Abstract
Three-dimensional (3D) organoids can serve as an in vitro platform to study cell–cell interactions, tissue development, and toxicology. Development of organoids with tissue architecture similar to testis in vivo has remained a challenge. Here, we present a microwell aggregation approach to establish multicellular 3D testicular organoids from pig, mouse, macaque, and human. The organoids consist of germ cells, Sertoli cells, Leydig cells, and peritubular myoid cells forming a distinct seminiferous epithelium and interstitial compartment separated by a basement membrane. Sertoli cells in the organoids express tight junction proteins claudin 11 and occludin. Germ cells in organoids showed an attenuated response to retinoic acid compared to germ cells in 2D culture indicating that the tissue architecture of the organoid modulates response to retinoic acid similar to in vivo. Germ cells maintaining physiological cell–cell interactions in organoids also had lower levels of autophagy indicating lower levels of cellular stress. When organoids were treated with mono(2-ethylhexyl) phthalate (MEHP), levels of germ cell autophagy increased in a dose-dependent manner, indicating the utility of the organoids for toxicity screening. Ablation of primary cilia on testicular somatic cells inhibited the formation of organoids demonstrating an application to screen for factors affecting testicular morphogenesis. Organoids can be generated from cryopreserved testis cells and preserved by vitrification. Taken together, the testicular organoid system recapitulates the 3D organization of the mammalian testis and provides an in vitro platform for studying germ cell function, testicular development, and drug toxicity in a cellular context representative of the testis in vivo.
Keywords: organoid, testicular morphogenesis
Prepubertal testicular cells undergo in vitro morphogenesis and give rise to spherical organoids with testes-specific tissue architecture.
Introduction
The testis is a complex multicellular environment. It is divided into two distinct compartments: the seminiferous tubules within which spermatogenesis occurs and the interstitial tissue where endocrine tissue is located, including Leydig cells [1]. In the immature testis, the seminiferous tubules are surrounded by a basement membrane and peritubular myoid cells, and contain spermatogonia and Sertoli cells [2]. Both Sertoli and peritubular myoid cells contribute to the production of the basement membrane: peritubular myoid cells produce laminin, collagen IV, and fibronectin, while Sertoli cells produce laminin and collagen IV [3]. Sertoli cells also form tight junctions as part of the blood testis barrier, which contributes to the immune privileged environment inside the seminiferous tubules [4], and control the fate of peritubular myoid cells and Leydig cells during development [5].
Spermatogenesis is a stem cell-driven system. During spermatogenesis in the mature testis, spermatogonial stem cells, located at the basement membrane, undergo several rounds of mitotic divisions, then enter meiosis and give rise to spermatocytes and subsequently haploid spermatids. Somatic cells in the interstitial tissue and seminiferous tubules produce a broad range of secreted factors such as GDNF, CSF1, FGF1, Wnt3a, Wnt6, and androgens such as testosterone that regulate the fate of spermatogonia [2, 6–8]. These cell–cell interactions are essential for testicular morphogenesis and function [9].
Established two-dimensional (2D) culture systems for testicular cells [10] do not reflect the complex multicellular interactions and intricate signaling found in vivo [11]. Production of fertilization-competent sperm in an organ culture system is described in mice [12], and testicular cells can form functional testis tissue de novo when ectopically grafted to mice [13–15]. However, the in vitro generation of three-dimensional (3D) testicular organoids with a testis-specific tissue architecture from self-organizing testicular single cells has remained elusive despite several attempted studies [16–19]. Baert and colleagues [17] reported generation of human testicular organoids using testicular cells from both adult and pre-pubertal human testes using a hanging transwell system but the organoids lacked testis-specific topography. Pendergraft et al. [18] reported generation of organoids from adult human testicular cells using a U-bottom plate. This approach required a large number of cells, and therefore used immortalized Sertoli and Leydig cell lines. It also involved use of decellularized human testis-derived extra cellular matrix (ECM), which is time consuming to prepare and limits practical application due to the scarce availability of human testes. A scalable testicular organoid system with clear and distinct seminiferous tubules and interstitial compartments is required to allow the study of germ cells in their native niche.
Methods such as hanging drop suspension culture may result in one or more organoids of variable size and shape, likely due to the slow rate of assembly from gravity-based settling on an arcuate surface. We previously employed a combination of centrifugal forced aggregation with microwells tapered to submicron dimensions to rapidly and reproducibly induce the formation of large numbers of size- and composition-controlled organoids, with uniformly spherical initial geometry [20–22].
To provide a consistent platform for comparative studies, we sought to establish a process applicable across a range of species. While the availability of donated human testicular material is limited, human organoids are of great interest for their utility in studies of human health. Macaques provide a valuable model closely related to humans [23–26], although both practical and ethical considerations can limit tissue supplies. With their greater availability and long history, mouse models have proved informative [10, 27, 28], although with substantial differences in scale and reproductive strategies, Discoveries in rodent models often fail to translate to humans [29–31]. Pigs have recently emerged as a widely used translational model for biomedical research. They are an outbred species that share many physiological similarities with humans [32, 33], and porcine testes are available in quantity as a by-product of agricultural production.
Here, we report uniform, reproducible formation of 3D testicular organoids with physiologically relevant tissue architecture, with evidence of germ cell and somatic cell association similar to testis in vivo. Our approach is applicable to fresh and cryopreserved testis cells, and to cells from pig, mouse, nonhuman primate, and human testis, giving it broad potential impact for the study of morphogenesis, cell–cell interactions, drug and toxicity screening, and regenerative medicine.
Methods
Isolation of porcine testicular cells
Testes from 1-week-old piglets were obtained from Sunterra Farms Ltd (Acme, Alberta, Canada), as by-product from castration of commercial pigs. Sourcing of testes was approved by the Animal Care Committee at the University of Calgary (UofC). Cells were harvested using a two-step enzymatic digestion process as previously described [34]. Briefly, testes were decapsulated and minced into ∼1–2 mm size pieces. The pieces were digested using collagenase IV (Sigma) (2 mg/mL), and cells were isolated using 0.25% trypsin–EDTA (Sigma) and DNase I (Sigma) (7 mg/mL) to obtain the starting cell population. All experiments were replicated using a minimum of three independently prepared cell suspensions.
Isolation of mouse testicular cells
Testes from P8-P10 mice were obtained by euthanasia (approved by the Animal Care Committee, the UofC) and digested using collagenase type IV in Hank's Balanced Salt Solution (HBSS, Gibco) for 15 min at 37°C. The tubules were then sedimented by centrifugation at 90 × g for 1.5 min and washed with HBSS. Individual cells were isolated using 0.25% trypsin–EDTA and DNase I (7 mg/mL) to obtain the starting cell population. All experiments were replicated using a minimum of three independently prepared cell suspensions.
Isolation of monkey and human testicular cells
Cells were isolated from 2-year-old rhesus macaque (n = 3), 6-month-old and 5-year-old human (n = 2) frozen testicular tissue using a two-step enzymatic digestion described previously [23, 24, 35, 36]. Macaque testis tissue was obtained with permission of the Institutional Animal Care and Use Committee, and human testis tissue was obtained through the University of Pittsburgh Health Sciences Tissue Bank and Center for Organ Recovery and Education (CORE) with approval from the University of Pittsburgh Committee for Oversight of Research and Clinical Training Involving Decedents (CORID protocol #686). Testicular tissue was digested using collagenase type IV for 5 min at 37°C on the shaker (250 rpm), then shaken vigorously and incubated for another 3+2 min at 37°C on the shaker. The tubules were then sedimented by centrifugation at 200 × g for 5 min and washed with HBSS. The tubules were digested with 0.25% trypsin/EDTA and DNase I. The suspension was triturated vigorously 3–5 times and incubated at 37°C for 5 min. The process was repeated in 5-min increments up to 15 min. The digestion was stopped by adding 10% fetal bovine serum, and the cells were strained through a 70-μm strainer (Becton Dickson). The cells were pelleted by centrifugation at 600 × g for 15 min.
