Abstract
Dengue virus (DENV) infection remains a major public health concern in many parts of the world, including Southeast Asia and the Americas. Sri Lanka experienced its largest dengue outbreak in 2017. Neurological symptoms associated with DENV infection have increasingly been reported in both children and adults. Here, we characterize DENV type 2 (DENV-2) strains, which were isolated from cerebrospinal fluid (CSF) and/or serum of patients with dengue encephalitis. Acute serum and CSF samples from each patient were subjected to dengue-specific non-structural protein 1 (NS1) antigen test, IgM and IgG enzyme-linked immunosorbent assay (ELISA), virus isolation, conventional and real-time polymerase chain reaction (PCR), and next-generation sequencing (NGS). Among the 5 dengue encephalitis patients examined, 4 recovered and 1 died. DENV-2 strains were isolated from serum and/or CSF samples of 3 patients. The highest viral genome levels were detected in the CSF and serum of the patient who succumbed to the illness. A phylogenetic tree revealed that the DENV-2 isolates belonged to a new clade of cosmopolitan genotype and were genetically close to strains identified in China, South Korea, Singapore, Malaysia, Thailand, and the Philippines. According to the NGS analysis, greater frequencies of nonsynonymous and synonymous mutations per gene were identified in the nonstructural genes. The full genomes of serum- and CSF-derived DENV-2 from the same patient shared 99.7% similarity, indicating that the virus spread across the blood-brain barrier. This is the first report to describe neurotropic DENV-2 using whole-genome analysis and to provide the clinical, immunological, and virological characteristics of dengue encephalitis patients during a severe dengue outbreak in Sri Lanka in 2017.
Introduction
Dengue is one of the most globally prevalent, arthropod-borne, viral diseases in humans [1]. The overall incidence of dengue, as well as the incidence of explosive dengue outbreaks, has increased dramatically over the last several years [2]. The causative agent, dengue virus (DENV), which includes four distinct, but closely related serotypes, belongs to the genus Flavivirus in the family Flaviviridae [3]. Transmitted by Aedes mosquitoes, dengue virus occurs primarily in tropical and subtropical areas of the world [4]. The infection causes a flu-like illness, and patients occasionally develop potentially lethal complications. The different degrees of dengue severity were re-categorized in 2009 by the World Health Organization (WHO) into dengue without warning signs (DwoWS), dengue with warning signs (DwWS), and severe dengue (SD) [5]. The annual incidence of dengue infections was estimated to be 400 million per year, of which approximately 96 million were clinically apparent [2]. Death occurs in about 2.5% of dengue-infected people [2, 3]. In recent years, there has been an increase in the number of reported cases of neurological manifestations associated with dengue infections. However, the precise incidence rate of neurological symptoms remains unclear [6]. Neurological signs were first reported in 1976 as atypical symptoms of dengue infection, and their incidence rates have varied from 0.5% to 20% in recent years [7, 8]. Neurological complications associated with DENV infection include encephalopathy (caused by hepatic failure or metabolic disorders), encephalitis (caused by direct viral invasion), neuromuscular complications (Guillain-Barre syndrome or transient muscle dysfunctions), and neuro-ophthalmic involvement [9]. In addition, other less common neurological features have been described as atypical manifestation of dengue infection. Dengue serotypes 2 and 3 are most commonly associated with neurological symptoms [10, 11]. Confirmed dengue cases with neurological manifestations have been confirmed by assessing the presence of the virus and/or antibody in the cerebrospinal fluid (CSF) [6, 12]. However, molecular and biological characterizations of neurotropic DENV strains have been extremely limited, despite their important roles in the neuropathogenesis of dengue.
In 2017, the largest dengue outbreak was reported in Sri Lanka, with over 185,000 clinical cases and at least 250 fatal cases [13]. The age distribution of infected individuals showed that many patients were young people (15–39 years age group) [13]. Atypical manifestations of DENV infection, i.e. dengue encephalitis, were reported during this outbreak. The aims of our study were to describe the neurotropic DENV-2 strains that we isolated from CSF and serum samples of pediatric and adult patients with dengue encephalitis during the severe dengue outbreak in Sri Lanka, in 2017 and to provide clinical, immunological, and virological characteristics of these patients.
