Abstract
SARS-CoV-2 emerged in 2019 and has become a major global pathogen in an astonishingly short period of time. The emergence of SARS-CoV-2 also has been notable due to its impacts on individuals residing within skilled nursing facilities (SNFs) such as rehabilitation centers and nursing homes. SNF residents tend to possess several risk factors for the most severe outcomes of SARS-CoV-2 infection, including advanced age and the presence of multiple comorbidities. Indeed, residents of long-term care facilities represent approximately 40 percent of US SARS-CoV-2 deaths. To assess the prevalence and incidence of SARS-CoV-2 among SNF workers, determine the extent of asymptomatic infection by SARS-CoV-2, and provide information on the genomic epidemiology of the virus within these unique care settings, we sampled workers weekly at five SNFs in Colorado using nasopharyngeal swabs, determined the presence of viral RNA and infectious virus among these workers, and sequenced 48 nearly complete genomes. This manuscript reports results from the first five to six weeks of observation. Our data reveal a strikingly high degree of asymptomatic infection, a strong correlation between RNA detection and the presence of infectious virus in NP swabs, persistent RNA in a subset of individuals, and declining incidence over time. Our data suggests that asymptomatic individuals infected by SARS-CoV-2 may contribute to virus transmission within the workplace.
Introduction
The COVID-19 pandemic has resulted in disproportionally high morbidity and mortality among residents in skilled nursing facilities (SNFs). As of June 2, 2020, the Centers for Medicare and Mediciaid Services reported over 30,000 deaths due to COVID-19 in long-term care facilities in the US, representing 42% of COVID-29-related US deaths (Nursing Home COVID-19 Public File Data.CMS.gov). In six states, deaths in long-term care facilities accounted for over 50% of all COVID-19 deaths (Delaware, Massachusetts, Oregon, Pennsylvania, Colorado, and Utah). The high burden of COVID-19 within SNFs is principally due to the risk profile of many residents, which includes advanced age and the presence of severe comorbidities (1). Accordingly, strategies to mitigate SARS-CoV-2 transmission to SNF residents have included restrictions on visitation, cessation of group activities and dining, and confinement to individual living quarters. While SNF residents are largely isolated, SNF employees are permitted to enter resident rooms provided they have passed a daily screening process for fever, COVID-19 respiratory symptoms or known exposure. However, a significant fraction of individuals infected with SARS-CoV-2, the causative agent of COVID-19, have a lengthy latency period prior to exhibiting COVID-19 symptoms, and many remain asymptomatic throughout the course of infection (2, 3). Pre-symptomatic and asymptomatic SNF workers are a potential source of unrecognized transmission within SNFs and are thus an attractive focus for interventions directed at suppressing transmission within these facilities.
To date, there have been no studies focused on longitudinal surveillance of asymptomatic workers within skilled nursing facilities. Therefore, we assessed SARS-CoV-2 infection among employees at five SNFs in Colorado. Workers were enrolled into the study and sampled by nasopharyngeal (NP) swab weekly for five or six consecutive weeks. Swabs were assayed for virus infection by qRT-PCR and plaque assay, and individuals with evidence of infection were instructed to self-quarantine for ten days. Using data on worker infection, site-specific prevalence at study onset and incidence rate over time was calculated. Viral genomes also were sequenced to assess viral genetic diversity within and between SNFs. Our results document a surprising degree of asymptomatic infection among healthy workers, and extreme variation in the prevalence and incidence of infections between different SNFs. We observed that the median number of consecutive positive weekly tests was two, indicating that RNA was present in the nasopharynx of most individuals for at least eight days, however some individuals had viral RNA in their nasopharynx for over five weeks. A small number of individuals had RNA reappear in the nasopharynx after apparent clearance. Sequencing studies lend limited support to the observation that transmission may occur within SNFs and, combined with the epidemiologic and other data provided here, highlight the importance of testing and removing positive workers from contact with vulnerable SNF residents. Data obtained from longitudinal surveillance studies such as this ongoing effort will provide crucial information about infectious disease transmission dynamics within complex workforces and inform best practices for preventing or mitigating future COVID-19 outbreaks within SNFs.
