Abstract
Human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes have many promising applications, including the regeneration of injured heart muscles, cardiovascular disease modeling, and drug cardiotoxicity screening. Current differentiation protocols yield a heterogeneous cell population that includes pluripotent stem cells and different cardiac subtypes (pacemaking and contractile cells). The ability to purify these cells and obtain well-defined, controlled cell compositions is important for many downstream applications; however, there is currently no established and reliable method to identify hiPSC-derived cardiomyocytes and their subtypes. Here, we demonstrate that second harmonic generation (SHG) signals generated directly from the myosin rod bundles can be a label-free, intrinsic optical marker for identifying hiPSC-derived cardiomyocytes. A direct correlation between SHG signal intensity and cardiac subtype is observed, with pacemaker-like cells typically exhibiting ~70% less signal strength than atrial- and ventricular-like cardiomyocytes. These findings suggest that pacemaker-like cells can be separated from the heterogeneous population by choosing an SHG intensity threshold criteria. This work lays the foundation for developing an SHG-based high throughput flow sorter for purifying hiPSC-derived cardiomyocytes and their subtypes.
Keywords: hiPSC-CMs, Pacemaking, Subtypes, Action potential, Second harmonic generation
SIGNIFICANCE STATEMENT
Current differentiation methods for generating human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) yield heterogeneous populations that remain a critical barrier for downstream applications where well-defined populations are required. Markers for identifying undesired populations such as teratoma-forming or arrhythmia-inducing cells are not currently available. We show a direct correlation between cardiac subtype and the intensity of second harmonic generation (SHG) signals generated from myosin filaments. We demonstrate SHG signals can be a label-free marker for eliminating undesired cell populations. These results highlight a novel method for using SHG signals to purify hiPSC-CMs.
Graphical Abstract
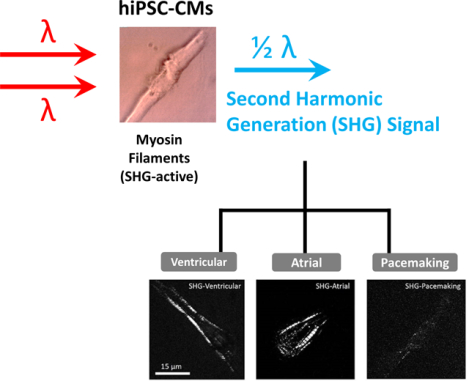
INTRODUCTION
Human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes are an ex-vivo source of heart cells to potentially treat heart disease, model cardiac cell function and disease, and assess drug cardiotoxicity [1]. Purified cell populations with defined compositions are highly desired for these applications. The removal of pluripotent stem cells is critical for human transplantation due to their potential for forming teratomas [2]. Moreover, the hiPSC-derived cardiomyocytes population itself is a mixture of pacemaker-, atrial-, and ventricular-like subtypes. The elimination of pacemaker-like cells and immature cells that spontaneously generate action potentials is particularly important to avoid irregular arrhythmic events after cardiac transplantation [3]. In drug screening, hiPSC-derived cardiomyocytes have an improved prediction of drug response if higher purity cells with more homogenous and desired cardiomyocyte subtype are used [4, 5].
Current hiPSC-derived cardiomyocytes differentiation protocols typically yield ~70% cardiomyocytes with ~15% of the cardiomyocytes being the pacemaking subtype [6–8]. Achieving higher-purity populations is challenging because there is no established purification method, largely due to the lack of a reliable marker that can be used to sort cells by phenotype, maturity, or subtype. Many methods have been explored (e.g. lentiviral transduction [9], molecular beacons [10, 11], size separation [12], metabolic selection [13], mitochondrial dye [14], surface markers [15]). These approaches rely on exogenous labels, viruses, or animal derived products that may complicate their use clinically and/or lack the specificity for achieving high-purity cell populations. None of these methods can identify cardiomyocytes by their subtype.
We previously reported that the SHG signal generated directly from myosin rod bundles can be a highly specific, label-free optical signal for identifying hiPSC-derived cardiomyocytes [16]. The high specificity is due to myosin filaments only being present in cardiomyocytes and absent in other types of cells. We further showed that hiPSC-derived cardiomyocytes of different maturities could also be discriminated by the SHG intensity due to their myosin development. In this study, we demonstrate that hiPSC-derived cardiomyocyte subtypes can be separated based on their SHG intensity due to the different levels of myosin filaments in atrial-, ventricular-, and pacemaker-like hiPSC-derived cardiomyocytes.