Materials and methods
Ethics statement
Ethical approvals for this study were provided by the Institution Ethical Committee on Medical Research and Review, General Hospital (Teaching) Kandy, Sri Lanka (THK/ERC/73/2017) and the Institute of Tropical Medicine Ethical Committee, Nagasaki University, Japan (180608200).
Sample collection
Paired serum and CSF samples used in this study were from five confirmed DEN patients between March 2017- January 2018. These patients included children (< 15 years old) and adults admitted to Teaching Hospital Kandy, Sri Lanka. Confirmation of dengue infection was based on the clinical findings supported by laboratory tests. Patients were classified as having DwoWS or DwWS or SD according to WHO 2009 guideline [5]. Informed Consent was obtained from patients or parents of children or their legal guardians prior to the collection of samples.
DENV IgM and IgG ELISA tests
Serological confirmation of DENV infection was done by an in-house DENV IgM capture ELISA and anti-DENV IgG indirect ELISA. NS1 antigen from serum samples was detected by using SD BIOLINE Dengue NS1 antigen (Ag) rapid test (Standard Diagnostic Inc., Korea). To confirm DENV infection, the in-house DENV IgM was performed following the procedure described previously [14, 15]. Optical density (OD) was read at 492nm and P/N (positive control or sample OD/ negative control OD) ratio ≥ 2 was considered as positive. To determine primary and secondary DENV infections, we used our in-house DENV IgG indirect ELISA [16] which had a high correlation with dengue hemagglutination inhibition (DEN HI) test, the gold standard recommended by the World Health Organization [17]. If the IgG titer was ≥ 29,000, infection was considered secondary, whereas a titer < 29,000 was considered primary [16].
Virus isolation in cell culture
To isolate DENV, serum and CSF samples at 10 ul volume each were inoculated onto Aedes albopictus clone mosquito cells (C6/36 E2) grown in flat culture tubes. Infected cells were incubated at 28°C for 7 days in Eagle’s Minimum Essential Medium supplemented with 2% fetal calf serum and 0.2 mM of non-essential amino acids [18]. The infected culture fluid (ICF) from each tube was collected, aliquoted and stored at -80°C until use. A second virus passage was done in tubes with fresh confluent cells which were incubated for one week with the same incubation conditions as the first passage. ICF from each tube was collected and processed as before.
RNA extraction and conventional RT-PCR
To test ICFs for the presence of DENV, RNA was extracted from them by using viral RNA mini kit (Quiagen, Hilden, Germany) according to the manufacturer’s instruction. Then, PrimeScript™ One Step RT-PCR Kit Ver.2 (Takara Bio Inc., Shiga, Japan) was used following the manufacturer’s instruction. A volume of 5 μl of RNA was used for conventional RT-PCR and amplification was done by using a total of 25 μl of reaction mixture consisting of 1 μl of enzyme mix, 13 μl of 2x buffer, 4 μl of nuclease water, 1 μl of 100 pmol forward and reverse primers with separate primer sets for the detection of DENV and determination of specific DENV serotypes [19–21].
Quantification of DENV genome levels
Viral RNA was directly extracted from 140 μl of patient serum and the same kit was used to extract RNA from ICF. A volume of 5 μl of RNA was used for quantitative real time RT-PCR (qRT-PCR) and amplification of the envelope gene was performed by using a total of 20 μl of reaction mixture consisting of 5 μl of Taqman master mix, 9 μl of nuclease water, 0.3 μl of 100 pmol forward and reverse primers, 0.4 μl of probe with DENV serotype specific primers of TaqMan Fast Virus 1-Step Master Mix (Life Technologies, CA,USA), following the protocol described in a previous report [22, 23]. The viral genome levels were expressed as log10 genome copies/ml.