Materials and Methods
Study sites
Five SNF facilities in Colorado were chosen to participate in the SARS-CoV-2 surveillance project. Weekly nasopharyngeal (NP) swabs were collected for a five to six week period on 454 consented individuals. Participants were asked to provide their job code but were otherwise de-identified to the investigators.
This study was reviewed and approved by the Colorado State University IRB under protocol number 20–10057H. Participants provided consent to participate in the study and were promptly informed of test results and, if positive, instructed to self-isolate for a period of ten days. Return to work also required absence of fever or other symptoms for the final three days of isolation.
Sample Collection
Nasopharyngeal swabs were performed by trained personnel at participating facilities on consented staff members. Swabs were placed in a 15ml conical tube containing 3ml viral transport media (Hanks Balanced Salt Solution w/ calcium and magnesium, w/o phenol red [HBSS; Fisher Scientific], 2% FBS [Atlas Biologicals], 50mg/ml gentamicin [VWR], 250ug/ml Amphotericin B/Fungizone [Gemini Bio]). Samples were returned to the laboratory on ice.
RNA Extraction
Tubes containing NP swabs were vortexed for 10sec, centrifuged at 1282 RCF for 5min at 4°C, and swabs removed. RNA was extracted using the Omega Mag-Bind Viral DNA/RNA 96 Kit with 200ul of input sample on a KingFisher Flex magnetic particle processor according to the manufacturers’ instructions.
qRT-PCR
One-step reverse transcription (RT) and PCR reaction was performed using the EXPRESS One-Step SuperScript qRT-PCR Kit (ThermoFisher Scientific) in a 20ul final reaction volume per the manufacturer’s instructions. Primer/probe sets for SARS-CoV-2 are as described elsewhere [(4) and CDC diagnostic testing guidelines: https://www.fda.gov/media/134922/download) and were obtained from IDT. A primer/probe set for human RNase P transcript served as a control for RNA quality (not shown). RNA standards for SARS-CoV-2 nucleocapsid (N) and envelope (E) were kindly provided by Dr. Nathan Grubaugh, and served as positive controls. 96-well PCR plates were prepared on ice and centrifuged at 1282 RCF for 2min at 4°C. Plates were run on a QuantStudio3 using the following cycling conditions: Reverse transcription at 50°C for 15 minutes, followed by a single inactivation step (95°C for 3 minutes); 40 cycles alternating between 95°C for 5 seconds and 60°C for 30 seconds completed the reaction. Specimens with a cycle threshold (CT) less than 38 were considered positive. Samples were initially screened with an N1 primer/probe set as described in the US CDC diagnostic guidelines. If a positive or inconclusive result was obtained, the sample was retested with both N2 and E primer/probe sets(4). Specimens positive by two or more primer sets were considered RNA positive for SARS-CoV-2.
Plaque assay for infectious virus
Plaque assays were performed on African Green Monkey Kidney (Vero) cells (ATCC CCL-81) according to standard methods (5). Briefly, 250 uL of qRT-PCR positive specimen was inoculated into nearly confluent cell monolayers. After incubation, cells were provided with a tragacanth semi-solid overlay, and fixed and stained after two days of incubation with 30% ethanol and 0.1% crystal violet. Plaques were counted manually.
Incidence Estimation
The rate at which workers acquired infections was estimated as the number of new infections per 100 workers per week at each facility and was estimated for weeks 2 through 6. A worker was classified as having an incident infection if they tested positive for the first time following a negative test one-week prior (or two weeks prior if they were not surveyed one-week prior) and if they had not previously tested positive for SARS-CoV-2 in our surveys. The population at risk includes all workers who had not yet been infected and with a negative test in the past week (or two weeks prior if not tested the prior week).