Characterization of testicular cells
The porcine testicular cells obtained after enzymatic digestion were fixed using 4% PFA/PBS and placed on slides by cytospin centrifugation. The slides were washed with PBS, blocked with CAS-Block, and incubated overnight with anti-GATA4 (GATA Binding Protein 4; 1:200 dilution; Santa Cruz Biotechnology) to identify Sertoli cells, anti-UCH-L1(Ubiquitin C-Terminal Hydrolase L1; 1:100 dilution; Abcam) to label spermatogonia, anti-α-SMA(alpha-Smooth Muscle Actin; 1:200 dilution; Abcam) to identify peritubular myoid cells, anti-Cytochrome P450(1:100; Bio-RAD) for Leydig cells, and anti-CD31(Cluster of differentiation 31, 1:100, Abcam) for endothelial cells (Supplementary Table S1). The slides were then incubated with secondary antibodies conjugated with Alexa Fluor 488 and 555. The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and the cells were analyzed using fluorescence microscopy. The relative percentages of germ cells and somatic cell types were performed by counting the cells with ImageJ software.
Generation of 3D testicular organoids in microwell culture
AggreWell 400 plates (STEMCELL Technologies Inc, Vancouver, Canada, cat# 34450) were prepared according to the manufacturer's instructions, washed once with 0.5 mL PBS, and 0.5 mL of organoid formation medium (Dulbecco Modified Eagle Medium F/12 supplemented with insulin 10 μg/mL, transferrin 5.5 μg/mL, selenium 6.7 ng/mL, 20 ng/mL epidermal growth factor, 1% Penicillin-Streptomycin) [37] was placed in the wells. Each plate was centrifuged at 2000 × g for 2 min to remove trapped air, 1.2 × 106 (1000 cell organoids) or 6 × 105 (500 cell organoids) fresh or frozen-thawed testicular cells suspended in 0.5 mL medium were placed into each well, and the plates were centrifuged at 500 × g for 5 min. The culture medium for the mouse samples was supplemented with 1:100 dilution of Matrigel. The forming organoids were maintained (1 mL medium/well) in microwell plates for 5 days at 37°C in 5% Co2 in air with half medium change every second day.
Characterization of organoids
The cultured organoids were collected at 5 days of culture and analyzed using confocal microscopy. Briefly, the organoids were fixed with 4% PFA/PBS and placed on slides by cytospin centrifugation. The slides were washed with PBS, blocked with CAS-Block, and incubated overnight with anti-GATA4 (1:200 dilution; Santa Cruz Biotechnology), anti-UCH-L1 (1:100 dilution; Abcam), anti-α-SMA (1:200 dilution; Abcam), anti-Cytochrome P450 (1:100; Bio-Rad), anti-Collagen IV (1:400 dilution; Abcam), anti-Laminin (1:400 dilution; Abcam), anti-Fibronectin (1:400 dilution; Abcam), anti-Occludin (1:100 dilution; Abcam), and anti-Claudin 11 (1:100 dilution; Thermo Fisher) (Supplementary Table S1). The slides were then incubated with secondary antibodies conjugated with Alexa Fluor 488 and 555. The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and the organoids were examined using a Leica TCS-SP8 confocal laser scanning microscope using the Leica Las X software.
Relative enrichment of germ cells
Testicular cells obtained after the enzymatic digestion were enriched for spermatogonia by differential adhesion culture as described [38]. A sample was fixed, and the percentage of germ cells was determined based on expression of UCH-L1. Enriched cells were then combined with the initial testicular cell population (5% germ cells) to obtain cell populations containing 15, 25, and 50% germ cells. These cell populations were seeded in microwells to produce organoids as previously described. Following 5 days of culture, organoids were collected, fixed, and stained with anti-UCH-L1 as described earlier. The number of germ cells attached to each organoid was counted.
Retinoic acid treatment
Testicular cell populations containing 25% germ cells were used to generate organoids (3D culture) as described above. For comparison, the same cell population was plated in a 24-well plate (2D culture). Both the 2D cell and 3D organoid cultures were maintained for 5 days. The samples were then exposed to 1 μM Retinoic acid (RA) for 48 h and were collected, fixed with 4% PFA/PBS, stained for Stra8 (Stimulated by Retinoic acid 8; 1:100 dilution, Abcam) and UCH-L1 (1:100 dilution, Abcam) (Supplementary Table S1). Using confocal microscopy, the numbers of Stra8+ve and UCHL1+ve cells were counted from both 2D and organoid cultures.
Comparison of autophagosome numbers between 2D and 3D cultures
Testicular cell populations containing 25% germ cells were used to generate organoids (3D culture) as described earlier. For comparison, the same cell population was plated in a 24-well plate (2D culture). Both the 2D and 3D cultures were maintained for 5 days. The samples were collected by pipetting, fixed with 4% PFA/PBS, and stained for LC3B (microtubule-associated protein 1A/1B-light chain 3; 1:100 dilution, Cell Signaling) and UCH-L1 (1:100 dilution, Abcam) (Supplementary Table S1). Using confocal microscopy and ImageJ software, the number of LC3B puncta of each germ cell was counted [38]. A total of 10 germ cells from 2D and 3D cultures each were analyzed.
MEHP treatment
Testicular organoids generated from 25% germ cell enriched testicular cells were exposed to 0.5, 1, and 1.5 μM MEHP (mono 2-ethylhexyl) phthalate in dimethyl sulfoxide (DMSO) for 12 h at day 5 of culture. The control group was treated with 1.5 μM DMSO for 12 h. The samples were processed and analyzed as above. A total of 10 germ cells from each treatment and control were analyzed.
Inhibition of primary cilia
Testicular cells from 1-week-old pigs were seeded into microwells, and 2 μM Ciliobrevin D dissolved in DMSO was added to the organoid formation medium. For the control group an equivalent amount of DMSO was added to the medium. The formation of testicular organoids was analyzed at day 5 using the inverted microscope. Organoids from the treatment and control groups were also stained with anti-GATA4 (1:200, Santa Cruz Biotechnology), anti-αSMA (1:200, Abcam), and anti-Collagen IV (1:400, Abcam) and analyzed using confocal microscopy.
Cryopreservation of porcine testicular organoids
After 5 days of culture, the organoids were vitrified using a vitrification protocol previously described [39]. Ethylene glycol containing 0.9% NaCl and 0.5M raffinose was used as the vitrification solution. The solution was diluted with PBS to prepare 12.5, 25, and 50% (v/v) vitrification solutions. The organoids were then immersed into 12.5 and 25% vitrification solutions at room temperature for 5 min and 50% vitrification solution in ice for 15 min. Finally, the organoids were transferred into cryovials with 100% vitrification solution and frozen in liquid nitrogen. For thawing, cryovials were removed from liquid nitrogen and thawed in ice water. Once the vitrification liquid became clear, the organoids were successively transferred into 50 and 25% vitrification solutions for 10 min each on ice. Lastly, the organoids were successively transferred to 12.5% vitrification solution and PBS at room temperature for 10 min each. The frozen-thawed organoids were then divided into two groups: one group was immediately fixed using 4%PFA/PBS, while the other one was cultured for 7 days at 37°C in 5% C02 in air with half medium change every second day. After 7 days of culture, the organoids were fixed using 4%PFA/PBS. Both groups of frozen-thawed organoids were analyzed by immunocytochemistry as described above.
Statistical analysis
All results reported are from at least three independent experiments performed with separately prepared starting cell populations. The data were analyzed by unpaired two-tailed t-tests to compare two groups, and one-way ANOVA for more than two groups with the Tukey multiple comparisons test. A value of P < 0.05 was set as the limit of statistical significance. All the statistical analyses were performed using the GraphPad Prism 7 software.