MATERIALS AND METHODS
Figure 1 illustrates the process we used to identify subtypes of individual hiPSC-derived cardiomyocytes and to quantify their total SHG intensity. Detailed descriptions of the methods for cell differentiation, sample preparation, optical measurements, and data analysis are provided in the Supporting Information.
Figure 1.
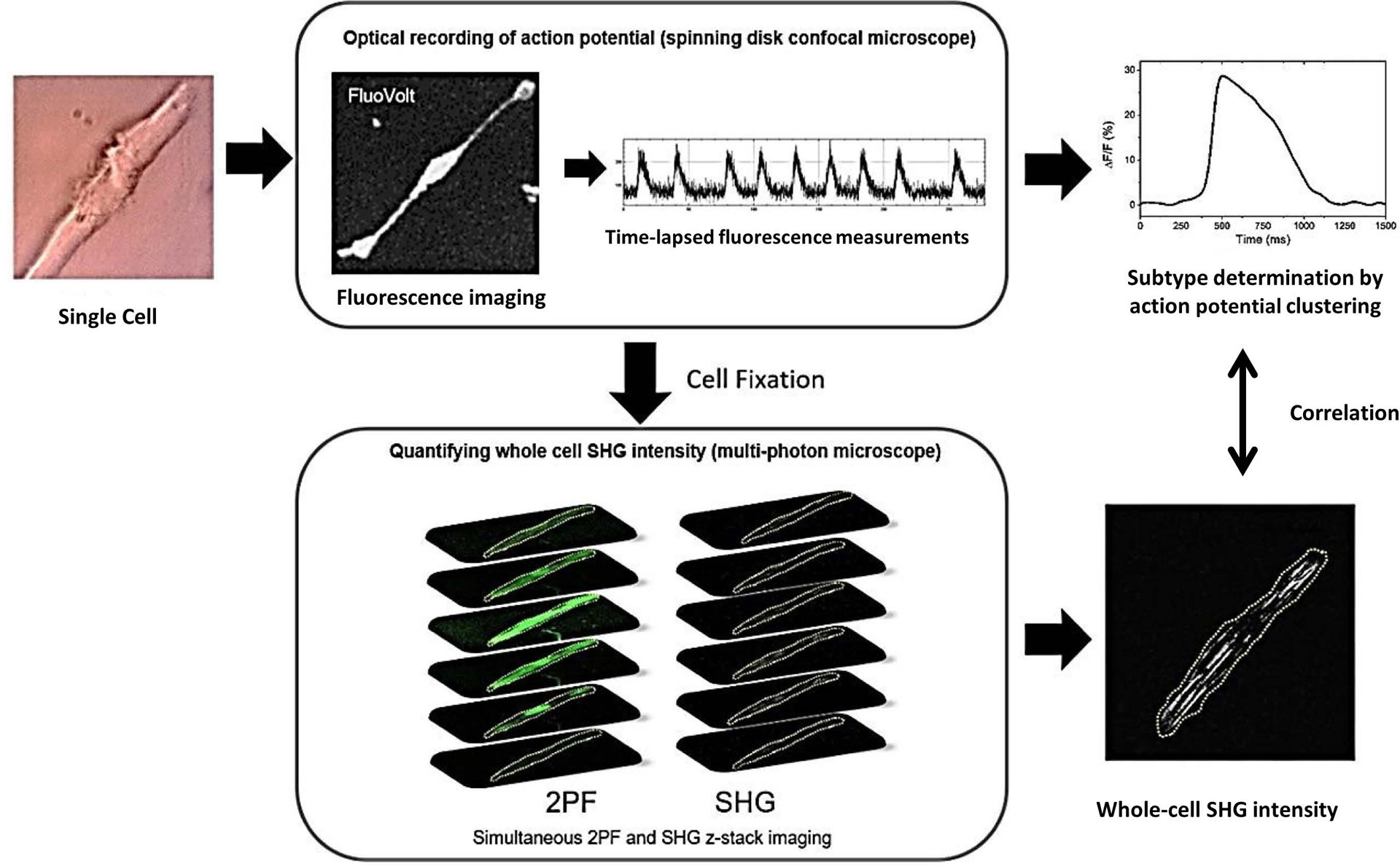
Diagram illustrating the method of identifying the subtype of individual hiPSC-derived cardiomyocytes and correlating it to their whole cell SHG intensity. Single cells (40 days in culture) were re-plated onto gridded glass dishes for 55–67 days for a total of 95–107 days post-differentiation (noted D95 in the manuscript). Cells were loaded with the voltage-sensitive dye FluoVolt. The spontaneous action potentials of individual cells were recorded with a spinning disk fluorescence microscope to measure the time-lapsed fluorescence intensity at ~100 frames per second for ~1 minute. The subtype was determined by the clustering analysis of the action potential (AP) profiles. Cells were then fixed and whole-cell SHG measurements were performed on a multi-photon microscope using a circularly polarized excitation beam. The whole-cell SHG intensity was obtained by performing z-stack imaging and summing the SHG signal intensity from all z-planes. The two-photon fluorescence (2PF) signal from the cell membrane was used as a guide to outline the cell boundary.