Next-generation sequencing library preparation for positive samples
Viral RNA from positive patient samples was prepared for next-generation sequencing. Briefly, cDNA was generated using SuperScript IV Reverse Transcriptase enzyme (Invitrogen) with random hexamers. PCR amplification was performed using ARTIC network (https://artic.network/)V2 tiled amplicon primers in two separate reactions by Q5 High-fidelity polymerase (NEB) essentially as previously described(6). First-round PCR products were purified using Ampure XP bead (Beckman Coulter). Libraries were prepared using the Nextera XT Library Preparation Kit (Illumina) according to manufacturer protocol. Unique Nextera XT i7 and i5 indexes for each sample were incorporated for dual indexed libraries. Indexed libraries were again purified using Ampure XP bead (Beckman Coulter). Final libraries were pooled and analyzed for size distribution using the Agilent High Sensitivity D1000 Screen Tape on the Agilent Tapestation 2200, final quantification was performed using the NEBNext® Library Quant Kit for Illumina® (NEB) according to manufacture protocol. Libraries were then sequenced on the Illumina MiSeq V2 using 2 × 250 paired end reads.
Deep sequencing analysis
Next-generation sequencing data were processed to generate consensus sequences for each viral sample. MiSeq reads were demultiplexed, quality checked by FASTQC, paired-end reads were processed to remove Illumina primers and quality trimmed with Cutadapt, duplicate reads were removed. Remaining reads were aligned to SARS-CoV-2 reference sequence by Bowtie2 (GenBank: MT020881.1). Alignments were further processed, and quality checked, using Geneious software, consensus sequences were determined and any gaps in sequences were filled in with the reference sequence or cohort specific consensus sequence. Consensus sequences were aligned in Geneious and a neighbor-joining tree generated with the Reference sequence as an outgroup and 1000 bootstrap replicates.
Results
SARS-CoV-2 prevalence and incidence in five SNFs
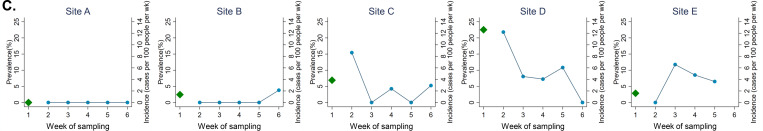
Employees at five SNFs throughout Colorado were tested weekly for SARS-CoV-2 viral RNA (vRNA) for a total of five or six weeks via NP swab. Staff included nursing, administrative, maintenance and other professions. A mean of 75 individuals per facility were tested weekly (range 29–115) with varying viral RNA levels within NP swabs (Fig. 1A). The percentage of NP swabs that tested positive for viral RNA each week varied considerably by facility, but showed a general downward trend over the course of the study period (Fig. 1B). SARS-CoV-2 infection prevalence during the first week of testing, and the incidence of infections in subsequent weeks also varied widely between facilities (Fig. 1C and Table A1). Staff at Site A remained uninfected throughout the entire six week study period. In contrast, 22.5% of workers at site D had prevalent infections at the start of the study and incidence was high initially (12.2 per 100 workers per week), declining over time. At site C, initial infection prevalence was lower (6.9%) and the incidence declined to zero by week 3. However, two facilities with low prevalence in week 1 (sites B and E) saw an increase in cases – including, at site B, incident infections detected after four weeks of no infections. Infections were observed in workers across all job types, including roles with typically high patient contact (e.g. nursing) and low patient contact (e.g., maintenance) (Table A2).
Figure 1. SARS-CoV-2 infection rates across five Colorado facilities.
