Abstract
Aging is a multifactorial process characterized by progressive changes in gut physiology and the intestinal mucosal immune system. These changes, along with alterations in lifestyle, diet, nutrition, inflammation and immune function alter both composition and stability of the gut microbiota. Given the impact of environmental influences on the gut microbiota, animal models are particularly useful in this field. To understand the relationship between the gut microbiota and aging in nonhuman primates, we collected fecal samples from 20 male and 20 female rhesus macaques (Macaca mulatta), across the natural macaque age range, for 16S rRNA gene analyses. Operational taxonomic units were then grouped together to summarize taxon abundance at different hierarchical levels of classification and alpha- and beta-diversity were calculated. There were no age or sex differences in alpha diversity. At the phylum level, relative abundance of Proteobacteria and Firmicutes and Firmicutes to Bacteriodetes ratio were different between age groups though significance disappeared after correction for multiple comparisons. At the class level, relative abundance of Firmicutes_Bacilli decreased and Proteobacteria_Alphaproteobacteria and Proteobacteria_Betaproteobacteria increased with each successively older group. Only differences in Firmicutes_Bacilli remained significant after correction for multiple comparisons. No sex differences were identified in relative abundances after correction for multiple comparisons. Our results are not surprising given the known impact of environmental factors on the gut microbiota.
Keywords: Monkey, Frailty, Animal model, Biological age
Within the mammalian gastrointestinal tract, there is a complex community of more than 100 trillion microbial cells (1) collectively known as the gut microbiota. The genomes associated with this community represent the gut microbiome. Overall interindividual variation in the gut microbiota is extremely large and is influenced by host genetic background, microbes, and environmental factors (2). Of the environmental factors, diet imposes a predominate and rapid effect (3). The gut microbiota is known to interact both with each other and the host and are associated with intestinal as well as systemic diseases. Compositional and functional changes of the human gut microbiome have been linked to many chronic metabolic diseases, such as malnutrition (4) and obesity (5), and there is increasing evidence that gut microbiota may influence metabolism and play a role in the onset of metabolic diseases (6).
Aging is a multifactorial process characterized by the progressive functional decline of the principal physiological systems, including gastrointestinal/digestive system and the intestinal mucosal immune system (7). These changes, along with concomitant alterations in life style, diet, nutrition, inflammation and immune function alter both the composition and stability of the human gut microbiota (8,9). While gut microbiotas of healthy adults are relatively stable even for decades (10), microbiota changes evident in older adults (8,11) are likely due to the effects of coexisting conditions and treatment thereof as well as age-related factors (12). In general, aging is associated with decreased intra-individual microbiota diversity and greater interindividual variation (2,8,12), though there is large variability in study results (13–15).
Animal model studies are particularly useful in situations such as these where the whole-body response is crucial and environmental effects are reasonably expected to impact results. Given the oversize impact of environmental influences on the gut microbiota and the degree to which the gut microbiota is involved in a multitude of body systems, animal models are particularly useful in this field. While rodent models are undoubtedly useful, they cannot recapitulate the human condition to the same degree as do nonhuman primate models such as the well-studied rhesus monkey (Macaca mulatta). Unlike in human studies, in nonhuman primate studies environment, dietary intake and medical history can be fully described, and studies can be designed to assure comprehensive subject monitoring and strict protocol adherence. At its most basic, studies in rhesus monkeys can be designed to eliminate or reduce the effect of lifestyle factors on the microbe community. In addition, unlike rodents, rhesus monkeys display patterns of eating and sleeping behavior that mirror those of humans, have a life span measured in decades, and develop and age in similar ways to humans. Furthermore, rhesus monkey share ~93% sequence identity with the human genome (16,17), and this similarity extends to numerous aspects of their anatomy, physiology, neurology, endocrinology, immunology, and behavior (18). While other nonhuman primate species serve as valuable models for studies of gut microbiota, the breadth of existing knowledge regarding captive rhesus macaques and aging rhesus macaques, in particular, make them the ideal animal model for studying aging and the gut microbiota.
Monkey gut microbiota has been explored in several contexts. For example, studies have explored the relationship between gut microbiota and obesity (19), diet type (20,21), diet and ecology (22), ecological plasticity (23), social structure (24,25), and impact of captivity (26). Unfortunately, few studies have been designed to explore the impact of aging on gut microbiota in nonhuman primate models. Here, we explore gut microbiota differences related to age and sex in rhesus macaques housed at the Wisconsin National Primate Research Center (WNPRC). We hypothesize that, as in humans, aged rhesus macaques will have decreased intra-individual microbiota diversity and greater interindividual variation compared with younger animals. We further hypothesize that there will be a sex difference in microbial composition. However, we acknowledge that it is possible that both age-related and sex-related differences will be attenuated by the tight environmental control under which our animals are maintained.