Full length viral genome amplification and phylogenetic analysis
Whole transcriptome libraries (Ion Total RNA-Seq Kit v2, Life Technologies, CA, USA) were synthesized by using RNA extracted from ICF. Sequencing was conducted by Next-Generation Sequencing (NGS), Ion Proton (Life Technologies, CA, USA). The low quality reads that had < 75% with quality score of < 20 were removed by FASTX-Toolkit Version (v) 0.0.14 from the input data file. Before and after the quality trimming, sequence quality was checked by FastQC v 0.11.8. For the de novo assembly, Trinity v 2.8.4 [24] was used and the sequence name was repaired by Seqkit v 10.0.1. while blastn v 2.7.1 [25] was used to the assembled de novo contig. Trimmed fastq data set were mapped by bwa v 0.7.17 [26] to the reference sequence chosen by blastn, and variants were detected by LoFreq v 2.1 3.1 [27], and Varscan v 2.4.3 [28]. From the output of Varscan, Samtools v 1.9 constructed the consensus dengue virus sequence. Data pre-processing was conducted according to the best practice workflow for GATK v 3.8.1 and Picard v 2.20. Dengue virus sequences in the International Nucleotide Sequence Database Consortium were collected and annotated with Entrez-edirect and annotated by Seqkit. The sequences of the full genome coding region were aligned by Mafft v 7.407 [29]. Maximum likelihood phylogenetic trees were constructed by Phyml v 3.2.0 [30]. Bootstrap values were obtained after 1000 replications. The substitution model was selected by jModelTest v 2.1.10 [31].
Statistical analysis
Data were analyzed by using SPSS for Windows, version 22.0 (IBM Corp., Armonk, NY). Categorical variables were presented as absolute number (n) and percentage (%) as appropriate. Chi-square test (or Fisher’s exact test) was used to determine association among the categorical variables. Comparison of continuous variables (ratio of non-synonymous vs synonymous mutations) was done by Mann-Whitney U test. An alpha level of 0.05 was used for all statistical analyses and a P-value less than 0.05 was considered statistically significant.
Results
Characteristics of patients with neurological manifestations
During the 2017 severe dengue outbreak in Sri Lanka, 295 patients suspected with dengue infection were admitted to the Kandy Teaching Hospital in Kandy, Sri Lanka. There were five patients who showed neurological manifestations. Paired serum and CSF samples were collected from these patients (2 children and 3 adults) who were diagnosed with SD (Table 1). All patients experienced the following symptoms: fever; joint, muscle and eye-socket pain; nausea; vomiting; headache; irritability and; photophobia. Of the 5 patients, 4 recovered and 1 died. The patient who died was a 19-year-old female. She had abdominal pain, liver enlargement, free fluid in the abdomen/pelvis, confusion, loss of consciousness, and generalized fits. She required ventilation. The 4 patients who recovered had signs and symptoms of SD, including neck stiffness, alteration of mental status, confusion, and liver enlargement. The CSF analysis of the samples from all 5 patients revealed low levels of white blood cells (WBCs) and protein, high glucose levels, and a high percentage of CSF/blood sugar, whereas, the serum samples showed low WBC counts and platelet levels (Table 1), leading to the diagnosis of leukopenia-associated thrombocytopenia in all patients. The detection of gram stain and bacterial and fungal cultures were negative for all CSF samples, and none of the CSF samples were contaminated with blood.
Table 1. Clinical features and laboratory findings for the 5 patients with neurological manifestations.