Results
Porcine testicular organoids have testes-specific cell associations
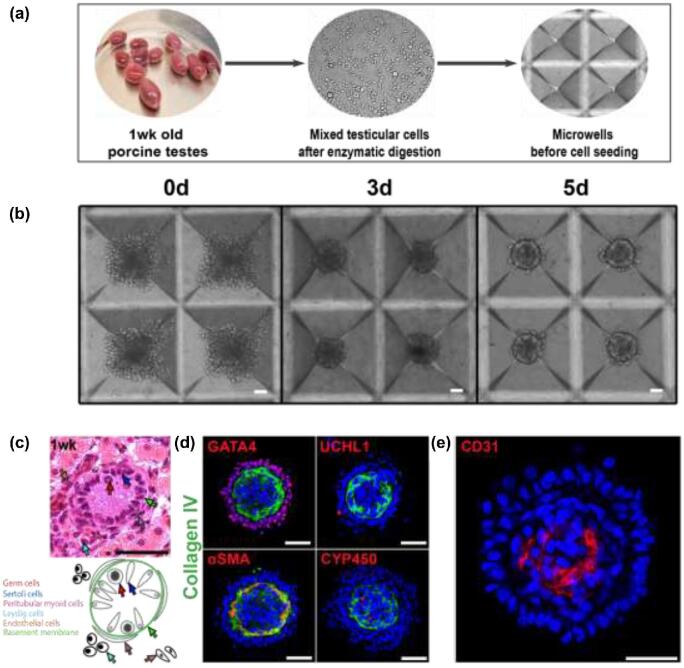
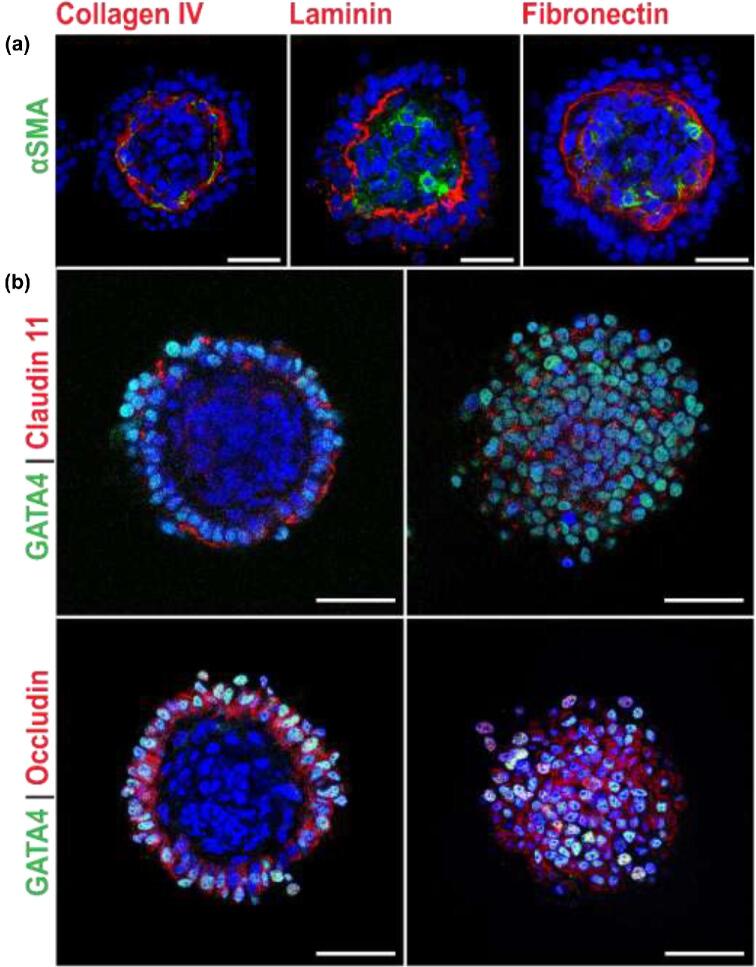
Porcine pre-pubertal testicular cells (containing 67.4% ± 0.45 GATA4+ve Sertoli cells [40], 4.5% ± 0.14 UCH-L1+ve germ cells [41–44], 1.9% ± 0.28 Cytohrome P450+ve Leydig cells [45], 3.6% ± 0.3 αSMA+ve peritubular myoid cells [46, 47], and 11.64% ± 0.82 CD31+ve endothelial cells [48]) (n = 3 separate starting cell populations) were seeded into microwells (1000 cells/microwell) and cultured for 5 days. Over the course of 4–5 days, clustered cells self-organized into spheroids (Figure 1a and b) with a diameter of 166.4 ± 4.15 μm (n = 3 experiments, 10 organoids analyzed from each), with a clearly delineated exterior and interior compartment (Figure 1d). UCHL1+ve germ cells and GATA4+ve Sertoli cells were located in the exterior compartment on a collagen IV+ve basement membrane (Figure 1d) located at the intercompartmental boundary. α-SMA+ve peritubular myoid cells were located in the interior compartment along the inside of this membrane; and Cytochrome P450+ve Leydig and CD31+ve endothelial cells were at the core of the interior compartment (Figure 1d and e). This two-compartment structure reflects the natural tissue organization in vivo, where Cytochrome P450+ve Leydig cells and αSMA+ve peritubular myoid cells are found in the interstitial compartment (Figure 1c); peritubular myoid cells are located in the interstitial compartment immediately adjacent to the basement membrane (Figure 1c); and UCHL1+ve germ cells and GATA4+ve Sertoli cells are in the tubule compartment on the opposite side of the basement membrane (Figure 1c). The intercompartment basement membrane in the organoids was positive for ECM proteins laminin and fibronectin (Figure 2a), indicative of peritubular myoid cell function [3], and tight junction proteins claudin 11 and occludin co-localized with GATA4+ve Sertoli cells (Figure 2b).
Figure 1.
Microwell-derived testicular organoids exhibit testis-specific tissue architecture with an inverted topography. (a) Schematic representation of cell harvesting and microwell seeding. (b) Porcine testicular cells at 0, 3, and 5 days of microwell culture. (c) Histological appearance (H&E) and schematic representation of 1-week-old pig testis. Specific cell types are indicated with corresponding arrows. (d) Immunofluorescence images of testicular organoids identifying specific cell types: Sertoli cells (GATA4), basement membrane (Collagen IV); germ cells (UCH-L1); peritubular myoid cells (α-SMA); Leydig cells (CYP450). (e) Immunofluorescence image of endothelial cells (CD31). Scale bars = 50 μm.
Figure 2.
Sertoli and peritubular myoid cells maintain key structural and functional features in organoids. (a) Sertoli cells and PMCs produce collagen IV, laminin, and fibronectin to form the basement membrane. (b) Sertoli cells express tight junction proteins claudin 11 (upper panel) and occludin (lower panel). Left panels show cross section of organoids, and right panels show a top down view of the same organoids. Scale bars = 50 μm.