A) Presence of viral RNA in all samples tested during study time period. Nasopharyngeal swabs were tested for the presence of SARS-CoV-2 N1 transcripts by qRT-PCR. CT represents PCR cycle threshold, n indicates number of samples tested each week. Dotted line indicates limit of detection for qRT-PCR. B) Percent of samples that tested positive for N1 weekly from each site. Calculated as number of samples with N1 CT<40, divided by total number of samples tested. C) Prevalence of SARS-CoV-2 among facility workers during the first week of surveillance (green triangles), and the incidence of new cases in following weeks (blue circles). Incident cases were defined as individuals with a new positive N1 RNA test who had tested negative for viral RNA one or two weeks prior, and had not previously tested positive for SARS-CoV-2. Not shown are prevalent infections among workers tested for the first time in week 2 (1 at site B, 3 at site C, 5 at site D, 1 at site E) or week 3 (1 at site C).
Infectious SARS-CoV-2 in nasopharyngeal swabs
All NP swabs with detectable SARS-CoV-2 N1 vRNA were assayed for N2- and E-containing viral transcripts and evaluated for the presence of infectious virus by plaque assay (Fig. 2). We observed high concordance between SARS-CoV-2 viral RNA regardless of genome region assayed (N1, N2 or E) (Fig. 2A). N1 viral RNA level was positively correlated with the amount of infectious virus (Fig. 2B) in swab material (least squares linear regression R2=0.7885), demonstrating the virus within these individuals is infectious.
Figure 2. Viral RNA and infectious virus.
Samples with detectable SARS-CoV-2 N1 RNA were evaluated for A) viral N2 and E-containing transcripts via qRT-PCR (CT represents PCR cycle threshold) and B) infectious virus via standard plaque assay (PFU = plaque forming units). Dashed lines represent limits of detection for each assay. B) Solid line represents least squares linear regression (R2=0.7885).
Levels of viral RNA tend to decline over the duration of infection and correspond to low levels of infectious virus
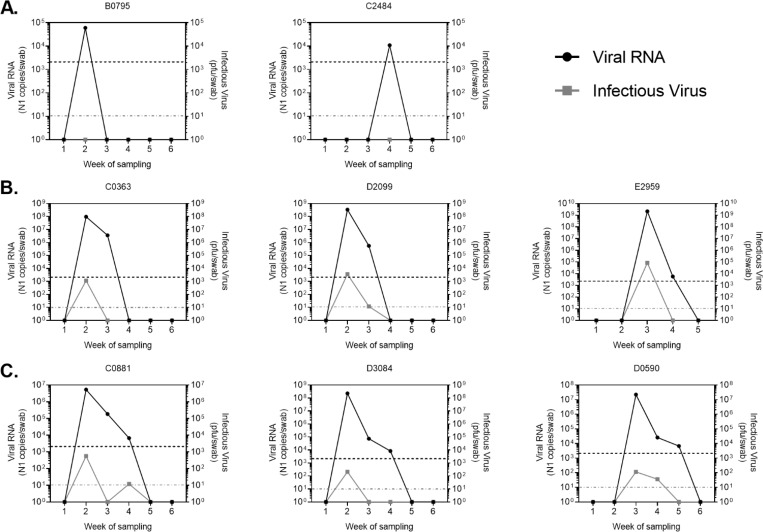
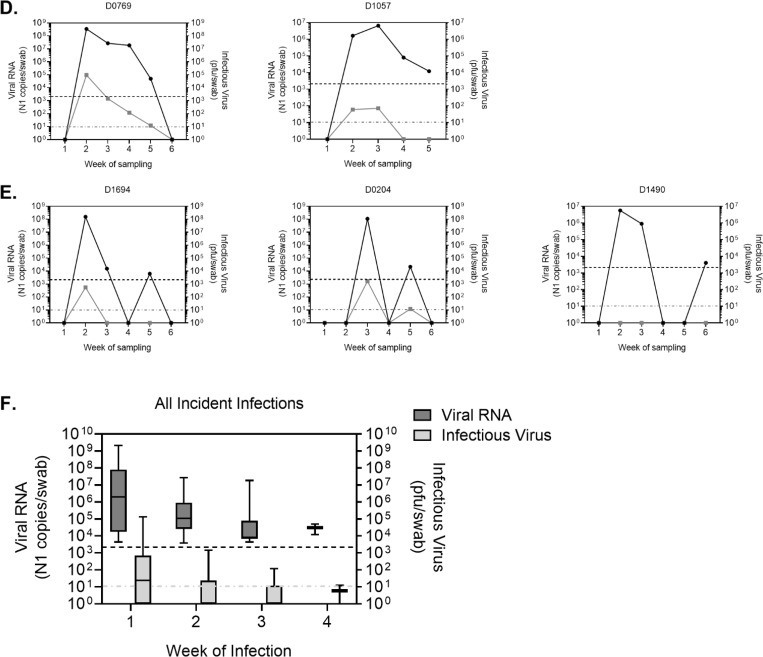
Within the study period, incident infections varied in length from one to four weeks (Fig. 3A–D), as determined by detection of viral RNA via qRT-PCR for the SARS-CoV-2 N1 gene. Levels of viral RNA were generally highest during the first week of infection and declined in subsequent weeks (Fig. 3F). Infectious virus was detected in individuals with high levels of viral RNA and also declined over the course of infection. In general, infectious virus was not detected in individuals with less than 100,000 N1 RNA copies/swab (Fig. 3 and 2B).
Figure 3: RNA and viral loads in individuals sampled repeatedly.