| Serum analysis | CSF analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Age (yr) /sex | Clinical classifi-cation* | Date of sample collection (mo-yr) | Clinical features | WBC (x103/mm) | Hematocrit (%) | platelet count (x103/mm) | WBC (x103/mm) | Protein (mg/dl) | Glucose (mg/dl) | CSF/blood sugar ratio (%) |
| 1 | 24/M | SD | Jun-17 | fever, muscle and joint pain, pain in eye socket, nausea, headache, vomiting, rash, irritability, lethargy, photophobia, neck stiffness, alternation of mental status, encephalitis | 2.7 | 35.7 | 47.8 | 15 | 67 | 124 | 97 |
| (80% neutrophils, 20% lymphocytes) | |||||||||||
| 2 | 19/F | SD | Aug-17 | flushing, gum bleeding, headache, vomiting, free fluid in abdomen/pelvis, abdominal pain, liver enlargement, photophobia, irritability, confusion, loss of consciousness, respiratory failure, generalized fits, encephalitis | 2.3 | 36.7 | 37.9 | 119 | 92.8 | - | - |
| 3 | 17/M | SD | Jul-17 | fever, muscle and joint pain, pain in eye socket, nausea, headache, vomiting, irritability, neck stiffness, alternation of mental status, encephalitis | 5.8 | 27.8 | 67.7 | 246 | 61.7 | 97.8 | 86.5 |
| (11% neutrophils, 89% lymphocytes) | |||||||||||
| 4 | 7/F | SD | Jun-17 | fever, muscle and joint pain, pain in eye socket, nausea, headache, vomiting, irritability, alternation of mental status, encephalitis | 3.8 | 29.9 | 113.9 | 42 | 42.1 | 132.1 | 78.9 |
| (83% neutrophils, 17%lymphocytes) | |||||||||||
| 5 | 10/F | SD | Nov-17 | fever, muscle and joint pain, pain in eye socket, nausea, headache, vomiting, liver enlargement, photophobia, irritability, confusion, alternation of mental status, encephalitis | 4.7 | 36.5 | 77.9 | 8 | 104 | 78.9 | 72 |
| (100% lymphocytes) | |||||||||||
* Clinical classification according to WHO 2009 criteria.
Virological and serological features of dengue patients with neurological manifestations
DENV-NS1 was detected in the serum of patients 1–4 and CSF of patient 2 (Table 2). Due to insufficient amount of CSF, NS1 Ag test was not performed for the three patients (patients 1, 3, and 4). Quantitative real-time PCR (qRT-PCR) was used to determine the viral genome levels of DENV-2 in the serum and CSF samples from all 5 patients; the highest viral genome levels were detected in the CSF and serum samples of the fatal case-patient (patient 2). DENV-2 isolates were identified in the serum and CSF samples of patients 1 and 2, and the serum sample of patient 3. For patients 4 and 5, virus isolation from their clinical samples failed (Table 2). DENV-2 was detected by conventional PCR in the brain tissue obtained at autopsy on patient 2, the patient who succumbed to the illness. The presence of DENV IgM was detected in the serum and CSF samples of the 4 patients, with the exception of patient 2. Out of the five patients, only one patient (patient 2) had primary infection, whereas all the other 4 patients had secondary infection (Table 2).
Table 2. Summary of serological and molecular diagnostic results for the 5 patients with neurological manifestations.
| Patient ID | Sample source | No. of days of illness | NS1 Antigen rapid test | Viremia level by real time RT-PCR (log10copies/ml) | Virus isolation | DENV conventional RT-PCR | NGS analysis | IgM ratioa | IgG titerb | Type of infection | Remark |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 252-Sc | 3 | + | 5.7 | + | DENV-2 | DENV-2 | 11.6 | 43,002 | Sec | recovery |
| 252-Cd | 5 | ND | 7.8 | + | DENV-2 | DENV-2 | 3.2 | 3,666 | |||
| 2 | 257-S | 5 | + | 8.9 | + | DENV-2 | DENV-2 | 0.5 | 294 | Pri | death |
| 257-C | 5 | + | 8.3 | + | DENV-2 | DENV-2 | 0.5 | 95 | |||
| Brain tissue (autopsy) | 7 | DENV-2 | |||||||||
| 3 | 256-S | 5 | + | 6.7 | + | DENV-2 | DENV-2 | 9.5 | 44,876 | Sec | recovery |
| 256-C | 5 | ND | 3.4 | - | - | - | 12.1 | 57,677 | |||
| 4 | 251-S | 3 | + | 4.2 | - | - | - | 11.5 | 72,099 | Sec | recovery |
| 251-C | 3 | ND | 6.3 | - | - | - | 4.5 | 8,760 | |||
| 5 | 1354-S | 10 | - | 3.6 | - | - | - | 11.8 | 60,405 | Sec | recovery |
| 1354-C | 10 | - | 3.9 | - | - | - | 2.2 | 16,183 |
ND: not done. Pri: primary infection. Sec: secondary infection.
a DENV IgM P/N ratio ≥ 2 considered as positive.
b DENV IgG titer ≥ 29,000 considered as secondary and < 29,000 as primary infection.
cS: serum
dC:CSF.
