Abstract
The structural and functional properties of G protein-coupled receptors (GPCRs) are often studied in a detergent micellar environment, but many GPCRs tend to denature or aggregate in short alkyl chain detergents. In our previous work [Lee, S., et al. (2016) J. Am. Chem. Soc.138, 15425–15433], we showed that GPCRs in alkyl glucosides were highly dynamic, resulting in the penetration of detergent molecules between transmembrane α-helices, which is the initial step in receptor denaturation. Although this was not observed for GPCRs in dodecyl maltoside (DDM, also known as lauryl maltoside), even this detergent is not mild enough to preserve the integrity of many GPCRs during purification. Lauryl maltose neopentylglycol (LMNG) detergents have been found to have significant advantages for purifying GPCRs in a native state as they impart more stability to the receptor than DDM. To gain insights into how they stabilize GPCRs, we used atomistic molecular dynamics simulations of wild type adenosine A2A receptor (WT-A2AR), thermostabilized A2AR (tA2AR), and wild type β2-adrenoceptor (β2AR) in a variety of detergents (LMNG, DMNG, OGNG, and DDM). Analysis of molecular dynamics simulations of tA2AR in LMNG, DMNG, and OGNG showed that this series of detergents exhibited behavior very similar to that of an analogous series of detergents DDM, DM, and OG in our previous study. However, there was a striking difference upon comparison of the behavior of LMNG to that of DDM. LMNG showed considerably less motion than DDM, which resulted in the enhanced density of the aliphatic chains around the hydrophobic regions of the receptor and considerably more hydrogen bond formation between the head groups. This contributed to enhanced interaction energies between both detergent molecules and between the receptor and detergent, explaining the enhanced stability of GPCRs purified in this detergent. Branched detergents occlude between transmembrane helices and reduce their flexibility. Our results provide a rational foundation to develop detergent variants for stabilizing membrane proteins.
G protein-coupled receptors (GPCRs) are heptahelical integral membrane proteins that play a critical role in cell signaling.1,2 The pivotal role that GPCRs play in intercellular communication and their pharmacological accessibility on the cell surface make them highly tractable drug targets, with 34% of current Food and Drug Administration-approved small molecule drugs targeting GPCRs.3 Although there have been more than 360 structures of GPCRs published in the past two decades, ∼81% of nonsensory human GPCRs have yet to be crystallized or had a structure determined by single-particle electron cryo-microscopy (cryo-EM). Thus, there is still a need for additional high-resolution GPCR structures, to gain greater insights into their precise mechanism of action and to facilitate rational drug design.3 The challenge in obtaining three-dimensional structures of GPCRs begins with the their overexpression and purification. Primarily, the major impediment to GPCR purification, or any membrane protein for that matter, is the lack of stability of GPCRs in a detergent solution during purification.4 Detergents such as dodecyl maltoside (DDM) and octyl thioglucoside (OTG) have been used for GPCR purification and crystallization, but only for stable GPCRs such as rhodopsin,5 A2AR,6 and the β2-adrenoceptor,7 or receptors that have been thermostabilized.8 Branched amphiphiles such as the maltose-neopentylgylcols (MNGs)9 have been shown to impart greater stability than detergents containing a single alkyl chain, and LMNG in particular has been very successful in GPCR purification. The neopentylglycols feature a central quaternary carbon atom, which places subtle restraints on conformational flexibility.10−12 Octylglucose neopentylglycol (OGNG) is the smallest neopentylglycol and was used to determine the structure of the β1-adrenoceptor8 and rhodopsin coupled to mini-Go.13
Lauryl maltose neopentyl glycol (LMNG), which consists of two maltose units in their hydrophilic domain and two n-dodecyl chains appended to a quaternary central carbon, has been used extensively to purify multiple GPCRs such as β2-adrenergic receptor (β2AR), opioid receptors, muscarinic receptors, and the neurotensin receptors.14−23 The receptors were then subsequently crystallized in a lipidic cubic phase, which strips the detergent away from the receptor when it is embedded in the monoolein bilayer.24 More recently, structures have been determined by single-particle cryo-EM of GPCR complexes in LMNG.25
Although LMNG has been used widely in GPCR purification, the mechanism by which it stabilizes GPCRs better than its counterpart DDM remains unclear. This understanding is essential for developing newer detergents for stabilizing less stable GPCRs for which there is no structural information. Previously, we used atomistic molecular dynamics (MD) simulations of the thermostabilized mutant of adenosine receptor A2AR in DDM, n-decyl β-d-maltoside (DM), n-nonyl β-d-glucoside (NG), and n-octyl β-d-glucoside (OG) to study the stability of GPCRs embedded in detergent micelles.26 We found that the short chain glucosides occlude less of the hydrophobic surface of the transmembrane region of GPCRs, compared to long chain glucosides or maltosides. OG shows high mobility in the micelle and destabilizes the GPCR through loss of helicity and interhelical packing interactions. In the study presented here, we used MD simulations to understand the stabilization conferred by the LMNG detergent on the thermostabilized mutant of human A2AR compared to DMNG and OGNG. We refer to the inactive state of the thermostabilized mutant of A2AR (StaR2),27 called tA2AR hereafter unless specified. We also studied the stability of the wild type human A2AR (WT-A2AR) and wild type human β2AR (WT-β2AR) in the branched detergent LMNG compared to its unbranched counterpart DDM. The WT-A2AR and WT-β2AR have been shown to be more stable in LMNG than in DDM.28 Our results show that LMNG stabilizes GPCRs better than DDM through more effective packing of its alkyl chains around the hydrophobic transmembrane region of the receptor. We also observed that LMNG forms bifurcated hydrogen bonds between its two polar head groups and the intracellular and extracellular regions of TM helices and loops. The bifurcated hydrogen bonds make the receptor less flexible. Both of these factors result in higher energies of interaction between LMNG molecules in the micelle and between LMNG and the receptor compared to those observed for DDM.