Methods
Animals
In order to assess the effects of age and sex on gut microbiota, we evaluated fecal microbiome in 40 healthy rhesus macaques housed at the WNPRC at the University of Wisconsin, Madison. An equal number of males and females were studied across four age groups: young, young adult, adult, and old. All females were known to be nonpregnant. Subject numbers, ages, and body weights are given in Table 1. Animals more than 18 years of age constitute the aged group as at this age rhesus macaques are known to exhibit age-related phenotypes such as sarcopenia (18). For reference, the overall median life span of rhesus monkeys at the WNPRC is 27 years, and the maximum life span is 40 years (27).
Table 1.
Animal Demographics
| Animals | Young | Young Adult | Adult | Old | p |
|---|---|---|---|---|---|
| Total number of animals | 10 | 10 | 10 | 10 | |
| Female* | 5 (5, 0) | 5 (4, 1) | 4 (3, 1) | 6 (2, 4) | |
| Male* | 5 (5, 0) | 5 (5, 0) | 6 (6, 0) | 4 (4, 0) | |
| Age (mean ± SD, years) | 4.5 ± 1.1a | 10.2 ± 0.6b | 16.2 ± 0.9c | 23.3 ± 3.6d | <.0001 |
| Female | 4.7 ± 1.2a | 10.2 ± 0.5b | 16.7 ± 0.7c | 24.8 ± 3.6d | <.0001 |
| Male | 4.3 ± 1.1a | 10.1 ± 0.7b | 15.9 ± 0.9c | 21.0 ± 2.4d | <.0001 |
| Age range (years) | 3.1–6.0 | 9.4–11.1 | 15.2–17.5 | 18.4–31.6 | |
| Female | 3.1–6.0 | 9.8–11.1 | 15.7–17.2 | 21.5–31.6 | |
| Male | 3.2–5.9 | 9.4–11.0 | 15.2–17.6 | 18.4–23.2 | |
| Body weight (kg) | 7.4 ± 1.8a | 11.0 ± 3.9ab | 11.2 ± 2.6b | 11.0 ± 3.4ab | .020 |
| Female | 6.8 ± 1.5 | 8.2 ± 2.0 | 9.8 ± 2.6 | 9.0 ± 1.8 | .154 |
| Male | 7.9 ± 2.2a | 13.7 ± 3.4b | 12.2 ± 2.3ab | 13.9 ± 3.1b | .013 |
| Body weight range (kg) | 5.1–11.0 | 5.6–19.1 | 7.4–16.6 | 7.0–17.0 | |
| Female | 5.1–8.5 | 5.6–10.3 | 7.4–12.9 | 7.0–11.2 | |
| Male | 5.2–11.0 | 10.6–19.1 | 9.6–16.6 | 9.9–17.0 |
Notes: For pair housed animals, only one animal per pair was included in analyses. Age and body weight are from the time of sample collection. p Values are from ANOVA, values with different superscripts within rows are significantly different by Tukey HSD (p < .05).
*Number of animals (number of animals singly housed, number of animals pair housed).
Animals were housed either singly or socially in standard primate enclosures and grouped into rooms with auditory and visual contact with other animals. Animal rooms were maintained at 21–26°C with ~50%−65% relative humidity. Artificial room lighting was automatically controlled to provide 12-hour light and dark periods. Animals had continuous access to municipal water and were fed standard primate chow (Teklad 2050, Envigo, Madison, WI) ad libitum. Animals had no antibiotic exposure within 2 months of sample collection. The protocol (G005726) for this study was approved by the Animal Care and Use Committee of the Office of the Vice Chancellor for Graduate Research and Education of the University of Wisconsin–Madison, an AAALAC-accredited program.