Manipulation of germ cell numbers in the organoids
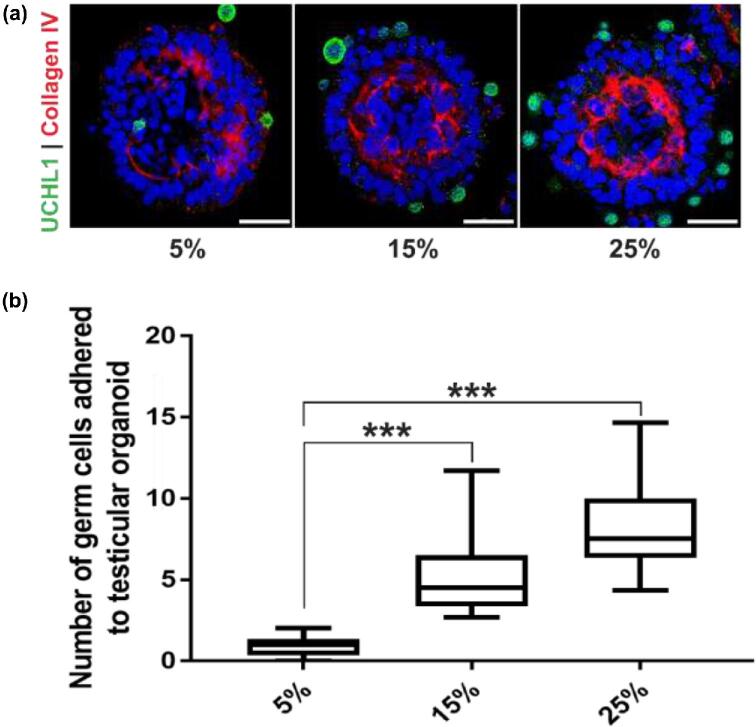
The number of germ cells on each organoid generated from a cell population containing 5% germ cells was low (0.9 ± 0.5; n = 3 experiments, 20 organoids each). To increase the number of germ cells adhered to the organoids, cell populations containing 15, 25, and 50% germ cells were used to generate organoids. Cell populations with 15 and 25% germ cells formed organoids with a topography consistent with the organoids from nongerm cell enriched cells, but with a higher number of germ cells on each organoid (5.1 ± 2.2, P < 0.001 for 15%; and 8.3 ± 2.8, P < 0.001 for 25% germ cells, n = 3 experiments, 20 organoids each, F = 60.45, df = 57) (Figure 3a and b). However, testicular cell populations with 50% germ cells failed to form any organoids presumably due to the lack of sufficient somatic cells to build the organoid scaffold.
Figure 3.
Number of germ cells on organoids can be increased. (a) Immunofluorescence images showing UCHL1+ve germ cells on organoids derived from 5, 15, and 25% germ cell enrichment in starting cell population. Scale bars = 50 μm. (b) Number of germ cells adhered to each organoid for 5, 15, and 25% germ cell enriched cell population. n = 3 experiments, mean ± SD, P < 0.001(***).
Germ cells in organoids exhibit an attenuated response to retinoic acid stimulation
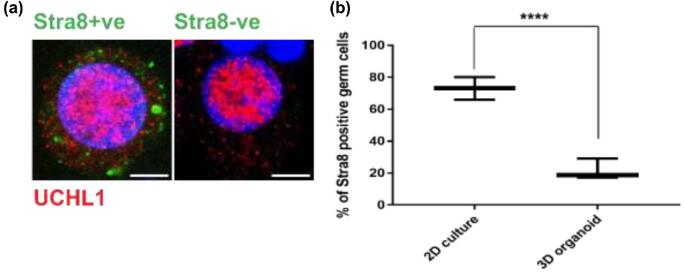
Retinoic acid, a biologically active form of retinol or vitamin A, is essential for germ cell differentiation [49], and responsiveness of mouse germ cells to RA is modulated by the testicular microenvironment [50]. Expression of Stra8 (stimulated by retinoid acid 8), an RA responsive element [51], was used as a readout for responsiveness of germ cells to RA stimulation. The numbers of Stra8+ve germ (UCHL1+ve) cells in 2D culture and 3D testicular organoid culture systems were counted. Mimicking the in vivo situation, the percentage of Stra8+ve germ cells in 3D testicular organoids was significantly lower (21.7 ± 6.5, n = 3 experiments, 50 germ cells each, P = 0.0008, t = 9.237, df = 4) compared to testicular cells in 2D culture (73.1 ± 7.1, n = 3, 100 germ cells each, P = 0.0008, t = 9.237, df = 4) using the same media conditions (Figure 4a and b).
Figure 4.
Germ cell response to retinoic acid is attenuated in organoids. (a) Representative immunofluorescence images of Stra8+ve (left panel) and Stra8−ve germ cells (right panel). Scale bars = 5 μm. (b) Number of Stra8+ve germ cells in 2D culture and 3D organoid culture. n = 3 experiments, mean ± SD, P < 0.001(***).
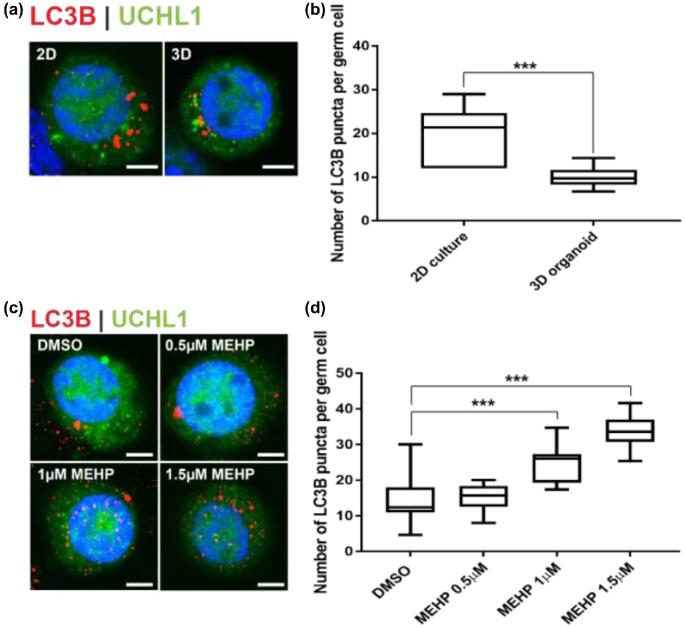
Germ cells in 3D organoids experience less cellular stress than in 2D culture
Autophagy, a process of cellular self-degradation, maintains cell homeostasis by clearing damaged organelles from cell cytoplasm [52]. Damaged organelles are engulfed by double-membrane vesicles known as autophagosomes. Autophagosomes then fuse with lysosomes for degradation and clearing [52, 53]. Germ cells respond to cellular stress by upregulating autophagy [38]. To compare the level of cellular stress in germ cells between 2D culture and 3D testicular organoid culture, the number of autophagosomes (LC3B+ve puncta) [38, 54] was counted in UCH-L1+ve germ cells in 2D culture and 3D organoids, respectively. Germ cells in 3D testicular organoids had a significantly lower number of autophagosomes (9.9 ± 2.4 puncta per germ cell, n = 3 experiments, 10 germ cells each, P = 0.0001, t = 4.86, df = 18), representative of the situation in vivo, compared to germ cells in 2D culture (20 ± 6.1 puncta per germ cell, n = 3, 10 germ cells each, P = 0.0001, t = 4.86, df = 18) using the same media conditions (Figure 5a and b).
Figure 5.
Germ cells experience less autophagy in organoids. (a) Representative images of autophagosomes (LC3B+ve puncta) in a UCHL1+ve single germ cell in 2D culture and 3D organoids. Scale bars = 5 μm. (b) Number of autophagosomes (LC3B puncta) per germ cells in 2D culture and 3D organoid culture. n = 3 experiments, 10 cells per group were analyzed, mean ± SD, P < 0.001(***). (c) Representative images of autophagosomes (LC3B+ve puncta) in a UCHL1+ve single germ cell on organoids exposed to four different concentrations of MEHP. Scale bars = 5 μm. (d) Number of LC3B puncta per germ cells for four different MEHP concentrations. n = 3 experiments, 10 cells per groups were analyzed, mean ± SD, P < 0.001(***).
Organoids are a model to study germ cell toxicity
To investigate the potential of the testicular organoid as a platform to study testicular toxicity, the organoids were exposed to 0.5, 1, and 1.5 μM of MEHP, an active metabolite of a commonly used plasticizer di-(2-ethylhexyl) phthalate (DEHP) that is implicated in reproductive toxicity [38, 55, 56]. The germ cells on the organoids were examined for levels of autophagy by the presence of LC3B+ve puncta. The autophagy levels increased with increasing doses of MEHP. There was no significant difference between the control (14.3 ± 6.67 puncta per germ cell, n = 3, 20 cells analyzed per replicate) and 0.5 μM MEHP (15.3 ± 3.6 puncta per germ cell, n = 3, 10 cells analyzed per replicate, P = 0.9732) treatments. But with higher doses of 1 (24.6 ± 5.2 puncta per germ cell, n = 3, 10 cells analyzed per replicate, P = 0.0002, F = 33.83, df = 40) and 1.5 μM (33.8 ± 4.6 puncta per germ cell, n = 3, 10 cells analyzed per replicate, P < 0.0001, F = 33.83, df = 40) MEHP treatment, there was a significant increase in levels of autophagy compared to the control (Figure 5c and d). These results were similar to other analyses for germ cells exposed to phthalate esters in vivo and in vitro [38]. No effect was observed on the tissue architecture of the organoids.
