Abstract
The lipid headgroup plays an important role in the association of polymers with lipid bilayer membranes. Herein, we report how a glycerol headgroup versus a choline headgroup affects the interaction of poly(ethylene oxide)-b-poly(propylene oxide) (PEO-PPO) block copolymers with lipid bilayer vesicles. Unilamellar vesicles composed of phosphatidylcholine and phosphatidylglycerol at various molar ratios were used as model membranes. The interactions between the block copolymers and lipid bilayers were quantified by pulsed-field gradient nuclear magnetic resonance (PFG-NMR) based on the distinctly different mobilities of free and bound polymers. All the investigated polymer species showed significantly higher binding with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG) liposomes than with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes, indicating stronger association with the glycerol headgroup compared to the choline headgroup. This effect did not become significant until the composition of mixed POPC/POPG liposomes contained more than 20 mol %POPG. A plausible explanation for the enhanced polymer binding with POPG invokes the role of hydrogen bonding between the glycerol headgroup and the ether moieties of the polymers.
Graphical abstract
INTRODUCTION
The major components of plasma membranes are phospholipids which form the bilayer scaffold separating the internal content of living cells from the exterior matrix.1,2 Phospholipids are amphiphilic molecules generally comprising two hydrocarbon chains and a polar headgroup consisting of a phosphate group and a functional group (e.g., choline, ethanolamine, serine, glycerol, and inositol).2 Headgroup chemistry, including electrostatic characteristics, can strongly affect the phospholipid membrane dipole potential,3 head-group conformations,4 phase transitions,5–7 raft formation,8,9 and interactions with drug molecules, polymers, peptides, and proteins.10–17 Triblock copolymers composed of poly(ethylene oxide) (PEO; A) and poly(propylene oxide) (PPO; B) in an A-B-A architecture are commercially known as Pluronics. They have shown great potential in cell membrane stabilization and permeabilization, depending on the hydrophilic-hydrophobic balance of the polymer.18–26 Exploring the influence of the lipid headgroup composition on the interactions of PPO-PEO block copolymers with lipid bilayers will enable a more comprehensive understanding of the mechanism of polymer-membrane association from a membrane perspective and help improve the design of model membrane systems.
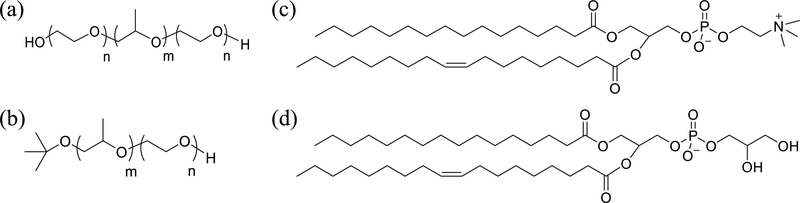
Herein, we report an investigation of the effect of lipid headgroup composition on polymer-membrane association, by quantifying the block copolymer binding to liposomes composed of phospholipids with two different types of headgroups. Narrowly distributed unilamellar liposomes were selected as model membranes, the preparation of which gives precise control over the lipid bilayer composition and size. Polymers involved in this study include commercial Pluronics and laboratory-synthesized diblock analogs, and their chemical structures are shown in Scheme 1a,b, respectively. Polymer binding to liposomes was determined by pulsed-field gradient nuclear magnetic resonance (PFG-NMR) based on the distinctly different diffusivities of free and bound polymers.27,28 Lipid headgroup composition was manipulated by mixing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Scheme 1c) with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG; Scheme 1d) at various molar ratios to compare the effect of glycerol headgroups with choline headgroups. Pure POPC liposomes were used as controls because phosphatidylcholine is one of the major components found in eukaryotic cell membranes and is widely used as the basic constituent in model membranes.29–32 POPG, on the other hand, is a relatively minor component in eukaryotic cell membranes and is mostly found in bacterial cell membranes.33 Nevertheless, given that POPG has the same phase transition temperature and almost the same chemical structure except for the headgroup as POPC, it can be considered as a simple model to study how the presence of net charge and hydroxyl groups affect membrane interactions with the copolymers. It has been reported that the net charge and hydroxyl groups carried by the headgroup of POPG play important roles in headgroup-headgroup interactions, which could change the orientation and conformation of the headgroups, as well as the area per lipid.34–37 The results obtained from this study demonstrate the critical role that lipid headgroup composition plays on polymer-membrane association and provides guidance to future studies using more complicated membrane compositions than a two-component membrane system.
Scheme 1.
Chemical Structures of (a) Pluronic, (b) a Diblock Polymer with a tert-Butyl Endgroup on the PPO Block, (c) Phosphatidylcholine, and (d) Phosphatidylglycerol
EXPERIMENTAL SECTION
Materials.
Pluronics F68, P84, P103, P105, F108, and F127 were provided by BASF. Ethylene oxide (EO, ≥99.5%), propylene oxide (PO, ≥99%), potassium, naphthalene, potassium tert-butoxide, n-butyllithium, 18-crown-6 ether, sodium chloride (NaCl, ≥99.0%), silver trifluoroacetate (AgTFA), and α-cyano-4-hydroxycinnamic acid (≥98%), were purchased from Sigma-Aldrich and used as received. POPC and POPG in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL) and used as received. Deuterium oxide (D2O, 99.9 atom % D) and chloroform-d (CDCl3, 99.8 atom % D + 0.05% V/V TMS) were purchased from Cambridge Isotope Laboratories, Inc. Ultrapure water with a resistivity of 18.2 MΩ·cm was obtained from a Millipore Direct Q-3 water system (EMD Millipore, Billerica, MA).
Polymer Synthesis and Characterization.
