Abstract
Hypoxia-inducible factor-1 (HIF-1), an important component of angiogenesis, is activated as a response to tumor hypoxia and facilitates tumor survival. Several case–control articles stressed the connection between lung cancer danger and HIF-1α gene polymorphism, but the conclusions were conflicting. Thus, this meta-analysis was carried out to assess the connection between HIF-1α gene polymorphisms (rs11549467, rs11549465, and rs2057482) and lung cancer risk.
PubMed, Embase, Cochrane Library, and Google Scholar were systematically searched up to November 1, 2018. The study quality was quantified by the c. The odds ratios (ORs) and 95% confidence intervals (CIs) were pooled in 5 genetic models for assessment under a fixed- or random-effect model. Subgroup analyses were carried out by ethnicity and genotype method. Sensitivity analysis and publication bias were tested. Five eligible articles were enrolled.
The rs11549467 significantly increased the lung cancer risk (OR [95% CI]: A vs G, 1.68 [1.03–2.76]; AA + AG vs GG, 1.70 [1.14–2.54]; AA vs GG, 1.59 [1.21–2.10]), whereas neither rs11549465 nor rs2057482 was related with the lung cancer risk. Subgroup analysis showed rs11549465 and rs11549467 increased lung cancer risk among Asians, but not whites. HIF-1α rs2057482 was unrelated to the risk of lung cancer in Asians and whites.
HIF-1α gene rs11549465 and rs11549467, but not rs2057482, increased the risk of lung cancer among Asians.
Keywords: HIF-1α, lung cancer, meta-analysis, polymorphisms
1. Introduction
Lung cancer is one of the primary occurring cancers worldwide and the leading cause of cancer-related death.[1] Lung cancer is caused primarily by smoking and endangered by other environmental factors, such as exposure to heavy metal, radiation, asbestos, and air pollution.[2] Nevertheless, only a small portion of the population exposed to these factors finally develop lung cancer, suggesting host factors also play important roles in lung carcinogenesis. General molecular genetic studies showed that lung cancer cells acquired multiple genetic and epigenetic changes in the DNA sequence, copy number, and aberrant promoter hypermethylation as a consequence of increasing genomic instability.[3,4] With the advent of next-generation sequencing and in-depth understanding into the molecular biology of lung cancer, sequencing of single-nucleotide polymorphisms (SNPs) may be pivotal in the personalized treatment of lung cancer.[5]
Hypoxia is an important environmental regulator of tumor angiogenesis and growth. Many of the adaptations to hypoxia are mediated by the activation of specific genes through the hypoxia-inducible factor (HIF).[6] HIF-1 is a hetero-dimer basic helix-loop-helix transcription factor that includes α and β subunits. HIF-1α is induced by hypoxia and contains 3 members, including HIF-1α and HIF-2α, 2 major functional members. HIF-1α is overexpressed in several cancers, such as colon, kidney, pancreas, esophagus, endometrial, prostate, breast, stomach, and lung cancers.[7–12] The target genes of HIF-1α are particularly relevant to cancers and encode proliferation/survival factors and angiogenic factors.[13] As such, variability in this protein may influence the individual risk to this disorder. Functional polymorphisms of HIF-1α gene can considerably affect lung carcinogenesis via increasing genomic instability, especially in adenocarcinomas.[14] The C2028T polymorphism in exon 12 and the dinucleotide repeat polymorphism in intron 13 of the HIF-1α gene both affect HIF-1α protein expression in lung cancer.[9] HIF-1α is a potential target for cancer chemo-sensitization, -therapy, and -prevention.[13,15,16] HIF-1α protein overexpression was associated with lymph node metastasis, and histological grade in breast cancer.[17] Low expression of HIF-1α was related with pathologic response to disease-free survival and 5-year overall survival in clinical stage II/III rectal cancer patients.[18] Variants of HIF-1α gene on the treatment response and long-term prognosis of lung cancer have been found. Reportedly, carriers of CC genotype of C1772T are more likely to be chemotherapeutic responders.[19] Lung cancer patients with high versus low HIF-1α expressions had a higher chance to be chemotherapeutic non-responders and CC genotype carriers have longer overall survival and progression-free survival.[19]