Fecal Sample Collection and Bacterial 16S rRNA Sequencing and Processing
After cage-washing in the morning, animals were separated from cage-mates (if applicable) and observed until fecal samples were produced. These samples were then collected from the drop-pan below each cage, subsampled into prepared sterile bead beating tubes, and stored at −80°C until processing by a bead beating protocol for DNA isolation as previously described (5,28,29). The variable region V4 amplicon of the bacterial 16S rRNA gene was amplified using barcoded primers (515F: 5′-GTGCCAGCMGCCGCGGTAA-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′) and sequencing was performed using quality-filtered paired end Illumina sequencing reads using the Illumina MiSeq platform (28,29). All sequencing was performed at the same time. 16S rRNA sequences were aligned and those with 97% similarity were clustered into operational taxonomic units (OTUs) using the UCLUST algorithm (30) using QIIME (Quantitative Insights Into Microbial Ecology, version 1.9.1) software package (31) and assigned the lowest possible taxonomic classifications from the Greengenes reference database (v13.8) (32) using a naive Bayesian classifier requiring an 80% confidence score. Samples were rarefied to 9,439 reads/sample with singletons removed.
OTUs with <0.0005% of total sequence reads were filtered out from the data set to account for sequencing errors. OTUs were then grouped together to summarize taxon abundance at different hierarchical levels of classification. Diversity between samples, or beta diversity (β-diversity) was measured by unweighted Unifrac analysis (33). Diversity of bacterial “species” in a given sample type, or the alpha diversity (α-diversity), was calculated based on the entire data set including all OTUs, using Shannon’s diversity matrix as implemented in QIIME (31). This matrix measures compositional complexity based both on evenness (relative abundance of difference OTUs/species and their even distribution in a sample) and richness (number of OTUs/species present in a sample) (34) of the microbial ecosystem. In order to visualize the dissimilarity matrix between all samples, we performed principal components analysis (PCoA) by QIIME and 3D PCoA plots were generated using EMPEROR (35).
Statistical Analysis
Data were analyzed by age group (combining sexes) and sex (combining age groups) separately. For each of the studies (age, sex), all data were evaluated for normality and homogeneity of variance. Age and body weight were compared by ANOVA with post hoc testing (Tukey HSD) or unpaired Student’s t-tests as appropriate. α-diversity was tested between groups by nonparametric Kruskal–Wallis tests or Mann–Whitney test as appropriate based on the number of groups. Differences in microbial DNA populations at the phylum, class, and species levels, across age and sex were tested by nonparametric Kruskal–Wallis or Mann–Whitney tests, as appropriate based on the number of groups. Initially, OTUs of interest were identified as those having a p < .05. Final determination of significance was established after post hoc correction for false discovery rate (FDR) using established techniques (36). Following comparative analyses, Spearman correlation analyses were performed between body weight and OTUs identified as differing in relative abundance by age or sex.