Organoids are a model to study testis development in vitro
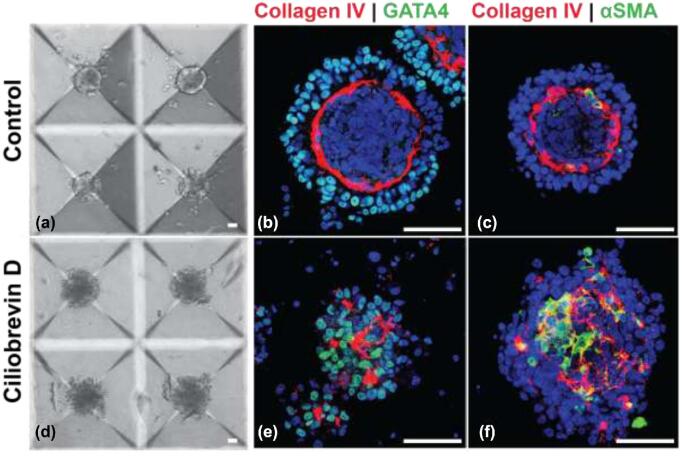
To investigate whether testicular organoids self-organize following developmentally relevant processes, we assessed the contribution of primary cilia. The primary cilium, an antenna-like organelle extending from the surface of the cell, is involved in several signal transduction pathways such as Wnt/β-catenin, Notch, Hippo, and Hedgehog signaling [57]. Primary cilia are present on somatic cells in the mammalian testis, and we previously demonstrated their critical role in normal testis development and morphogenesis [37, 58]. When primary cilia on testicular somatic cells were chemically ablated using 5 μM Ciliobrevin D, a small molecule inhibitor of primary cilium, organoid development was arrested (Figure 6). This finding is consistent with our previous observations on seminiferous tubule formation [37].
Figure 6.
Chemical ablation of primary cilia inhibits morphogenesis. (a–c) Control cells, (d–f) Ciliobrevin D treated cells after day 5. (b, c, e, f) Sertoli cells (GATA4), peritubular myoid cells (α-SMA), basement membrane (Collagen IV). Scale bars = 50 μm.
Organoids can form with lower numbers of cells
A total of 1000 cells/microwell were initially used for the formation of organoids, which translated to 1.2 × 106 cells per well. This cell number can be high for applications where the number of available cells is limited. To evaluate generation of organoids with lower cell numbers, organoids were formed using 6 × 105, 3 × 105, 15 × 104, and 75 × 103 cells which translated to 500, 250, 125, and 60 cells seeded in each microwell. The organoids generated with 500, 250, and 125 cells were topographically similar to 1000 cell organoids (collagen IV+ve basement membrane with GATA4+ve Sertoli and UCH-L1+ve germ cells on the outside of the basement membrane and Cytochrome P450+ve Leydig cells and αSMA+ve peritubular myoid cells on the inside (Figure 7)). However, 60 cells did not form organoids with distinct seminiferous epithelium and interstitium.
Figure 7.
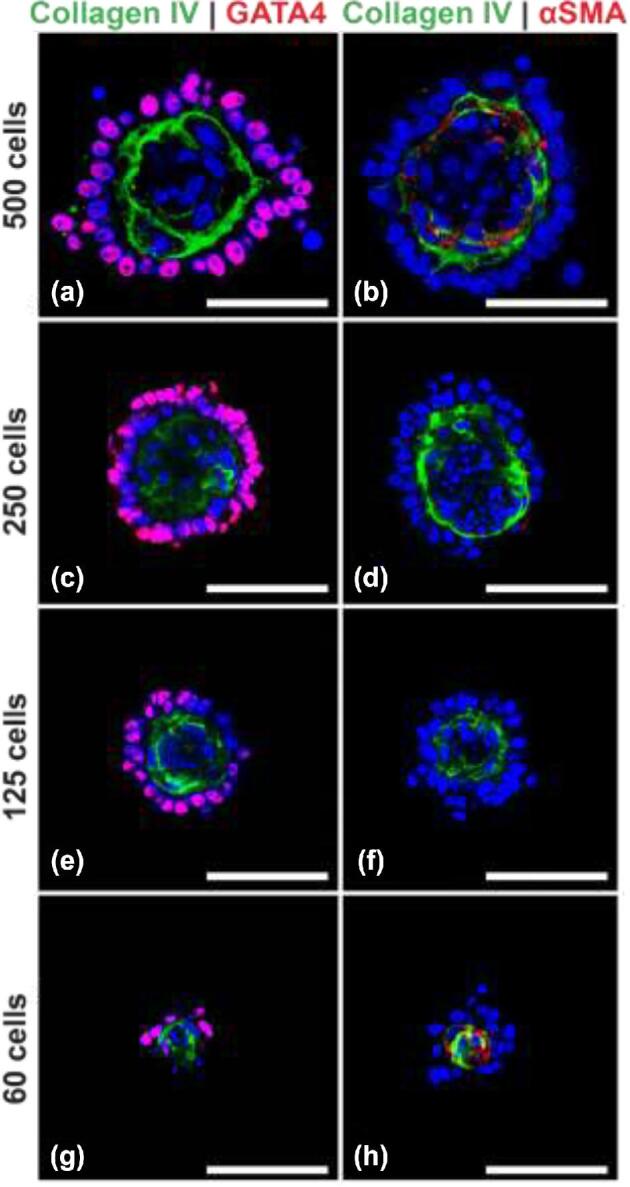
Organoids can be generated with 125 cells each. (a,b) Organoids with 500 cells. (c,d) Organoids with 250 cells. (e,f) Organoids with 125 cells. (g, h) Organoids with 60 cells showing lack of distinct seminiferous epithelium and interstitial compartment. Scale bars = 50 μm.
Organoids can be generated from cryopreserved cells and can be cryopreserved after formation
Fresh testicular cells are not always available, particularly when human patients or rare and endangered species are involved. To evaluate if organoids can be derived from frozen-thawed cells, cryopreserved testicular cells were thawed, and used to generate both 500 and 1000 cell organoids. The organoids generated from frozen-thawed cells exhibited a tissue architecture similar to organoids derived from fresh cells (Figure 8a and b). A cryopreservation strategy is also required for clinical and practical applications of testicular organoids [39]. To evaluate if cryopreservation of organoids has any deleterious effect on the testis-specific tissue architecture, testicular organoids were frozen by vitrification, thawed, and analyzed (immediately and after 7 days of culture) by immunofluorescence. Despite lower GATA4 immunoreactivity and a more diffuse αSMA distribution, the cryopreserved organoids seem to retain the delineated interior-interstitial and exterior-seminiferous compartments (Figure 8c and d).
Figure 8.
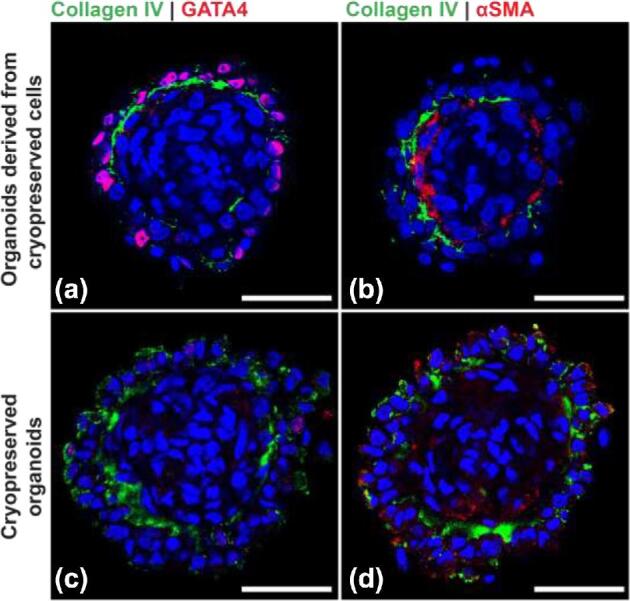
Organoids can be generated from cryopreserved cells and can be cryopreserved post formation. (a, b) Organoids generated with cryopreserved cells. (c, d) Cryopreserved organoids stained after thawing. Scale bars = 50 μm.