RESULTS AND DISCUSSION
The subtype of each hiPSC-derived cardiomyocyte was first determined by its action potential (AP) profile through the clustering analysis, an established alternative method to patch clamp for subtype classification [17–20]. The AP of a ventricular-like cell shows a more prominent plateau phase than that of an atrial-like and pacemaker-like cell (Figs. 2, A1-A3). Moreover, ventricular-like cells have an average AP duration (APD50, action potential duration at 50% repolarization) near 324±20 ms, which is longer than that of atrial-like cells (166±4 ms) and pacemaker-like cells (171±8 ms). The average membrane voltage change of pacemaker-like cells is ~15% (ΔF/F0, the maximum change in fluorescence), while contractile CMs have a higher amplitude of ~21%. See graphs in Fig. S1 in Supporting Information. Among all 200 cells that were sampled, pacemaker-like hiPSC-derived cardiomyocytes represent the smallest population at 10%, while ventricular-like and atrial-like cells account for 44% and 46%, respectively. This subtype distribution is comparable to previously published reports [8, 21–23].
Figure 2.
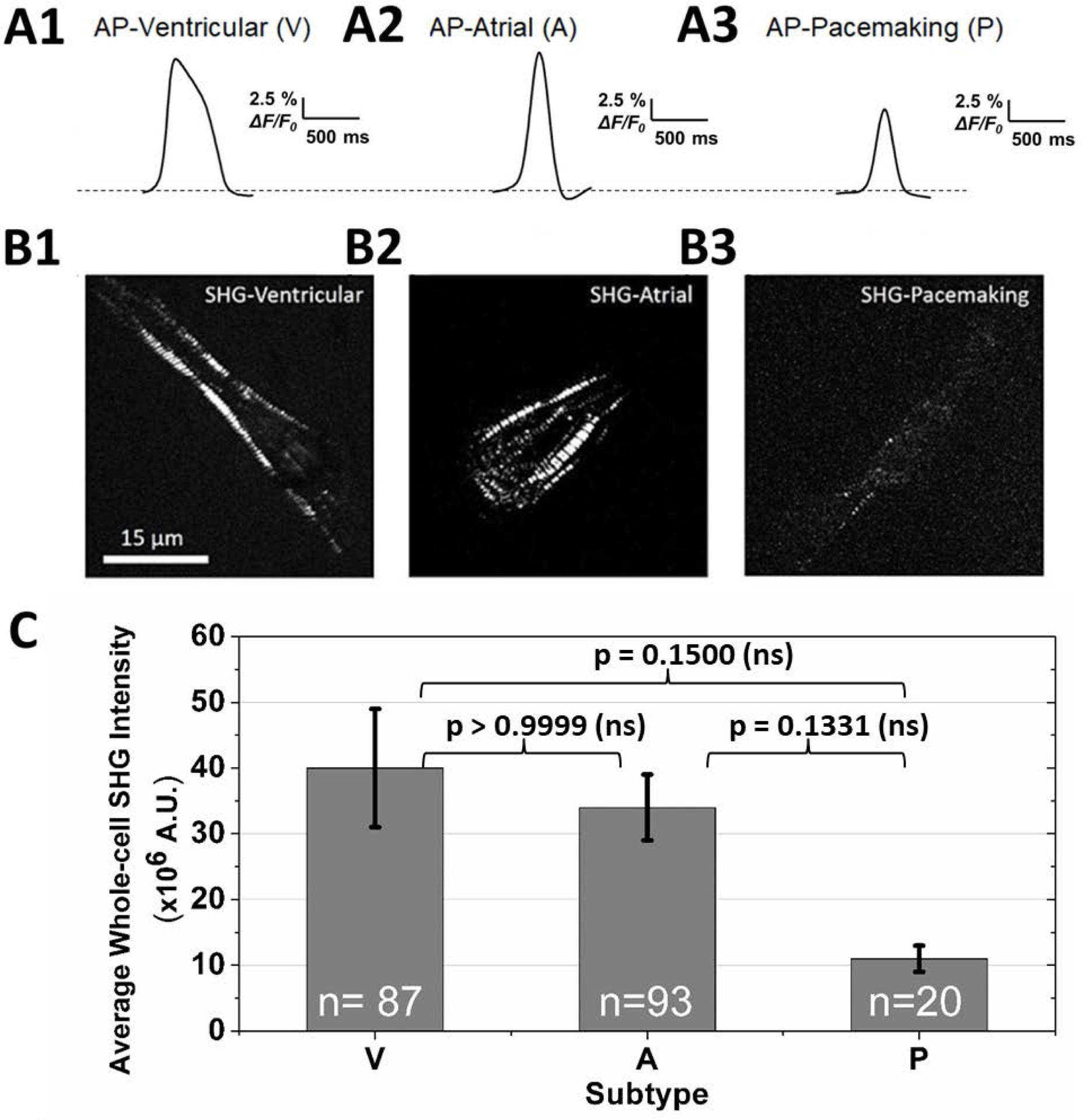
(A) Representative action potential profiles used to classify the cardiomyocyte subtype of each cell (V: ventricular-like; A: atrial-like, P: pacemaking-like hiPSC-CMs). (B) Representative whole-cell SHG images show that the contractile cells exhibit more organized sarcomeric striation patterns, while pacemaker-like cells typically have a lower SHG signal intensity from less organized sarcomeric structures. (C) Average whole-cell SHG intensities for each cardiac subtype. The SHG signal strength of pacemaker-like cells is ~70% less than the contractile cells. The average whole-cell SHG intensity ± standard error for ventricular-like, atrial-like, and pacemaking-like cells