Viral RNA levels as determined by qRT-PCR amplification of N1 gene (left y-axis) and infectious virus as determined by plaque assay (right y-axis) for a subset of subjects. Viral RNA was detectable for one (A), two (B), three (C) or four (D) consecutive weeks. E) Representative individuals in which viral RNA was detected again after a period of negative tests. F) Average viral RNA levels and infectious virus (+/− SD) for all incident infections during first, second, third and fourth week of consecutive infection. Viral RNA and infectious virus levels were calculated per nasopharyngeal swab. Letters at the beginning of graph titles indicate site of origin. Black dotted line is the limit of detection for viral RNA. Grey dotted line is the limit of detection for infectious virus. pfu, plaque forming units.
Six individuals exhibited two positive tests, separated by a period of negative tests (Fig. 3E). In these individuals, initial infections were typically followed by a period of 1–2 weeks during which viral RNA was undetectable. Viral RNA was then detected a second time, usually for just one week. These resurgences in viral RNA were normally associated with no, or very low levels of infectious virus. RNA quality was evaluated for the interim negative tests and was found to be within acceptable parameters (not shown).
SARS-CoV-2 sequencing
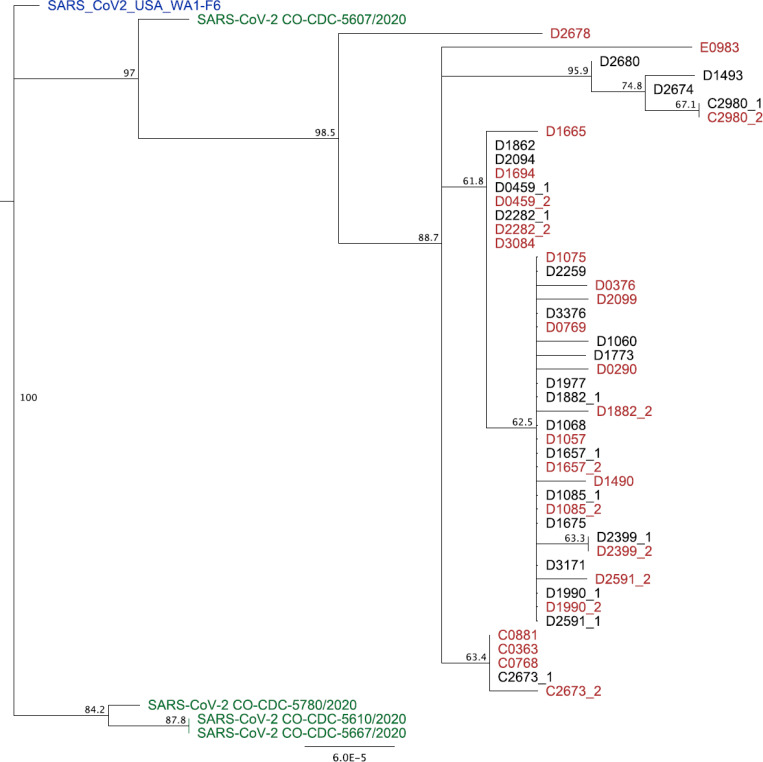
48 partial genome sequences were obtained over the first two weeks of observation. Mean genome coverage was 29,268nt (range = 27,656 to 29,831) and mean coverage depth was 621 reads per position (range = 376 – 2,138). Gaps in sequencing alignment due to ARTIC V2 primer incompatibilities were filled in with the reference strain MT020881.1 pending additional sequencing. Once complete, these sequences will be deposited into NCBI. The resulting NJ tree obtained from these 48 sequences were aligned to a reference strain from early in the US outbreak and to four strains collected from Colorado. The tree was reasonably clearly resolved into a number of clusters with moderate bootstrap support (i.e. greater than 50%). These included two major clusters that were composed exclusively of sequences obtained from individuals employed at the same SNF (Fig. 4). Thirty-six sequences derived from 31 individuals from Site D formed a single cluster apparent in the lower part of the tree. Five sequences from four individuals from Site C similarly clustered in our preliminary analysis. In contrast, the remaining seven sequences from six individuals did not tend to associate with others from the same facility. Three different facilities are represented in this group of sequences. Finally, we sequenced SARS-CoV-2 from ten individuals on two successive weeks. In general, sequences from the same individuals were identical to, or very closely related to, those collected previously from that individual (e.g. C2980_1 and C2980_2). Some evidence for mutation accumulation was detected in, for example, C2673_1 and C2573_2, as well as D1882_1 and D1882_2.