Results
Impact of Age
Demographic data for the 40 animals evaluated in this study are presented in Table 1. Per the study design, age was different across the four groups (young, young adult, adult, old). Body weight in females did not differ across the age groups but in the males, animals in the young group weighed less than those in the young adult and old groups leading to an overall difference in body weight between the age groups.
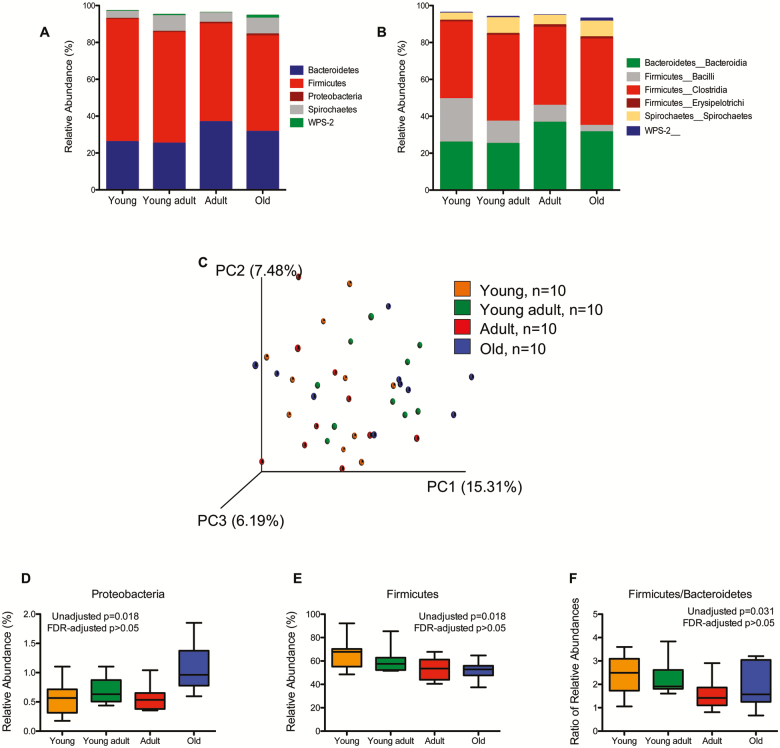
Figure 1 represents the overall composition of the fecal microbiomes in young, young adult, adult, and old at the phylum (Figure 1A) and class (Figure 1B) taxonomic level. PCoA revealed no differences in overall fecal microbiota community structure as determined by unweighted UniFrac (Figure 1C). At the phylum level, relative abundance of Proteobacteria (Figure 1D) and Firmicutes (Figure 1E) and ratio of Firmicutes to Bacteriodetes (F/B ratio) were significantly different between age groups though significance did not remain after correction for multiple comparisons by FDR. At the class level (Supplementary Table 1), relative abundance of Firmicutes_Bacilli decreased with each successively older group (p = .002) and relative abundance of Proteobacteria_Alphaproteobacteria and Proteobacteria_Betaproteobacteria increased with each successively older group (p = .038 and p = .012, respectively). Only differences in Firmicutes_Bacilli remained significant after correction for multiple comparisons by FDR. A total of 19 different species from the phyla Bacteroidetes, Firmicutes, and Proteobacteria were significantly different between age groups though none remained significant after correction for multiple comparisons by FDR (Supplementary Table 2). No correlations were found between body weight and OTUs identified as differing in relative abundance by age (data not shown).
Figure 1.
Taxonomic distribution of fecal microbiome of healthy young (3.1–6.0 years of age), young adult (9.4–11.1 years of age), adult (15.2–17.5 years of age), and old (18.4–31.6 years of age) rhesus macaques by phylum (A) and class (B). Individual phyla and classes are included in the graph only if the average abundance in at least one of the age groupings exceeded 1%. (C) Comparison of fecal microbiome ß-diversity (unweighted UniFrac) between different rhesus macaque age groups. (D–F) Microbial DNA populations differentially expressed between young (orange), young adult (green), adult (red), and old (blue) rhesus macaques at the phylum level. None remain significant after correction for multiple comparisons via false discovery rate. Box and whiskers represent the range of observed values.
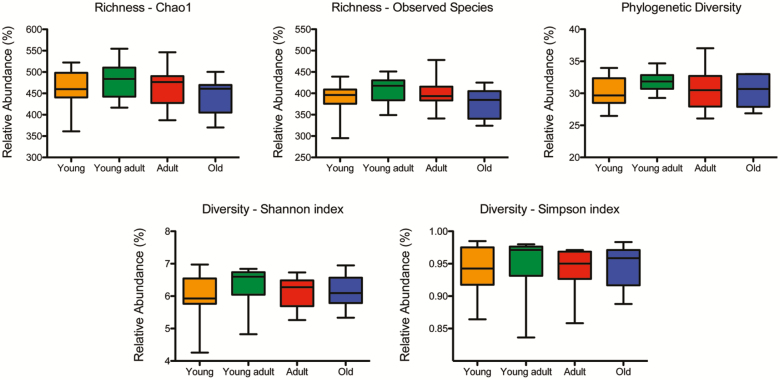
Figure 2 depicts α-diversity measurements, including richness (chao1 and observed species), phylogenetic diversity, and overall sample diversity measured according to Shannon and Simpson metrics. Measures of α-diversity did not differ between age groups.
Figure 2.
Comparison of α-diversity of the fecal microbiome between healthy young (orange), young adult (green), adult (red), and old (blue) rhesus macaques. Five indices were used to represent the richness (chao1, observed species), phylogenetic diversity, and sample diversity (shannon and simpson indices). Box whiskers indicate the range of observed values.