are 40±9, 34±5, and 11±2 million A.U., respectively. A Kruskal-Wallis test was used to assess the significance of the difference in SHG intensities between subtypes. P-values were greater than 0.05, indicating the differences were non-significant (ns). The distribution of SHG intensity for all cells of each subtype is shown in the histograms in Figure 3.
The contractile cells (i.e. ventricular-like, atrial-like) on average exhibit a higher SHG intensity than pacemaker-like cells (Figs. 2B–2C; V: 40-, A: 34-, P: 11-million counts). For contractile cells, the SHG signals originate from relatively periodic, organized sarcomere patterns in the cells, while pacemaker-like cells have lower intensity counts and the signal is distributed across the cell from a less-organized myosin network (Figs. 2, B1-B3). On average, contractile cells generate >3.5 times more signal than pacemaker-like cells.
A Kruskal-Wallis test performed on the results in Figure 2C indicate that the SHG intensities are not statistically different between the three groups. However, more detailed information of the cell-to-cell variability of the SHG intensity for each subtype can be obtained by plotting histograms from the three cell groups (Fig. 3). All pacemaker-like cells have signal intensity less than 30×106 counts. In contrast, the contractile cells have a wider distribution: the atrial-like and ventricular-like populations have cells that exhibit SHG intensities up to ~200×106 and ~500×106 counts, respectively. Many of these contractile cells, however, also exhibit low SHG signal intensities. These cells tend to be smaller and have a partially developed, disordered myofilament network, which is in stark contrast to the ordered, well-developed myosin network of adult contractile cardiomyocytes (Supporting Information Fig. S2) that emit strong SHG signals.
