells, Peritubular cells, Macrophage, Perivascular cells (adult) | – | Sertoli cells, pertibular myoid cells, blood/endothelial cells, macrophages, Leydig cells (adult) Peritubular myoid cells, Leydig cells, blood/endothelial cells (neonatal) | Sertoli cells, peritubular myoid cells, Leydig cells, testicular macrophages (normal or OA and NOA patiants) |
| Validation methods | IHC | smFISH | IHC | IHC | IHC, Flow cytometory | IHC, ISH, NOA |
| scRNA-seq Chemistry/Method | full length sequence (Fluidigm C1) | 3′ sequencing and barcoding (10x Genomics) | full length sequence (Fluidigm C1), 3′ sequencing and barcoding (10x Genomics) | full length sequence | 3′ sequencing and barcoding (10x Genomics) | 3′ sequencing (SMART-seq2) |
| Replication | Replication unclear | 3 biological replicates from healthy men and 2 replicates from infants (each with 2 techinical replicates) | 9 x C1 adult human spermatogonia, 3 x 10x Genomics adult steady-state spermatogenesis (unselected), 2 x Adult StaPut Spermatocytes, 2 x Adult StaPut Round Spermatids | 2 biological replicates | One replicate each from neonatal day 2 ITGA6 enriched or unfractionated and neonatal day 7 ITGA6 enriched or unfractionated, 2 adult replicates, each with ITGA6 enriched and unfractionated. | 2 replicates from healthy men with obstructive azoospermia, 1 replicate from man with nonobstructive azoospermia (NOA) |
| GEO number | GSE92280 | GSE120508 | GSE108970, GSE108974, GSE108977, GSE109049, GSE109033, and GSE109037 | None | GSE124263 | GSE106487 |
Bioinformatic analysis
Perhaps an equally important consideration in the generation of single-cell transcriptomes of spermatogenic cells are the methods used to analyze the data and draw conclusions about the underlying biology of the system. The bioinformatics processing of scRNA-seq data is not trivial and can be undertaken in a variety of ways using different algorithms, assumptions, and parameters, in unbiased and biased manners, and with varying degrees of extrapolation to draw biological inferences. In general, the data are simplified for presentation/visualization using dimensionally reduction strategies like as Principal Component Analysis (PCA) and t-Distributed Stochastic Neighbor Embedding (t-SNE) and cells are grouped into clusters (e.g. k-means) on the basis of variably expressed genes among all cells in the datasets (Figure 2A) [67]. Differentially expressed genes (DEGs) can then be determined between cell clusters (Figure 2B) to facilitate pathway analyses or trajectory analysis in pseudotime to dynamically evaluate cellular relationships (Figure 2C) [67]. Seurat (https://satijalab.org/seurat/) [73] is one software package that can be used to perform these analyses and has been employed in almost all of the scRNA-seq studies of spermatogenic cells [63–65, 74–81]. Likewise, Monocle (http://cole-trapnell-lab.github.io/monocle-release/) [82–84] has been utilized in several of these same studies to perform pseudotime analysis [65, 74, 75, 77, 79, 85–88].
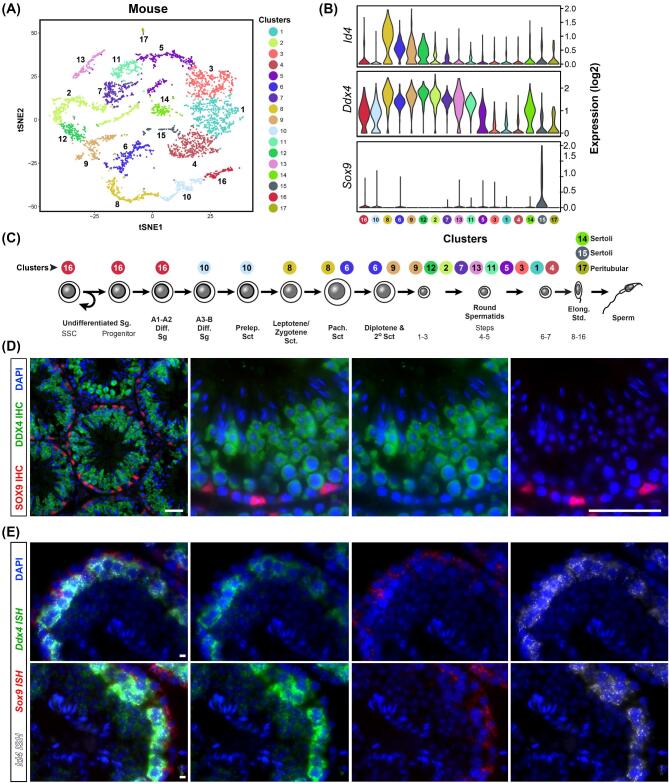
Figure 2.
Validation methods of scRNA-seq. (A) tSNE projection of scRNA-seq data from unselected adult mouse spermatogenic cells (from Hermann et al., 2018). Unbiased clusters indicate cell populations with significantly distinct transcriptomes. (B) Violin plots show expression of three key marker genes, Id4 (marks a subset of undifferentiated spermatogonia and many spermatocytes), Ddx4 (germ cell marker—expressed at low levels in spermatogonia and relatively higher levels in spermatocytes & spermatids), and Sox9 (Sertoli cell marker). (C) Spermatogenesis lineage diagram showing alignment of cell clusters with known cell types (from Hermann et al., 2018) (D) Adult mouse testis sections immunostained for SOX9 (red) and DDX4 (green) counterstained with DAPI (blue). Scale bar = 50μm. (E) smFISH triple-labeling using the RNAScope approach and adult mouse testis sections for Id4 mRNA (white), Sox9 mRNA (red), Ddx4 mRNA (green) and counterstained with DAPI. Scale bar = 5 μm. A, B and C are reprinted and adapted from Cell Reports 25(6) Hermann et al., “The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids,” pages 1650–1667.e8, Copyright 2018, with permission from Elsevier. This figure is available in color at Biology of Reproduction online.
Validation
Following establishment and analysis of single-cell transcriptomes, it is essential to confirm the reliability of the analysis with independent approaches. Tissue-based approaches are amongst the most popular and can help recapitulate the spatial expression patterns lost during tissue dissociation. Tissue validation includes low-throughput methods like protein immunostaining (Figure 2D) and single-molecule RNA fluorescence in situ hybridization (smFISH, Figure 2E), as well as high-throughput approaches like a sequential in situ hybridization (seqFISH) [89–92]. However, when applied to the testis, these tissue-based validation approaches are not created equal. Consider that during spermatogenesis, post-transcriptional gene regulation may disturb the correlation between mRNA and protein, making immunostaining potentially less informative than smFISH or seqFISH [93]. Nearly all of the testicular scRNA-seq datasets produced to date have utilized some form of tissue-based validation (Tables 1 and 2). Many alternative approaches to validate scRNA-seq datasets, including combined marker-based cell sorting combined with batch qRT-PCR/RNA-seq, cross-validation with other scRNA-seq methods, and use of gene knockout or knock-down, have been also been utilized with success (Tables 1 and 2).
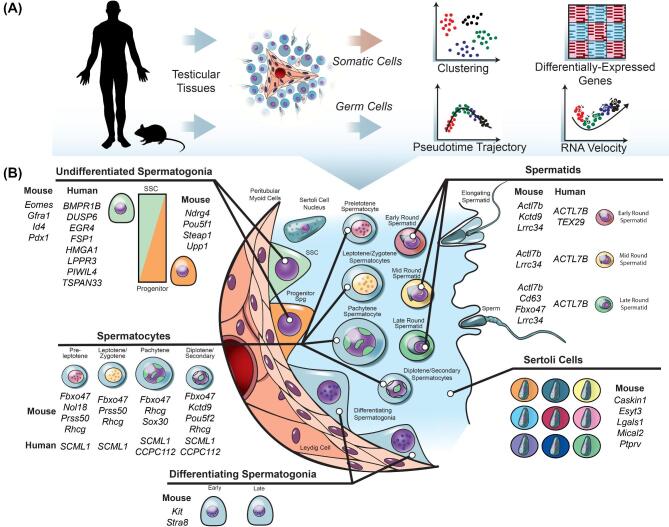
The newfound accessibility of scRNA-seq methods within the spermatogenesis research community and the relative ease with which resulting data can be analyzed have led to a flood of scRNA-seq studies reporting results from various testis cell populations of mice and humans. Consequently, all spermatogenic cell types can be recognized and separated into at least 11 different major populations based on validated unique expression of numerous genes and Sertoli cells are divisible into at least 9 different subpopulations (Figure 3). For the remainder of this review, we will discuss each of these studies grouped thematically by: (1) developing mouse testis during the first-wave of spermatogenesis, (2) steady-state adult mouse spermatogenesis, and (3) human spermatogenesis.