Impact of Sex
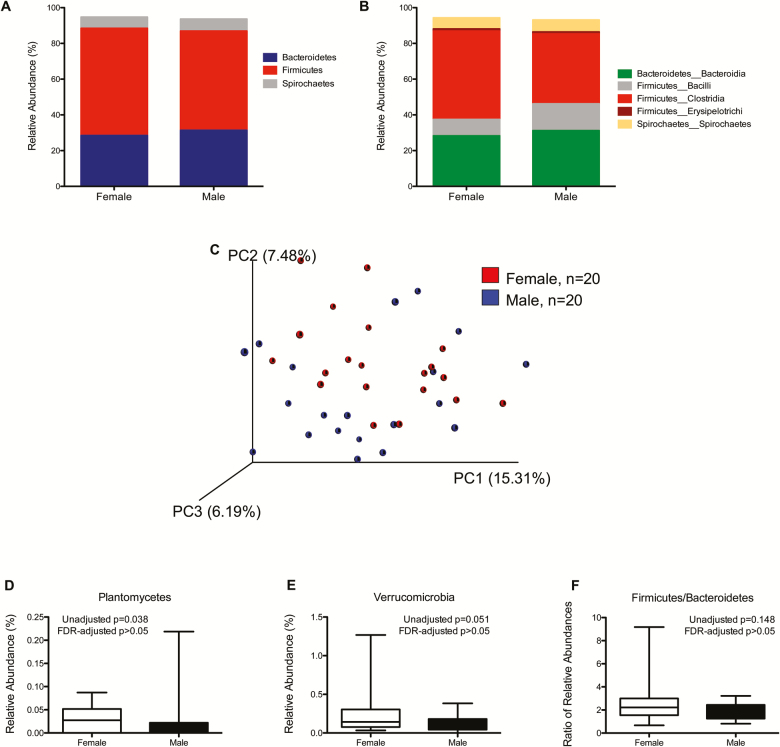
We evaluated a total of 20 males and 20 females. There were no differences in age between males and females. As expected for this sexually dimorphic species, males weighed more (p = .0006) than females.
Figure 3 represents the overall composition of the fecal microbiomes in females and males at the phylum (Figure 3A) and class (Figure 3B) taxonomic level. PCoA revealed no differences in overall fecal microbiota community structure as determined by unweighted UniFrac (Figure 3C). At the phylum level, relative abundance of Plantomycetes (Figure 3D) and Verrucomicrobia (Figure 3E) were significantly different between males and females though significance did not remain after correction for multiple comparisons by FDR. The F/B ratio of was not different between males and females. At the class level (Supplementary Table 3), relative abundance of Firmicutes_Clostridia and Verrucomicrobia_Verruco-5 were lower in males than females though significance disappeared after correction for multiple comparisons by FDR. A total of 23 different species from the phyla Actinobacteria, Bacteroidetes, Elusimicrobia, Firmicutes, Plantomycetes, and Verrucomicrobia were significantly different between males and females though none remained significant after correction for multiple comparisons by FDR (Supplementary Table 4). No correlations were found between body weight and OTUs identified as differing in relative abundance between males and females (data not shown).
Figure 3.
Taxonomic distribution of fecal microbiome of healthy female and male rhesus macaques by phylum (A) and class (B). Individual phyla and classes are included in the graph only if the average abundance in at least one of the sex groupings exceeded 1%. (C) Comparison of fecal microbiome ß-diversity (unweighted UniFrac) between female and male rhesus macaques. (D–F) Microbial DNA populations differentially expressed between female (red) and male (blue) rhesus macaques at the phylum level. None remain significant after correction for multiple comparisons via false discovery rate. Box and whiskers represent the range of observed values.
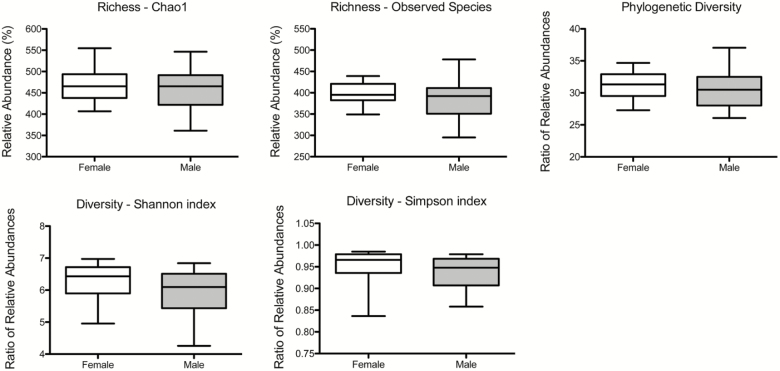
Figure 4 depicts α-diversity measurements, including richness (chao1 and observed species), phylogenetic diversity, and overall sample diversity measured according to Shannon and Simpson metrics. Measures of α-diversity did not differ between females and males.
Figure 4.