Results
Measurement of the Apparent Tm for the A2AR Thermostable Mutant of the Inactive State in Branched Detergent Micelles
As described in Methods, we measured the apparent melting temperature of tA2AR bound to the antagonist ZM241385 in three detergent micelles, namely, LMNG, DMNG, and OGNG. It was important to use the thermostabilized version of A2AR for these studies, because the wild type A2AR is not stable in very short chain detergents such as OGNG and therefore an accurate apparent melting temperature (Tm) would not have been possible to measure. We observed that tA2AR is more stable in LMNG than in DMNG or OGNG, with the respective apparent Tm values being 44.2 ± 0.2, 33.9 ± 0.2, and 24.2 ± 0.6 °C, respectively. The measurements of stability for β2AR were extracted from literature.9
For studying the mechanism of stability of tA2AR and β2AR in the branched detergent micelles, atomistic MD simulations were performed on receptors embedded in LMNG, DMNG, OGNG, and DDM micelles (see Figure 1A for the structures of the detergents). The simulations were started from the antagonist-bound inactive state of the two receptors, namely, the antagonist ZM241385 bound to A2AR [Protein Data Bank (PDB) entry 3PWH]27 and antagonist carazolol-bound β2AR (PDB entry 2RH1).29 We used 192 molecules of DDM and 96 molecules of LMNG, DMNG, and OGNG to build the micelles (see Methods for more details).
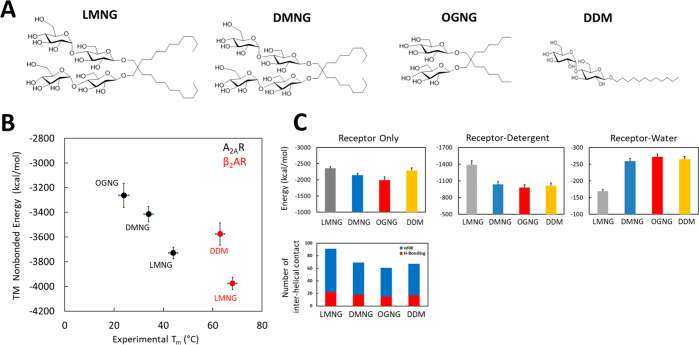
Figure 1.
Stability of the tA2AR–detergent complexes. (A) Chemical structures of the detergents used in this study. (B) Comparison of the calculated nonbond energy of the transmembrane (TM) region of tA2AR (black) and WT-β2AR (red) in different detergent micelles to the measured apparent Tm. (C) Total enthalpic stability (ΔH) of tA2AR. The total nonbond energy of the TM regions of tA2AR is shown, including the energy of receptor–detergent interactions and receptor–water interactions, averaged over the MD trajectories. The total number of sustained (>50% of the MD simulation snapshots) interhelical hydrogen bonds and van der Waals interactions of tA2AR in different detergent micelles is depicted in the bottom panel.
Receptor–Detergent Complex Geometry and Stability of the Receptor–Detergent Complex
After assembly of the receptor–detergent complex (RDC) using CHARMM GUI,30 RDC was equilibrated and subjected to five MD simulations, 250 ns each. To validate the MD simulations, we compared the experimentally measured Tm to the calculated total energy (enthalpic contribution only) of the RDCs in the transmembrane regions, from the MD trajectories that reflect its stability (Figure 1B). The total energy of the RDC includes the receptor nonbond energy, receptor–detergent interaction energy, and receptor–water interaction energy for the transmembrane (TM) region of the receptors.
The ordering of the calculated total energy of tA2AR and WT-β2AR in LMNG, OGNG, DMNG, and DDM is similar to that of the measured Tm, suggesting that the RDC models are good for understanding the stability of GPCRs in detergents.26 The internal energy (enthalpy) of tA2AR is 370 kcal/mol better in LMNG than in OGNG. The total internal energy of WT-β2AR is 116 kcal/mol lower in LMNG than in DDM. It should be noted that this is just the enthalpy of the folded state of the receptors. The apparent Tm of β2AR in DMNG and OGNG has not been published, so we could not include them in our analysis. The calculated nonbond energies of the receptor alone and receptor–detergent interactions (Figure 1C and Figure S1A) contribute significantly to favor the RDC stability of both tA2AR and WT-β2AR in LMNG compared to OGNG. We calculated the total number of interhelical hydrogen bonds and van der Waals contacts that are sustained (over 50% of the MD snapshots) over the course of MD simulations in tA2AR and WT-β2AR in LMNG, DMNG, and OGNG (Figure 1C and Figure S1B). The number of interhelical H-bonds (colored red) and van der Waals contacts (colored blue) within both tA2AR and WT-β2AR are higher in the LMNG micelle than in DMNG, OGNG, and DDM micelles.
Shape of the Receptor–Detergent Complex
We analyzed the shape of the receptor–micelle complex and calculated its eccentricity. As shown in Figure 2A and Figure S2B, the representative structure of the most populated conformation ensemble for tA2AR and β2AR micelle complexes forms oblate spheroids.31,32 LMNG forms the most oblate spheroid compared to DMNG and OGNG as shown in Figure S2A. Figure 2B and Figure S2C show the population density distribution of the eccentricity of the spheroids for both tA2AR and WT-β2AR, respectively. The data for WT-β2AR simulation are shown in Figure S2.
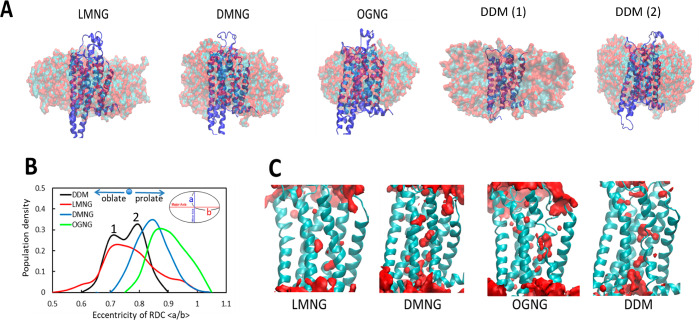
Figure 2.