Diblock polymers tPPO14-PEO46 and tPPO29-PEO68 were synthesized by anionic polymerization as described previously,27 where the subscripts represent the average number of repeat units in the PPO and PEO blocks and “t” signifies a tert-butyl end group on the PPO block, which originates from the tert-butyl potassium initiator used for the PPO synthesis. The number-average molecular weight (Mn) and dispersity (Đ) of commercial triblock Pluronics and laboratory-synthesized diblocks were characterized by matrix-assisted laser desorption ionization (MALDI) mass spectroscopy (AB SCIEX TOF/TOF 5800). The weight fraction of PEO (wPEO) was determined from the molar ratios of PPO to PEO measured by 1H NMR spectroscopy. Details of the characterization procedures and the corresponding results were reported in our previous studies;27,28 data relevant to the current study are summarized in Table 1.
Table 1.
Polymer Characterization
| Mna (g/mol) | Đa | wPEOb | NPOc | NEOc | |
|---|---|---|---|---|---|
| F68 | 8200 | 1.01 | 0.81 | 27 | 75 |
| F108 | 15,900 | 1.01 | 0.82 | 49 | 148 |
| F127 | 13,200 | 1.01 | 0.72 | 64 | 108 |
| P103 | 5200 | 1.02 | 0.33 | 60 | 19 |
| P105 | 6500 | 1.02 | 0.50 | 56 | 37 |
| P84 | 4200 | 1.03 | 0.42 | 42 | 20 |
| tPPO14-PEO46 | 2900 | 1.03 | 0.70 | 14 | 46 |
| tPPO29-PEO68 | 4700 | 1.04 | 0.63 | 29 | 68 |
Determined by MALDI mass spectroscopy.
Determined from molar ratios by 1H NMR spectroscopy.
Liposome Preparation.
Liposomes were prepared using an extrusion method described elsewhere.27–29,38–40 Briefly, the bicomponent liposomes were prepared by mixing POPC with POPG in chloroform at a specific ratio, followed by evaporating the organic solvent under nitrogen. The resulting dry lipid thin films of various compositions were then hydrated with D2O or 150 mM NaCl salt solution in D2O at 37 °C for an hour. Vortex was applied every 5 min to facilitate the formation of large multilamellar liposomes. The final unilamellar liposome solution was obtained by repeated extrusion through a polycarbonate membrane with 100 nm diameter pores using an Avanti Mini-Extruder assembly. The hydrodynamic radius Rh and size dispersity of the liposomes were characterized by dynamic light scattering (DLS), as described previously.28
PFG-NMR Measurements.
Polymer-lipid bilayer association was evaluated by quantifying the polymer binding to the liposomes via PFG-NMR as described in our previous studies.27,28 The experiments were conducted on a Bruker AVANCE III 500 MHz NMR spectrometer with a 5 mm broadband fluorine observe probe. A stimulated pulse sequence named “ledbpgp2s” (longitudinal eddy current delay experiment using bipolar gradients acquired in two dimensions) was applied, and a series of 1D 1H spectra were recorded corresponding to increasing gradient strength G (from 2 to 95% of the maximum strength). The echo-attenuated intensity I observed from each 1D 1H spectrum can be correlated to G based on the following equation
| (1) |
where I0 is the intensity at zero gradient strength, γ is the gyromagnetic ratio of 1H (42.6 MHz/T), δ is the length of the gradient pulse (δ = 5 ms), and Δ is the diffusion time (Δ = 300, 500, and 700 ms for polymer-liposome mixtures and Δ = 700 ms for pure polymer solutions). The diffusion coefficient D can be extracted from the slope of a linear fit to eq 1. Polymer diffusivity was obtained from the attenuated intensity of the PEO peak because of its strong signal at zero gradient strength. The decay curves of POPG liposomes were also obtained, using the 1H signal of POPG alkyl chains because the 1H signal of the headgroup can be barely observed.
In polymer-liposome mixtures, the binding of a fraction of the polymers to the liposomes gives rise to two distinct diffusivities of polymers, that is, free and bound polymers (Dfree and Dbound, respectively). The diffusivity of the bound polymers is about an order of magnitude slower than that of the free polymers, assuming that the bound polymers have the same diffusivity as the liposomes and that the liposome diffusivity changes little after polymer binding. These two distinct polymer diffusivities result in a bi-exponential signal decay profile, which can be expressed by
| (2) |
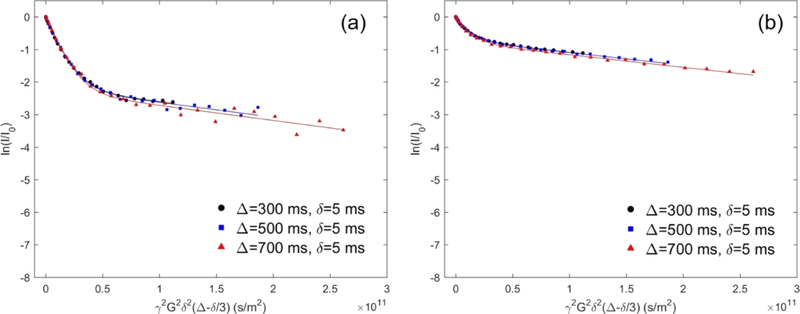
where f bound is the molar percentage of the bound polymers. The fraction of bound polymers can be obtained from the biexponential fit to the signal decay profile of polymers in the presence of liposomes (see Figure 1 for an example). Note that fbound is the only fitting parameter because Dfree and Dbound can be directly extracted from the initial and final slopes of the decay profile, respectively. Additionally, as also reported in our previous studies, molecular exchange of block copolymer between the bound and free states is negligible based on the close overlap of the three decay profiles at different diffusion time points, as illustrated in Figure 1. All samples were measured at 27 °C with a polymer concentration of 0.2 or 1 mg/mL and a liposome concentration of 5 mM.