Figure 3.
Analyses and outcomes of scRNA-seq data from mouse and human spermatogenic cells. (A) Cells from human and mouse testicular tissue have been used for separate analyses of testicular germ and somatic cells at the single-cell level (Tables 1 & 2). In general, the analysis pipelines of various studies involve unbiased cell clustering, detection of differentially expressed genes (DEGs) between clusters, and pseudotime trajectory analysis to infer developmentally-regulated gene expression changes. (B) In summary, from the various scRNA-seq studies of spermatogenic cells, at least 11 distinct types of spermatogenic cells have been reported, distinguished by DEGs and 9 types of Sertoli cells. Noted are examples of validated genes corresponding to these various cell types. This figure is available in color at Biology of Reproduction online.
First-wave mouse spermatogenesis
Investigating spermatogonia in the adult testis during steady-state spermatogenesis is technically complicated due to the relative rarity of spermatogonia. To address this issue and study developmental questions such as the origin of SSCs, many have resorted to studying the biologically-enriched spermatogonia found in the early postnatal mouse testis. In 2015, the first study to profile gene expression at the single-cell level among postnatal male germ cells was published [93]. This study examined expression of a 172-gene panel by qRT-PCR among 584 mouse first-wave spermatogonia and hypothesized that observed subsets of cells that exhibited gene expression difference correlate with functionally distinct subgroups [93]. Speculation that a spermatogonial subset at postnatal day 6 (P6) was enriched for SSCs was buoyed by knowledge that the ID4 was a candidate SSC-specific marker [23, 28, 94] and transplantation of cultured ID4-EGFP + spermatogonia definitively demonstrated that SSCs were exclusively found within the Id4 expressing fraction [23]. Still, it was only in subsequent studies where this concept was borne out with evidence that a novel marker (TSPAN8) which differentially labeled these subsets correlated with enriched transplantable SSC numbers [34]. This initial foray into the single-cell mRNA characterization of postnatal male germ cells was limited by profiling expression of only a hand-picked subset of genes. Subsequent experiments reported by Song and colleagues in 2016 examined the complete transcriptomes of nearly 200 sorted ID4-EGFP + mouse spermatogonia at two time points (P3 and P7) [95] (Table 1). Heterogeneity was evident among these spermatogonia in expression of more than 1000 genes that pointed to a subset of spermatogonia likely to populate the SSC pool [95]. Liao et al., examined the transcriptomes of 71 sorted P5.5 OCT4-GFP + /KIT-spermatogonia and reported CD87 as a stem cell marker which they posited was involved in the initial establishment of SSC pool [86] (Table 1). Makino et al., examined the transcriptomes of 80 prospermatogonia at P1.5 and 96 spermatogonia at two time points (P3.5 and P5.5) and reported that DEC2 regulates the maintenance of SSCs via direct inhibition of Sohlh1 expression [88] (Table 1). Earlier in testis development, Neirijnck et al., reported results from 483 prospermatogonia at embryonic day (E) 16.5, although no in-depth analysis was performed since the subject of the study was the developing testicular soma [76] (Table 1). Together, these data may point to important distinguishing features which allow recognition of future SSCs as they arise during the prospermatogonia to spermatogonia transition, a poorly understood process which is thought to begin as early as E18.5 in mice [96].
More recently, 10x Genomics analyses have been applied to profile spermatogenic cells during postnatal testis development across the first wave without cell selection (Table 1). Specifically, Hermann et al., examined 3466 unselected P6 testis cells [77], Grive et al., examined 1200–2500 cells each from P6, P14, P18, P25, and P30 mouse testes [80], and Ernst et al., examined cells from P5, P10, P15, P20, P25, P30, and P35 mouse testes [97] (Table 1). Beyond simply the sequential emergence of spermatogenic cells across the first wave of spermatogenesis, which were defined manually based on expression of key cell type-specific markers, Grive and colleagues observed dynamic changes in the transcriptomes of spermatogenic cells as the first wave progressed (P6, P14, P18, P25, and P30) [80]. For instance, Asrgl1 was highly expressed in spermatogonia from P6 mice, but decreased as the first wave proceeded. Reciprocally, the genes encoding ATM and RAD51 exhibited progressively higher expression among pachytene spermatocytes as the first wave progressed. Ernst and colleagues also observed dynamic changes in the transcriptomes of spermatogenic cells as the first wave progressed (P5, P10, P15, P20, P25, P30, and P35) [97]. For instance, as the first wave progressed, Prss50 and Pou5f2 exhibited progressively higher expression from pre-leptotene to zygotene spermatocytes and among diplotene spermatocyte, respectably.
Uniquely, Hermann and colleagues focused only on P6 spermatogonia and compared unselected spermatogonia to sorted subsets that were enriched or depleted for transplantable SSCs [77] (Table 1). This study selected the most epifluorescent (ID4-EGFPbright) spermatogonia from mice bearing the Id4-Egfp transgene, which are highly enriched for transplantable SSCs, along with those GFP + spermatogonia with low epifluorescence (ID4-EGFPdim) which have little SSC activity [98]. Parallel 10x Genomics and Fluidigm C1 analyses were performed of sorted P6 ID4-EGFPbright and ID4-EGFPdim spermatogonia in order to directly correlate functional SSCs with single-cell phenotypes. SSCs expressed known markers (Id4, Gfra1, Etv5), novel signaling intermediates (Tcl1, Dusp6) as well as, unexpectedly, genes involved in autophagy (Figure 3B) [99, 100]. Reciprocally, first-wave progenitor and differentiating spermatogonia were characterized by activation of the cell cycle, a metabolic shift to oxidative phosphorylation, and could be recognized by a new marker, NDRG4 (Figure 3B) [77]. Together, these single-cell analyses of spermatogenic cells during the first-wave of mouse spermatogenesis have resolved new subsets of spermatogonia and defined the dynamic nature of spermatogenic initiation.
Steady-state adult mouse spermatogenesis
By far, the most abundant of type spermatogenesis single-cell datasets are those that examined cells from adult mouse testes [74, 75, 77, 80, 81, 85, 97] (Table 1). In common among the Hermann, Grive, Lukassen, Green, and Ernst studies is analysis of data from spermatogenic cells that were not selected for any particular cell type, which allows for confidence in the relative proportions of cells as well as the low likelihood that selection bias leads to erroneous loss of rare or transitional cell types (Figure 1A). These datasets shared a striking common feature—the clear demonstration that spermatogenic cell phenotypes existing on a continuum, rather than distinct subgroups separated by large transcriptome changes (Figure 2A). That is, the heterogeneity within and between spermatogenic cell types is so high that the borders between cell types become blurred. Notably, one of the rationales for profiling unselected cells is to prevent selection bias which could exclude rare unique/novel cell populations. Yet, collectively, identification of novel cell subsets has not been an outcome of these experiments.
Green et al., uniquely supplemented their spermatogenesis profiling with selected cell populations that were found to be relatively infrequent in their datasets [75]. Specifically, this compensated for low abundance of spermatogonia and Sertoli cells, allowing more comprehensive evaluation of those cell types, but with the drawback that altered cell proportions influence the statistics underlying trajectory (pseudotime) modeling. These data indicated the presence of transient cellular states from spermatogonia to early spermatocytes, in vivo. Moreover, expression of genes related to RNA splicing and RNA binding proteins, early meiotic genes, chromatin remodeling and epigenetic modifiers was distinct among cellular subsets. Importantly, this study also highlighted data indicating that Sertoli cells contained high levels of mRNAs that are typically thought to be expressed only by round spermatids (e.g. Prm2). SmFISH demonstrated that round spermatids, but not Sertoli cells, transcribe Prm2, and Prm2 transcripts found in Sertoli cells arise from engulfment of spermatid residual bodies in which Prm2 was highly abundant [75].