Comparison of α-diversity of the fecal microbiome between healthy female and male rhesus macaques. Five indices were used to represent the richness (chao1, observed species), phylogenetic diversity, and sample diversity (shannon and simpson indices). Box whiskers indicate the range of observed values. No significant differences were observed.
Discussion
The main purpose of this study was to understand the effect of aging on the fecal microbiota in rhesus monkeys. We found no differences between our age groups in α-diversity. At the phylum level, we found higher levels of proteobacteria in old monkeys and decreasing levels of Firmicutes with increasing age as seen in humans (8). We also found an age-related difference in the F/B ratio. Although unlike in many human studies, these differences did not remain significant after statistical correction for multiple correlations. The only age-related difference that remained significant was decreased Firmicutes_Bacilli with increasing age group which is contrary to what has been found in humans (8). We additionally examined our data for sex effects. Contrary to recent reports from human studies (37,38), we found no differences between male and female monkeys that remained significant after correction for multiple comparisons. However, the relatively small group sizes, particularly in the older ages and given the expected inter-individual variation, should be taken into account.
It is not clear why results in this study disagree with findings from human studies. It is possible that we did not see more age-related alterations in gut microbiota due to our level of control over diet and environment and the fact that we restricted our sampling to clinically healthy individuals. Firmicutes_Bacilli is known to participate in dietary and drug metabolism. Perhaps the decreased dietary variety and general lack of polypharmacy in our population may in part explain why our results do not mimic those in humans.
When studying aging microbiota it can not be ignored that older individuals often have a variety of comorbidities that are known to impact the gut microbiota. This likely leads to the fact that, as seen with the ELDERMET cohort, the microbiota of older adults is highly variable between individuals and becomes more diverse and variable with age (11). This large variation can be explained by a host of external factors, such as increasing gut dysbiosis with age together with the use of antibiotics, lack of nutrition (12), overall health status, living situation (13), and increased inflammatory status (8,39). A major benefit of exploring the gut microbiota in an animal model is the ability to control external factors which may explain the lack of an age-related effect on interindividual diversity in our study.
Within individuals, gut microbial diversity declines with age and its function in metabolism and regulation of the immune system is reduced. This gives the chance for opportunistic pathogens to invade and inflame the gut giving rise to various diseases (12). In a study of centenarians from Northern Italy, Biagi and colleagues found lower species diversity in the centenarians compared with younger adults along with specific changes within Firmicutes and enrichment of Proteobacteria which contains many opportunistic bacteria (8). These specific changes match what we observed in our aging study.
There is large variability in results from human studies exploring age-related differences in gut microbiota with some studies showing increased diversity and others showing decreased diversity (13). This lack of consensus is likely a result of the fact that gut microbiota diversity is inversely correlated with biological age (ie, overall health or frailty) as opposed to chronological age (14). Specifically, as biological age increases, overall gut microbiota richness decreases, while some microbial taxa associated with unhealthy aging emerge (15). This relationship with biological versus chronological age raises the question of appropriate study design. Are the longest-lived individuals (ie, super-centenarians) able to somehow counteract the negative effects of aging? Is it possible that the aging process is somehow different in these individuals; is successful aging different from normal aging? Our use of all clinically healthy animals for this study may have somewhat curtailed potential differences between biological and chronological age.
Studies specifically addressing the effect of aging on the gut microbiome in nonhuman primates are limited. The loss of mucosal barrier function with increasing age in old world monkeys has been documented (vervet: (40,41); baboon: (42)). The two vervet monkey studies (40,41) additionally assessed the fecal microbiome. As in our study, no age-related differences were found in either vervet monkey study. Similar to our study, these studies were also likely hampered by inclusion of a limited number of animals in the old group. Furthermore, the vervet studies included only females and animals were moved to new housing shortly before sampling began. Therefore, it is important to note that while our results are different from those in human studies, they do recapitulate the overall lack of difference in microbiome with aging found in another old world monkey species, the vervet monkey.