Phylogenetic analysis
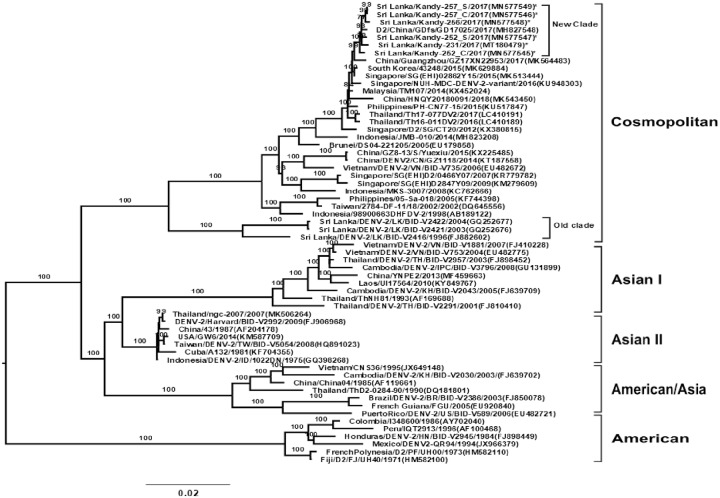
By performing an NGS analysis, the complete genome of DENV-2 was determined from the culture fluids of cells inoculated with the patients’ serum and CSF samples. To compare the genetic background of DENV-2 strains, we compared the sequences of the neurotropic DENV-2 isolates from this study with a closely related reference sequence of DENV-2 (Sri Lanka/Kandy/231-2017/GenBank acc. no. MT180479) which was isolated from serum of non-encephalitis patient who was admitted in the same Kandy hospital during the 2017 dengue outbreak. We determined the genetic relationship of these 5 isolates, the MT 180479 strain and the other 54 DENV-2 strains that have been isolated from geographically diverse areas including Sri Lanka by performing a phylogenetic analysis on their complete genome sequences. The complete genome sequences of the other DENV-strains came from Genbank (Fig 1). The DENV-2 isolates including MT 180479 strain belonged to a new cosmopolitan genotype clade when compared with previously identified DENV-2 isolates (GQ252677, GQ 252676), which circulated in Sri Lanka during 2003 and 2004. The DENV-2 isolates identified in this study were genetically close (99%) to isolates from China, South Korea, Singapore, Malaysia, Thailand, and the Philippines.
Fig 1. Phylogenetic tree of DENV-2 based on the whole genome.
It shows the relationship of the 54 virus strains from different sources, the MT 180479 strain from Sri Lanka, and the 5 isolates (*) of DENV-2 in this study. The 5 isolates in this study and the MT 180479 formed a new clade whereas the strains previously circulating in Sri Lanka belonged to an old clade. The representative strains of each genotype obtained from GenBank are named by country-origin, strain name, year of isolation, and GenBank accession number.
Amino acid variability analysis
To characterize viral amino acid differences by an NGS strategy, the amino acid sequences of the DENV-2 isolates were aligned. The sequence alignment revealed 99% identity among the isolates. Compared with MT180479 strain, amino acid substitutions were identified in 165 sites throughout the complete coding region of the 5 DENV-2 isolates in this study (Table 3). Across the complete genome, the number of positions with variant frequencies > 1% revealed 149 synonymous and 16 nonsynonymous sites among DENV isolates in this study (S1 Table). The variant frequencies > 50% indicated that these variants became predominant in the particular patient (Table 3). The 30 synonymous and 4 nonsynonymous mutations were shared among DENV isolates from the CSF and serum of patients. The fifteen synonymous and one nonsynonymous (K1400R) mutations were found in the virus particles from both serum and CSF of patient 1. The thirty synonymous and three nonsynonymous (S643N, K1400R, V1779I) were shared by the virus particles from serum and CSF of patient 2.