Hermann et al., performed side-by-side comparisons of unselected spermatogenic cell datasets with enriched populations of spermatogonia, spermatocytes, and spermatids in order to validate the gene expression signatures obtained for various cell types [77]. Among the immunostaining validated novel markers, KCTD9 was found to be expressed in spermatocytes and early round spermatids, RHCG was expressed in spermatocytes, and ACTL7B was expressed in round spermatids (Figure 3B). Exclusive to this study, sorted subsets of spermatogonia based on ID4-EGFPbright and ID4-EGFPdim were examined with both 10x Genomics and Fluidigm C1 platform and distinguished adult SSCs from progenitor spermatogonia based on correlation between single-cell expression signatures and functional transplantation results. Similar to first-wave spermatogonia, SSC-containing subsets expressed known SSC markers, but a unique subsets exhibited unique features that may point to heterogeneity among SSCs (Figure 3B). Progenitor spermatogonia exhibited enhanced cell cycle gene expression and differentiating spermatogonia appeared to undergo a metabolic shift to oxidative phosphorylation.
La et al., selectively examined Plzf-mCherry + undifferentiated spermatogonia and observed a very similar continuum profile to others’ observations with unselected steady-state spermatogonia [85]. This experiment led the to the observation that Pdx1 mRNA was co-expressed by cells expressing SSC markers, which led to their discovery that PDX1 is expressed by a subset of undifferentiated spermatogonia and may participate in cell state regulation in concert with signals from the niche (Figure 3B).
In the Lukassen et al., study, a unique analysis of sex chromosome gene expression during spermatogenesis demonstrated differences in X- and Y-linked gene transcription after meiosis [74]. As expected, sex chromosome transcription was silenced among spermatocytes due to meiotic sex chromosome inactivation (a feature others have also observed and noted from their data). Here, the authors noted that Y-linked transcript levels preferentially decreased before protamine expression initiated, which was distinct from the later decline in X-linked and autosomal transcripts.
As noted above, mammalian spermatogenesis proceeds asynchronously. Thus, a likely fundamental contributor to the apparent continuum of spermatogenic gene expression signatures and lack of distinct novel/rare spermatogenic cell types, is the inherent asynchrony in spermatogenesis across the entirety of a given testis [101]. To address this, spermatogenesis can be synchronized by suppressing spermatogonial differentiation from P2-P8 with WIN18 446 followed by initiating spermatogonial differentiation with a bolus of RA at P9 [47]. Chen and colleagues used this approach and examined a total 1136 cells obtained from synchronized mouse testes at 20 time points varying from 10 to 540 h after RA induction [64]. Examination of mouse testes with synchronized spermatogenesis demonstrated that there is still underlying heterogeneity among spermatogenic cells independent of spermatogenic synchrony [64]. Still, these analyses were informative by providing a window into the underpinnings of specific cellular events during spermatogenesis. For instance, since Fbxo47 was found to be uniquely expressed at the time of meiotic recombination and synaptonemal complex formation, the authors examined Fbxo47 knockout testes and found defective meiotic recombination and infertility (Figure 3B) [64]. During spermiogenesis, differential expression of Cd63 between early (step 1–2, CD63high) and late (steps 7–8, CD63−) round spermatids was validated through the significantly higher efficiency of blastocyst development with CD63– round spermatids (via ROSI) than CD63high round spermatids (Figure 3B). Lastly, Sox30 was found to be highly expressed in pachytene spermatocytes and round spermatids but not in somatic cells. Consequently, evaluation of Sox30 knockout testis and ChIP-seq for SOX30 demonstrated the repertoire of SOX30 regulates genes involved in spermatid differentiation including Chd5, Hils1, and Sun5 (Figure 3B) [64]. Importantly, these results also suggested that common mechanisms underlie the first-wave of spermatogenesis and the steady-state spermatogenesis.
The average sequence similarity among protein-coding genes between mice and humans is 85% [102], and since mice are relatively easy to maintain and manipulate genetically, they have become the most popular mammalian model for studying human health and disease. Although the scRNA-seq datasets from mouse spermatogenic cells may provide insights into human spermatogenesis biology given the high degree of conservation in spermatogenesis across taxa [77, 78], there may be important differences in the underlying biology between species [103]. For instance, mouse spermatogonia can be expanded exponentially and maintained indefinitely in vitro [104, 105], yet similarly robust culture conditions for human spermatogonia have yet be established. Thus, molecular and cellular mechanisms responsible for human spermatogonial maintenance likely have unique contributors not found in mice.
Human spermatogenesis
Of particular interest due to the potentially high translational significance, five studies have examined various spermatogenic cell types from human testes at the single-cell level: Guo et al., Hermann et al., Sohni et al. and Wang et al. (Table 2). Three of these studies examined unselected spermatogenic cells from adult testes [65, 77, 78], one focused exclusively on sorted adult spermatogonia [87] and the most recent compared selected cells from testes of human infant and adults [79] (Table 2). In a similar fashion to the mouse datasets noted above, a common theme was to identify cell types based on expression of key markers, typically extrapolated from mice, but without the benefit of functional and genetic validation. Also like results from mice, human single-cell spermatogenic cell transcriptomes largely appeared to be arranged in a continuum with considerable heterogeneity within and between spermatogenic cell types.
Guo et al., isolated human undifferentiated and differentiating spermatogonia by FACS using SSEA4 + or KIT + selection, respectively, and profiled a small number of cells that met a stringent quality threshold (60 SSEA4 + spermatogonia and 32 KIT + spermatogonia) [87]. Data from this study supported the conclusion that upregulation of genes related to the FGF signaling pathway, which is known to be involved in SSC self-renewal in mice [106–108], occurred specifically in human spermatogonia with a more primitive phenotype. Similarly, these data revealed upregulation of WNT pathway related genes and BMP pathway related genes in less differentiated human spermatogonia, which mirrors the SSC to progenitor transition in mice (Figure 3B) [33, 87, 109, 110].
Guo et al., expanded these analyses to profile adult and infant human testis cells without cell section and validated a variety of putative spermatogonial markers (FGFR3, PHGDH, TCF3, KI67, MAGEAB1, PIWIL4, PPP1R36, and TSPAN33) at the protein level using immunostaining (Figure 3B) [78]. Similar to prior results [87], data from this study also supported the conclusion that reciprocal upregulation of genes related to the FGF and WNT signaling pathways in the most primitive and advanced spermatogonia, respectively. Notably, this study also introduced a novel approach, RNA velocity analysis [111, 112] (Figure 3A), which uses integrated analysis of levels of mature spliced mRNA (steady-state) and unspliced (nascent) mRNA, to identify dynamic relationships among adult human spermatogonia. This analysis of human spermatogonia predicted the possibility that more advanced spermatogonia (state 2, KIT+, MKI67+) may revert to a more primitive state (state 1, GFRA1+, ID4+, ETV5+), which aligns with the controversial clone fragmentation model of mouse SSC renewal (Figure 4A) [24, 26]. Ultimately, confirmation of this predicted dynamic relationship between human spermatogonia will be dependent on lineage tracing methodologies that, at this point, do not exist for human spermatogenesis.
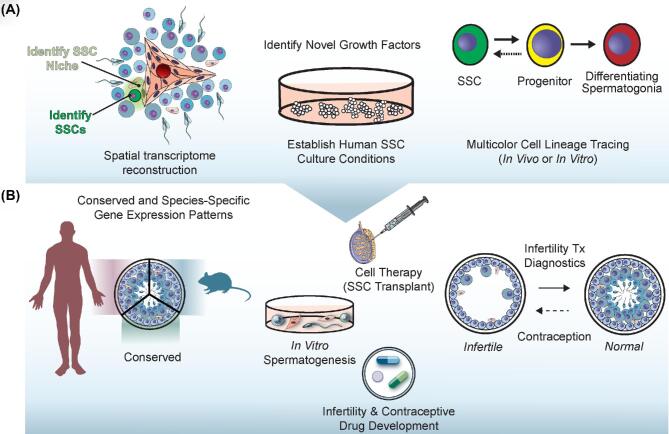
Figure 4.
Future advancements in spermatogenesis built on the single-cell foundation. (A) Spatial reconstitution of single-cell transcriptomes for SSCs may allow prospective localization of SSCs and their cognate niches in the testis. These same data may help identify key growth factor ligand-receptor combinations to facilitate establishment of human SSC culture conditions. Lastly, novel markers of SSCs, progenitors and differentiating spermatogonia could be used to drive transgenic reporters for lineage tracing to confirm assumed cellular relationships. (B) The accumulated knowledge from scRNA-seq of human and mouse testes will enable identification of species-specific and conserved gene expression patterns underlying spermatogenesis. These data will undoubtedly help advance diagnosis and treatment of male infertility as well as new modes for male contraception. Directly emanating from the single-cell data and underlying such translational outcomes would be development of new drugs to solve male infertility and achieve contraception as well as better characterization of cells to be used therapeutically (e.g. SSC transplant) or production and characterization of gametes in a dish.