Representative structures of tA2AR–detergent complexes extracted from the MD simulations. (A) The hydrophobic tail of the detergent is shown as cyan spheres, and the hydrophilic atoms in the head group are colored red. (B) Calculated eccentricity of tA2AR in four different micelle systems defined as the ratio of the short axis to the long axis (a/b). The two peaks in the bimodal distribution of the eccentricity of the tA2AR–DDM complex are denoted by numerals 1 and 2 (black curve). Representative structures (Methods) of tA2AR embedded in DDM corresponding to these two peaks are shown in panel A as DDM (1) and DDM (2). (C) Water density representation within 3.5 Å of tA2AR during MD simulations. Volumetric density maps were contoured by iso-surface treatment by Volmap of VMD software.33
The eccentricity of the RDC of both tA2AR and WT-β2AR in DDM shows two peaks. The peak closer to that of LMNG shows a more oblate structure (DDM 1 in Figure 2A), and the other that is farther away shows a more spherical shape (DDM 2 in Figure 2A). The bimodal distribution of the eccentricity in DDM indicates that the RDC is more flexible in DDM than in other detergents (black curve, Figure 2B). The results of the energy analysis and eccentricity taken together show that although the RDC spheroids in DDM are similar to those in LMNG they are not as tightly packed as in LMNG. Our previous study of A2AR showed that they form “oblate” spheroids in DM and OG.26,34−36Figure 2C shows water density near the TM regions of tA2AR, showing fewer waters in the LMNG system than in the OGNG system, highlighting the tighter packing in the LMNG–A2AR complex. The corresponding results for WT-β2AR are shown in Figure S2D.
Conformational Heterogeneity and Receptor Dynamics
The conformational heterogeneity of the receptor and the stability of its structure when inserted in a detergent micelle were investigated by analyzing the MD simulations. Snapshots of tA2AR and WT-β2AR were clustered according to the structural deviation from the crystal structure using the root-mean-square deviation in Cα atom coordinates and percentage helicity retained in the TM α-helices during the dynamics.
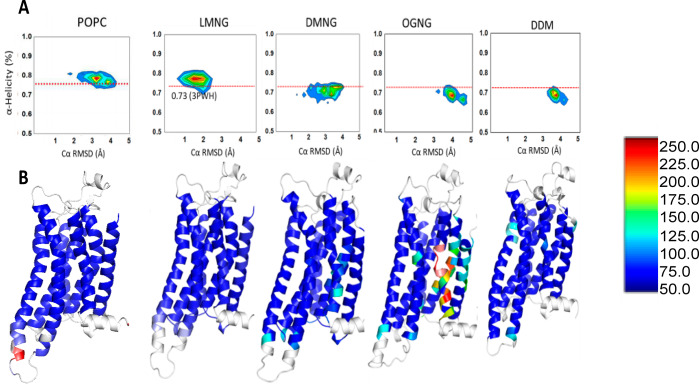
Both of these properties indicate the extent of unfolding of the receptor structure in the detergent micelle. Both tA2AR and β2AR exhibit less unraveling of the receptor structures in LMNG micelles than in DMNG, OGNG, and DDM micelles (Figure 3A and Figure S3A). Interestingly, the structural ensemble of tA2AR in its lipid bilayer environment [represented by the palmitoyloleoylphosphatidylcholine (POPC) bilayer] shows high helicity just like LMNG. Figure 3B and Figure S3B show the flexibility of each amino acid in tA2AR and WT-β2AR, respectively, as a heat map on the structures. The flexibility is quantified by the root-mean-square fluctuation (RMSF) from the average structure extracted from the MD simulations. Both tA2AR and WT-β2AR in the OGNG micelle show considerable deviations from the crystal structure, especially in TM2 and TM7. This is similar to what we observed in our previous study of the effects of DDM, DM, and OG on GPCR structure, where the harsher the detergent, the greater the deviation from the crystal structure and the lower the α-helicity of the TM regions.26 Each of the TM regions in tA2AR maintains a stable α-helix in DDM and DM simulations but shows a relatively low helicity (50–70%) in OG.
Figure 3.
Conformational heterogeneity of the inactive state of tA2AR in the POPC lipid bilayer, LMNG, DMNG, OGNG, and DDM. (A) Distributions of the average helicity of all TM region residues and RMSD (root-mean-square deviation) of each MD snapshot of tA2AR. Simulations of tA2AR (PDB entry 3PWH) were performed in POPC, LMNG, DMNG, OGNG, and DDM micelles. The red dotted lines in the figure show the average helicity of the crystal structure of tA2AR. (B) Residue-based thermal B-factor calculated from the RMSF (root-mean-square fluctuation) of tA2AR from the MD simulations in POPC, LMNG, DMNG, OGNG, and DDM detergent complexes shown as a heat map. The loop regions and helix 8 colored white have been omitted from the representation for the sake of clarity.
Packing of the Detergent Micelle around the Receptor
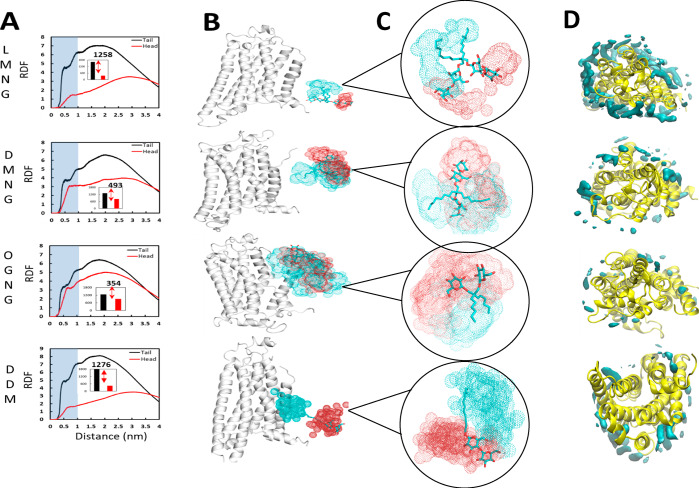
To understand the details of the detergent packing around the receptor, we calculated the distribution of the density of the head group and tail group of the detergent molecules in the micelles around tA2AR and β2AR using a radial distribution function for each detergent (Figure 4A and Figure S4A). The densities of the hydrophobic tail group of both LMNG and its unbranched counterpart DDM, are high within 10 Å of the receptor compared to those of DMNG and OGNG. The density of the polar head group peaks around 25–30 Å in LMNG and DDM for both tA2AR and β2AR. In contrast, the head and the groups show similar densities at 10 Å in OGNG. This is due to tumbling of the detergent molecules in the micelle. The spatial distribution of a selected detergent molecule shows that the LMNG molecule is less mobile in the micelle than are other detergents (Figure 4B and Figure S4B). Individual LMNG detergent molecules diffuse relatively slowly, and thus, the position of a given molecule varies little with time (Figure 4C). In contrast, molecules of OGNG tumble and thus are flexible within the micelle (Figure 4C). Tumbling of LMNG or DMNG was not observed within the simulation time. Figure 4D and Figure S4C show the volumetric density distribution of any tail carbon within 4 Å of a residue in the receptor. In the LMNG micelle, there is a higher density of the hydrophobic aliphatic “tail” groups being closer to the receptor than in DMNG, OGNG, and DDM. Thus, LMNG forms a stiffer micelle around both tA2AR and WT-β2AR, providing the hydrophobic coverage of the TM regions of the receptor.