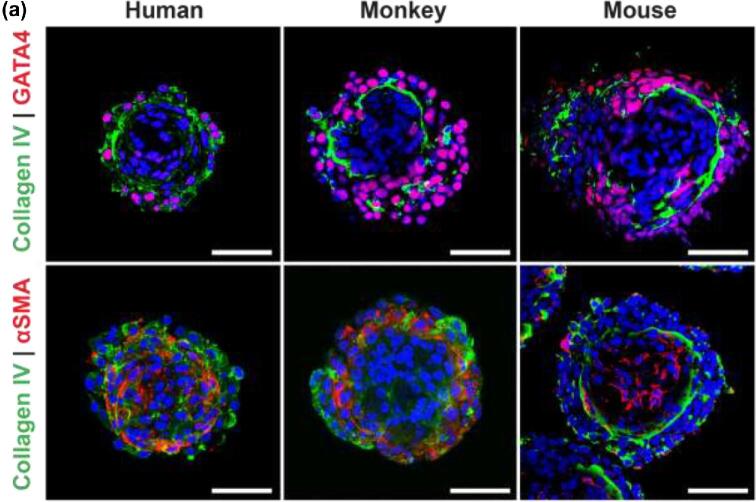
Organoids can form from mouse, human, and monkey testicular cells
To validate testicular organoid formation across mammalian species, organoids were generated using 1.2 × 106 fresh mouse, 1.2 × 106 frozen-thawed monkey, and 6 × 105 frozen thawed human (Figure 9) testicular cells. These organoids showed relatively similar cell associations to the porcine testicular organoids: GATA4+ve Sertoli cells and UCHL1+ve germ cells were located on one side of the Collagen IV+ve basement membrane, while Cytochrome P450+ve Leydig cells and the αSMA+ve peritubular myoid cells were mostly localized inside the organoids (Figures 9 and 10a,b). In human and monkey organoids, the αSMA staining pattern appears to be more diffuse (Figures 1d and 9) compared to porcine organoids. This is most likely due to the use of frozen-thawed testicular cells. Interestingly, mouse cells failed to produce organoids without supplementation of the medium with Matrigel, which may indicate a lower ECM production rate of mouse testicular cells compared to larger species such as pigs and primates.
Figure 9.
Testicular organoids form with similar organization across species. (a) Left panel, human organoids. Middle panel, monkey organoids and right panel, mouse organoids. Scale bars = 50 μm.
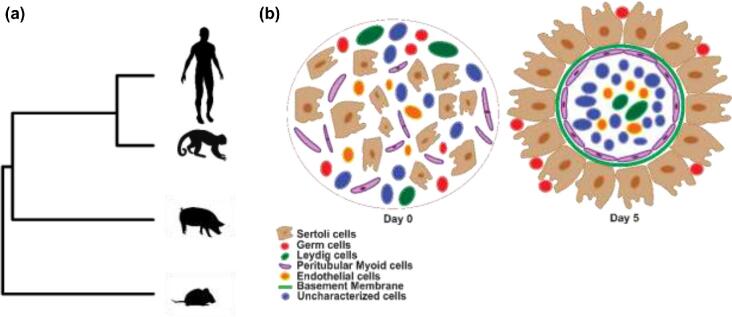
Figure 10.
Summary. (a) Phylogenetic relationship of donor species. (b)Schematic representation of organoid formation.
Discussion
Interest in 3D organoids [59, 60] as an intermediate system between in vivo animal models and more conventional 2D in vitro culture models has surged. The development of organoids from various organs paves the way for the study of development and for high-throughput drug and toxicity screening [61–64]. In this study, we report for the first-time generation of organotypic testicular organoids from porcine, murine, human, and primate testicular cells.
The microwell centrifugal forced-aggregation approach allows for the reproducible generation of 3D organoids of defined size and composition [21, 65, 66]. Testicular organoids formed from primary pre-pubertal testis cells with a well-defined interstitial compartment and seminiferous epithelium, separated by a basement membrane similar to testes tissue architecture in vivo. This high level of structural organization was not seen in organoids previously reported from rodent or human cells [16–19], possibly due to the interference by added ECM proteins and/or use of pubertal and adult cells, which display limited morphogenic capacity and survival in xenografts [67]. Pendergraft et al. generated adult human testicular organoids using immortalized testicular somatic cells capable of androgen production [18]. The organoids reported here have been entirely generated from primary cells, which are more reflective of in vivo conditions as immortalized cells often differ genetically and phenotypically from their tissue of origin [68, 69]. Our organoids were formed from pre-pubertal testis cells, which do not yet produce androgens. It will be interesting to determine if they can be further matured to activate this pathway as well. Given their dual-compartment architecture reflective of the native testes, the organoids reported here promise to be an important tool for the study of cell–cell interactions and testicular morphogenesis.
Although microwell-derived testicular organoids have testis-specific cell associations, the overall tissue architecture is “inside-out” compared to the native structures. The seminiferous epithelium is located at the surface, while the interstitial compartment is located inside the organoid enclosed by the basement membrane, resulting in a physiologically appropriate relationship between fluid spaces (lumen/culture medium) and the various cell types. This is potentially mediated by the differential adhesion and motility properties of different testicular cell types [70, 71]. The cell–cell interaction among the interstitial cells (Leydig and endothelial cells) may be faster than the cell–cell interaction among the cells of the seminiferous epithelium. As a result, the interstitial core is generated at the onset of organoid formation with the seminiferous compartment forming around it. This inside-out morphology is anticipated to be beneficial for studies of germ cells—for example, cultured germ cells that have undergone genetic manipulation could be readily transplanted on pre-built niches by centrifugation on top of pre-formed organoids and studied for downstream effects.
Although RA signaling is essential for spermatogenesis [51], a subpopulation of germ cells can resist RA signaling and maintain testis homeostasis. Lord and colleagues recently reported that germ cells in situ have a higher resistance to RA stimulation compared to germ cells in 2D testicular cell culture [50]. Interestingly, the mere presence of testicular somatic cells is not enough to protect germ cells from RA signaling, rather the cells must be part of a testis-specific architecture [50]. Our study supports a significant, reduced response to RA stimulation in germ cells cultured on 3D organoids compared to cell populations cultured without testis-specific cell associations. This indicates that germ cell–somatic cell interactions in the organoids are functionally representative of the situation in vivo.
Autophagy, a process of cellular self-degradation, plays an important role in cell homeostasis [72, 73]. Germ cells in situ have lower levels of autophagy compared to isolated germ cells in culture, and when cultured germ cells are reintegrated to the testicular microenvironment the level of autophagy returns to basal levels (Valenzuela-Leon and Dobrinski, unpublished). The multicellular tissue architecture in the 3D organoid mimics physiological environment in situ compared to 2D culture, since germ cells in the 3D organoids exhibit less autophagy than germ cells in 2D culture. Notably, while exposure to phthalate esters increased the autophagic response in germ cells in organoids as previously reported for isolated germ cells, the response was attenuated in germ cells exposed in organoids. Exposure to 0.5 μM MEHP increased autophagic levels of germ cells in 2D culture [38], yet failed to induce autophagy in germ cells on 3D organoids. Hence, 3D organoid systems that mimic in vivo physiology is a more representative in vitro alternative to 2D cell cultures that are now largely used due to high-throughput applicability and reduced need for animal experimentation [74].