Figure 3.
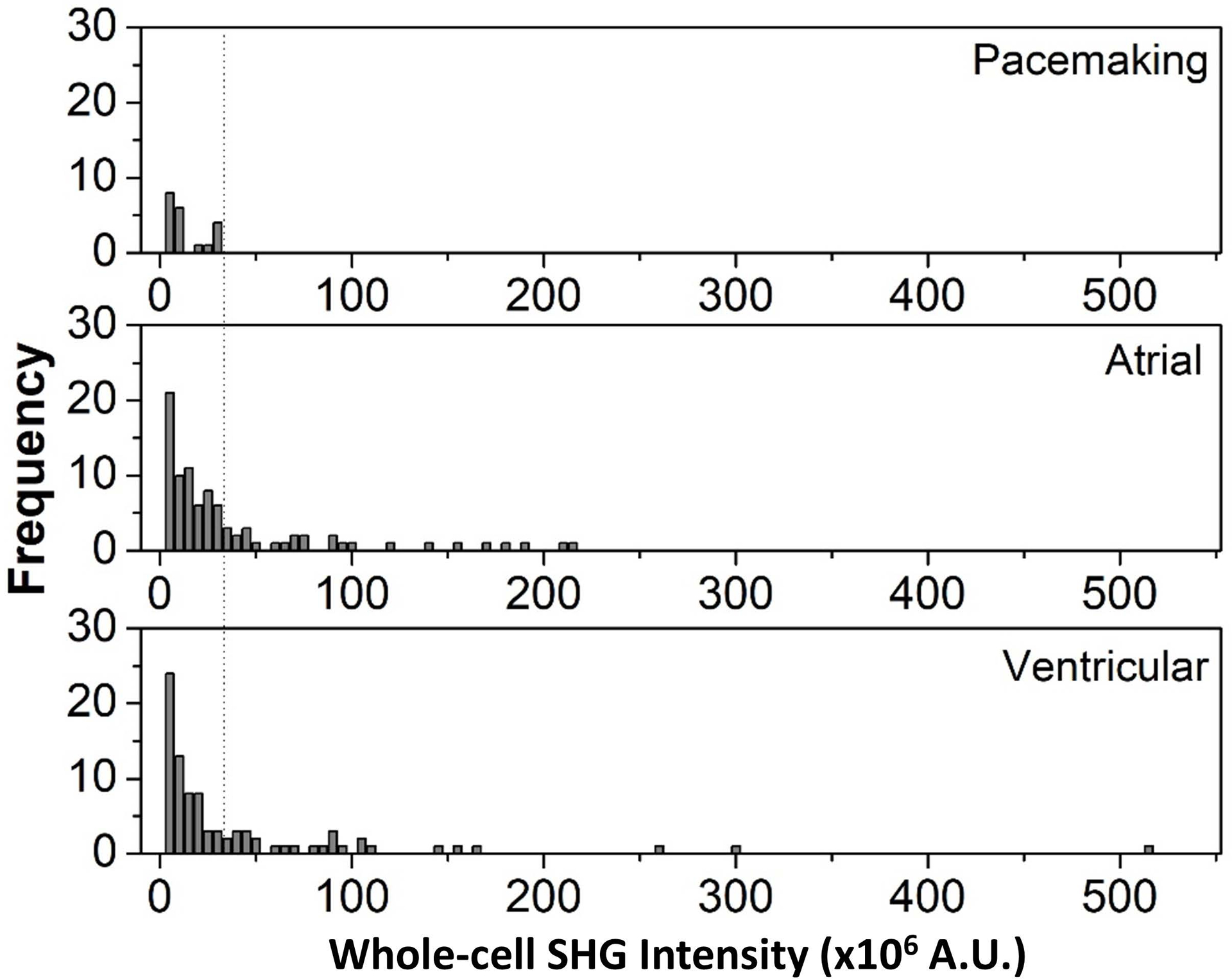
Histogram plots of whole-cell SHG intensity for each cardiomyocyte subtype. The dashed line indicates an SHG signal threshold of 30×106 counts that can be used to separate pacemaker-like cells from the other cells.