Figure 1.
Experimental and fitted echo decay curves of the protons from PEO for 0.2 mg/mL P105 in the presence of 5 mM liposome at 27 °C with Δ = 300, 500, 700 ms and with fixed δ = 5 ms. The value of f bound in eq 2 was fit to the data. (a) POPC liposome and (b) POPG liposome solution in 150 mM NaCl.
RESULTS AND DISCUSSION
Liposome Size Characterization.
The size and dispersity of the liposomes composed of POPC and POPG are summarized in Table 2. An NaCl solution at physiologic concentration (150 mM) was used as the solvent instead of pure D2O in order to screen electrostatic interactions between anionic POPG lipids, as discussed in more detail below. The size of neat POPC liposomes remained the same after switching the solvent from D2O to the salt solution. The liposome size decreases slightly as the molar percentage of POPG increases in the POPC/POPG lipid bilayer. However, the dispersity of the liposomes was not affected as the values of the reduced second cumulant μ2/Γ2 determined by DLS in all cases remained around 0.1. Note also that the liposome diffusivity remains 1 order of magnitude lower than that of the free polymers despite the minor reduction in the liposome size.
Table 2.
Characterization of the Liposome Size by DLS
| POPG molar percentage (%) | Rha (nm) | μ2/Γ2 at 90° |
|---|---|---|
| 0b | 70 | 0.04 |
| 0 | 70 | 0.08 |
| 20 | 66 | 0.08 |
| 40 | 66 | 0.11 |
| 60 | 61 | 0.07 |
| 80 | 62 | 0.08 |
| 100 | 62 | 0.10 |
Rh measured by DLS has approximately 5% uncertainty.
Measured in D2O. All the other samples were measured in 150 mM NaCl solution in D2O.
Effect of the Lipid Headgroup.
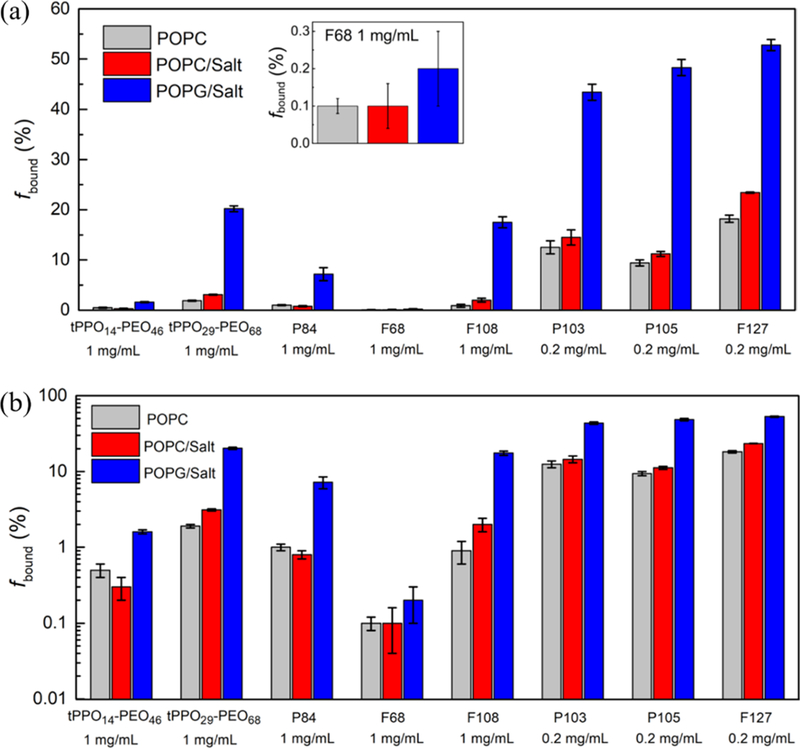
The headgroup effect was investigated by comparing block copolymer binding to POPC liposomes containing a choline group and POPG liposomes with a glycerol group. Salt solutions containing 150 mM NaCl in D2O were used instead of pure D2O to stabilize the liposomes by screening the electrostatic interactions between the negatively charged headgroups of POPG. Additional remarks about this effect can be found at the end of this subsection. Polymer binding to POPC liposomes in D2O, to POPC liposomes in 150 mM NaCl, and to POPG liposomes in 150 mM NaCl is displayed in Figure 2a; the same data plotted on a logarithmic scale are shown in Figure 2b. Values of the binding percentage and related parameters used in fitting the data are summarized in Table S2. In order to assess whether adding salt makes a difference, the polymer binding to POPC liposomes in D2O (gray bars) is compared to that in 150 mM NaCl (red bars). Some polymers show slightly more binding with POPC lipid bilayers in salt solution than in D2O. The presence of salt can lead to a slight decrease in the CMC,41–44 which may result in a stronger drive for the hydrophobic block of the polymers to form a micellar core or insert into the hydrophobic bilayer interior, rather than interacting with water molecules.27 It is to be noted that the physiologic salt concentration used is not high enough to induce micellization, so the polymers still remain as free chains, despite the polymer concentration moving slightly closer to the CMC.42–44 This point is supported by the linearity of the PFG-NMR profiles of free polymers in salt solution, as shown in Figure S1, which illustrates the echo decay curves of F127 in D2O and in 150 mM NaCl. The two linear fits give virtually identical diffusivities, indicating that the presence of 150 mM NaCl does not induce polymer micellization. Therefore, changing the solvent from D2O to 150 mM NaCl slightly increases the polymer binding to POPC lipid bilayers, but the effect is not significant, especially considering the error bars associated with the measurement of polymer binding. Here, we note that P103, P105, and F127 were measured at a lower concentration (i.e., 0.2 mg/mL instead of 1 mg/mL) because of lower CMCs compared to those of other polymers that have been evaluated.45,46 The polymer concentration was kept below the CMC because the presence of micelles would result in a third value of polymer diffusivity (i.e., Dmicelle), in addition to Dfree and Dbound, and thereby complicate the data analysis.47
Figure 2.