This study has several important limitations. We characterized only fecal-derived populations. It is known that throughout the length of the gut different regions harbor distinct microbial communities that are adapted to local conditions, such as tissue structure, host secretions, pH, and oxygen concentration. In this study, because we used easily accessible feces, we focus only on the distal part of the large intestine as a proxy for the entire GI tract. Furthermore, less than 10,000 reads/sample is low. At the time of sample analysis, this was the best technology that was reasonably available to us. We appreciate that this may have impacted species richness; however, we believe our depth of sequencing is adequate to infer relative abundance of members of the microbial community. Any future analyses of microbiome would use a higher number of reads. Although we have excellent diet and environmental control over our animals, there are differences in diet, treatment exposures, and health that were unavoidable. An advantage is that we have complete medical, treatment, and experimental histories available so we can utilize this information to provide better understanding of our data, and in a larger study, we would be able to use this information to segregate our data. Finally, some of the animals in the old group exceed median survival age for this species with some as old as the human equivalent of 90+ years of age. It cannot be ignored that these individuals may in fact represent “optimal agers” that have some degree of survivor benefit and therefore do not accurately represent normal aging. Moving forward, larger studies in this species including frailty and microbiome assessments will help clarify the relationship between biological age and gut microbiota and solidify the rhesus monkey as a model for integrating mechanisms of aging.
In conclusion, although our results do not recapitulate those from human studies, they do match the findings from what are, to the best of our knowledge, the only other studies that have explored the effect of aging on gut microbiota in captive old world monkeys. Given the abundance of information supporting the overwhelming impact of diet, health, and environment on gut microbiota composition, it is not surprising that under our well-controlled conditions we saw limited age effects and no sex differences. This does not detract from the use of the rhesus monkey model for explorations involving the gut microbiota. In fact, moving forward, larger studies in this species including frailty and microbiome assessments will help clarify the relationship between biological age and gut microbiota and solidify the rhesus monkey as a model for integrating mechanisms of aging.
Supplementary Material
Acknowledgments
Thank you to the animal care, veterinary, and research staff at the WNPRC. Additional thanks to Federico Rey, Chrissi Illgen, and Robert Kerby from the Department of Bacteriology, University of Wisconsin, Madison for initial sample preparation and analysis. J.A. and S.T.B. participated in study design, sample collection, and manuscript preparation. E.J.L. and W.J.V. performed data analysis and interpretation and participated in manuscript preparation. T.W.B. provided guidance on study design and data analysis and interpretation and participated in manuscript preparation. R.J.C. designed the study and had overall oversight of the study and manuscript preparation.
Funding
This work was supported by the National Institutes of Health (R01AG07831, P01AG011915, R01AG040178, R01AG047358, P51RR000167, P51OD011106, and UL1TR003096). This publication was made possible in part by the National Institutes of Health, National Center for Research Resources/Office of Research Infrastructure and Policy grants (P51RR000167 and P51OD011106) to the Wisconsin National Primate Research Center, University of Wisconsin–Madison. This publication was made possible by the University of Alabama at Birmingham Center for Clinical and Translational Science (UL1TR003096) from the National Center for Advancing Translational Sciences of the National Institutes of Health.
Conflict of Interest
None reported.
References
- 1. Zheng X, Wang S, Jia W. Calorie restriction and its impact on gut microbial composition and global metabolism. Front Med. 2018;12:634–644. doi: 10.1007/s11684-018-0670-8 [DOI] [PubMed] [Google Scholar]
- 2. Lu M, Wang Z. Microbiota and aging. Adv Exp Med Biol. 2018;1086:141–156. doi: 10.1007/978-981-13-1117-8_9 [DOI] [PubMed] [Google Scholar]
- 3. Chassaing B, Vijay-Kumar M, Gewirtz AT. How diet can impact gut microbiota to promote or endanger health. Curr Opin Gastroenterol. 2017;33:417–421. doi: 10.1097/MOG.0000000000000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Subramanian S, Huq S, Yatsunenko T, et al. . Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. . A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ussar S, Griffin NW, Bezy O, et al. . Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mabbott NA, Kobayashi A, Sehgal A, Bradford BM, Pattison M, Donaldson DS. Aging and the mucosal immune system in the intestine. Biogerontology. 2015;16:133–145. doi: 10.1007/s10522-014-9498-z [DOI] [PubMed] [Google Scholar]