Recently, accumulating evidence showed that HIF-1α gene polymorphisms may contribute to lung cancer development.[14,20–23] Many studies have attached importance to the following SNPs: rs11549465, rs11549467, rs2057482, rs10873142, and rs41508050. The rs11549465 (1744C > T and Pro582Ser), rs11549467 (1762G > A and Ala588Thr), and rs41508050 (1253C > T and Thr418IIe) are located in the exon region of HIF-1α gene and have many alternative names. The HIF-1α expression or activity may be affected in carriers with the genotypes of the above SNPs. The rs10873142 (1029–145C>T and 213+14141T>C) polymorphism is located in the intron region of HIF-1α, whereas rs2057482 (1960A>G and 45T>C) is located in the 3’-UTR region of HIF-1α. The rs10873142 may have linkage disequilibrium with another potentially functional variants or be closely linked to susceptibility gene. Genetic variations in the 3’-UTR may affect the binding of miRNA to its target mRNA and thereby confer the susceptibility to lung cancer. However, the findings of the relationship between these polymorphisms and lung cancer risk remain controversial or inconclusive, due to the limited sample sizes, clinical heterogeneity, and different ethnic populations. Thus, we performed this meta-analysis including all eligible case-controlled studies to investigate whether HIF-1α gene polymorphisms were associated with the risk of lung cancer.
2. Material and methods
In this meta-analysis based on the published studies, we followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses.[24] This study did not need either informed consent of the patients or ethical approval.
2.1. Search strategy
Relevant research was systematically searched on PubMed, Embase, Cochrane Library, and Google Scholar until November 1, 2018. The relevant keywords and search strategy were as follows: “Hypoxia-inducible factor or HIF,” “polymorphisms or mutation or variants,” and “lung cancer or lung carcinoma.” These terms were combined differently in the search. Moreover, the reference lists of original studies were searched manually for additional literature. All the eligible studies were checked carefully to prevent overlapping datasets, and only published studies were included.
2.2. Inclusion and exclusion criteria
Inclusion criteria were: studies with case–control design, assessment of potential relationship between HIF-1α gene polymorphism and lung cancer risk, and enough data for computation of odds ratios (ORs) and 95% confidence interval (CI).
Exclusion criteria were: review, meta-analysis, letter, case report, abstract only, and absence of controls.
2.3. Data extraction
Data were extracted from all the eligible studies by 2 investigators independently according to the inclusion criteria, including surname of first author, year of publication, ethnicity, genotype method, source of controls (SOC), and numbers of cases and controls for HIF-1α genotypes. Any conflicting evaluation was settled after discussion with the third reviewer.
2.4. Statistical analysis
The available data from each study were analyzed on STATA 11.0 (STATA Corporation, College Station, TX). The pooled statistic data were analyzed using the fixed-effects model, but a random-effects model was used in case of P < .1 in the heterogeneity test.[25–27] Results were expressed as ORs for dichotomous data, and 95% CIs were also calculated. P < .05 indicated the pooled OR was significant.[28]I2 was used to test the between-study heterogeneity. Potential publication bias was assessed by Egger and Begg linear regression tests.[29] The effect on the test of heterogeneity and the stability of the overall results were investigated through sensitivity analysis by omitting each study in turn.
3. Results
3.1. Study characteristics
The primary search returned 38 articles, of which 11 duplications were deleted. Then 10 studies were removed after title and abstract screening. Of the 17 remaining articles, 3 without sufficient data, 7 meta-analyses, and 2 with different study design were deleted. The whole process of article inclusion is shown in Figure 1. Finally, only 5 studies[14,20–23] involving 2067 cases and 2055 controls were enrolled in this meta-analysis. Tables 1 and 2 summarize the main characteristics of the included articles. Two ethnicities were included: Asian (n = 4) and white (n = 1). The studies were population-based[23] or hospital-based.[14,20–22] The sample sizes ranged from 141 to 1096. The results of Hardy-Weinberg equilibrium (HWE) in the controls and Newcastle-Ottawa scale (NOS) scores are also shown in Table 1. One study about rs2057482[23] and 1 about rs11549467[22] are inconsistent with HWE. The NOS scores of all 5 studies are >5 points, which suggests a high quality.