(a) Polymer binding with POPC liposomes in D2O (gray), with POPC liposomes in 150 mM NaCl D2O solution (red), and with POPG liposomes in 150 mM NaCl D2O solution (blue). The inset shows a zoom-in view of the binding of F68. (b) Data replotted on a logarithmic scale.
The effect of a choline versus a glycerol headgroup was probed by quantifying the polymer binding to POPC liposomes (red bars) and to POPG liposomes (blue bars) in 150 mM NaCl, as shown in Figure 2. NMR decay curves of P105 are shown in Figure 1a,b as representative examples of block copolymer binding to POPC and POPG liposomes in 150 mM NaCl, respectively. The polymer signal intensity in the second linear region is much higher in the presence of POPG liposomes versus POPC liposomes, indicating that P105 has more association with the POPG bilayer membrane. Similarly, all the other polymer species that were evaluated, except F68, show significantly higher binding with POPG than POPC, as the blue bars are much taller than the red bars in Figure 2. In the case of F68, both POPG and POPC lipid bilayers show very low binding and there is no statistically significant difference between the two lipids. This weak association between F68 and lipid bilayers can be attributed to the relatively short hydrophobic PPO blocks and large content of hydrophilic PEO blocks (ca. 80 wt %).27 The NMR decay curves of all the other polymer species in addition to P105 are shown in Figure S2.
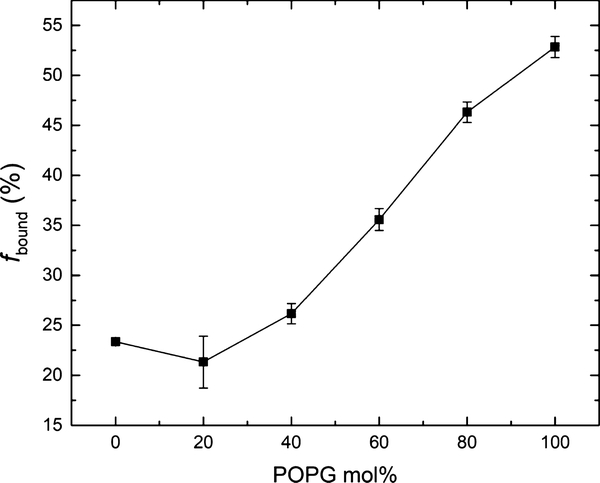
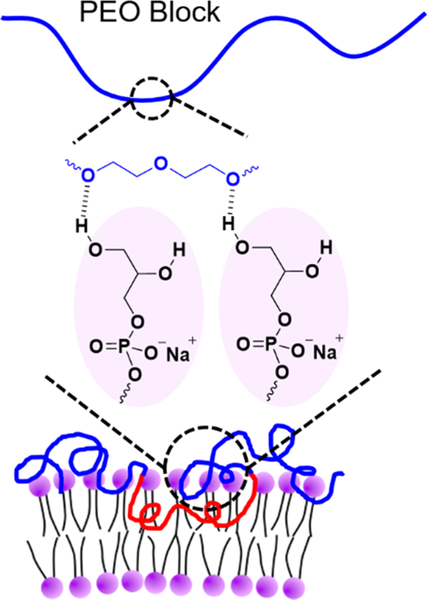
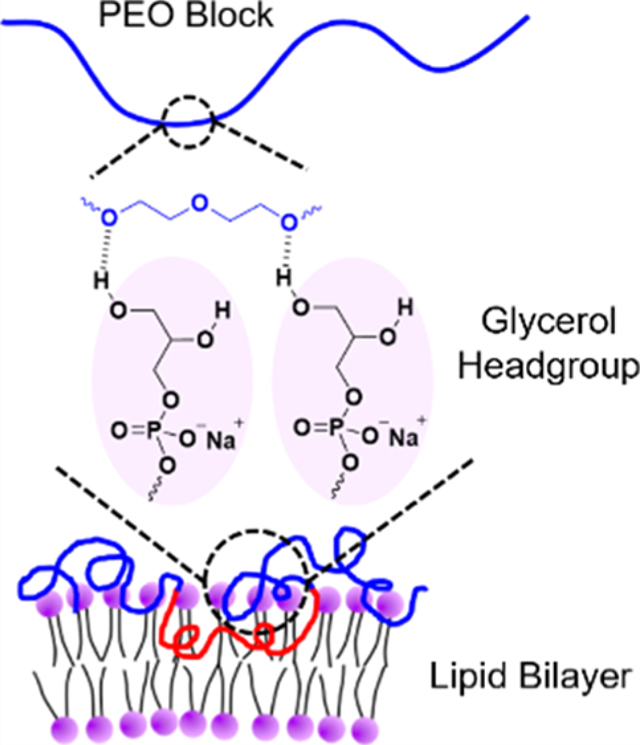
Next, we investigated how polymer binding varied with increasing POPG content in the lipid bilayer. F127 was selected as a representative polymer to illustrate binding as a function of the molar percentage of POPG in POPC/POPG bicomponent liposomes in 150 mM NaCl (Figure 3). Binding of F127 at each POPG molar percentage is summarized in Table S3, and the corresponding echo decay curves are shown in Figure S3. Figure 3 shows that the binding of F127 increases as the POPG content increases, with the headgroup effect becoming significant when the bicomponent lipid bilayer contains more than 20 mol % POPG. A possible reason that the glycerol group of POPG enhances polymer binding is hydrogen bonding. Earlier work in the literature demonstrated that POPG lipid bilayers can form strong intermolecular hydrogen bonds within the hydrophilic headgroup layer because of the presence of the hydroxyl groups.34–36 It was also found that POPG can form hydrogen bonds with specific sites on some proteins, resulting in tight binding between the lipid headgroup and the protein.48 Additionally, it was suggested that ether oxygen in PEO favors hydrogen bonding with the surrounding hydroxyl groups.49–53 In this context, we suggest that the headgroup of POPG could form hydrogen bonds with PPO-PEO block polymers, where the hydroxyl groups function as hydrogen bonding donors, while the ether oxygen of the PEO or PPO blocks are potential hydrogen bonding acceptors. A schematic of this proposed mechanism is shown in Figure 4. The fact that the headgroup effect is not significant until the bilayer contains more than 20 mol % POPG could be because multiple hydrogen bonds need to be formed between one polymer molecule and two or more glycerol groups in order to produce enhanced polymer binding. We speculate that there is a threshold POPG content in the lipid bilayer, below which the lipid bilayer is unable to form enough hydrogen bonds with the polymers because of the lack of glycerol groups, and thereby shows little increase of polymer binding.