Hermann et al., using a nearly identical strategy to their mouse studies, completed parallel assessments of unselected human spermatogenic cells with enriched populations of spermatogonia (FACS), spermatocytes (StaPut) and spermatids (StaPut) in order to establish gene expression signatures for various cell types [77]. Both Fluidigm C1 and 10x Genomics analyses were employed and exemplary markers were validated at the protein level using immunostaining methods, including DUSP6, which is expressed in undifferentiated spermatogonia, and ACTL7B, which is expressed in round spermatids (Figure 3B). Moreover, the parallel examination of mouse and human cells supported robust interspecies comparisons and permitted extrapolation of the identity of human SSCs from their functionally defined mouse counterparts. Importantly, one outcome of this study was demonstration of both conserved and unique transcriptome features across the spermatogenic lineage between mice and humans (Figure 4B).
The Sohni et al., study placed a particular emphasis on undifferentiated spermatogonia and differentiating spermatogonia in adult and infant human testes [79]. A new cell surface marker, LPPR3, was identified which appears to mark a more restrictive population of spermatogonia than FGFR3, and therefore, may be a more selective SSC surface marker than FGFR3 (Figure 3B). Given the absence of a functional assay for human SSCs, this will be a difficult hypothesis to support experimentally, but the identification of new markers provides additional tools for future basic and translational studies. In the infant testis, Sohni and colleagues identified two germ cell populations, one which exhibits gene expression features that are more similar to primordial germ cells (PGCs) and another that appears to be intermediate between PGCs and adult spermatogonia in pseudotime. Lastly, they report that UTF1, ETV4, and PIM2 are markers of the population with PGC-like features and validate these results with immunostaining.
Last, but not least, Wang et al., uniquely compared steady-state spermatogenic cells from fertile men to testis cells from a man with non-obstructive azoospermia [65]. This comparison confirmed the specificity of single-cell transcriptomes and identified that apoptosis related genes are upregulated and the accumulation of γH2AX was observed in testicular somatic cells from NOA patient. These phenomena would be potential drivers of male infertility. Along with immunostaining studies, they used smFISH validation of HMGA1 transcripts to show specific labeling in undifferentiated and differentiating spermatogonia, BMPR1B in putative SSCs, and TEX29 in spermatids (Figure 3B).
Given the genetic variability between individual men, the inability to carefully control developmental timing, and underlying reasons for tissue availability (disease, trauma), there may be particular value to future combined re-analysis of the varied human spermatogenic single-cell data sets. Such analyses may help to distinguish between sample-dependent and cell type-dependent gene expression variation that is not evident with only the few samples used for each individual study. As the field moves forward with these data, such integrated analyses, including those leveraging new data types (e.g. ATAC-Seq, DNA methylome, histone modifications, etc.), may help to boil these complex and somewhat isolated datasets down into transformative tools for studying the biology of human spermatogenesis (Figure 4).
Testicular somatic cells
In the testis, growth factors and morphogen signals emanate from somatic cells and play important roles instructing spermatogenic development, including driving the balance between SSC self-renewal and differentiation [10, 14–16, 30, 32, 104, 105, 107, 113–124]. For instance, GDNF produced from testicular somatic cells (Sertoli cells, peritubular myoid cells) regulates SSC self-renewal via the MAPK and AKT signaling pathways [30, 104, 105, 107, 113–120, 125, 126]. Transgenic overexpression of GDNF causes the accumulation of undifferentiated spermatogonia, while the deletion of genes encoding Gdnf or its receptor components (Gfra1 & Ret) depletes undifferentiated spermatogonia [114, 116, 117, 120]. But, the timing of GDNF stimulation along the seminiferous epithelial cycle, as well as the spatial location of GDNF-producing cells (e.g. the SSC niche) are not well understood [27, 115, 127]. Therefore, characterization of testicular somatic cells at the single-cell level may reveal unique characteristics that define spatially and temporally resolved subsets that point to key spermatogenic regulators. Notably, though, several of the unselected spermatogenic single-cell transcriptome datasets demonstrated only a minor contribution from testicular somatic cells identified on the basis of somatic cell markers [64, 65, 74, 77–80]. The limited representation of somatic cells located in the testicular interstitium between seminiferous tubules (e.g. Leydig cells, peritubular myoid cells, macrophages, endothelial cells, etc.) is typical for 2-step enzymatic digestion protocols used for seminiferous tubule cell preparation since these cells are lost during the collagenase treatment prior to tryptic digestion of seminiferous tubules. Further, adult Sertoli cells were also underrepresented, but not absent, in both human and mouse seminiferous datasets because these cells are well known to be highly sensitive to the harsh tryptic digestion. Green et al., overcame this limitation by specifically selecting Sertoli cells and interstitial cells with immunological and genetic lineage tracing methods to supplement the lack of somatic cells [75]. They then performed in-depth analysis of these somatic cells and reported four type of Sertoli cells, validated using smFISH for a panel of genes, including Ptprv, Meical2, Caskin1, Esyt3, and Lgals1, to assign these cell types to groups of stages in the cycle of the seminiferous epithelium (Figure 3B). It is likely that future analyses extending from similar data may reveal distinct somatic cell programs that could be spatially traced to specific testicular locations to address unsolved questions related to the SSC niche and overarching somatic instruction of the spermatogenic program (Figure 4A).
Conclusions
Human birth rates are declining in both the developing and developed world [128, 129] and male infertility affects more than 5% of the world population. This problem is only expected to worsen in developed countries as the generational time increases due to social factors [130]. Conversely, there is a desperate need for effective contraception strategies to address the global need for population control [131]. Together, these two significant problems illustrate the need to obtain a better understanding of the fundamental biological processes driving sperm production in the testis. A considerable body of mechanistic knowledge about spermatogenesis has emerged from the study of rodents, which has been extrapolated to humans. Yet, this is only sometimes effective and valid—consider the simple difference that the duration of mouse spermatogenesis is 35 days while human spermatogenesis takes 62 days [22]. Therefore, human and model species (e.g. mice) can and should be examined in parallel.
High-resolution profiling of gene expression signatures across both rodent and human spermatogenesis can arm the research community with data to begin solving infertility and executing effective male contraception (Figure 4B). In this review, we highlighted the available scRNA-Seq datasets generated from mouse and human testicular somatic and germ cells and pointed to particularly notable features of each that focus their utility for future investigation (Figures 3 and 4A, Table 1). We also note specific advances in knowledge that emerged from these analyses (Figure 3) and highlight that for humans, the combination analysis of these datasets may help to refine the conserved cellular states and underlying gene sets that mark each. After considering each dataset in isolation, a logical extension is to ask about consistency and variation between investigations, which includes cross-platform, cross-laboratory, and cross-species comparisons that introduce technical, logistical and conceptual challenges that still have yet to be adequately addressed. One step towards using these data synergistically was the recent upgrade to the ReproGenomics platform (http://rgv.genouest.org/app/#/) which now enables web-based dataset comparisons [132, 133], and indeed, scRNA-Seq data produced from a variety of groups have already been included in this resource [64, 65, 74, 75, 77, 85, 95].
In the immediate future, there is a strong possibility that these single-cell transcriptome datasets may find utility in diagnosis of male infertility and discovery of underlying etiologies (Figure 4B). Indeed, Hermann et al., defined specific subsets of three genes that can be used to uniquely recognize 11 different spermatogenic cell types in both human and mouse testes [77]. Theoretically, such gene panels could be used as diagnostic tools to determine the precise point at which the spermatogenic lineage is blocked (Figure 4B). Establishing the efficacy of such a diagnostic approach would require empirical tests akin to the comparison made by Wang et al., between profiles of fertile and infertile testis cells (Figure 4B, [65] Table 1).
Generating single-cell transcriptomes of spermatogenic cells to this point has necessitated the dissociation of testis tissue into its component cellular constituents. While this allows maximal resolution among individual unit cells, the process of dissociation necessarily removes key spatial information linking transcriptomes back to the cellular locations in the testis. This is a key challenge that has prompted many to perform validation experiments that establish spatial profiles for individual mRNAs, panels of few mRNAs or their encoded proteins using tissue sections to reestablish spatial context (Figures 2 and 4B). While largely informative, these approaches necessarily involve low-dimensionality profiling under current technological constraints that limit the ability to relate scRNA-seq results back to the spatial organization of the testis. Transcriptome-wide spatial mRNA profiling using tissue sections is possible [134, 135], but the resolution is limited to ∼200μm, which is roughly the diameter of a normal mouse seminiferous tubule, and thus, has insufficient resolution to define cellular transcriptomes in space. Correcting the loss of spatial information to achieve high-resolution spatial transcriptomics in the testis will be a key advancement moving forward for understanding testis biology (Figure 4A).