Table 3. Synonymous and nonsynonymous variant (>1%) alleles shared among the DENV-2 isolates in this study.
| Sample IDa | Feature | Nucleo tide position | Amino acid position | Ref: Alleleb | ALT Allelec | Frequency (%) | Changed | Sample IDa | Feature | Nucleo tide position | Amino acid position | Ref: Alleleb | ALT Allelec | Frequency (%) | Changed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 252-C | C | 276 | 60 | A | G | 98.4 | Syn-P | 257-C | NS3 | 4647 | 1517 | A | G | 99.4 | Syn-K |
| 252-S | C | 276 | 60 | A | G | 100.0 | Syn-P | 257-S | NS3 | 4647 | 1517 | A | G | 98.4 | Syn-K |
| 257-C | C | 276 | 60 | A | G | 98.4 | Syn-P | 256-S | NS3 | 4647 | 1517 | A | G | 100.0 | Syn-K |
| 257-S | C | 276 | 60 | A | G | 98.5 | Syn-P | ||||||||
| 256-S | C | 276 | 60 | A | G | 100.0 | Syn-P | 252-C | NS3 | 5182 | 1696 | T | C | 99.0 | Syn-L |
| 252-S | NS3 | 5182 | 1696 | T | C | 98.3 | Syn-L | ||||||||
| 257-C | C | 370 | 92 | C | T | 99.1 | Syn-L | 257-C | NS3 | 5182 | 1696 | T | C | 96.0 | Syn-L |
| 257-S | C | 370 | 92 | C | T | 100.0 | Syn-L | 257-S | NS3 | 5182 | 1696 | T | C | 98.8 | Syn-L |
| 256-S | NS3 | 5182 | 1696 | T | C | 100.0 | Syn-L | ||||||||
| 252-C | pr | 678 | 194 | T | C | 98.8 | Syn-C | ||||||||
| 252-S | pr | 678 | 194 | T | C | 100.0 | Syn-C | 252-C | NS3 | 5406 | 1770 | A | G | 95.7 | Syn-A |
| 257-C | pr | 678 | 194 | T | C | 99.4 | Syn-C | 252-S | NS3 | 5406 | 1770 | A | G | 100.0 | Syn-A |
| 257-S | pr | 678 | 194 | T | C | 99.4 | Syn-C | 257-C | NS3 | 5406 | 1770 | A | G | 98.7 | Syn-A |
| 256-S | pr | 678 | 194 | T | C | 97.7 | Syn-C | 257-S | NS3 | 5406 | 1770 | A | G | 99.3 | Syn-A |
| 256-S | NS3 | 5406 | 1770 | A | G | 100.0 | Syn-A | ||||||||
| 257-C | M | 879 | 261 | A | G | 100.0 | Syn-T | ||||||||
| 257-S | M | 879 | 261 | A | G | 100.0 | Syn-T | 257-C | NS3 | 5431 | 1779 | G | A | 100.0 | V-I |
| 256-S | M | 879 | 261 | A | G | 100.0 | Syn-T | 257-S | NS3 | 5431 | 1779 | G | A | 98.9 | V-I |
| 257-C | E | 1785 | 563 | T | C | 98.3 | Syn-L | 252-C | NS3 | 5523 | 1809 | C | T | 98.9 | Syn-D |
| 257-S | E | 1785 | 563 | T | C | 98.4 | Syn-L | 252-S | NS3 | 5523 | 1809 | C | T | 100.0 | Syn-D |
| 256-S | E | 1785 | 563 | T | C | 100.0 | Syn-L | 257-C | NS3 | 5523 | 1809 | C | T | 100.0 | Syn-D |
| 257-S | NS3 | 5523 | 1809 | C | T | 99.1 | Syn-D | ||||||||
| 257-C | E | 2024 | 643 | G | A | 99.4 | S-N | 256-S | NS3 | 5523 | 1809 | C | T | 100.0 | Syn-D |