Figure 1.
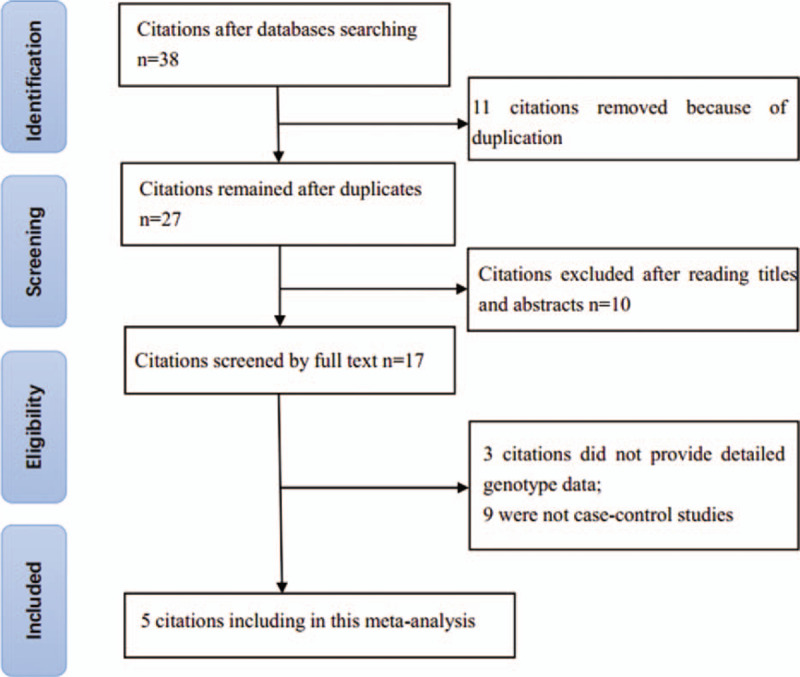
Flowchart of the literature search and selection.
Table 1.
Characteristics of included studies.
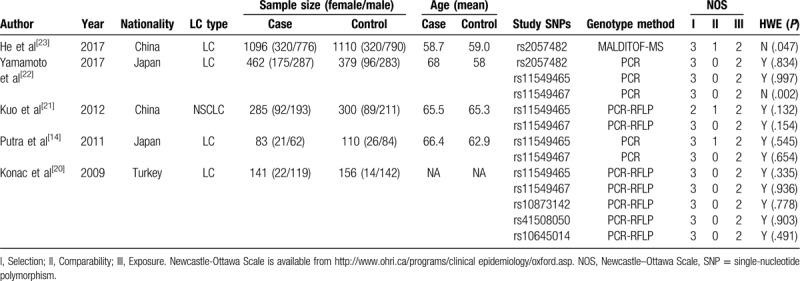
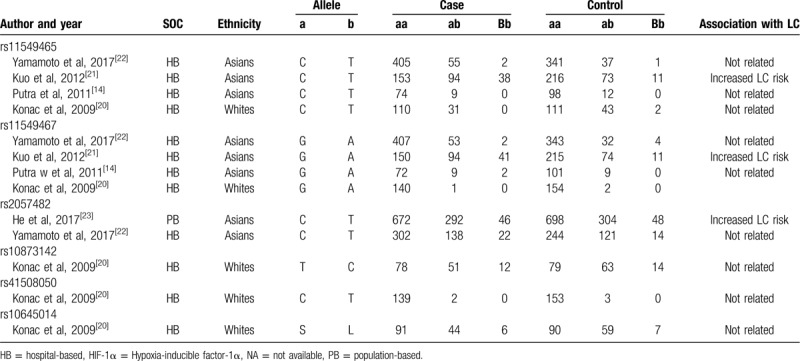
Table 2.
Genotype distributions of HIF-1α polymorphisms in the included studies.