Inferences and predictions emanating from analyses of single-cell transcriptomes using pseudotime can be very powerful for identifying novel cell states and their relationships. However, those inferences are limited by the static nature of the underlying measurements of steady-state mRNA abundance. The field has been introduced to the potential power of cutting-edge analyses like RNA velocity analysis to provide empirical evidence of cell relationships [78, 111, 112]. Wide application of this strategy would help identify unique gene sets that could be used to build lineage tracing panels and execute toxin-dependent lineage ablation to validate cell lineage relationships, in vivo. Likewise, leveraging CRISPR/Cas9 to delete key novel target [136, 137], along with CRISPR activation [138–142], CRISPR inhibition [143, 144], and CRISPR epigenetic modification [145–153], which will define the molecular circuitry controlling cell fate in the spermatogenic lineage.
Mouse spermatogonia can be maintained indefinitely and robustly amplified in culture [104, 105] and this has provided a tremendous resource for basic studies of spermatogonial biology because of amenability for genetic modification, factor treatment studies, and biochemical experimentation. Progress culturing human spermatogonia (potentially including SSCs) has also been reported [154–158], but a similarly robust culture system for human spermatogonia has yet to be developed, likely due to absence of some key growth factor or other constituent necessary for human spermatogonial propagation and survival in vitro. Consequently, progress advancing our knowledge of human spermatogonial biology has remained slow. Clinically, a human spermatogonial culture system may also provide an unlimited source of germ cells for therapeutic use via transplantation [159, 160]. Therefore, combinatorial analyses of human and mouse spermatogenic and testicular somatic single-cell datasets provides opportunities to define reciprocal factor-receptor combinations that may point to missing constituents that improve spermatogonial survival and propagation in the dish (Figure 4).
Moreover, there has been considerable interest paid to in vitro gametogenesis, where the entirety of germ cell development, from their initial specification during early embryogenesis through gamete formation, is modeled in a dish starting with pluripotent stem cells (PSCs) [161, 162]. Using embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) to sequentially produce PGC like cells (PGCLCs) has been accomplished in both mouse and human [163–173], and mouse PGCLCs are fully capable of producing sperm and offspring upon transplantation into host testes [163]. It remains unclear, however, whether their human counterparts are similarly functional due to a lack of functional tests dictated by appropriate ethical boundaries. Likewise, in vitro mouse spermatogenesis using organ culture of testis from neonatal and adult in vitro has been established [174–178]. Curiously, GSC-like cells (GSCLCs) from PGCLCs exhibit transplantable SSC ability [170], but these cells are also considered to be epigenetically abnormal. These are just a few examples of in vitro systems modeling spermatogenic development that would benefit from comparison to the existing datasets discussed here that constitute references of the normal spermatogenic lineage (Figure 4B). As scRNA-seq becomes more commonplace, cheaper and expected as a characterization tool, such comparisons would provide a wealth of information about the extent of variation among cells produced in vitro, as well as their comparability to their normal in vivo counterparts.
In conclusion, the flurry of new transcriptome studies examining mouse and human spermatogenic cells at the single-cell level have provided a wealth of new information to advance our understanding of spermatogenic biology (Figures 3 and 4). The impact of this work is already being realized through advancements in identification of specific cell types, including the elusive human SSC, and new appreciation of the extent of heterogeneity among spermatogenic cells and how that relates to their development. New bioinformatics and wet bench techniques already available and on the horizon will allow the field to build on these observations by solving the limitations of static observations and spatial remapping to bring these data full-circle to truly advance our foundational knowledge. Ultimately, a more advanced understanding of the biology of spermatogenesis will help the field work towards developing new diagnoses for male infertility, deciphering their etiologies, devising new treatment modalities, and conversely, cultivate new methods for male contraception to maximize human health (Figure 4).
Acknowledgment
Notes
Edited by Dr. P. Jeremy Wang, MD, PhD, University of Pennsylvania
Author Biographical
 Dr. Brian P. Hermann is an Associate Professor at the University of Texas at San Antonio. He received his PhD in 2005 under the direction of Dr. Leslie Heckert from the University of Kansas in the Department of Molecular and Integrative Physiology. He then received postdoctoral training in the laboratory of Dr. Kyle Orwig at the University of Pittsburgh in the Magee-Womens Research Institute and the Center for Research in Reproductive Physiology. He started his lab at the University of Texas at San Antonio in 2011 with support from an NIH K99/R00 Career Development Award. The Hermann lab studies spermatogonial stem cells (SSCs), which are adult stem cells responsible for sperm production throughout life in mammals and are essential for male fertility. Dr. Hermann's primary research interests are uncovering the mechanisms regulating SSC fate, understanding how SSCs are specified during testis development, and identification of potential SSC-based therapeutic strategies to address male infertility. His lab utilizes mouse and non-human primate models and human testicular tissue to tackle these objectives. More recently, the Hermann lab has been among the pioneering groups employing single-cell transcriptomics to address questions about heterogeneity among spermatogenic cells, including SSCs. Data from such experiments are helping to uncover novel pathways involved in SSC fate control and are revolutionizing the understanding of spermatogenic development.
Dr. Brian P. Hermann is an Associate Professor at the University of Texas at San Antonio. He received his PhD in 2005 under the direction of Dr. Leslie Heckert from the University of Kansas in the Department of Molecular and Integrative Physiology. He then received postdoctoral training in the laboratory of Dr. Kyle Orwig at the University of Pittsburgh in the Magee-Womens Research Institute and the Center for Research in Reproductive Physiology. He started his lab at the University of Texas at San Antonio in 2011 with support from an NIH K99/R00 Career Development Award. The Hermann lab studies spermatogonial stem cells (SSCs), which are adult stem cells responsible for sperm production throughout life in mammals and are essential for male fertility. Dr. Hermann's primary research interests are uncovering the mechanisms regulating SSC fate, understanding how SSCs are specified during testis development, and identification of potential SSC-based therapeutic strategies to address male infertility. His lab utilizes mouse and non-human primate models and human testicular tissue to tackle these objectives. More recently, the Hermann lab has been among the pioneering groups employing single-cell transcriptomics to address questions about heterogeneity among spermatogenic cells, including SSCs. Data from such experiments are helping to uncover novel pathways involved in SSC fate control and are revolutionizing the understanding of spermatogenic development.