These results suggest that by selecting an SHG intensity threshold criteria (i.e. 30×106 counts in this case), it is possible to purge potential arrhythmia-inducing pacemaker-like cells from a population of hiPSC-derived cardiomyocytes and retrieve a pure population of contractile cells. The tradeoff, however, is that a fraction of contractile cells would also be lost. We estimated that about 66% of the contractile cells would be discarded along with all pacemaker-like cells. About 30% of the total cell population, comprised of only contractile cells, can be recovered with this technique (Fig. 4).
Figure 4.
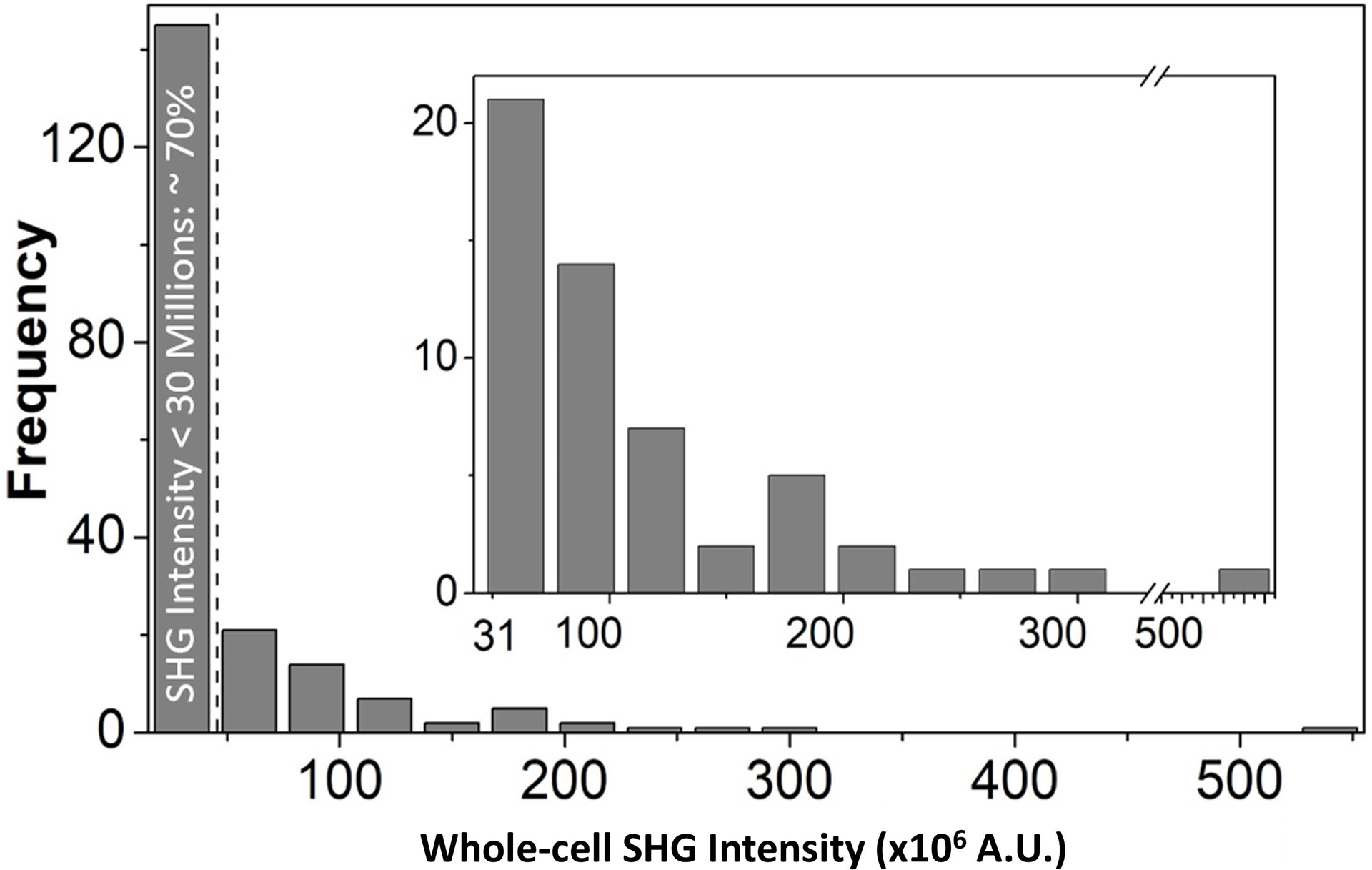
Histogram of the whole-cell SHG intensity with a 30×106 intensity increment. The dashed line indicates the 30×106 intensity threshold that will filter ~70% of cells, including all pacemaker-like cells. The remaining 30% of gated cells (inset plot) are all atrial- and ventricular-like cells.