Figure 4. Phylogenetic analysis of 48 SARS-CoV-2 nearly complete genomes collected during the first two weeks of observation.
Neighbor-Joining tree constructed using Tamura-Nei distance model including both transitions and transversions in Geneious Prime. Numbers at the nodes indicate bootstrap confidence based on 1000 replicates. Distance matrix was computed, and the tree was visualized, in Geneious Prime. Letters at the beginning of taxon names indicate site of origin. “_1” and “_2” indicate that sequences were derived from the same individual, with “_1” collected in week 1 and “_2” collected in week 2 of sampling. Reference sequences and four Colorado-derived sequences were obtained from NCBI.
Discussion
SNFs, including nursing homes, residential treatment facilities and other long-term care providers are increasingly recognized as key venues for SARS-CoV-2 transmission due to the vulnerable populations that tend to inhabit them. Due to their disproportionate contribution to the burden of COVID-19 mortality, they also represent an attractive target for surveillance testing and interventions that may include removing SARS-CoV-2 positive staff from the workplace. Therefore, we longitudinally sampled asymptomatic workers at five SNFs in Colorado to determine the proportion of workers at these facilities who had SARS-CoV-2 RNA in their nasopharynx, and continued weekly testing as they self-isolated for ten days. Return to work also required absence of fever for the final three days of isolation, without antipyretic use. Individuals who continued to test positive after two weeks were notified and recommended to continue self-isolation until a negative test result was returned. Our data clearly demonstrate the potential for large numbers of workers at SNFs to be asymptomatically infected and for the concentration of infections to vary widely across facilities. One facility never had a single worker test positive, while otfhers had up 22.5% of workers with SARS-CoV-2 RNA during the first week of surveillance. Infections varied considerably over time. The steady declines in the incidence of infections in staff in the two facilities with the highest initial infection prevalence is encouraging and hints at the potential impact of worker screening programs. However, the detection of incident infections at facility B, after four weeks of negative tests underscores the on-going threat of infections in worker populations. Notably, participation in our sampling scheme was high, with approximately 85% of workers from each facility being sampled each week. These results clearly demonstrate that asymptomatically infected workers may be common in particular SNFs.