References
- 1. Zuckerman S. The number of oocytes in the mature ovary. Recent Prog Horm Res 1951; 6:63–109. [Google Scholar]
- 2. Tilly JL. Commuting the death sentence: How oocytes strive to survive. Nat Rev Mol Cell Biol 2001; 2:838–848. [DOI] [PubMed] [Google Scholar]
- 3. Perez GI, Robles R, Knudson CM, Flaws JA, Korsmeyer SJ, Tilly JL. Prolongation of ovarian lifespan into advanced chronological age by Bax-deficiency. Nat Genet 1999; 21:200–203. [DOI] [PubMed] [Google Scholar]
- 4. Canning J, Takai Y, Tilly JL. Evidence for genetic modifiers of ovarian follicular endowment and development from studies of five inbred mouse strains. Endocrinology 2003; 144:9–12. [DOI] [PubMed] [Google Scholar]
- 5. Gosden RG, Laing SC, Felicio LS, Nelson JF, Finch CE. Imminent oocyte exhaustion and reduced follicular recruitment mark the transition to acyclicity in aging C57BL/6J mice. Biol Reprod 1983; 28:255–260. [DOI] [PubMed] [Google Scholar]
- 6. Ryu BY, Orwig KE, Oatley JM, Avarbock MR, Brinster RL. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells 2006; 24:1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang X, Ebata KT, Robaire B, Nagano MC. Aging of male germ line stem cells in mice1. Biol Reprod 2006; 74:119–124. [DOI] [PubMed] [Google Scholar]
- 8. De Rooij DG. Stem cells in the testis. Int J Exp Pathol 1998; 79:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiarini-Garcia H, Russell LD. High-resolution light microscopic characterization of mouse spermatogonia1. Biol Reprod 2001; 65:1170–1178. [DOI] [PubMed] [Google Scholar]
- 10. de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21:776–798. [PubMed] [Google Scholar]
- 11. Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. Histological and histopathological evaluation of the testis. Clearwater, FL: Cache River Press; 1990. [Google Scholar]
- 12. Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140:5894–5900. [DOI] [PubMed] [Google Scholar]
- 13. Yoshinaga K, Nishikawa S, Ogawa M, Hayashi SI, Kunisada T, Fujimoto T, Nishikawa SI. Role of c-kit in mouse spermatogenesis - identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113:689–699. [DOI] [PubMed] [Google Scholar]
- 14. Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Develop Biol 2015; 397:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Endo T, Freinkman E, de Rooij DG, Page DC. Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc Natl Acad Sci USA 2017; 114:E10132–E10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro1. Biol Reprod 2008; 78:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36:653–659. [DOI] [PubMed] [Google Scholar]
- 18. Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 2010; 142:468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 2016; 143:1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mutoji KN, Hermann BP. Defining the Phenotype and Function of Mammalian Spermatogonial Stem Cells. In: Oatley JM, Griswold MD (eds.), The Biology of Mammalian Spermatogonia; New York, NY: Springer; 2017: 67–90. [Google Scholar]
- 21. Fawcett DW. The mammalian spermatozoon. Dev Biol 1975; 44:394–436. [DOI] [PubMed] [Google Scholar]
- 22. Hermann BP, Sukhwani M, Hansel MC, Orwig KE. Spermatogonial stem cells in higher primates: are there differences from those in rodents? Reproduction 2010; 139:479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev 2014; 28:1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, Simons BD, Yoshida S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 2014; 14:658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helsel AR, Yang QE, Oatley MJ, Lord T, Sablitzky F, Oatley JM. ID4 levels dictate the stem cell state in mouse spermatogonia. Development 2017; 144:624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 2010; 328:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell 2007; 12:195–206. [DOI] [PubMed] [Google Scholar]
- 28. Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice1. Biol Reprod 2011; 85:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun F, Xu Q, Zhao D, Degui Chen C. Id4 marks spermatogonial stem cells in the mouse testis. Sci Rep 2015; 5:17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takashima S, Kanatsu-Shinohara M, Tanaka T, Morimoto H, Inoue K, Ogonuki N, Jijiwa M, Takahashi M, Ogura A, Shinohara T. Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports 2015; 4:489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133:1495–1505. [DOI] [PubMed] [Google Scholar]
- 32. Ikami K, Tokue M, Sugimoto R, Noda C, Kobayashi S, Hara K, Yoshida S. Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development 2015; 142:1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tokue M, Ikami K, Mizuno S, Takagi C, Miyagi A, Takada R, Noda C, Kitadate Y, Hara K, Mizuguchi H, Sato T, Taketo MM et al.. SHISA6 confers resistance to differentiation-promoting wnt/β-catenin signaling in mouse spermatogenic stem cells. Stem Cell Reports 2017; 8:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mutoji K, Singh A, Nguyen T, Gildersleeve H, Kaucher AV, Oatley MJ, Oatley JM, Velte EK, Geyer CB, Cheng K, McCarrey JR, Hermann BP. TSPAN8 expression distinguishes spermatogonial stem cells in the prepubertal mouse testis. Biol Reprod 2016; 95:117–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat 1956; 99:391–413. [DOI] [PubMed] [Google Scholar]
- 36. Lahn BT, Tang ZL, Zhou JX, Barndt RJ, Parvinen M, Allis CD, Page DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Nat Acad Sci 2002; 99:8707–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laiho A, Kotaja N, Gyenesei A, Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS ONE 2013; 8:e61558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ellis PJI, Furlong RA, Wilson A, Morris S, Carter D, Oliver G, Print C, Burgoyne PS, Loveland KL, Affara NA. Modulation of the mouse testis transcriptome during postnatal development and in selected models of male infertility. Mol Human Reprod 2004; 10:271–281. [DOI] [PubMed] [Google Scholar]
- 39. Lehti MS, Sironen A. Formation and function of sperm tail structures in association with sperm motility defects. Biol Reprod 2017; 97:522–536. [DOI] [PubMed] [Google Scholar]
- 40. Griswold MD. Spermatogenesis: the commitment to meiosis. Physiological Rev 2016; 96:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoshida S. Open niche regulation of mouse spermatogenic stem cells. Dev Growth Differ 2018; 60:542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiological Rev 1972; 52:198–236. [DOI] [PubMed] [Google Scholar]
- 43. Clermont Y, Leblond CP. Differentiation and renewal of spermatogonia in the monkey, Macacus rhesus. Am J Anat 1959; 104:237–273. [DOI] [PubMed] [Google Scholar]
- 44. Clermont Y. The cycle of the seminiferous epithelium in man. Am J Anat 1963; 112:35–51. [DOI] [PubMed] [Google Scholar]
- 45. Amann RP. The cycle of the seminiferous epithelium in humans: A need to revisit? J Androl 2008; 29:469–487. [DOI] [PubMed] [Google Scholar]
- 46. Nihi F, Gomes MLM, Carvalho FAR, Reis AB, Martello R, Melo RCN, Almeida FRCL, Chiarini-Garcia H. Revisiting the human seminiferous epithelium cycle. Hum Reprod 2017; 32:1170–1182. [DOI] [PubMed] [Google Scholar]
- 47. Hogarth CA, Evanoff R, Mitchell D, Kent T, Small C, Amory JK, Griswold MD. Turning a Spermatogenic Wave into a Tsunami: Synchronizing Murine Spermatogenesis Using WIN 18,4461. Biol Reprod 2013; 88:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hogarth CA, Arnold S, Kent T, Mitchell D, Isoherranen N, Griswold MD. Processive Pulses of Retinoic Acid Propel Asynchronous and Continuous Murine Sperm Production1. Biol Reprod 2015; 92:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci USA 2015; 112:E2347–E2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Rooij DG. Organization of the seminiferous epithelium and the cycle, and morphometric description of spermatogonial subtypes (rodents and primates). In: Oatley JM, Griswold MD (eds.), The Biology of Mammalian Spermatogonia; New York, NY: Springer; 2017: 3–20. [Google Scholar]
- 51. Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis1. Biol Reprod 2004; 71:319–330. [DOI] [PubMed] [Google Scholar]
- 52. Bellvé AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 1993; 225:84–113. [DOI] [PubMed] [Google Scholar]
- 53. Bellvé AR, Millette CF, Bhatnagar YM, O’Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 1977; 25:480–494. [DOI] [PubMed] [Google Scholar]
- 54. Romrell LJ, Bellvé AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. Dev Biol 1976; 49:119–131. [DOI] [PubMed] [Google Scholar]
- 55. Bellvé AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse: isolation and morphological characterization. J Cell Biol 1977; 74:68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc Res Tech 2010; 73:241–278. [DOI] [PubMed] [Google Scholar]
- 57. Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell 2015; 58:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 2018; 50:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peng X, Wu J, Brunmeir R, Kim SY, Zhang Q, Ding C, Han W, Xie W, Xu F. TELP, a sensitive and versatile library construction method for next-generation sequencing. Nucleic Acids Res 2015; 43:e35–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J et al.. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative analysis of single-cell RNA sequencing methods. Molecular Cell 2017; 65:631–643.e4. [DOI] [PubMed] [Google Scholar]
- 62. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA et al.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y et al.. Mapping the mouse cell atlas by microwell-seq. Cell 2018; 173:1307. [DOI] [PubMed] [Google Scholar]