- 8. Biagi E, Nylund L, Candela M, et al. . Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li H, Qi Y, Jasper H. Preventing age-related decline of gut compartmentalization limits microbiota dysbiosis and extends lifespan. Cell Host Microbe. 2016;19:240–253. doi: 10.1016/j.chom.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faith JJ, Guruge JL, Charbonneau M, et al. . The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claesson MJ, Cusack S, O’Sullivan O, et al. . Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(suppl 1):4586–4591. doi: 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vemuri R, Gundamaraju R, Shastri MD, et al. . Gut microbial changes, interactions, and their implications on human lifecycle: an ageing perspective. Biomed Res Int. 2018;2018:4178607. doi: 10.1155/2018/4178607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. An R, Wilms E, Masclee AAM, Smidt H, Zoetendal EG, Jonkers D. Age-dependent changes in GI physiology and microbiota: time to reconsider? Gut. 2018;67:2213–2222. doi: 10.1136/gutjnl-2017-315542 [DOI] [PubMed] [Google Scholar]
- 14. Claesson MJ, Jeffery IB, Conde S, et al. . Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 15. Kim S, Jazwinski SM. The gut microbiota and healthy aging: a mini-review. Gerontology. 2018;64:513–520. doi: 10.1159/000490615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhesus Macaque Genome Sequencing and Analysis Consortium; Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–2 34. doi: 10.1126/science.1139247 [DOI] [PubMed] [Google Scholar]
- 17. Zimin AV, Cornish AS, Maudhoo MD, et al. . A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colman RJ. Non-human primates as a model for aging. Biochim Biophys Acta Mol Basis Dis. 2018;1864(9 Pt A):2733–2741. doi: 10.1016/j.bbadis.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koo BS, Hwang EH, Kim G, et al. . Evaluation of fecal microbiomes associated with obesity in captive cynomolgus monkeys (Macaca fascicularis). J Vet Sci. 2019;20:e19. doi: 10.4142/jvs.2019.20.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ardeshir A, Narayan NR, Méndez-Lagares G, et al. . Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6:252ra120. doi: 10.1126/scitranslmed.3008791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagpal R, Wang S, Solberg Woods LC, et al. . Comparative microbiome signatures and short-chain fatty acids in mouse, rat, non-human primate, and human feces. Front Microbiol. 2018;9:2897. doi: 10.3389/fmicb.2018.02897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amato KR, Mallott EK, McDonald D, et al. . Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 2019;20:201. doi: 10.1186/s13059-019-1807-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trosvik P, Rueness EK, de Muinck EJ, Moges A, Mekonnen A. Ecological plasticity in the gastrointestinal microbiomes of Ethiopian Chlorocebus monkeys. Sci Rep. 2018;8:20. doi: 10.1038/s41598-017-18435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amaral WZ, Lubach GR, Proctor A, Lyte M, Phillips GJ, Coe CL. Social influences on Prevotella and the gut microbiome of young monkeys. Psychosom Med. 2017;79:888–897. doi: 10.1097/PSY.0000000000000454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2:e1500997. doi: 10.1126/sciadv.1500997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clayton JB, Vangay P, Huang H, et al. . Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. 2016;113:10376–10381. doi: 10.1073/pnas.1521835113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colman RJ, Anderson RM. Nonhuman primate calorie restriction. Antioxid Redox Signal. 2011;14:229–239. doi: 10.1089/ars.2010.3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kreznar JH, Keller MP, Traeger LL, et al. . Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep. 2017;18:1739–1750. doi: 10.1016/j.celrep.2017.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vogt NM, Kerby RL, Dill-McFarland KA, et al. . Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 2017;7:13537. doi: 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2017;2:e00073–17. doi: 10.1128/mSphereDirect.00073-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDonald D, Price MN, Goodrich J, et al. . An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1 [DOI] [PubMed] [Google Scholar]
- 35. Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 37. Dominianni C, Sinha R, Goedert JJ, et al. . Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10:e0124599. doi: 10.1371/journal.pone.0124599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saraswati S, Sitaraman R. Aging and the human gut microbiota—from correlation to causality. Front Microbiol. 2014;5:764. doi: 10.3389/fmicb.2014.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bischoff SC. Microbiota and aging. Curr Opin Clin Nutr Metab Care. 2016;19:26–30. doi: 10.1097/MCO.0000000000000242 [DOI] [PubMed] [Google Scholar]
- 40. Mitchell EL, Davis AT, Brass K, et al. . Reduced intestinal motility, mucosal barrier function, and inflammation in aged monkeys. J Nutr Health Aging. 2017;21:354–361. doi: 10.1007/s12603-016-0725-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson QN, Wells M, Davis AT, et al. . Greater microbial translocation and vulnerability to metabolic disease in healthy aged female monkeys. Sci Rep. 2018;8:11373. doi: 10.1038/s41598-018-29473-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tran L, Greenwood-Van Meerveld B. Age-associated remodeling of the intestinal epithelial barrier. J Gerontol A Biol Sci Med Sci. 2013;68:1045–1056. doi: 10.1093/gerona/glt106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.