3D organoids from primary cells [75] such as the microwell-derived testicular organoids reported here are invaluable for studying organogenesis in vitro; they can be formed with a high degree of repeatability and conservation of organ-specific cell architecture. We provided proof of principle that chemical ablation of primary cilia on testicular somatic cells plays a role in testicular development [37, 58], and prevents formation of organoids. Small molecule inhibitors or gene editing combined with the organoid culture system therefore will help identify regulatory mechanisms involved in testicular morphogenesis.
To summarize, we report high-throughput generation of testicular organoids with a consistent testis-specific architecture from pig, mouse, and primate cells. These organoids support cell type specific functions and can serve as a platform for studying testicular function and morphogenesis and allow for rapid toxicity screening on testicular tissues.
Supplementary Material
Notes
Conference presentation: Presented in part at the 50th Annual Meeting of the Society for the Study of Reproduction, 13–16 July 2017, Washington D.C., USA, 2017 Alberta Children's Hospital Research Institute Symposium, Calgary, AB and 17th Annual Alberta Biomedical Engineering Conference, 21–23 October 2016, Banff, AB, Canada.
Footnotes
† Grant support: This work was supported by NIH/NICHD HD091068-01 to Dr Ina Dobrinski.
Supplementary data
Supplementary Table S1. List of all antibodies used in the study.
Competing interests
ID is a member of the scientific advisory board of Recombinetics, Inc. M.U. is an inventor of the microwell system employed in this work and has a financial interest in it. The authors declare that there are no other competing interests associated with their contribution to this manuscript.
References
- 1. De Felici M, Dolci S. From testis to teratomas: a brief history of male germ cells in mammals. Int J Dev Biol 2013; 57(2-4):115–121. [DOI] [PubMed] [Google Scholar]
- 2. Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev 2012; 92(2):577–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richardson LL, Kleinman HK, Dym M. Basement membrane gene expression by sertoli and peritubular myoid cells in vitro in the rat. Biol Reprod 1995; 52(2):320–330. [DOI] [PubMed] [Google Scholar]
- 4. Zhao S, Zhu W, Xue S, Han D. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol 2014; 11(5):428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rebourcet D, O'Shaughnessy PJ, Pitetti JL, Monteiro A, O’Hara L, Milne L, Tsai YT, Cruickshanks L, Riethmacher D, Guillou F, Mitchell RT, van’t Hof R et al.. Sertoli cells control peritubular myoid cell fate and support adult Leydig cell development in the prepubertal testis. Development 2014; 141(10):2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 2009; 138(1):151–162. [DOI] [PubMed] [Google Scholar]
- 7. Oatley MJ, Racicot KE, Oatley JM. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol Reprod 2011; 84(4):639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takase HM, Nusse R.. Paracrine Wnt/beta-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc Natl Acad Sci USA 2016; 113(11):E1489–E1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skinner MK, Norton JN, Mullaney BP, Rosselli M, Whaley PD, Anthony CT. Cell-cell interactions and the regulation of testis function. Ann NY Acad Sci 1991; 637 (1):354–363. [DOI] [PubMed] [Google Scholar]
- 10. Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69(2):612–616. [DOI] [PubMed] [Google Scholar]
- 11. Abbott A. Biology's new dimension. Nature 2003; 424(6951):870–872. [DOI] [PubMed] [Google Scholar]
- 12. Sato T, Katagiri K, Gohbara A, Inoue K, Ogonuki N, Ogura A, Kubota Y, Ogawa T. In vitro production of functional sperm in cultured neonatal mouse testes. Nature 2011; 471(7339):504–507. [DOI] [PubMed] [Google Scholar]
- 13. Honaramooz A, Megee SO, Rathi R, Dobrinski I. Building a testis: formation of functional testis tissue after transplantation of isolated porcine (Sus scrofa) testis cells. Biol Reprod 2007; 76(1):43–47. [DOI] [PubMed] [Google Scholar]
- 14. Dores C, Rancourt D, Dobrinski I. Stirred suspension bioreactors as a novel method to enrich germ cells from pre-pubertal pig testis. Andrology 2015; 3(3):590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kita K, Watanabe T, Ohsaka K, Hayashi H, Kubota Y, Nagashima Y, Aoki I, Taniguchi H, Noce T, Inoue K, Miki H, Ogonuki N et al.. Production of functional spermatids from mouse germline stem cells in ectopically reconstituted seminiferous tubules. Biol Reprod 2007; 76(2):211–217. [DOI] [PubMed] [Google Scholar]
- 16. Alves-Lopes JP, Soder O, Stukenborg JB. Testicular organoid generation by a novel in vitro three-layer gradient system. Biomaterials 2017; 130:76–89. [DOI] [PubMed] [Google Scholar]
- 17. Baert Y, De Kock J, Alves-Lopes JP, Söder O, Stukenborg J-B, Goossens E. Primary human testicular cells self-organize into organoids with testicular properties. Stem Cell Rep 2017; 8(1):30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pendergraft SS, Sadri-Ardekani H, Atala A, Bishop CE. Three-dimensional testicular organoid: a novel tool for the study of human spermatogenesis and gonadotoxicity in vitro. Biol Reprod 2017; 96(3):720–732. [DOI] [PubMed] [Google Scholar]
- 19. Strange DP, Zarandi NP, Trivedi G, Atala A, Bishop CE, Sadri-Ardekani H, Verma S. Human testicular organoid system as a novel tool to study Zika virus pathogenesis. Emerg Microbes Infect 2018; 7(1):82–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Razian G, Yu Y, Ungrin M. Production of large numbers of size-controlled tumor spheroids using microwell plates. J Vis Exp 2013; 81:50665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One 2008; 3(2):e1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Y, Gamble A, Pawlick R, Pepper AR, Salama B, Toms D, Razian G, Ellis C, Bruni A, Gala-Lopez B, Lu JL, Vovko H et al.. Bioengineered human pseudoislets form efficiently from donated tissue, compare favourably with native islets in vitro and restore normoglycaemia in mice. Diabetologia 2018; 61(9):2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hermann BP, Sukhwani M, Lin CC, Sheng Y, Tomko J, Rodriguez M, Shuttleworth JJ, McFarland D, Hobbs RM, Pandolfi PP, Schatten GP, Orwig KE. Characterization, cryopreservation, and ablation of spermatogonial stem cells in adult rhesus macaques. Stem Cells 2007; 25(9):2330–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hermann BP, Sukhwani M, Salati J, Sheng Y, Chu T, Orwig KE. Separating spermatogonia from cancer cells in contaminated prepubertal primate testis cell suspensions. Hum Reprod 2011; 26(12):3222–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mclachlan RI, O’Donnell L, Meachem SJ, Stanton PG, de K, Pratis K, Robertson DM. Hormonal regulation of spermatogenesis in primates and man: insights for development of the male hormonal contraceptive. J Androl 2002; 23(2):149–162. [PubMed] [Google Scholar]
- 26. Schmid N, Stöckl JB, Flenkenthaler F, Dietrich KG, Schwarzer JU, Köhn FM, Drummer C, Fröhlich T, Arnold GJ, Behr R, Mayerhofer A. Characterization of a non-human primate model for the study of testicular peritubular cells-comparison with human testicular peritubular cells. Mol Hum Reprod 2018; 24(8):401–410. [DOI] [PubMed] [Google Scholar]
- 27. Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133(8):1495–1505. [DOI] [PubMed] [Google Scholar]
- 28. Fayomi AP, Orwig KE.. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res 2018; 29:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM et al.. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA 2013; 110(9):3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. González R, Dobrinski I.. Beyond the mouse monopoly: studying the male germ line in domestic animal models. ILAR J 2015; 56(1):83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gutierrez K, Dicks N, Glanzner WG, Agellon LB, Bordignon V. Efficacy of the porcine species in biomedical research. Front Genet 2015; 6:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vodicka P, Smetana K Jr, Dvoránková B, Emerick T, Xu YZ, Ourednik J, Ourednik V, Motlík J. The miniature pig as an animal model in biomedical research. Ann N Y Acad Sci 2005; 1049 (1):161–171. [DOI] [PubMed] [Google Scholar]