It may be possible to improve the recovery rate of contractile cells by optimizing parameters such as differentiation time, re-plating time, substrate stiffness, or electrical stimulation of cells to mature myosin development and increase SHG signal strength [24, 25]. For example, we performed the same experiment on D53 cells post-differentiation that have a shorter re-plating time (i.e. 10 days after re-plating) and compared the results to the D95 cells post-differentiation (SI-Fig. S3). We observed similar overall trends in the SHG intensities of the three cardiac subtypes indicating good data reproducibility. Also, if the same SHG intensity threshold is used, the number of contractile cells that would fall within the pacemaker-like cell population slightly decreases. As such, for D53 cells, ~60% of the contractile cells would be discarded. HiPSC-derived contractile cardiomyocytes are known to exhibit a decrease in frequency of automaticity over time in culture, which can adversely affect myofilament development without an external electrical stimulus [23, 26]. Therefore, single plated contractile cardiomyocytes, as in D95 cardiomyocytes, may have loss of myofilaments with long-term culture. The higher whole-cell SHG signal counts in D53 hiPSC-derived contractile cardiomyocytes is presumably due to these cells retaining their contractile structure before further reduction of automaticity frequency. As maturation strategies for hiPSC-derived contractile cardiomyocytes improve [23], the separation between pacemaker and contractile phenotype may be more distinct, since it is known that pacemaking cells have underdeveloped and poorly organized myofilaments compared to contractile cells in vivo, in humans and other animals [27]. Hence, the SHG signal intensities, which depend on myosin filament content, will be more distinct for these two cell groups, enabling improved contractile cell yield and reduced numbers of discarded false-negative contractile cardiomyocytes.
CONCLUSION
We demonstrate that, in addition to being able to identify hiPSC-derived cardiomyocytes [16], SHG signals can be also used as a label-free optical marker to remove pacemaker-like cells from a heterogeneous population. By selecting a proper SHG intensity threshold criteria, as is commonly done in flow cytometry, cells that might form teratomas (i.e. undifferentiated hiPSCs with no SHG signal) or induce arrhythmias (i.e. pacemaker-like cells with a low SHG intensity) can be identified in a label-free, noninvasive manner. While we used a confocal microscope in this study to accurately quantify whole cell SHG signal intensities, we are actively working on integrating SHG into a microfluidic sorting system for analyzing individual hiPSC-derived cardiomyocytes prepared as suspension cells using collagenase [16]. SHG has the potential to be a novel nonlinear optical modality in flow cytometry [28] for high-throughput purification of hiPSC-derived cardiomyocytes to obtain sorted cell populations with well-defined cell compositions that can be used in disease modeling, drug screening, and stem cell based therapies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from National Science Foundation (1827611) (PI: Chan) and California Institute for Regenerative Medicine (DISC2-10120) (PI: Lieu). C.-W. Chang acknowledges support from the T32 Training Program in Basic and Translational Cardiovascular Science established by National Heart, Lung and Blood Institute (T32 HL086350). We thank Prof. Nipavan Chiamvimonvat’s laboratory for providing the adult mouse cardiomyocytes.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST:
D.K.L. serves as a consultant for Novoheart Ltd. The other authors indicate no potential conflicts of interest.