Figure 4.
LMNG micelle showing tight packing around the receptor. (A) Radial distribution function (RDF) plot for the density for either the head group (red) or tail group (black) of the detergents as a function of distance from tA2AR in LMNG, DMNG, OGNG, and DDM. The inset and corresponding number are the RDF area difference from tail to head group within 1.0 nm (10 Å). (B) Spatial distribution (SDF) plots of the detergent molecules for LMNG, DMNG, OGNG, and DDM detergents near tA2AR from the most populated ensemble. The initial position of the detergent molecules is shown in stick representation, and the resultant spatial distributions are shown as dots: cyan (end carbon in the tail group) and red (the ether oxygen atom of the first sugar ring). (C) Close-up of the SDFs from panel B. (D) Volumetric density of the tail carbon within 4 Å of each residue in the tA2ARs. See Figure S4 for data on β2AR simulations.
Effect of Branching in LMNG Compared to Its Unbranched Counterpart DDM
The thermostable mutant of the inactive state of A2AR does not show a difference in thermostability in LMNG and DDM. However, Ashok et al. showed that the wild type A2AR shows a higher thermostability in LMNG by 11 °C compared to that in DDM. To understand the difference between the branched chain detergent LMNG and its counterpart DDM (see Figure S5A), we performed simulations on the WT-A2AR in LMNG and DDM.28
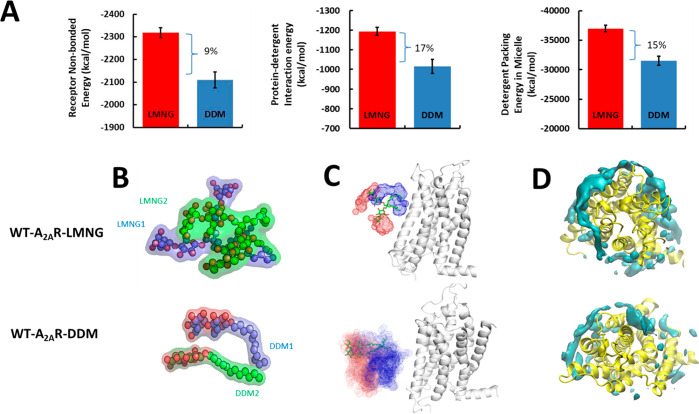
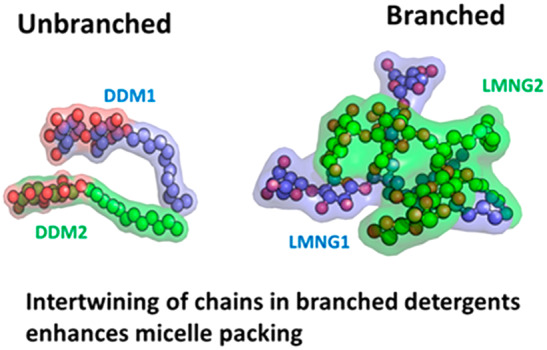
The receptor–detergent interaction energy as well as the detergent–detergent packing energy within the RDC is more favorable by 15–17% as shown in Figure 5A for the WT-A2AR in LMNG than in DDM. The enthalpic gain in receptor packing energy comes from enhanced interhelical van der Waals contacts (Figure S5B). The LMNG detergent molecules show intertwining of the polar head groups as shown in Figure 5B, making multiple hydrogen bonds between the polar head groups. The number of hydrogen bonds among the polar head groups in LMNG compared to DDM is higher as seen from the time series plot of the number of hydrogen bonds in Figure S5C. This enthalpic gain in the hydrogen bonds between the head groups in LMNG is absent in DDM because it is more mobile compared to the branched LMNG. The restricted mobility of LMNG compared to DDM within the RDC complex is evident from the density distribution of its atoms over the course of the MD simulations of a typical LMNG and DDM (Figure 5C) and also from the RMSF of each detergent molecule (Figure S5D). Additionally, the density of the hydrophobic contacts between the tail groups of LMNG and the TM region of WT-A2AR is higher in LMNG than in DDM (Figure 5D and Figure S5E). The restricted mobility and favorable packing of detergent molecules among the LMNG molecules come from the strong hydrogen bonding potential among the head groups. Thus, branching the two mobile hydrophobic alkyl chains in DDM through a central carbon as in LMNG reduces the entropy of the RDC and the extent of packing of the detergents within the micellar particle. This leads to improved stability of WT-A2AR in LMNG compared to that in DDM.
Figure 5.
Effect of branched tail groups in LMNG compared to their unbranched counterpart DDM on the stability of the wild type A2AR (WT-A2AR). (A) Average nonbond interaction energy between the WT-A2AR and LMNG or DDM molecules averaged across the MD simulation trajectories. Also shown is the average nonbond interaction energy between the detergent molecules in LMNG and DDM receptor complexes. (B) Internal structure of a representative LMNG and DDM molecule in the WT-A2AR–LMNG and WT-A2AR–DDM RDCs. The two chains in LMNG or the two monomers of DDM are colored differently, and the oxygen atoms of the hydrophilic head group are colored red. (C) Spatial distribution plots of a typical LMNG or DDM molecule near the WT-A2AR during the MD simulations. The head group is colored red, and tail groups are colored blue. (D) Volumetric density of tail carbon atoms of detergent within 4 Å of each WT-A2AR.