3.2. Quantitative synthesis
For HIF-1α gene rs11549465, no significant connection was found by any of the 5 genetic models (OR [95% CI]: T vs C, 1.23 [0.69–2.20], P = .489; TT + TC vs CC, 1.23 [0.71–2.13], P = .466; TT vs TC + CC, 1.93 [0.43–8.66], P = .388; TT vs CC, 1.93 [0.35–10.53], P = .452; TC vs CC, 1.19 [0.78–1.82], P = .426, Table 3). However, the stratification analysis revealed that rs11549465 increased the risk of lung cancer among Asians (OR [95% CI]: 1.47 (1.07–2.02), P = .016, Fig. 2).
Table 3.
Meta-analysis of the association between HIF-1α polymorphisms and LC risk.
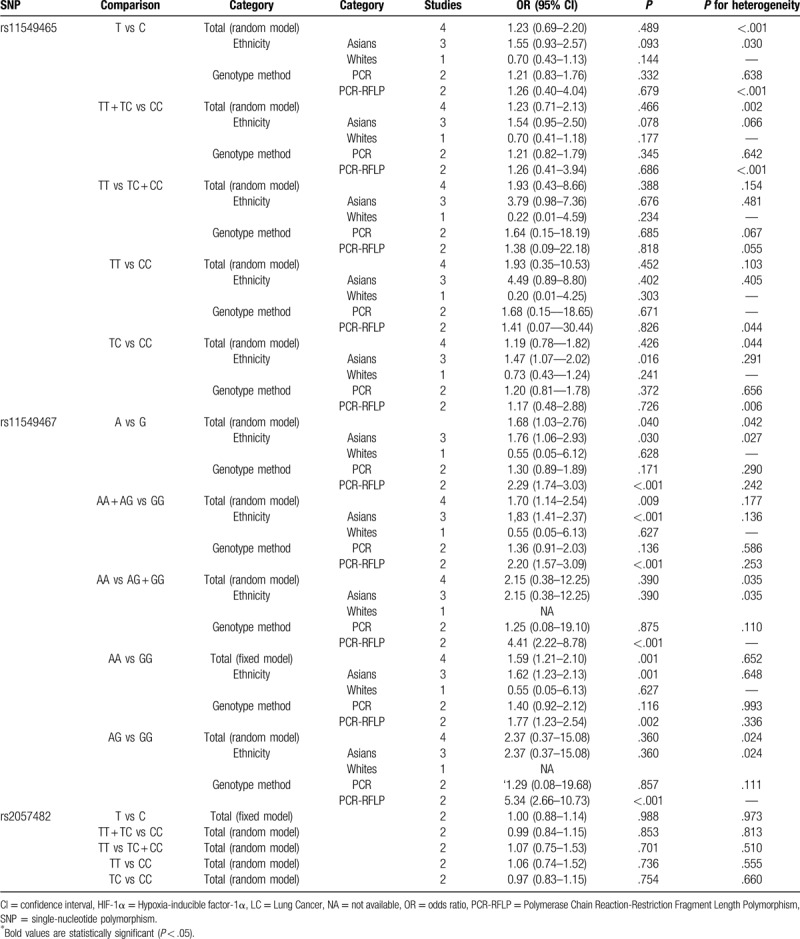
Figure 2.