DATA AVAILABILITY STATEMENT:
The data supporting the findings are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Oikonomopoulos A, Kitani T, Wu JC. Pluripotent Stem Cell-Derived Cardiomyocytes as a Platform for Cell Therapy Applications: Progress and Hurdles for Clinical Translation. Molecular therapy : the journal of the American Society of Gene Therapy. 2018;26:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelakanti RV, Kooreman NG, Wu JC. Teratoma formation: a tool for monitoring pluripotency in stem cell research. Current protocols in stem cell biology. 2015;32:4A 8 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida SO, Skelton RJ, Adigopula S, et al. Arrhythmia in stem cell transplantation. Cardiac electrophysiology clinics. 2015;7:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawatou M, Masumoto H, Fukushima H, et al. Modelling Torsade de Pointes arrhythmias in vitro in 3D human iPS cell-engineered heart tissue. Nat Commun. 2017;8:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretti A, Bellin M, Welling A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. [DOI] [PubMed] [Google Scholar]
- 6.Burridge PW, Keller G, Gold JD, et al. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell stem cell. 2012;10:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burridge PW, Matsa E, Shukla P, et al. Chemically defined generation of human cardiomyocytes. Nature methods. 2014;11:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yechikov S, Copaciu R, Gluck JM, et al. Same-Single-Cell Analysis of Pacemaker-Specific Markers in Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Subtypes Classified by Electrophysiology. Stem Cells. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woods NB, Muessig A, Schmidt M, et al. Lentiviral vector transduction of NOD/SCID repopulating cells results in multiple vector integrations per transduced cell: risk of insertional mutagenesis. Blood. 2003;101:1284–1289. [DOI] [PubMed] [Google Scholar]
- 10.Wile BM, Ban K, Yoon YS, et al. Molecular beacon-enabled purification of living cells by targeting cell type-specific mRNAs. Nature protocols. 2014;9:2411–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyduk E, Knoll E, Heyduk T. Molecular beacons for detecting DNA binding proteins: mechanism of action. Analytical biochemistry. 2003;316:1–10. [DOI] [PubMed] [Google Scholar]
- 12.Xu C, Police S, Rao N, et al. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circulation research. 2002;91:501–508. [DOI] [PubMed] [Google Scholar]
- 13.Tohyama S, Hattori F, Sano M, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell stem cell. 2013;12:127–137. [DOI] [PubMed] [Google Scholar]
- 14.Hattori F, Chen H, Yamashita H, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nature methods. 2010;7:61–66. [DOI] [PubMed] [Google Scholar]
- 15.Dubois NC, Craft AM, Sharma P, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature biotechnology. 2011;29:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awasthi S, Matthews DL, Li RA, et al. Label-free identification and characterization of human pluripotent stem cell-derived cardiomyocytes using second harmonic generation (SHG) microscopy. Journal of biophotonics. 2012;5:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yechikov S, Copaciu R, Gluck JM, et al. Same-Single-Cell Analysis of Pacemaker-Specific Markers in Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Subtypes Classified by Electrophysiology. Stem cells. 2016;34:2670–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takaki T, Inagaki A, Chonabayashi K, et al. Optical Recording of Action Potentials in Human Induced Pluripotent Stem Cell-Derived Cardiac Single Cells and Monolayers Generated from Long QT Syndrome Type 1 Patients. Stem Cells Int. 2019;2019:7532657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedut S, Seminatore-Nole C, Lamamy V, et al. High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. Am J Physiol Heart Circ Physiol. 2016;311:H44–53. [DOI] [PubMed] [Google Scholar]
- 20.Biendarra-Tiegs SM, Secreto FJ, Nelson TJ. Addressing Variability and Heterogeneity of Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Adv Exp Med Biol. 2019. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Guo L, Fiene SJ, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301:H2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He JQ, Ma Y, Lee Y, et al. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–39. [DOI] [PubMed] [Google Scholar]
- 23.Lieu DK, Fu JD, Chiamvimonvat N, et al. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Arrhythm Electrophysiol. 2013;6:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro AJ, Ang YS, Fu JD, et al. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A. 2015;112:12705–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engler AJ, Carag-Krieger C, Johnson CP, et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartiani L, Bettiol E, Stillitano F, et al. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. [DOI] [PubMed] [Google Scholar]
- 27.Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47:658–687. [DOI] [PubMed] [Google Scholar]
- 28.Collier BB, Awasthi S, Lieu DK, et al. Non-Linear Optical Flow Cytometry Using a Scanned, Bessel Beam Light-Sheet. Sci Rep-Uk. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirish P, Ledford HA, Timofeyev V, et al. Action Potential Shortening and Impairment of Cardiac Function by Ablation of Slc26a6. Circulation. Arrhythmia and electrophysiology 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