Branched Detergents Reduce the Flexibility of Intracellular Regions of the Wild Type A2AR Similar to POPC in the Bilayer
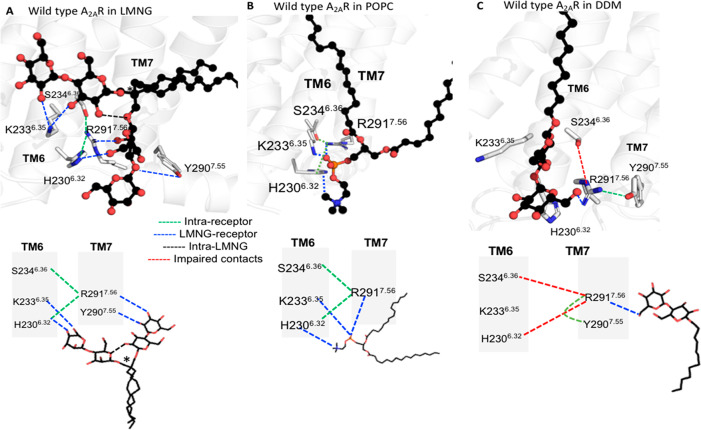
During the MD simulations of WT-A2AR in LMNG, we observed the two polar groups of LMNG form persistent bifurcated hydrogen bonds with polar residues in the intracellular regions of the helices and loops (Figure 6A and Figure S6). The bifurcated hydrogen bonds between TM helices reduce the flexibility of the receptor in the intracellular regions. This also facilitates the formation of favorable packing interactions within the receptor. A two-dimensional schematic of the hydrogen bond patterns is also shown for the sake of clarity in Figure 6A. The intramolecular hydrogen bonds between the head groups in LMNG (shown as black dotted lines in Figure 6A) are essential for keeping the two arms of the LMNG molecule in the same plane for making bifurcated hydrogen bonds with the receptor. Such bifurcated hydrogen bonds were also formed by the phosphate and choline head groups in the POPC lipid during MD simulations of WT-A2AR in the POPC bilayer (Figure 6B). These MD simulation trajectories in the POPC bilayer were taken from our previous work.37 However, such bifurcated hydrogen bonds were absent in the unbranched DDM detergents (Figure 6C). Similar bifurcated hydrogen bonds were identified in the intra- and extracellular regions of the receptor as shown in Figure S6.
Figure 6.
Branched detergent LMNG forms bifurcated hydrogen bonds between TM helices and loops in WT-A2AR. WT-A2AR is shown in white cartoon representation, and the detergents and POPC are shown in ball-and-stick representation (oxygen atoms of the polar head groups shown as red spheres). The hydrogen bonds within the receptor are shown as green dotted lines, those between LMNG and the receptor as blue dotted lines, those within the LMNG molecule as black dotted lines, and those that are broken as red dotted lines. (A) Bifurcated hydrogen bonds formed by the two polar head groups of LMNG. The two-dimensional schemes of the hydrogen bond patterns are also shown (bottom). The quaternary carbon atom of LMNG is denoted with an asterisk. (B) Bifurcated hydrogen bonds formed by the POPC head group and WT-A2AR in the lipid bilayer. The two-dimensional (2D) scheme of the hydrogen bonds is also shown (bottom). (C) Hydrogen bonds between DDM and the receptor and the 2D scheme (bottom). The Ballesteros–Weinstein (BW) residue numbering scheme is shown as superscripts.
Discussion
The class of novel branched neopentylglycol of amphiphiles has been shown to stabilize multiple GPCRs much more robustly than its unbranched counterparts.8 LMNG is a widely used member of the MNG series with a central carbon that connects the two chains of a single chain counterpart detergent DDM. Although LMNG has been used widely, the basis of how it stabilizes the GPCR structures compared to two molecules of DDM is unknown. Such knowledge is critical to the further design of detergents to solubilize >80% of GPCRs for which structures have not been determined.3 Here we have used MD simulations to study the stabilization forces in the receptor–detergent complexes of two GPCRs A2AR and β2AR. We have studied the thermostable mutant and the wild type of A2AR and wild type β2AR in four different detergents, namely, LMNG, DMNG, OGNG, and DDM.
The receptor–LMNG complex shows favorable energetics compared to those of the other three detergents studied here (DMNG, OGNG, and DDM). Although the internal nonbond energy of the receptors is similar in all four detergent complexes, the interaction energy of the receptor transmembrane region with the detergents is more favorable in LMNG than in the other detergents. The difference in the packing of the detergent molecules within the receptor–detergent complex is the major cause of the differences in stability between the LMNG complex compared to the other three detergents. The short chain detergent OGNG is flexible and tumbles within the complex, as previously observed for its single-chain counterpart OG.26 This leads to increased flexibility within the complex and therefore weaker interactions of the detergent with the receptors. All of the results for LMNG, DMNG, and OGNG are qualitatively similar to those of our previous study on DDM, DM, NG, and OG.26 This indicates that the introduction of the central quaternary carbon in the neopentylglycol detergent series did not fundamentally alter the basic properties of their behavior. Thus, the smaller the head group and the shorter the aliphatic tail, the more the detergent tumbles, the more motion there is in the receptor, and the less stable the receptor is.
LMNG is essentially two DDM molecules fused through a central quaternary carbon. The fusing of the chains in LMNG constrains the polar atoms in the maltoside head group to be near each other, thus favoring the formation of sustained hydrogen bonds between the head groups in the receptor–detergent complex. This favorable enthalpic effect is reduced in DDM because the two chains are free to move and hence the hydrogen bonds between the head groups are not sustained. Thus, we have shown that LMNG forms a tightly packed micelle around the receptor with a high degree of coverage of the hydrophobic surface of the transmembrane regions of the receptors by their hydrophobic tail groups compared to other members of the series, namely, DMNG and OGNG, and its single-chain counterpart DDM. This tight packing of the detergent molecules among themselves stabilizes the receptor conformations, promoting conformational homogeneity and enhancing the interhelical packing interactions in the GPCRs.
The receptor flexibility at the edge of TM helices and loops on the extracellular and intracellular sides is reduced by LMNG compared to DDM. This is due to the two polar head groups of LMNG forming bifurcated hydrogen bonds across TM helices and loops. This also facilitates new polar and hydrophobic contacts between adjacent TM helices. Such stabilizing features were also observed in the POPC bilayer and not in DDM. Thus, the edges of TM helices remain flexible in DDM. We posit that this could be a reason why GPCRs show a higher activity in DDM than in LMNG.