Because qRT-PCR detects viral RNA, not infectious virus, it may be that RNA-positive workers are not infectious to others, despite high levels of viral RNA. This could be attributable to the presence of free RNA (i.e. RNA that is not packaged into virus particles) or to antibodies within the mucosa that neutralize virus infectivity. Therefore, we tested NP swab samples for the presence of infectious virus via plaque assay. Importantly, we found that viral RNA was strongly positively correlated with infectious virus. In samples with high levels of viral RNA (N1 CT<30), infectious virus tended to be present, whereas lower viral RNA levels often had undetectable levels of infectious virus. Because plaque assays have lower sensitivity than qRT-PCR, it is unsurprising that samples with fewer than ~1000 RNA copies tended to have undetectable levels of infectious virus. Moreover, our data supports the observation that asymptomatic workers can harbor high levels of infectious virus within their mucosa and may therefore contribute to transmission of SARS-CoV-2.
The longitudinal design of this study permitted characterization of asymptomatic individuals over time, including several who were vRNA and/or plaque assay positive for one, two, three or four consecutive weeks. We also observed individuals who were vRNA positive, then negative, then again became vRNA positive. While it is possible that these individuals were re-infected with SARS-CoV-2 after clearing their initial infection, we find that unlikely(7). Instead, this phenomenon may be due to host factors that led to suppression of viral replication in the nasopharynx, or an NP swab that failed to capture virus. It is also unlikely that the intervening negative tests in these individuals were due to poor RNA quality, because all samples were tested for human RNase P (CDC diagnostic guidelines) and had comparable levels across all samples. Sequencing of the viruses from these individuals, will help determine the likelihood of re-infection versus host factor activity.
Viral RNA and infectious virus levels tended to peak on the first week of infection and decline thereafter, with a few exceptions (Fig. 3C & D, individuals C0881 and D1057). Some individuals however had infectious virus for multiple consecutive weeks. These data highlight the heterogeneity in human SARS-CoV-2 infection, and the need to further understand host and viral factors that permit varying lengths of infection, often in the complete absence of symptoms.
Sequencing of virus genomes also provided insights into SARS-CoV-2 transmission in our study population. Our data encompasses a sample of 48 genomes obtained during the first two weeks of observation (Site D is most highly represented because it had the highest number of SARS-CoV-2 cases during the first two weeks). Sequences from our study were compared to a strain sequenced during the early phase of the COVID-19 outbreak in the US, and to the four other SARS-CoV-2 sequences currently available from Colorado. The most notable feature of the phylogenetic tree is the fairly clear and consistent clustering of virus sequences by facility. This type of clustering could be due to transmission within staff at the facility, or from a shared community source outside of the workplace. For example, it may be that workers at these facilities socialize frequently outside of work or reside in close proximity, and that transmission occurred during non-work-related activities. Sampling in the workplace would therefore represent the distribution of genomes in the community and not work-related transmission. While we cannot rule out this possibility, it seems more likely transmission occurred within the workplace. Community transmission seems more likely to produce clusters that are not associated with a given facility, which is not what we have observed most prominently in this data thus far. Our sequencing results therefore are consistent with workplace transmission of SARS-CoV-2, but we cannot rule out the possibility that transmission occurred elsewhere. Additional data on the degree of viral genetic diversity in the larger community would add significant power to our ability to discriminate between these two non-mutually exclusive scenarios.