Figure 3.
Polymer binding percentage of 0.2 mg/mL F127 as a function of POPG molar percentage in the POPC/POPG lipid bilayer in 150 mM NaCl.
Figure 4.
Schematic of possible hydrogen bonding between a PEO block and glycerol headgroups of POPG in the lipid bilayers.
In an attempt to verify the proposed binding mechanism, Raman spectroscopy was employed to probe hydrogen bonding between the polymers and the POPG headgroups. However, because of the limitations on the signal resolution, we were unable to obtain definitive evidence of hydrogen bonding. These experiments and associated discussion are present in the Supporting Information.
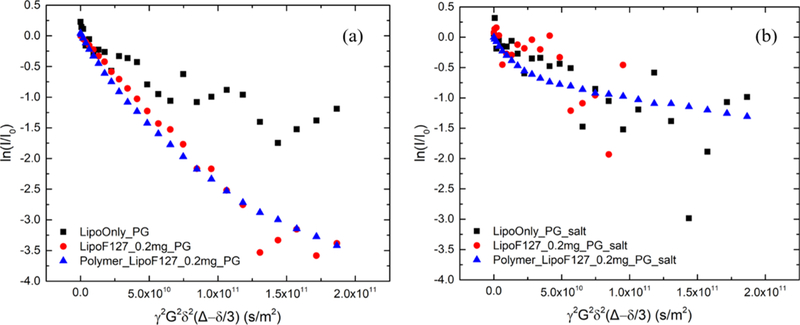
Additionally, PFG-NMR was employed to show that using 150 mM NaCl instead of pure D2O can stabilize the liposomes by screening the electrostatic interactions between the negatively charged headgroups of POPG. In pure D2O (Figure 5a), the PFG-NMR decay profile for the POPG liposomes after mixing with polymers (red circles) decays much more rapidly than that of the neat liposomes (black squares), notwithstanding the noisy signal obtained with the neat liposomes. Faster diffusion and the nonlinear decay profile of the liposomes in the presence of polymers suggest that the liposomes are disrupted into smaller vesicles or aggregates with high dispersity. In the absence of salt, the repulsive interactions between the POPG headgroups become significant, which we speculate makes the liposomes unstable and vulnerable to disturbances induced by polymer association. We attribute the highly disperse diffusivity of the copolymers in the presence of liposomes, as evidenced by the nonlinearity of the PFG-NMR decay (blue triangles), to polymer association with the disrupted polydisperse liposomes. In contrast, as shown in Figure 5b, the bi-exponential character of the polymer decay curves in the presence of liposomes is recovered with a 150 mM NaCl solution. The plateau region (i.e., final slope of the polymer biexponential decay) indirectly indicates that the integrity of the liposome structure is retained. Additional indirect evidence of liposome integrity is that liposome diffusion, after mixing with polymers (red circles), as shown in Figure 5b, no longer resembles that in Figure 5a which is a much faster decay, although a quantitative diffusion coefficient could not be clearly resolved because of a noisy signal. We attribute the noisy signal of the liposomes to the broad NMR peak because of the poor solubility of the hydrocarbon chains of lipid in water, which significantly reduces the signal-to-noise ratio.
Figure 5.
Echo decay curves of 5 mM neat POPG liposomes (black squares), 5 mM POPG liposomes in the presence of 0.2 mg/mL F127 (red circles), and 0.2 mg/mL F127 in the presence of 5 mM POPG liposome (blue triangles) at 27 °C in (a) D2O and (b) 150 mM NaCl D2O solution. The decay curves of POPG liposomes were obtained using the 1H signal of POPG alkyl chains because the 1H signal of the headgroup is barely observable. The noisy signal of liposomes is attributed to the broad signal peak because of poor solubility of alkyl chains in water, which significantly decreases the signal-to-noise ratio.