It is not possible to know all of the factors that contribute to the difference in thermal stability of the receptor–detergent complexes with a single alkyl chain and branched alkyl chain detergent. However, we have identified that the presence of a central quaternary carbon in branched micelles like MNGs would allow less flexibility of the detergent alkyl chain relative to single-chain DDM. This restricted mobility of detergents would reduce the dynamic nature of the receptor–detergent complex, resulting in enhanced stability. We also find that the double sugar rings are essential to reduce the flexibility and tumbling of detergents in the complex. It will be interesting to investigate the effect of the central carbon on micellar properties in the context of various detergent architectures.
Methods
Experimental Measurement of Tm of tA2AR in Detergents
The receptor was expressed in HEK293T cells by transient transfection (GeneJuice, Merck), maintained in culture in DMEM with 10% FBS (Sigma-Aldrich) for 36 h at 37 °C, and the thermostability assessed by measuring binding of 100 nM [3H]ZM241385 (American Radiolabeled Chemicals) for 60 min at 4 °C and then for 30 min at a range of temperatures. The excess and unbound radioligand were separated using gel filtration mini-columns following solubilization (∼10 × 106 cells) in 800 μL of buffer [50 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 100 nM [3H]ZM241385] containing either 2% LMNG, 2% DMNG, or 3% OGNG (all detergents sourced from Anatrace) for 1 h at 4 °C, and then lysates cleared by centrifugation at 16000g for 15 min. Fifty microliter portions of lysate samples were heated at different temperatures for 30 min and applied to 300 μL of gel slurry pre-equilibrated in solubilization buffer and packed prior to addition of lysate. The bound ligand fractions were separated from the free ligand by centrifugation at 200g for 5 min, and the elutions measured for radioactivity by the addition of 180 μL of a liquid scintillant; the levels of retained radioligand were then determined using a liquid scintillation counter. The Tm values measured in this work are listed in Table S1.
Systems Used for Molecular Dynamics Simulations
We used atomistic molecular dynamics (MD) simulations to investigate the effect of various detergents on GPCR stability, which has a significant impact on membrane protein extraction, solubilization, and purification. For this study, we considered three detergents, OGNG, DMNG, and LMNG,8,39,40 to examine their effects on two class A GPCRs (human β2AR and human adenosine A2AR). We performed simulations on the antagonist-bound inactive state of the thermostabilized mutant of human adenosine tA2AR, wild type A2AR, and the antagonist-bound inactive state of wild type β2AR in four different detergents as detailed below. We performed five independent velocity MD simulations, 250 ns each, totaling 1.25 μs for each receptor–detergent complex (Table S2).38
Construction of Receptor–Detergent Complex Structures
The Micelle Builder module41 in CHARMM-GUI30 was used to build the starting structure of the receptor–detergent complexes for three detergents (OGNG, DMNG, and LMNG). We used 96 monomers of OGNG, DMNG, and LMNG to surround the GPCRs simulated on the basis on an experimental study, which concluded that the number of LMNG monomers required to shield the hydrophobic region of a membrane receptor was half of the number of DDM monomers.9 We used 192 monomers of DDM in our previous study to construct a receptor–detergent complex for human A2AR (class A GPCR) that recapitulated the experimental properties of the receptor–detergent micelle.26 We chose LMNG, DMNG, and OGNG to understand how the difference in head and tail length affects the GPCR stability despite having the same number of detergent monomers in the receptor–detergent complex systems. For simulations in a membrane, the tA2AR structures was solvated in explicit 128 lipid (POPC). These simulations were performed in our previous work.26 We used these simulation trajectories from our previous work.
Building of the Receptor–Micelle Complex
All of the MD simulations were performed using the GROMACS package42 with the GROMOS force field.43 The initial coordinates of A2AR with antagonist ZM241385 bound and β2AR with antagonist carazolol bound were taken from PDB entries 3PWH(27) and 2RH1,29 respectively. The A2AR thermostable mutant in the inactive state contains eight mutations (A54L2.52, T88A3.36, R107A3.55, K122A4.43, L202A5.63, L235A6.37, V239A6.41, and S277A7.42). The A2AR and β2AR were inserted into each of the three detergent micelles built as described above. The receptor–detergent complex was constructed by inserting the receptor into hollow micelles. The partial atomic charges for each ligand are the ESP charges calculated using the HF-631G** method as implemented in the Jaguar program of the Schrödinger suite.44 The bonded and nonbonded parameters of the ligands were obtained using the web utility PRODRG.45 Each of the prepared structures was minimized in energy using the steepest descent (SD) method in GROMACS. We retained all of the crystal waters and added counterions to neutralize each system. We used the SPC force field for the waters in the simulations. Each system was replicated and assigned with different initial velocities to generate five independent simulations, resulting in a total of 30 simulations.
MD Simulation Protocol
GROMACS ver. 2016 was used for all MD simulations in this study. The receptor–detergent system and solvent waters were independently coupled to a temperature bath with a relaxation time of 0.2 ps.46 The pressure was calculated using a molecular virial and held constant by weak coupling to a pressure bath with a relaxation time of 0.5 ps. For all of the equilibration simulations, the receptor was positionally restrained, and the simulations were performed at constant pressure (NPT). The bond lengths and geometry of the water molecules were constrained using the SHAKE algorithm.47 For equilibration of the receptor–detergent complexes, the atoms of the protein were positionally restrained using a harmonic restraining force with a force constant of 10000 kJ mol–1 nm–1 during the 1 ns equilibration at 310 K. In this step, the water molecules and detergent could be moved to optimize their packing around the receptor. The system was further equilibrated using the NPT ensemble, while the force constant of the restraining force was set to 2100 kJ mol–1 nm–1 and reduced to zero stepwise each 2.5 ns. At this point, the pressure coupling was switched on. We performed an additional 5 ns of simulations without restraints before the production runs. Five independent simulations each to 250 ns were performed with different starting velocities. The details of the methods used for calculating properties calculated from MD simulation trajectories are given in the Supporting Information.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00183.
Details of analysis methods, data tables, and supporting figures (PDF)
Author Present Address
# S.L.: PharmCADD, 221, 17, APEC-ro, Haeundae-gu, Busan, Republic of Korea.
Author Present Address
∥ S.J.: Quantumzyme LLP, 110/8 1st Floor, Lal Bagh Main Rd, Krishnappa Layout, Bengaluru, Karnataka 560027, India.