| 257-S | E | 2024 | 643 | G | A | 98.0 | S-N | ||||||||
| 257-C | NS3 | 5676 | 1860 | C | T | 100.0 | Syn-L | ||||||||
| 257-C | E | 2095 | 667 | C | T | 100.0 | Syn-L | 257-S | NS3 | 5676 | 1860 | C | T | 98.8 | Syn-L |
| 257-S | E | 2095 | 667 | C | T | 100.0 | Syn-L | 256-S | NS3 | 5676 | 1860 | C | T | 100.0 | Syn-L |
| 256-S | E | 2095 | 667 | C | T | 96.6 | Syn-L | ||||||||
| 252-C | NS3 | 6174 | 2026 | C | A | 100.0 | Syn-A | ||||||||
| 257-C | E | 2388 | 764 | C | A | 97.2 | Syn-V | 252-S | NS3 | 6174 | 2026 | C | A | 100.0 | Syn-A |
| 257-S | E | 2388 | 764 | C | A | 96.0 | Syn-V | 257-C | NS3 | 6174 | 2026 | C | A | 100.0 | Syn-A |
| 257-S | NS3 | 6174 | 2026 | C | A | 100.0 | Syn-A | ||||||||
| 252-C | NS1 | 2901 | 935 | C | T | 99.2 | Syn-F | 256-S | NS3 | 6174 | 2026 | C | A | 100.0 | Syn-A |
| 252-S | NS1 | 2901 | 935 | C | T | 97.8 | Syn-F | ||||||||
| 257-C | NS1 | 2901 | 935 | C | T | 97.4 | Syn-F | 252-C | NS4A | 6510 | 2138 | C | T | 99.0 | Syn-L |
| 257-S | NS1 | 2901 | 935 | C | T | 97.0 | Syn-F | 252-S | NS4A | 6510 | 2138 | C | T | 100.0 | Syn-L |
| 256-S | NS1 | 2901 | 935 | C | T | 100.0 | Syn-F | 257-C | NS4A | 6510 | 2138 | C | T | 100.0 | Syn-L |
| 257-S | NS4A | 6510 | 2138 | C | T | 100.0 | Syn-L | ||||||||
| 257-C | NS1 | 3327 | 1077 | T | C | 100.0 | Syn-T | 256-S | NS4A | 6510 | 2138 | C | T | 100.0 | Syn-L |
| 257-S | NS1 | 3327 | 1077 | T | C | 100.0 | Syn-T | ||||||||
| 256-S | NS1 | 3327 | 1077 | T | C | 100.0 | Syn-T | 252-S | NS4A | 6603 | 2169 | A | G | 100.0 | Syn-K |
| 257-C | NS4A | 6603 | 2169 | A | G | 99.4 | Syn-K | ||||||||
| 252-C | NS1 | 3459 | 1121 | T | C | 100.0 | Syn-V | 257-S | NS4A | 6603 | 2169 | A | G | 99.0 | Syn-K |
| 257-C | NS1 | 3459 | 1121 | T | C | 98.7 | Syn-V | 256-S | NS4A | 6603 | 2169 | A | G | 100.0 | Syn-K |
| 257-S | NS1 | 3459 | 1121 | T | C | 99.1 | Syn-V | ||||||||
| 256-S | NS1 | 3459 | 1121 | T | C | 88.9 | Syn-V | 257-C | 2Ke | 6801 | 2235 | T | C | 100.0 | Syn-L |
| 257-S | 2K | 6801 | 2235 | T | C | 99.1 | Syn-L | ||||||||
| 252-C | NS2A | 3534 | 1146 | A | G | 100.0 | Syn-L | 256-S | 2K | 6801 | 2235 | T | C | 97.0 | Syn-L |
| 252-S | NS2A | 3534 | 1146 | A | G | 100.0 | Syn-L | ||||||||
| 257-C | NS2A | 3534 | 1146 | A | G | 99.4 | Syn-L | 252-C | NS4B | 6900 | 2268 | T | C | 100.0 | Syn-S |
| 257-S | NS2A | 3534 | 1146 | A | G | 100.0 | Syn-L | 252-S | NS4B | 6900 | 2268 | T | C | 100.0 | Syn-S |