CONCLUSIONS
We have investigated the effect of the lipid headgroup on the interactions of PPO-PEO block copolymers with lipid bilayers. Polymer binding to POPC liposomes and to POPG liposomes dispersed in 150 mM NaCl solution was studied in order to ascertain the roles of glycerol versus choline headgroups. PFG-NMR was employed to determine polymer binding based on the quantitative measurement of the relative amounts of the free and bound polymer. All the selected polymer species showed higher binding to POPG liposomes than to POPC liposomes. Binding of F127 as a function of the mole fraction of POPG in POPC/POPG bicomponent liposomes revealed that the enhanced binding effect becomes substantial when the lipid bilayer contains more than 20 mol % POPG. Enhanced polymer association with POPG is tentatively attributed to hydrogen bonding between the glycerol groups in POPG and the ether oxygen atoms in the polymers. These findings suggest that other lipids containing headgroups relevant to mammalian cells, such as phosphatidylserine, may play a role in the physiological action of PPO-PEO block copolymers.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Institutes of Health (grants HL122323; AR071349). We acknowledge Karen J. Haman for synthesizing the diblock polymers and Mihee Kim for useful discussions. We also acknowledge Letitia J. Yao for helping with the NMR experiments. NMR instrumentation was supported by the Office of the Vice President of Research, College of Science and Engineering, and the Department of Chemistry at the University of Minnesota. The Raman measurements were conducted in the Characterization Facility, University of Minnesota, with help from Bing Luo.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.0c00553.
Polymer characterization; summary of polymer binding to 5 mM POPC liposomes in D2O, to POPC liposomes in 150 mM NaCl D2O solution, and to POPG liposomes in 150 mM NaCl D2O solution; NMR decay curves of 0.2 mg/mL F127 in D2O and in 150 mM NaCl D2O solution; summary of polymer binding of 0.2 mg/mL F127 to 5 mM POPC/POPG liposomes at various POPG molar percentages in the lipid bilayer in 150 mM NaCl D2O solution; echo decay curves of polymers in the presence of 5 mM POPC and POPG liposomes in 150 mM NaCl D2O solution; echo decay curves of 0.2 mg/mL F127 in the presence of 5 mM POPC/POPG liposomes with various POPG molar percentage in 150 mM NaCl D2O solution; and results and discussion of Raman spectroscopy (PDF)
The authors declare no competing financial interest.
J.M.M. is on the scientific advisory board of and holds zero value equity shares in Phrixus Pharmaceuticals Inc., a company developing novel therapeutics for heart failure and DMD, and this is actively managed by the UMN Office of Institutional Compliance.
Contributor Information
Wenjia Zhang, Department of Chemical Engineering and Materials Science, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Joseph M. Metzger, Department of Integrative Biology and Physiology, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Benjamin J. Hackel, Department of Chemical Engineering and Materials Science, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Frank S. Bates, Department of Chemical Engineering and Materials Science, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Timothy P. Lodge, Department of Chemical Engineering and Materials Science and Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States.
REFERENCES
- (1).Shrestha R; Anderson CM; Cardenas AE; Elber R; Webb LJ Direct Measurement of the Effect of Cholesterol and 6-Ketocholestanol on the Membrane Dipole Electric Field Using Vibrational Stark Effect Spectroscopy Coupled with Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 3424–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yeagle PL The Lipids of Biological Membranes The Membranes of Cells; Academic Press: New York, 1993; pp 27–56. [Google Scholar]
- (3).Starke-Peterkovic T; Clarke RJ Effect of Headgroup on the Dipole Potential of Phospholipid Vesicles. Eur. Biophys. J 2009, 39, 103–110. [DOI] [PubMed] [Google Scholar]
- (4).Yeagle PL; Hutton WC; Huang C-H; Martin RB Phospholipid Head-Group Conformations; Intermolecular Interactions and Cholesterol Effects. Biochemistry 1977, 16, 4344–4349. [DOI] [PubMed] [Google Scholar]
- (5).Garidel P; Blume A. Miscibility of Phospholipids with Identical Headgroups and Acyl Chain Lengths Differing by Two Methylene Units: Effects of Headgroup Structure and Headgroup Charge. Biochim. Biophys. Acta, Biomembr 1998, 1371, 83–95. [DOI] [PubMed] [Google Scholar]
- (6).Cevc G. Effect of Lipid Headgroups and (Nonelectrolyte) Solution on the Structural and Phase Properties of Bilayer Membranes. Ber. Bunsen Ges. Phys. Chem 1988, 92, 953A–961. [Google Scholar]
- (7).Nagle JF Theory of Lipid Monolayer and Bilayer Phase Transitions: Effect of Headgroup Interactions. J. Membr. Biol 1976, 27, 233–250. [DOI] [PubMed] [Google Scholar]