Author Present Address
⊥ N.R.: Antibody Discovery/Protein Engineering at Oxford Genetics Ltd, Harrow, United Kingdom.
Author Contributions
N.V. and C.G.T. conceived the ideas and designed the experiments. S.L., S.G. and S.J. executed the MD simulation work and performed the analysis. N.R. performed thermostability assays. S.L., S.G., N.V., N.R., and C.G.T. wrote the paper.
This work was funded by National Institutes of Health Grants R01-GM097261 and R01-GM117923 to N.V. Work on GPCRs in C.G.T.’s lab is funded by the Medical Research Council (MC_U105197215) and an ERC Advanced Grant (EMPSI 339995).
The authors declare no competing financial interest.
Supplementary Material
References
- Rosenbaum D. M.; Rasmussen S. G.; Kobilka B. K. (2009) The structure and function of G-protein-coupled receptors. Nature 459 (7245), 356–363. 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J.; Deupi X.; Lebon G.; Tate C. G.; Schertler G. F.; Babu M. M. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494 (7436), 185–194. 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Hauser A. S.; Attwood M. M.; Rask-Andersen M.; Schioth H. B.; Gloriam D. E. (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discovery 16 (12), 829–842. 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C. G. (2010) Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol. Biol. 601, 187–203. 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- Palczewski K.; Kumasaka T.; Hori T.; Behnke C. A.; Motoshima H.; Fox B. A.; Le Trong I.; Teller D. C.; Okada T.; Stenkamp R. E.; Yamamoto M.; Miyano M. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289 (5480), 739–745. 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Carpenter B.; Nehme R.; Warne T.; Leslie A. G.; Tate C. G. (2016) Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536 (7614), 104–107. 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. G.; Choi H. J.; Rosenbaum D. M.; Kobilka T. S.; Thian F. S.; Edwards P. C.; Burghammer M.; Ratnala V. R.; Sanishvili R.; Fischetti R. F.; Schertler G. F.; Weis W. I.; Kobilka B. K. (2007) Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450 (7168), 383–387. 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Warne T.; Serrano-Vega M. J.; Baker J. G.; Moukhametzianov R.; Edwards P. C.; Henderson R.; Leslie A. G.; Tate C. G.; Schertler G. F. (2008) Structure of a beta1-adrenergic G-protein-coupled receptor. Nature 454 (7203), 486–491. 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae P. S.; Rasmussen S. G.; Rana R. R.; Gotfryd K.; Chandra R.; Goren M. A.; Kruse A. C.; Nurva S.; Loland C. J.; Pierre Y.; Drew D.; Popot J. L.; Picot D.; Fox B. G.; Guan L.; Gether U.; Byrne B.; Kobilka B.; Gellman S. H. (2010) Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods 7 (12), 1003–1008. 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R. W. (1992) Flexible Molecules with Defined Shape - Conformational Design. Angew. Chem., Int. Ed. Engl. 31 (9), 1124–1134. 10.1002/anie.199211241. [DOI] [Google Scholar]
- McQuade D. T.; Quinn M. A.; Yu S. M.; Polans A. S.; Krebs M. P.; Gellman S. H. (2000) Rigid Amphiphiles for Membrane Protein Manipulation. Angew. Chem., Int. Ed. 39 (4), 758–761. . [DOI] [PubMed] [Google Scholar]
- Chae P. S.; Wander M. J.; Bowling A. P.; Laible P. D.; Gellman S. H. (2008) Glycotripod amphiphiles for solubilization and stabilization of a membrane-protein superassembly: importance of branching in the hydrophilic portion. ChemBioChem 9 (11), 1706–1709. 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J.; Pamula F.; Nehme R.; Muhle J.; Weinert T.; Flock T.; Nogly P.; Edwards P. C.; Carpenter B.; Gruhl T.; Ma P.; Deupi X.; Standfuss J.; Tate C. G.; Schertler G. F. X. (2018) Crystal structure of rhodopsin in complex with a mini-Go sheds light on the principles of G protein selectivity. Sci. Adv. 4 (9), eaat7052 10.1126/sciadv.aat7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. G.; Choi H. J.; Fung J. J.; Pardon E.; Casarosa P.; Chae P. S.; Devree B. T.; Rosenbaum D. M.; Thian F. S.; Kobilka T. S.; Schnapp A.; Konetzki I.; Sunahara R. K.; Gellman S. H.; Pautsch A.; Steyaert J.; Weis W. I.; Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469 (7329), 175–180. 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D. M.; Zhang C.; Lyons J. A.; Holl R.; Aragao D.; Arlow D. H.; Rasmussen S. G.; Choi H. J.; Devree B. T.; Sunahara R. K.; Chae P. S.; Gellman S. H.; Dror R. O.; Shaw D. E.; Weis W. I.; Caffrey M.; Gmeiner P.; Kobilka B. K. (2011) Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature 469 (7329), 236–240. 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A. C.; Hu J.; Pan A. C.; Arlow D. H.; Rosenbaum D. M.; Rosemond E.; Green H. F.; Liu T.; Chae P. S.; Dror R. O.; Shaw D. E.; Weis W. I.; Wess J.; Kobilka B. K. (2012) Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482 (7386), 552–556. 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K.; Kruse A. C.; Asada H.; Yurugi-Kobayashi T.; Shiroishi M.; Zhang C.; Weis W. I.; Okada T.; Kobilka B. K.; Haga T.; Kobayashi T. (2012) Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482 (7386), 547–551. 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A.; Kruse A. C.; Kobilka T. S.; Thian F. S.; Mathiesen J. M.; Sunahara R. K.; Pardo L.; Weis W. I.; Kobilka B. K.; Granier S. (2012) Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature 485 (7398), 321–326. 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier S.; Manglik A.; Kruse A. C.; Kobilka T. S.; Thian F. S.; Weis W. I.; Kobilka B. K. (2012) Structure of the delta-opioid receptor bound to naltrindole. Nature 485 (7398), 400–404. 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. F.; Noinaj N.; Shibata Y.; Love J.; Kloss B.; Xu F.; Gvozdenovic-Jeremic J.; Shah P.; Shiloach J.; Tate C. G.; Grisshammer R. (2012) Structure of the agonist-bound neurotensin receptor. Nature 490 (7421), 508–513. 