- 33. Bode G, Clausing P, Gervais F, Loegsted J, Luft J, Nogues V, The utility of the minipig as an animal model in regulatory toxicology. J Pharmacol Toxicol Methods 2010; 62(3):196–220. [DOI] [PubMed] [Google Scholar]
- 34. Sakib S, Dores C, Rancourt D, Dobrinski I. Use of stirred suspension bioreactors for male germ cell enrichment, In: Turksen K. (ed.) Bioreactors in Stem Cell Biology: Methods and Protocols. New York, NY: Springer; 2016:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, Chu T, Sanfilippo JS, Orwig KE. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest 2013; 123(4):1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, Chu T, Marshall GR, Orwig KE. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril 2014; 102(2):566–580.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dores C, Alpaugh W, Su L, Biernaskie J, Dobrinski I. Primary cilia on porcine testicular somatic cells and their role in hedgehog signaling and tubular morphogenesis in vitro. Cell Tissue Res 2017; 368(1):215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valenzuela-Leon P, Dobrinski I.. Exposure to phthalate esters induces an autophagic response in male germ cells. Environ Epigenet 2017; 3(3):dvx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeng W, Snedaker AK, Megee S, Rathi R, Chen F, Honaramooz A, Dobrinski I. Preservation and transplantation of porcine testis tissue. Reprod Fertil Dev 2009; 21(3):489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen S-R, Tang JX, Cheng JM, Li J, Jin C, Li XY, Deng SL, Zhang Y, Wang XX, Liu YX. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 2015; 6(35):37012–37027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Devi L, Pawar RM, Makala H, Goel S. Conserved expression of ubiquitin carboxyl-terminal esterase L1 (UCHL1) in mammalian testes. Indian J Exp Biol 2015; 53(5):305–312. [PubMed] [Google Scholar]
- 42. Goel S, Mahla RS, Suman SK, Reddy N, Imai H. UCHL-1 protein expression specifically marks spermatogonia in wild bovid testis. Eur J Wildl Res 2011; 57(3):663–667. [Google Scholar]
- 43. Luo J, Megee S, Dobrinski I. Asymmetric distribution of UCH-L1 in spermatogonia is associated with maintenance and differentiation of spermatogonial stem cells. J Cell Physiol 2009; 220(2):460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo J, Megee S, Rathi R, Dobrinski I. Protein gene product 9.5 is a spermatogonia-specific marker in the pig testis: application to enrichment and culture of porcine spermatogonia. Mol Reprod Dev 2006; 73(12):1531–1540. [DOI] [PubMed] [Google Scholar]
- 45. Payne AH. Hormonal regulation of cytochrome P450 enzymes, cholesterol side-chain cleavage and 17 alpha-hydroxylase/C17-20 lyase in Leydig cells. Biol Reprod 1990; 42(3):399–404. [DOI] [PubMed] [Google Scholar]
- 46. Holt WV, Waller J, Moore A, Jepson PD, Deaville R, Bennett PM. Smooth muscle actin and vimentin as markers of testis development in the harbour porpoise (Phocoena phocoena). J Anat 2004; 205(3):201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tung PS, Fritz IB.. Characterization of rat testicular peritubular myoid cells in culture: alpha-smooth muscle isoactin is a specific differentiation marker. Biol Reprod 1989; 42(2):351–365. [DOI] [PubMed] [Google Scholar]
- 48. Lertkiatmongkol P, Liao D, Mei H, Hu Y, Newman PJ. Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr Opin Hematol 2016; 23(3):253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140(12):5894–5900. [DOI] [PubMed] [Google Scholar]
- 50. Lord T, Oatley MJ, Oatley JM. Testicular architecture is critical for mediation of retinoic acid responsiveness by undifferentiated spermatogonial subtypes in the mouse. Stem Cell Rep 2018; 10(2):538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Endo T, Freinkman E, de Rooij DG, Page DC. Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc Natl Acad Sci USA 2017; 114(47):E10132–E10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levine B, Kroemer G.. Autophagy in the pathogenesis of disease. Cell 2008; 132(1):27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12 (S2):1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol 2008; 445: 77–88. [DOI] [PubMed] [Google Scholar]
- 55. Howdeshell KL, Rider CV, Wilson VS, Gray LE Jr. Mechanisms of action of phthalate esters, individually and in combination, to induce abnormal reproductive development in male laboratory rats. Environ Res 2008; 108(2):168–176. [DOI] [PubMed] [Google Scholar]
- 56. Rodriguez-Sosa JR, Bondareva A, Tang L, Avelar GF, Coyle KM, Modelski M, Alpaugh W, Conley A, Wynne-Edwards K, França LR, Meyers S, Dobrinski I. Phthalate esters affect maturation and function of primate testis tissue ectopically grafted in mice. Mol Cell Endocrinol 2014; 398(1-2):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wheway G, Nazlamova L, Hancock JT. Signaling through the primary cilium. Front Cell Dev Biol 2018; 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ou Y, Dores C, Rodriguez-Sosa JR, van der Hoorn FA, Dobrinski I. Primary cilia in the developing pig testis. Cell Tissue Res 2014; 358(2):597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009; 459(7244):262–265. [DOI] [PubMed] [Google Scholar]
- 60. Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 2015; 21(3):256–262. [DOI] [PubMed] [Google Scholar]
- 61. Forsythe SD, Devarasetty M, Shupe T, Bishop C, Atala A, Soker S, Skardal A. Environmental toxin screening using human-derived 3D bioengineered liver and cardiac organoids. Front Public Health 2018; 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, Martins TJ, Pippin JW. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 2018; 22(6):929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jabs J, Zickgraf FM, Park J, Wagner S, Jiang X, Jechow K, Kleinheinz K, Toprak UH, Schneider MA, Meister M, Spaich S, Sütterlin M. Screening drug effects in patient‐derived cancer cells links organoid responses to genome alterations. Mol Syst Biol 2017; 13(11):955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 2017; 144(6):958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kabiri M, Kul B, Lott WB, Futrega K, Ghanavi P, Upton Z, Doran MR. 3D mesenchymal stem/stromal cell osteogenesis and autocrine signalling. Biochem Biophys Res Commun 2012; 419(2):142–147. [DOI] [PubMed] [Google Scholar]
- 66. Schepers A, Li C, Chhabra A, Seney BT, Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 2016; 16(14):2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schlatt S, Honaramooz A, Ehmcke J, Goebell PJ, Rübben H, Dhir R, Dobrinski I, Patrizio P. Limited survival of adult human testicular tissue as ectopic xenograft. Hum Reprod 2006; 21(2):384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics 2009; 8(3):443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J Proteome Res 2006; 5(4):862–878. [DOI] [PubMed] [Google Scholar]
- 70. Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci USA 2009; 106(35):14890–14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moreno-Ruiz P, Arluzea J, Silván U, Díez-Torre A, Andrade R, Bonilla Z, Díaz-Núñez M, Silió M, Aréchaga J. Testis peritubular myoid cells increase their motility and express matrix-metalloproteinase 9 (MMP-9) after interaction with embryonal carcinoma cells. Andrology 2016; 4(1):111–120. [DOI] [PubMed] [Google Scholar]
- 72. Ryter SW, Cloonan SM, Choi AM. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells 2013; 36(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yin J, Ni B, Tian ZQ, Yang F, Liao WG, Gao YQ. Regulatory effects of autophagy on spermatogenesis. Biol Reprod 2017; 96(3):525–530. [DOI] [PubMed] [Google Scholar]
- 74. DelRaso NJ, In vitro methodologies for enhanced toxicity testing. Toxicol Lett 1993; 68(1-2):91–99. [DOI] [PubMed] [Google Scholar]
- 75. Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015; 160(1-2):324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.