- (8).Bakht O; Pathak P; London E. Effect of the Structure of Lipids Favoring Disordered Domain Formation on the Stability of Cholesterol-Containing Ordered Domains (Lipid Rafts): Identification of Multiple Raft-Stabilization Mechanisms. Biophys. J 2007, 93, 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Niu S-L; Litman BJ Determination of Membrane Cholesterol Partition Coefficient Using a Lipid Vesicle-Cyclodextrin Binary System: Effect of Phospholipid Acyl Chain Unsaturation and Headgroup Composition. Biophys. J 2002, 83, 3408–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhao L; Feng S-S Effects of Lipid Chain Unsaturation and Headgroup Type on Molecular Interactions between Paclitaxel and Phospholipid within Model Biomembrane. J. Colloid Interface Sci 2005, 285, 326–335. [DOI] [PubMed] [Google Scholar]
- (11).Ding L; Chi EY; Schanze KS; Lopez GP; Whitten DG Insight into the Mechanism of Antimicrobial Conjugated Polyelectrolytes: Lipid Headgroup Charge and Membrane Fluidity Effects. Langmuir 2010, 26, 5544–5550. [DOI] [PubMed] [Google Scholar]
- (12).mstedt AA; Wessman P; Ringstad L; Edwards K; Malmsten M. Effect of Lipid Headgroup Composition on the Interaction between Melittin and Lipid Bilayers. J. Colloid Interface Sci 2007, 311, 59–69. [DOI] [PubMed] [Google Scholar]
- (13).Hädicke A; Blume A. Binding of the Cationic Peptide (KL) 4 K to Lipid Monolayers at the Air-Water Interface: Effect of Lipid Headgroup Charge, Acyl Chain Length, and Acyl Chain Saturation. J. Phys. Chem. B 2016, 120, 3880–3887. [DOI] [PubMed] [Google Scholar]
- (14).Ishitsuka Y; Pham DS; Waring AJ; Lehrer RI; Lee KYC Insertion Selectivity of Antimicrobial Peptide Protegrin-1 into Lipid Monolayers: Effect of Head Group Electrostatics and Tail Group Packing. Biochim. Biophys. Acta, Biomembr 2006, 1758, 1450–1460. [DOI] [PubMed] [Google Scholar]
- (15).Heller WT; He K; Ludtke SJ; Harroun TA; Huang HW Effect of Changing the Size of Lipid Headgroup on Peptide Insertion into Membranes. Biophys. J 1997, 73, 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Middleton ER; Rhoades E. Effects of Curvature and Composition on α-Synuclein Binding to Lipid Vesicles. Biophys. J 2010, 99, 2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Elmore DE; Dougherty DA Investigating Lipid Composition Effects on the Mechanosensitive Channel of Large Conductance (MscL) Using Molecular Dynamics Simulations. Biophys. J 2003, 85, 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yasuda S; Townsend D; Michele DE; Favre EG; Day SM; Metzger JM Dystrophic Heart Failure Blocked by Membrane Sealant Poloxamer. Nature 2005, 436, 1025–1029. [DOI] [PubMed] [Google Scholar]
- (19).Houang EM; Haman KJ; Kim M; Zhang W; Lowe DA; Sham YY; Lodge TP; Hackel BJ; Bates FS; Metzger JM Chemical End Group Modified Diblock Copolymers Elucidate Anchor and Chain Mechanism of Membrane Stabilization. Mol. Pharm 2017, 14, 2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Houang EM Copolymer-Based Membrane Stabilizers for Duchenne Muscular Dystrophy; University of Minnesota, 2016. [Google Scholar]
- (21).Kim M; Haman KJ; Houang EM; Zhang W; Yannopoulos D; Metzger JM; Bates FS; Hackel BJ PEO-PPO Diblock Copolymers Protect Myoblasts from Hypo-Osmotic Stress In Vitro Dependent on Copolymer Size, Composition, and Architecture. Biomacromolecules 2017, 18, 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Venne A; Li S; Mandeville R; Kabanov A; Alakhov V. Hypersensitizing Effect of Pluronic L61 on Cytotoxic Activity, Transport, and Subcellular Distribution of Doxorubicin in Multiple Drug-Resistant Cells. Cancer Res. 1996, 56, 3626–3629. [PubMed] [Google Scholar]
- (23).Valle JW; Lawrance J; Brewer J; Clayton A; Corrie P; Alakhov V; Ranson M. A Phase II, Window Study of SP1049C as First-Line Therapy in Inoperable Metastatic Adenocarcinoma of the Oesophagus. J. Clin. Oncol 2004, 22, 4195. [Google Scholar]
- (24).Batrakova EV; Kabanov AV Pluronic Block Copolymers: Evolution of Drug Delivery Concept from Inert Nanocarriers to Biological Response Modifiers. J. Controlled Release 2008, 130, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kabanov AV; Batrakova EV; Alakhov VY Pluronic® Block Copolymers as Novel Polymer Therapeutics for Drug and Gene Delivery. J. Controlled Release 2002, 82, 189–212. [DOI] [PubMed] [Google Scholar]
- (26).Alakhov V; Klinski E; Li S; Pietrzynski G; Venne A; Batrakova E; Bronitch T; Kabanov A. Block Copolymer-Based Formulation of Doxorubicin. From Cell Screen to Clinical Trials. Colloids Surf., B 1999, 16, 113–134. [Google Scholar]
- (27).Zhang W; Haman KJ; Metzger JM; Hackel BJ; Bates FS; Lodge TP Quantifying Binding of Ethylene Oxide-Propylene Oxide Block Copolymers with Lipid Bilayers. Langmuir 2017, 33, 12624–12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhang W; Coughlin ML; Metzger JM; Hackel BJ; Bates FS; Lodge TP Influence of Cholesterol and Bilayer Curvature on the Interaction of PPO-PEO Block Copolymers with Liposomes. Langmuir 2019, 35, 7231–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wang J-Y; Marks J; Lee KYC Nature of Interactions between PEO-PPO-PEO Triblock Copolymers and Lipid Mem-branes: (I) Effect of Polymer Hydrophobicity on Its Ability to Protect Liposomes from Peroxidation. Biomacromolecules 2012, 13, 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang J-Y; Chin J; Marks JD; Lee KYC Effects of PEO-PPO-PEO Triblock Copolymers on Phospholipid Membrane Integrity under Osmotic Stress. Langmuir 2010, 26, 12953–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang J; Segatori L; Biswal SL Probing the Association of Triblock Copolymers with Supported Lipid Membranes Using Microcantilevers. Soft Matter 2014, 10, 6417–6424. [DOI] [PubMed] [Google Scholar]