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A. C.; Ring A. M.; Manglik A.; Hu J.; Hu K.; Eitel K.; Hubner H.; Pardon E.; Valant C.; Sexton P. M.; Christopoulos A.; Felder C. C.; Gmeiner P.; Steyaert J.; Weis W. I.; Garcia K. C.; Wess J.; Kobilka B. K. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504 (7478), 101–106. 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A. M.; Manglik A.; Kruse A. C.; Enos M. D.; Weis W. I.; Garcia K. C.; Kobilka B. K. (2013) Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature 502 (7472), 575–579. 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. S.; Aricescu A. R. (2014) Crystal structure of a human GABAA receptor. Nature 512 (7514), 270–275. 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M.; Cherezov V. (2009) Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4 (5), 706–731. 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Nafria J.; Tate C. G. (2019) Cryo-EM structures of GPCRs coupled to Gs, Gi and Go. Mol. Cell. Endocrinol. 488, 1–13. 10.1016/j.mce.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Lee S. B.; Mao A.; Bhattacharya S.; Robertson N.; Grisshammer R.; Tate C. G.; Vaidehi N. (2016) How Do Short Chain Nonionic Detergents Destabilize G-Protein-Coupled Receptors?. J. Am. Chem. Soc. 138 (47), 15425–15433. 10.1021/jacs.6b08742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore A. S.; Robertson N.; Errey J. C.; Ng I.; Hollenstein K.; Tehan B.; Hurrell E.; Bennett K.; Congreve M.; Magnani F.; Tate C. G.; Weir M.; Marshall F. H. (2011) Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure 19 (9), 1283–1293. 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok Y.; Nanekar R.; Jaakola V. P. (2015) Defining thermostability of membrane proteins by western blotting. Protein Eng., Des. Sel. 28 (12), 539–542. 10.1093/protein/gzv049. [DOI] [PubMed] [Google Scholar]
- Cherezov V.; Rosenbaum D. M.; Hanson M. A.; Rasmussen S. G.; Thian F. S.; Kobilka T. S.; Choi H. J.; Kuhn P.; Weis W. I.; Kobilka B. K.; Stevens R. C. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318 (5854), 1258–1265. 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29 (11), 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Oliver R. C.; Lipfert J.; Fox D. A.; Lo R. H.; Doniach S.; Columbus L. (2013) Dependence of micelle size and shape on detergent alkyl chain length and head group. PLoS One 8 (5), e62488 10.1371/journal.pone.0062488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J.; Columbus L.; Chu V. B.; Lesley S. A.; Doniach S. (2007) Size and shape of detergent micelles determined by small-angle X-ray scattering. J. Phys. Chem. B 111 (43), 12427–12438. 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD-Visual Molecular Dynamics. J. Mol. Graphics 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Rouse S. L.; Sansom M. S. (2015) Interactions of lipids and detergents with a viral ion channel protein: molecular dynamics simulation studies. J. Phys. Chem. B 119 (3), 764–772. 10.1021/jp505127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy C.; Auvray X.; Petipas C.; Rico-Lattes I.; Lattes A. (1997) Anomeric effects on the structure of micelles of alkyl maltosides in water. Langmuir 13 (15), 3965–3967. 10.1021/la9604285. [DOI] [Google Scholar]
- Das M.; Du Y.; Mortensen J. S.; Ramos M.; Ghani L.; Lee H. J.; Bae H. E.; Byrne B.; Guan L.; Loland C. J.; Kobilka B. K.; Chae P. S. (2019) Trehalose-Cored Amphiphiles for Membrane Protein Stabilization: Importance of the Detergent Micelle Size in GPCR Stability. Org. Biomol. Chem. 17, 3249–3257. 10.1039/C8OB03153C. [DOI] [PubMed] [Google Scholar]
- Lee S.; Bhattacharya S.; Grisshammer R.; Tate C.; Vaidehi N. (2014) Dynamic Behavior of the Active and Inactive States of the Adenosine A2A Receptor. J. Phys. Chem. B 118, 3355–3365. 10.1021/jp411618h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. Y.; Kim T. H.; Manglik A.; Alvares R.; Kobilka B. K.; Prosser R. S. (2012) Role of Detergents in Conformational Exchange of a G Protein-coupled Receptor. J. Biol. Chem. 287 (43), 36305–36311. 10.1074/jbc.M112.406371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellosalo J.; Kajander T.; Kogan K.; Pokharel K.; Goldman A. (2012) The structure and catalytic cycle of a sodium-pumping pyrophosphatase. Science 337 (6093), 473–476. 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]
- Frick A.; Eriksson U. K.; de Mattia F.; Oberg F.; Hedfalk K.; Neutze R.; de Grip W. J.; Deen P. M.; Tornroth-Horsefield S. (2014) X-ray structure of human aquaporin 2 and its implications for nephrogenic diabetes insipidus and trafficking. Proc. Natl. Acad. Sci. U. S. A. 111 (17), 6305–6310. 10.1073/pnas.1321406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.; Jo S.; Lee H. S.; Klauda J. B.; Im W. (2013) CHARMM-GUI micelle builder for pure/mixed micelle and protein/micelle complex systems. J. Chem. Inf. Model. 53 (8), 2171–2180. 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. (2008) GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 4 (3), 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Oostenbrink C.; Villa A.; Mark A. E.; van Gunsteren W. F. (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 25 (13), 1656–1676. 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Bochevarov A.; Harder E.; Hughes T. F.; Greenwood J. R.; Braden D. A.; Philipp D. M.; Rinaldo D.; Halls M. D.; Zhang J.; Friesner R. A. (2013) Jaguar: A High-Performance Quantum Chemistry Software Program with Strengths in Life and Material Sciences. Int. J. Quantum Chem. 113 (18), 2110–2142. 10.1002/qua.24481. [DOI] [Google Scholar]
- Schüttelkopf A.; van Aalten D. M. F. (2004) PRODRG: A Tool for High-Throughput Crystallography of Protein-Ligand Complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60 (8), 1355–1963. 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; Vangunsteren W. F.; Dinola A.; Haak J. R. (1984) Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 81 (8), 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23 (3), 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.