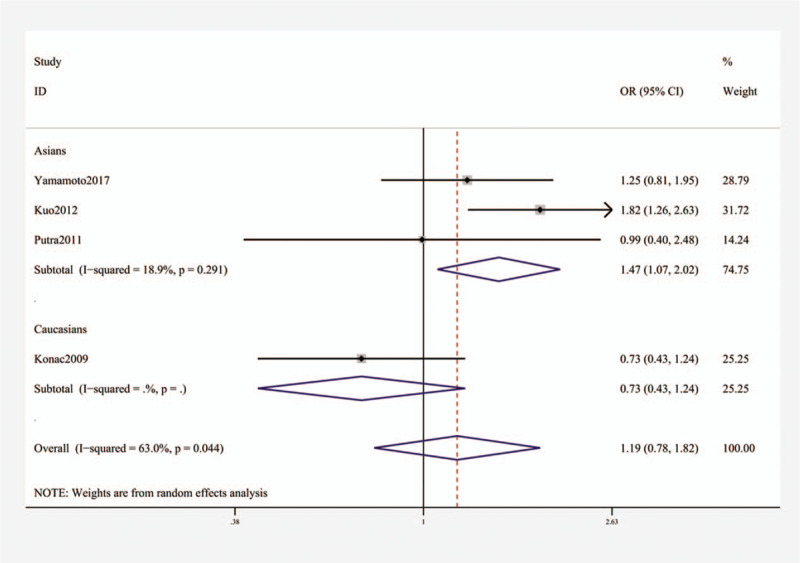
Stratification analysis by ethnicity showing OR for the association between the rs11549465 polymorphism and lung cancer risk (TC vs CC). OR = odds ratio.
For rs11549467, a significant relationship with higher risk of lung cancer was found (OR [95% CI]: A vs G, 1.68 [1.03–2.76], P = .040; AA + AG vs GG, 1.70 (1.14–2.54], P = .009; AA vs GG, 1.59 [1.21–2.10], P = .001, Table 3 and Fig. 3). Subgroup analysis by ethnicity uncovered substantial connection in Asians (OR [95% CI]: A vs G, 1.76 (1.06–2.93), P = .030; AA + AG vs GG, 1.83 [1.41–2.37], P < .001; AA vs GG, 1.62 (1.23–2.13), P = 0.001, Table 3 and Fig. 4), but not in whites.
Figure 3.
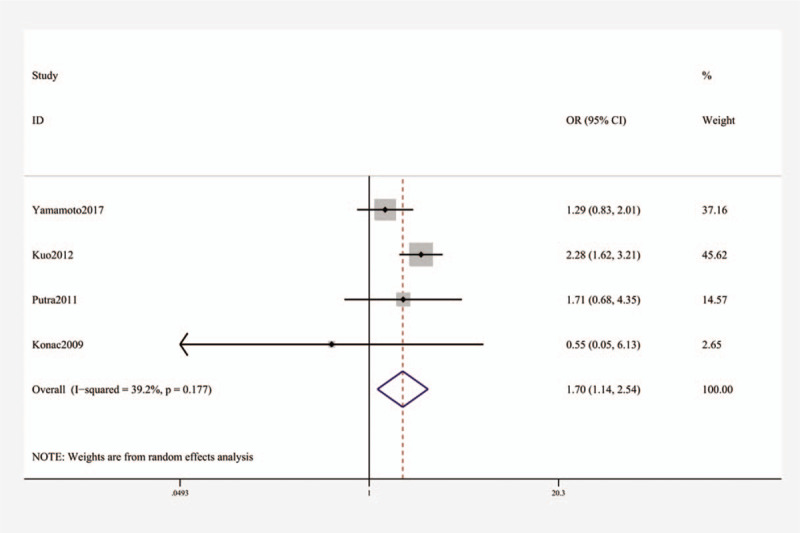
Forest plot showing OR for the associations between the rs11549467 polymorphism and lung cancer risk (AA + AG vs GG). OR = odds ratio.
Figure 4.
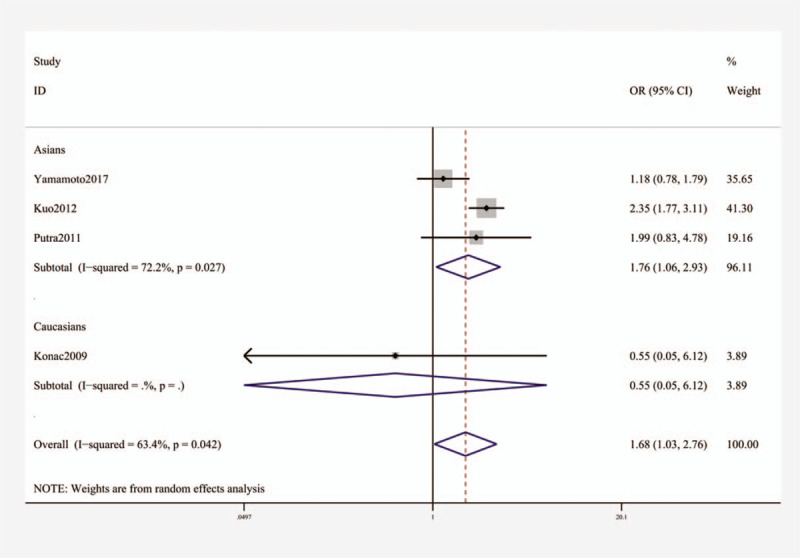
Stratification analysis by ethnicity showing OR for the association between the rs11549467 polymorphism and lung cancer risk (A vs G). OR = odds ratio.