Overall, our study highlights the high SARS-CoV-2 infection rates within asymptomatic individuals at a high-transmission risk/spread setting. Identifying, and removing these infected and infectious individuals from the facility, provides a way to reduce transmission and potential outbreaks. While our work focused on skilled nursing facilities, this approach could be applied to other high-risk settings (correctional facilities, factories, etc.).
Acknowledgements
This work was supported by funds donated by the Colorado State University Colleges of Health and Human Sciences, Veterinary Medicine and Biomedical Sciences, Natural Sciences, and Walter Scott, Jr. College of Engineering, and the Colorado State University Columbine Health Systems Center for Healthy Aging. KQ was supported by a fellowship from the National Institute of Allergy and Infectious Diseases, National Institutes of Health under grant number F32AI150123-01. The authors also gratefully acknowledge the participation of the workers in the facilities that participated in this study, without which it could not have been completed.
Appendix
Table A1.
The prevalence of infections at enrollment and the incidence of infections over time, by site.
| Site | Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | |
|---|---|---|---|---|---|---|---|
| # tested | Prevalence (%) | Incidence* | Incidence* | Incidence* | Incidence* | Incidence* | |
| A | 86 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| B | 82 | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 2.1 |
| C | 29 | 6.9 | 8.6 | 0.0 | 2.4 | 0.0 | 2.9 |
| D | 111 | 22.5 | 12.2 | 4.5 | 4.1 | 6.1 | 0.0 |
| E | 70 | 2.86 | 0.0 | 6.6 | 4.8 | 3.6 | . |
Incidence is estimated as the number of new infections per week per 100 workers. A worker was classified as having an incident infection if it was their first positive test and they had a negative test one week prior (or two weeks prior if not tested one week prior).
Table A2.
The distribution of infections by job code among workers at long-term care facilities.
| Job code | Num tested | % positive* |
|---|---|---|
| Nursing | 140 | 25.0 |
| Housekeeping | 90 | 8.9 |
| Administration | 41 | 14.6 |
| Dietary | 29 | 24.1 |
| Activities | 15 | 40.0 |
| Maintenance | 10 | 50.0 |
| Social services | 10 | 10.0 |
| Therapy | 10 | 10.0 |
| Other | 6 | 16.7 |
Analysis looks at the percent of workers that tested positive at least once during the 5–6 week study period. Analysis is limited to the four sites where COVID-19 was detected (B, C, D, E).
References Cited
- 1.Gorges RJ, Sanghavi P, Konetzka RT. 2019. A National Examination Of Long-Term Care Setting, Outcomes, And Disparities Among Elderly Dual Eligibles. Health Aff (Millwood) 38:1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, Wang M. 2020. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA doi: 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Hu Z, Yi Y, Shen H. 2020. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 63:706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders D, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weger-Lucarelli J, Duggal NK, Bullard-Feibelman K, Veselinovic M, Romo H, Nguyen C, Ruckert C, Brault AC, Bowen RA, Stenglein M, Geiss BJ, Ebel GD. 2016. Development and Characterization of Recombinant Virus Generated from a New World Zika Virus Infectious Clone. J Virol doi: 10.1128/JVI.01765-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, du Plessis L, Liu Z, Hill V, Kang M, Lin H, Sun J, Francois S, Kraemer MUG, Faria NR, McCrone JT, Peng J, Xiong Q, Yuan R, Zeng L, Zhou P, Liang C, Yi L, Liu J, Xiao J, Hu J, Liu T, Ma W, Li W, Su J, Zheng H, Peng B, Fang S, Su W, Li K, Sun R, Bai R, Tang X, Liang M, Quick J, Song T, Rambaut A, Loman N, Raghwani J, Pybus OG, Ke C. 2020. Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 181:997–1003 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victor Okhuese A. 2020. Estimation of the Probability of Reinfection With COVID-19 by the Susceptible-Exposed-Infectious-Removed-Undetectable-Susceptible Model. JMIR Public Health Surveill 6:e19097. [DOI] [PMC free article] [PubMed] [Google Scholar]