- 64. Chen Y, Zheng Y, Gao Y, Lin Z, Yang S, Wang T, Wang Q, Xie N, Hua R, Liu M, Sha J, Griswold MD et al.. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res 2018; 28:879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang M, Liu X, Chang G, Chen Y, An G, Yan L, Gao S, Xu Y, Cui Y, Dong J, Fan X, Hu Y et al.. Single-Cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 2018; 23:599–614.e4. [DOI] [PubMed] [Google Scholar]
- 66. Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH et al.. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 2017; 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lafzi A, Moutinho C, Picelli S, Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc 2018; 13:2742–2757. [DOI] [PubMed] [Google Scholar]
- 68. Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res/Fundamental Mol Mechan Mutagenesis 1993; 290:193–200. [DOI] [PubMed] [Google Scholar]
- 69. Bloom JD. Estimating the frequency of multiplets in single-cell RNA sequencing from cell-mixing experiments. PeerJ 2018; 6:e5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, Marioni JC, Teichmann SA. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 2015; 17:471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A, Teichmann SA. Power analysis of single-cell RNA-sequencing experiments. Nat Methods 2017; 14:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tung PY, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, Gilad Y. Batch effects and the effective design of single-cell gene expression studies. Sci Rep 2017; 7:39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015; 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lukassen S, Bosch E, Ekici AB, Winterpacht A. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Sci Rep 2018; 8:6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, Tallquist MD, Li JZ et al.. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq. Dev Cell 2018; 46:651–667.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Neirijnck Y, Kühne F, Mayère C, Pavlova E, Sararols P, Foti M, Atanassova N, Nef S. Tumor suppressor PTEN regulates negatively sertoli cell proliferation, testis size, and sperm production in vivo. Endocrinology 2019; 160:387–398. [DOI] [PubMed] [Google Scholar]
- 77. Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, Gildersleeve H, Lehle JD, Mayo M, Westernströer B, Law NC, Oatley MJ et al.. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Reports 2018; 25:1650–1667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guo JT, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie XC, Guo YX, Takei Y, Yun JN, Cai L, Kim R, Carrell DT et al.. The adult human testis transcriptional cell atlas. Cell Res 2018; 28:1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sohni A, Tan K, Song HW, Burow D, de Rooij DG, Laurent L, Hsieh TC, Rabah R, Hammoud SS, Vicini E, Wilkinson MF. The neonatal and adult human testis defined at the single-cell level. Cell Reports 2019; 26:1501–1517.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grive KJ, Hu Y, Shu E, Grimson A, Elemento O, Grenier JK, Cohen PE. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet 2019; 15:e1007810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jung M, Wells D, Rusch J, Ahmed S, Marchini J, Myers S, Conrad D. Unified single-cell analysis of testis gene regulation and pathology in 5 mouse strains. BioRxiv 2018: (doi: 10.1101/393769). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017; 14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017; 14:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. La HM, Mäkelä JA, Chan AL, Rossello FJ, Nefzger CM, Legrand JMD, De Seram M, Polo JM, Hobbs RM. Identification of dynamic undifferentiated cell states within the male germline. Nat Commun 2018; 9:2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liao J, Ng SH, Luk ACS, Suen AHC, Qian Y, Tu J, Fung JCL, Tang NLS, Feng B, Chan WY, Fouchet P, Lee TL. Revealing cellular and molecular transitions in neonatal germ cell differentiation using single cell RNA sequencing. Development 2019; 146 pii: dev174953. doi: 10.1242/dev.174953. [DOI] [PubMed] [Google Scholar]
- 87. Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, Zhu X, Li R et al.. Single-Cell RNA-Seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 2017; 20:891–892. [DOI] [PubMed] [Google Scholar]
- 88. Makino Y, Jensen NH, Yokota N, Rossner MJ, Akiyama H, Shirahige K, Okada Y. Single cell RNA-sequencing identified Dec2 as a suppressive factor for spermatogonial differentiation by inhibiting Sohlh1 expression. Sci Rep 2019; 9:6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 2014; 11:360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 2016; 92:342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shah S, Takei Y, Zhou W, Lubeck E, Yun J, Eng CL, Koulena N, Cronin C, Karp C, Liaw EJ, Amin M, Cai L. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 2018; 174:363–376.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ji N, van Oudenaarden A. Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook 2012:1–16. doi: 10.1895/wormbook.1.153.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB, McCarrey JR. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia1. Biol Reprod 2015; 92:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hermann BP, Phillips BT, Orwig KE. The elusive spermatogonial stem cell marker? Biol Reprod 2011; 85:221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Song HW, Bettegowda A, Lake BB, Zhao AH, Skarbrevik D, Babajanian E, Sukhwani M, Shum EY, Phan MH, Plank TM, Richardson ME, Ramaiah M et al.. The homeobox transcription factor RHOX10 drives mouse spermatogonial stem cell establishment. Cell Reports 2016; 17:149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pui HP, Saga Y. Gonocytes-to-spermatogonia transition initiates prior to birth in murine testes and it requires FGF signaling. Mechan Dev 2017; 144:125–139. [DOI] [PubMed] [Google Scholar]
- 97. Ernst C, Eling N, Martinez-Jimenez CP, Marioni JC, Odom DT. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat Commun 2019; 10:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Helsel AR, Oatley MJ, Oatley JM. Glycolysis-optimized conditions enhance maintenance of regenerative integrity in mouse spermatogonial stem cells during long-term culture. Stem Cell Reports 2017; 8:1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhao XL, Xu WJ, Wu JJ, Zhang D, Abou-Shakra A, Di L, Wang ZX, Wang LY, Yang F, Qiao ZD. Nicotine induced autophagy of Leydig cells rather than apoptosis is the major reason of the decrease of serum testosterone. Int J Biochem Cell Biol 2018; 100:30–41. [DOI] [PubMed] [Google Scholar]
- 100. Huang XY, Liao W, Huang YH, Jiang MJ, Chen JJ, Wang MX, Lin H, Guan SB, Liu J. Neuroprotective effect of dual specificity phosphatase 6 against glutamate-induced cytotoxicity in mouse hippocampal neurons. Biomed Pharmacother 2017; 91:385–392. [DOI] [PubMed] [Google Scholar]
- 101. Davis JC, Snyder EM, Hogarth CA, Small C, Griswold MD. Induction of spermatogenic synchrony by retinoic acid in neonatal mice. Spermatogenesis 2013; 3:e23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J et al.. Initial sequencing and comparative analysis of the mouse genome. Nature 2002; 420:520–562. [DOI] [PubMed] [Google Scholar]
- 103. Perlman RL. Mouse models of human disease: an evolutionary perspective. Evol Med Public Health 2016; 2016:170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells1. Biol Reprod 2003; 69:612–616. [DOI] [PubMed] [Google Scholar]
- 105. Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Nat Acad Sci 2004; 101:16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kaucher AV, Oatley MJ, Oatley JM. NEUROG3 is a critical downstream effector for STAT3-regulated differentiation of mammalian stem and progenitor spermatogonia1. Biol Reprod 2012; 86:164, 161-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development 2012; 139:1734–1743. [DOI] [PubMed] [Google Scholar]
- 108. Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Nat Acad Sci 2006; 103:9524–9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Takase HM, Nusse R. Paracrine Wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc Natl Acad Sci USA 2016; 113:E1489–E1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pellegrini M, Grimaldi P, Rossi P, Geremia R, Dolci S. Developmental expression of BMP4/ALK3/SMAD5 signaling pathway in the mouse testis: a potential role of BMP4 in spermatogonia differentiation. J Cell Sci 2003; 116:3363–3372. [DOI] [PubMed] [Google Scholar]
- 111. Svensson V, Pachter L. RNA velocity: molecular kinetics from single-cell RNA-Seq. Molecular Cell 2018; 72:7–9. [DOI] [PubMed] [Google Scholar]
- 112. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE et al.. RNA velocity of single cells. Nature 2018; 560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lee J, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Kimura T, Nakano T, Ogura A, Shinohara T. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 2007; 134:1853–1859. [DOI] [PubMed] [Google Scholar]
- 114. Jijiwa M, Kawai K, Fukihara J, Nakamura A, Hasegawa M, Suzuki C, Sato T, Enomoto A, Asai N, Murakumo Y, Takahashi M. GDNF-mediated signaling via RET tyrosine 1062 is essential for maintenance of spermatogonial stem cells. Genes Cells 2008; 13:365–374. [DOI] [PubMed] [Google Scholar]
- 115. Uchida A, Kishi K, Aiyama Y, Miura K, Takase HM, Suzuki H, Kanai-Azuma M, Iwamori T, Kurohmaru M, Tsunekawa N, Kanai Y. In vivo dynamics of GFRα1-positive spermatogonia stimulated by GDNF signals using a bead transplantation assay. Biochem Biophy Res Commun 2016; 476:546–552. [DOI] [PubMed] [Google Scholar]
- 116. Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H et al.. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287:1489–1493. [DOI] [PubMed] [Google Scholar]
- 117. Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate1. Biol Reprod 2006; 74:314–321. [DOI] [PubMed] [Google Scholar]