The sensitivity analysis showed that after eliminating the study by Kuo et al, rs11549467 was still associated with the risk of lung cancer, but rs11549465 polymorphism was related with increased risk for lung cancer in the heterozygous model.
No substantial relationship was identified between rs2057482 and lung cancer risk (Table 3).
3.3. Publication bias
The Begg funnel plot did not reveal any evident dissymmetry (Fig. 5), which indicated absence of publication bias.
Figure 5.
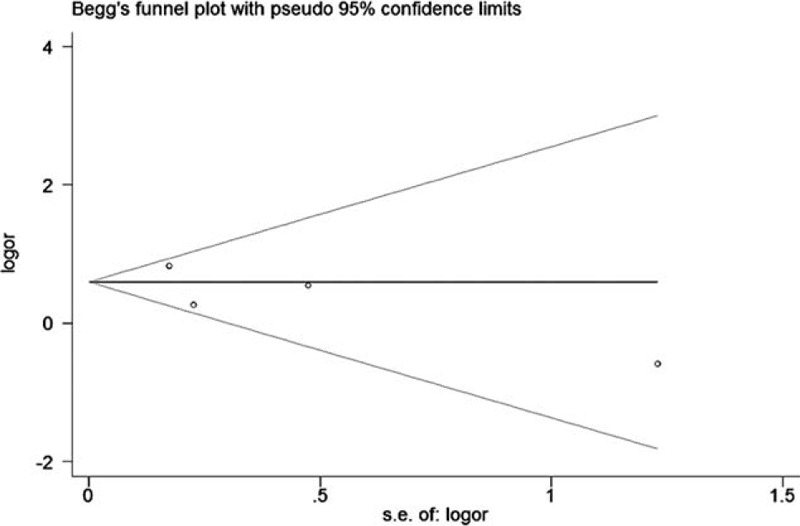
Begg tests between the rs11549465 polymorphism and lung cancer (T vs C).
4. Discussion
This meta-analysis showed HIF-1α gene rs11549465 and rs11549467 were both associated with increased risk for lung cancer among Asians, but not among whites.
Recently, several studies explored the associations between and HIF-1α gene polymorphisms and lung cancer risk, but yielded contradictory results. A study from Turkey firstly evaluated whether HIF-1α gene polymorphisms conferred susceptibility to lung cancer and found HIF-1α gene polymorphisms did not relate to the risk of lung cancer.[20] Additionally, no significant association was found between the genotypes and clinopathological characteristics of the lung cancer cases.[20] Later, 2 studies from Japan replicated negative findings[14,22] like the Turkish study.[20] However, a study from China consisting of 285 non-small cell lung cancer cases and 300 controls showed that HIF-1α gene rs11549465 and rs11549467 were associated with increased risk for lung cancer, but reported no relationship between HIF-1α rs11549465 or rs11549467 and the severity of lung cancer.[21] Another Chinese study with 1096 cases and 1110 controls found HIF-1α rs2057482 polymorphism was associated with increased risk for lung cancer.[23] Due to the conflicting findings of the above studies, several meta-analyses were conducted,[30–33] which all showed HIF-1α rs11549465 and rs11549467 can both increase the risk of lung cancer.[30–33] However, they only included 2 studies with limited sample size (405 cases and 481 controls), which greatly affected the relationship between gene SNPs and lung cancer risk. Thus, we think their conclusions are not trustworthy. In this meta-analysis, we enrolled 5 studies containing 2067 cases and 2055 controls[14,20–23] and found HIF-1α gene rs11549465 was not related to lung cancer susceptibility, which is inconsistent with previous meta-analyses. But our subgroup analysis of ethnicity showed this SNP conferred susceptibility to lung cancer. Furthermore, we showed lung cancer risk was related to HIF-1α rs11549467, but not to HIF-1α gene rs2057482, which has not been investigated before.