- (32).Evans WH; Hardison WG Phospholipid, Cholesterol, Polypeptide and Glycoprotein Composition of Hepatic Endosome Subfractions. Biochem. J 1985, 232, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Dowhan W. Molecular Basis for Mmebrane Phospholipid Diversity: Why Are There So Many Lipids? Annu. Rev. Biochem 1997, 66, 199–232. [DOI] [PubMed] [Google Scholar]
- (34).Elmore DE Molecular Dynamics Simulation of a Phosphatidylglycerol Membrane. FEBS Lett. 2006, 580, 144–148. [DOI] [PubMed] [Google Scholar]
- (35).Dickey A; Faller R. Examining the Contributions of Lipid Shape and Headgroup Charge on Bilayer Behavior. Biophys. J 2008, 95, 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhao W; Roǵ T; Gurtovenko AA; Vattulainen I; Karttunen M. Atomic-Scale Structure and Electrostatics of Anionic Palmitoyloleoylphosphatidylglycerol Lipid Bilayers with Na+ Counterions. Biophys. J 2007, 92, 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).rka N; Holland BW; Gray CG; Tomberli B; Katsaras J. Scattering Density Profile Model of POPG Bilayers As Determined by Molecular Dynamics Simulations and Small-Angle Neutron and X-Ray Scattering Experiments. J. Phys. Chem. B 2012, 116, 232–239. [DOI] [PubMed] [Google Scholar]
- (38).Olson F; Hunt CA; Szoka FC; Vail WJ; Papahadjopoulos D. Preparation of Liposomes of Defined Size Distribution by Extrusion through Polycarbonate Membranes. Biochim. Biophys. Acta, Biomembr 1979, 557, 9–23. [DOI] [PubMed] [Google Scholar]
- (39).MacDonald RC; MacDonald RI; Menco BPM; Takeshita K; Subbarao NK; Hu L.-r. Small-Volume Extrusion Apparatus for Preparation of Large, Unilamellar Vesicles. Biochim. Biophys. Acta, Biomembr 1991, 1061, 297–303. [DOI] [PubMed] [Google Scholar]
- (40).Hope MJ; Bally MB; Webb G; Cullis PR Production of Large Unilamellar Vesicles by a Rapid Extrusion Procedure. Characterization of Size Distribution, Trapped Volume and Ability to Maintain a Membrane Potential. Biochim. Biophys. Acta, Biomembr 1985, 812, 55–65. [DOI] [PubMed] [Google Scholar]
- (41).Patel K; Bahadur P; Guo C; Ma JH; Liu HZ; Yamashita Y; Khanal A; Nakashima K. Salt Induced Micellization of Very Hydrophilic PEO-PPO-PEO Block Copolymers in Aqueous Solutions. Eur. Polym. J 2007, 43, 1699–1708. [Google Scholar]
- (42).Mata JP; Majhi PR; Guo C; Liu HZ; Bahadur P. Concentration, Temperature, and Salt-Induced Micellization of a Triblock Copolymer Pluronic L64 in Aqueous Media. J. Colloid Interface Sci 2005, 292, 548–556. [DOI] [PubMed] [Google Scholar]
- (43).Jain NJ; George A; Bahadur P. Effect of Salt on the Micellization of Pluronic P65 in Aqueous Solution. Colloids Surf., A 1999, 157, 275–283. [Google Scholar]
- (44).Pandit N; Trygstad T; Croy S; Bohorquez M; Koch C. Effect of Salts on the Micellization, Clouding, and Solubilization Behavior of Pluronic F127 Solutions. J. Colloid Interface Sci 2000, 222, 213–220. [DOI] [PubMed] [Google Scholar]
- (45).Batrakova E; Lee S; Li S; Venne A; Alakhov V; Kabanov A. Fundamental Relationships between the Composition of Pluronic Block Copolymers and Their Hypersensitization Effect in MDR Cancer Cells. Pharm. Res 1999, 16, 1373–1379. [DOI] [PubMed] [Google Scholar]
- (46).Alexandridis P; Holzwarth JF; Hatton TA Micellization of Poly(Ethylene Oxide)-Poly(Propylene Oxide)-Poly(Ethylene Oxide) Triblock Copolymers in Aqueous Solutions: Thermodynamics of Copolymer Association. Macromolecules 1994, 27, 2414–2425. [Google Scholar]
- (47).Kim M; Vala M; Ertsgaard CT; Oh S-H; Lodge TP; Bates FS; Hackel BJ Surface Plasmon Resonance Study of the Binding of PEO-PPO-PEO Triblock Copolymer and PEO Homopolymer to Supported Lipid Bilayers. Langmuir 2018, 34, 6703–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yeagle PL Lipid-Protein Interactions in Membranes The Membranes of Cells; Academic Press: New York, 1993; pp 291–334. [Google Scholar]
- (49).Chang L-C; Chang Y-Y; Gau C-S Interfacial Properties of Pluronics and the Interactions between Pluronics and Cholesterol/ DPPC Mixed Monolayers. J. Colloid Interface Sci 2008, 322, 263–273. [DOI] [PubMed] [Google Scholar]
- (50).Mpofu P; Addai-Mensah J; Ralston J. Investigation of the Effect of Polymer Structure Type on Flocculation, Rheology and Dewatering Behaviour of Kaolinite Dispersions. Int. J. Miner. Process 2003, 71, 247–268. [Google Scholar]
- (51).Spitzer M; Sabadini E; Loh W. Entropically Driven Partitioning of Ethylene Oxide Oligomers and Polymers in Aqueous/Organic Biphasic Systems. J. Phys. Chem. B 2002, 106, 12448–12452. [Google Scholar]
- (52).Blandamer MJ; Fox MF; Powell E; Stafford JW A Viscometric Study of Poly(Ethylene Oxide) in T-butyl Alcohol/Water Mixtures. Die Makromol. Chem 1969, 124, 222–231. [Google Scholar]
- (53).Anselmo AG; Sassonia RC; Loh W. Thermodynamics of the Partitioning of Poly(Propylene Oxide) between Aqueous and Chlorinated Organic Phases Compared to Poly(Ethylene Oxide) and Other Hydrophilic Polymers. J. Phys. Org. Chem 2006, 19, 780–785. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.