- 118. Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282:25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. He Z, Jiang J, Kokkinaki M, Golestaneh N, Hofmann MC, Dym M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells 2008; 26:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sada A, Hasegawa K, Pin PH, Saga Y. NANOS2 acts downstream of glial cell line-derived neurotrophic factor signaling to suppress differentiation of spermatogonial stem cells. Stem Cells 2012; 30:280–291. [DOI] [PubMed] [Google Scholar]
- 121. Abé K, Eto K, Abé S. Epidermal growth factor mediates spermatogonial proliferation in newt testis. Reprod Biol Endocrinol 2008; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Simon L, Ekman GC, Tyagi G, Hess RA, Murphy KM, Cooke PS. Common and distinct factors regulate expression of mRNA for ETV5 and GDNF, Sertoli cell proteins essential for spermatogonial stem cell maintenance. Experim Cell Res 2007; 313:3090–3099. [DOI] [PubMed] [Google Scholar]
- 123. Yang QE, Kim D, Kaucher A, Oatley MJ, Oatley JM. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J Cell Sci 2013; 126:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 2009; 136:1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tadokoro Y, Yomogida K, Ohta H, Tohda A, Nishimune Y. Homeostatic regulation of germinal stem cell proliferation by the GDNF/FSH pathway. Mechan Dev 2002; 113:29–39. [DOI] [PubMed] [Google Scholar]
- 126. Chen LY, Willis WD, Eddy EM. Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc Natl Acad Sci USA 2016; 113:1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kitadate Y, Jörg DJ, Tokue M, Maruyama A, Ichikawa R, Tsuchiya S, Segi-Nishida E, Nakagawa T, Uchida A, Kimura-Yoshida C, Mizuno S, Sugiyama F et al.. Competition for mitogens regulates spermatogenic stem cell homeostasis in an open niche. Cell Stem Cell 2019; 24:79–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lutz W, O’Neill BC, Scherbov S. DEMOGRAPHICS: enhanced: Europe's population at a turning point. Science 2003; 299:1991–1992. [DOI] [PubMed] [Google Scholar]
- 129. Jain AK, Ross JA. Fertility differences among developing countries: are they still related to family planning program efforts and Social Settings? IPSRH 2012; 38:015–022. [DOI] [PubMed] [Google Scholar]
- 130. de Kretser DM. Determinants of male health: the interaction of biological and social factors. Asian J Androl 2010; 12:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jafarzadeh-Kenarsari F, Ghahiri A, Habibi M, Zargham-Boroujeni A. Exploration of infertile couples' support requirements: a qualitative study. Int J Fertility Sterility 2015; 9:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Darde TA, Sallou O, Becker E, Evrard B, Monjeaud C, Le Bras Y, Jégou B, Collin O, Rolland AD, Chalmel F. The ReproGenomics Viewer: an integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res 2015; 43:W109–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Darde TA, Lecluze E, Lardenois A, Stévant I, Alary N, Tüttelmann F, Collin O, Nef S, Jégou B, Rolland AD, Chalmel F. The ReproGenomics viewer: a multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics 2019; btz047, 10.1093/bioinformatics/btz047. [DOI] [PubMed] [Google Scholar]
- 134. Salmén F, Ståhl PL, Mollbrink A, Navarro JF, Vickovic S, Frisén J, Lundeberg J. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat Protoc 2018; 13:2501–2534. [DOI] [PubMed] [Google Scholar]
- 135. Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III Cutaneous Malignant Melanoma. Cancer Res 2018; 78:5970–5979. [DOI] [PubMed] [Google Scholar]
- 136. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun 2018; 9:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013; 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 2013; 23:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015; 160:339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N et al.. Comparison of Cas9 activators in multiple species. Nat Methods 2016; 13:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS et al.. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell 2016; 167:233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 2016; 167:219–232.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016; 44:5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biology Open 2016; 5:866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, Hatada I. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 2016; 34:1060–1065. [DOI] [PubMed] [Google Scholar]
- 150. Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 2015; 12:401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015; 33:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cano-Rodriguez D, Gjaltema RA, Jilderda LJ, Jellema P, Dokter-Fokkens J, Ruiters MH, Rots MG. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 2016; 7:12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kwon DY, Zhao YT, Lamonica JM, Zhou Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat Commun 2017; 8:15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, Hovingh S, de Reijke TM, de la Rosette JJ, van der Veen F, de Rooij DG, Repping S et al.. Propagation of human spermatogonial stem cells in vitro. JAMA 2009; 302:2127–2134. [DOI] [PubMed] [Google Scholar]
- 155. Wu X, Schmidt JA, Avarbock MR, Tobias JW, Carlson CA, Kolon TF, Ginsberg JP, Brinster RL. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Nat Acad Sci 2009; 106:21672–21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. He Z, Kokkinaki M, Jiang J, Dobrinski I, Dym M. Isolation, characterization, and culture of human spermatogonia1. Biol Reprod 2010; 82:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lim JJ, Sung SY, Kim HJ, Song SH, Hong JY, Yoon TK, Kim JK, Kim KS, Lee DR. Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Prolif 2010; 43:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Smith JF, Yango P, Altman E, Choudhry S, Poelzl A, Zamah AM, Rosen M, Klatsky PC, Tran ND. Testicular niche required for human spermatogonial stem cell expansion. Stem Cells Transl Med 2014; 3:1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hermann BP, Orwig KE. Translating spermatogonial stem cell transplantation to the clinic. In: Orwig KE, Hermann BP (eds.), Male germline stem cells: developmental and regenerative potential, New York, NY: Springer; 2011: 227–253. [Google Scholar]
- 160. Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell 2012; 11:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ginsburg M, Snow MHL, McLaren A. Primordial germ-cells in the mouse embryo during gastrulation. Development 1990; 110:521-&. [DOI] [PubMed] [Google Scholar]
- 162. Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature 2002; 418:293–300. [DOI] [PubMed] [Google Scholar]
- 163. Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011; 146:519–532. [DOI] [PubMed] [Google Scholar]
- 164. Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 2012; 338:971–975. [DOI] [PubMed] [Google Scholar]
- 165. Hayashi K, Saitou M. Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nat Protoc 2013; 8:1513–1524. [DOI] [PubMed] [Google Scholar]
- 166. Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature 2013; 501:222–226. [DOI] [PubMed] [Google Scholar]
- 167. Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z et al.. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013; 504:282–286. [DOI] [PubMed] [Google Scholar]
- 168. Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015; 160:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, Nakamura S, Sekiguchi K et al.. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 2015; 17:178–194. [DOI] [PubMed] [Google Scholar]
- 170. Ishikura Y, Yabuta Y, Ohta H, Hayashi K, Nakamura T, Okamoto I, Yamamoto T, Kurimoto K, Shirane K, Sasaki H, Saitou M. In vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Reports 2016; 17:2789–2804. [DOI] [PubMed] [Google Scholar]
- 171. Irie N, Surani MA. Efficient induction and isolation of human primordial germ ell-like cells from competent human pluripotent stem cells. Methods Mol Biol 2017; 1463:217–226. [DOI] [PubMed] [Google Scholar]
- 172. Yamashiro C, Sasaki K, Yabuta Y, Kojima Y, Nakamura T, Okamoto I, Yokobayashi S, Murase Y, Ishikura Y, Shirane K, Sasaki H, Yamamoto T et al.. Generation of human oogonia from induced pluripotent stem cells in vitro. Science 2018; 362:356–360. [DOI] [PubMed] [Google Scholar]
- 173. Hayashi K, Hikabe O, Obata Y, Hirao Y. Reconstitution of mouse oogenesis in a dish from pluripotent stem cells. Nat Protoc 2017; 12:1733–1744. [DOI] [PubMed] [Google Scholar]
- 174. Sato T, Katagiri K, Gohbara A, Inoue K, Ogonuki N, Ogura A, Kubota Y, Ogawa T. In vitro production of functional sperm in cultured neonatal mouse testes. Nature 2011; 471:504–507. [DOI] [PubMed] [Google Scholar]
- 175. Sato T, Katagiri K, Kubota Y, Ogawa T. In vitro sperm production from mouse spermatogonial stem cell lines using an organ culture method. Nat Protoc 2013; 8:2098–2104. [DOI] [PubMed] [Google Scholar]
- 176. Sato T, Katagiri K, Kojima K, Komeya M, Yao M, Ogawa T. In vitro spermatogenesis in explanted adult mouse testis tissues. PLoS ONE 2015; 10:e0130171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Komeya M, Kimura H, Nakamura H, Yokonishi T, Sato T, Kojima K, Hayashi K, Katagiri K, Yamanaka H, Sanjo H, Yao M, Kamimura S et al.. Long-term ex vivo maintenance of testis tissues producing fertile sperm in a microfluidic device. Sci Rep 2016; 6:21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Sanjo H, Komeya M, Sato T, Abe T, Katagiri K, Yamanaka H, Ino Y, Arakawa N, Hirano H, Yao T, Asayama Y, Matsuhisa A et al.. In vitro mouse spermatogenesis with an organ culture method in chemically defined medium. PLoS ONE 2018; 13:e0192884. [DOI] [PMC free article] [PubMed] [Google Scholar]