This meta-analysis has several potential limitations. First, the sample sizes and the number of included studies were not large enough, especially for subgroup analyses, which may decrease the power and robust of this meta-analysis. Secondly, some unpublished studies may be omitted, although our results showed no significant publication bias. Thirdly, subgroup analyses of age, sex, or smoking were not addressed due to limited data. Fourthly, only white and Asian populations were included. In addition, the 4 Asian studies were from China (n = 2) and Japan (n = 2), indicating a clear regional publication bias. This becomes significant because both ethnicity and geographical location profoundly affect the lung cancer risk. Thus, other ethnic groups should be explored. Fifthly, only 1 study with only 141 cases and 156 controls was focused on whites, indicating the positive results regarding whites should be interpreted with caution. Lastly, gene–gene or gene–environment interactions were not analyzed because of data insufficiency.
5. Conclusion
HIF-1α gene rs11549465 and rs11549467 polymorphisms are both associated with increased risk for lung cancer among Asians. HIF-1α rs2057482 polymorphism is not associated with the risk of lung cancer. These findings should be validated in other ethnicities.
Author contribution
Conceptualization: X.S.G. and Y.K.J.; Methodology: X.S.G. and Y.K.J.; Software and data analysis: X.S.G.; Validation: Y.K.J.; Writing - original draft preparation: X.S.J.; Writing - review and editing: Y.K.J.
Footnotes
Abbreviations: CI = confdence interval, HIF-1α = hypoxia-inducible factor-1α, HWE = Hardy–Weinberg equilibrium, NOS = Newcastle–Ottawa Scale, OR = odds ratio, SNP = single-nucleotide polymorphism.
How to cite this article: Xu S, Ying K. Association between HIF-1α gene polymorphisms and lung cancer: a meta-analysis. Medicine. 2020;99:24(e20610).
The authors declare there are no sources of funding to be acknowledged.
Availability of data and materials: Not applicable.
The authors report no conflicts of interests.
All data generated or analyzed during this study are included in this published article (and its supplementary information files);
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e1S–29S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sato M, Shames DS, Gazdar AF, et al. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007;2:327–43. [DOI] [PubMed] [Google Scholar]
- [4].Sun S, Schiller JH, Spinola M, et al. New molecularly targeted therapies for lung cancer. J Clin Invest 2007;117:2740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen K, Zhou YX, Li K, et al. A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal Bioanal Chem 2016;408:4371–7. [DOI] [PubMed] [Google Scholar]
- [6].Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev 2000;14:1983–91. [PubMed] [Google Scholar]
- [7].Talks KL, Turley H, Gatter KC, et al. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol 2000;157:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bos R, Zhong H, Hanrahan CF, et al. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst 2001;93:309–14. [DOI] [PubMed] [Google Scholar]
- [9].Koukourakis MI, Papazoglou D, Giatromanolaki A, et al. C2028T polymorphism in exon 12 and dinucleotide repeat polymorphism in intron 13 of the HIF-1alpha gene define HIF-1alpha protein expression in non-small cell lung cancer. Lung Cancer 2006;53:257–62. [DOI] [PubMed] [Google Scholar]
- [10].Tzao C, Lee SC, Tung HJ, et al. Expression of hypoxia-inducible factor (HIF)-1alpha and vascular endothelial growth factor (VEGF)-D as outcome predictors in resected esophageal squamous cell carcinoma. Dis Markers 2008;25:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Koukourakis MI, Giatromanolaki A, Skarlatos J, et al. Hypoxia inducible factor (HIF-1a and HIF-2a) expression in early esophageal cancer and response to photodynamic therapy and radiotherapy. Cancer Res 2001;61:1830–2. [PubMed] [Google Scholar]
- [12].Amankwah EK, Sellers TA, Park JY. Gene variants in the angiogenesis pathway and prostate cancer. Carcinogenesis 2012;33:1259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721–32. [DOI] [PubMed] [Google Scholar]
- [14].Putra AC, Tanimoto K, Arifin M, et al. Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology 2011;16:796–802. [DOI] [PubMed] [Google Scholar]
- [15].Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol 2011;4:62–77. [DOI] [PubMed] [Google Scholar]
- [16].Kizaka-Kondoh S, Inoue M, Harada H, et al. Tumor hypoxia: a target for selective cancer therapy. Cancer Sci 2003;94:1021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim HO, Jo YH, Lee J, et al. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep 2008;20:1181–7. [PubMed] [Google Scholar]
- [18].Lin S, Lai H, Qin Y, et al. Thymidine phosphorylase and hypoxia-inducible factor 1-alpha expression in clinical stage II/III rectal cancer: association with response to neoadjuvant chemoradiation therapy and prognosis. Int J Clin Exp Pathol 2015;8:10680–8. [PMC free article] [PubMed] [Google Scholar]
- [19].Wu F, Zhang J, Liu Y, et al. HIF1alpha genetic variants and protein expressions determine the response to platinum based chemotherapy and clinical outcome in patients with advanced NSCLC. Cell Physiol Biochem 2013;32:1566–76. [DOI] [PubMed] [Google Scholar]
- [20].Konac E, Dogan I, Onen HI, et al. Genetic variations in the hypoxia-inducible factor-1alpha gene and lung cancer. Exp Biol Med (Maywood) 2009;234:1109–16. [DOI] [PubMed] [Google Scholar]
- [21].Kuo WH, Shih CM, Lin CW, et al. Association of hypoxia inducible factor-1alpha polymorphisms with susceptibility to non-small-cell lung cancer. Transl Res 2012;159:42–50. [DOI] [PubMed] [Google Scholar]
- [22].Yamamoto Y, Kiyohara C, Ogata-Suetsugu S, et al. Association between genetic polymorphisms involved in the hypoxia-inducible factor pathway and lung cancer risk: a case-control study in Japan. Asia Pac J Clin Oncol 2017;13:234–42. [DOI] [PubMed] [Google Scholar]
- [23].He F, Qi Q, Li X, et al. [Association of indoor air pollution, single nucleotide polymorphism of HIF-1alpha gene with susceptibility to lung cancer in Han population in Fujian Province]. Zhongguo Fei Ai Za Zhi 2017;20:149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 2009;62:1006–12. [DOI] [PubMed] [Google Scholar]
- [25].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [27].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [28].Yang H, Hu Z, Zhuang C, et al. Association between the polymorphisms of CALM1 gene and osteoarthritis risk: a meta-analysis based on observational studies. Biosci Rep 2018;38:BSR20181128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006;295:676–80. [DOI] [PubMed] [Google Scholar]
- [30].Liao SH, Liu WZ, Liu T, et al. Potential signaling pathway of hypoxia-inducible factor in lung cancer and its gene polymorphism with lung cancer risk. J Recept Signal Transduct Res 2015;35:233–7. [DOI] [PubMed] [Google Scholar]
- [31].Li Y, Li C, Shi H, et al. The association between the rs11549465 polymorphism in the hif-1alpha gene and cancer risk: a meta-analysis. Int J Clin Exp Med 2015;8:1561–74. [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou Y, Lin L, Wang Y, et al. The association between hypoxia-inducible factor-1 alpha gene G1790A polymorphism and cancer risk: a meta-analysis of 28 case-control studies. Cancer Cell Int 2014;14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang X, Zhu HC, Zhang C, et al. HIF-1alpha 1772 C/T and 1790 G/A polymorphisms are significantly associated with higher cancer risk: an updated meta-analysis from 34 case-control studies. PLoS One 2013;8:e80396. [DOI] [PMC free article] [PubMed] [Google Scholar]