| 256-S | NS2A | 3534 | 1146 | A | G | 100.0 | Syn-L | 257-C | NS4B | 6900 | 2268 | T | C | 100.0 | Syn-S |
| 257-S | NS4B | 6900 | 2268 | T | C | 96.1 | Syn-S | ||||||||
| 252-C | NS2A | 3597 | 1167 | T | C | 99.4 | Syn-F | 256-S | NS4B | 6900 | 2268 | T | C | 100.0 | Syn-S |
| 252-S | NS2A | 3597 | 1167 | T | C | 100.0 | Syn-F | ||||||||
| 257-C | NS2A | 3597 | 1167 | T | C | 100.0 | Syn-F | 252-C | NS4B | 8028 | 2644 | T | C | 97.6 | Syn-N |
| 257-S | NS2A | 3597 | 1167 | T | C | 99.4 | Syn-F | 252-S | NS4B | 8028 | 2644 | T | C | 96.8 | Syn-N |
| 256-S | NS2A | 3597 | 1167 | T | C | 100.0 | Syn-F | 257-C | NS4B | 8028 | 2644 | T | C | 96.8 | Syn-N |
| 257-S | NS4B | 8028 | 2644 | T | C | 100.0 | Syn-N | ||||||||
| 252-C | NS2A | 3624 | 1176 | T | C | 93.9 | Syn-S | 256-S | NS4B | 8028 | 2644 | T | C | 100.0 | Syn-N |
| 252-S | NS2A | 3624 | 1176 | T | C | 100.0 | Syn-S | ||||||||
| 257-C | NS2A | 3624 | 1176 | T | C | 92.1 | Syn-S | 257-C | NS4B | 8325 | 2743 | C | T | 20.0 | Syn-Y |
| 257-S | NS2A | 3624 | 1176 | T | C | 96.5 | Syn-S | 257-S | NS4B | 8325 | 2743 | C | T | 23.1 | Syn-Y |
| 256-S | NS2A | 3624 | 1176 | T | C | 100.0 | Syn-S | ||||||||
| 257-C | NS4B | 8946 | 2950 | G | A | 99.2 | Syn-E | ||||||||
| 252-C | NS2B | 4295 | 1400 | A | G | 100.0 | K-R | 257-S | NS4B | 8946 | 2950 | G | A | 66.2 | Syn-E |
| 252-S | NS2B | 4295 | 1400 | A | G | 100.0 | K-R | 256-S | NS4B | 8946 | 2950 | G | A | 58.3 | Syn-E |
| 257-C | NS2B | 4295 | 1400 | A | G | 98.5 | K-R | ||||||||
| 257-S | NS2B | 4295 | 1400 | A | G | 100.0 | K-R | 252-C | NS4B | 9189 | 3031 | G | A | 100.0 | Syn-T |
| 256-S | NS2B | 4295 | 1400 | A | G | 100.0 | K-R | 252-S | NS4B | 9189 | 3031 | G | A | 99.4 | Syn-T |
| 257-C | NS4B | 9189 | 3031 | G | A | 99.4 | Syn-T | ||||||||
| 252-C | NS2B | 4488 | 1464 | C | T | 100.0 | Syn-A | 257-S | NS4B | 9189 | 3031 | G | A | 100.0 | Syn-T |
| 252-S | NS2B | 4488 | 1464 | C | T | 100.0 | Syn-A | 256-S | NS4B | 9189 | 3031 | G | A | 100.0 | Syn-T |
| 257-C | NS2B | 4488 | 1464 | C | T | 99.2 | Syn-A | ||||||||
| 257-S | NS2B | 4488 | 1464 | C | T | 100.0 | Syn-A | 257-C | NS4B | 9399 | 3101 | T | C | 99.4 | Syn-N |
| 256-S | NS2B | 4488 | 1464 | C | T | 100.0 | Syn-A | 257-S | NS4B | 9399 | 3101 | T | C | 100.0 | Syn-N |
| 256-S | NS4B | 9399 | 3101 | T | C | 100.0 | Syn-N | ||||||||
| 257-C | NS3 | 4587 | 1497 | C | T | 99.2 | Syn-A | ||||||||
| 257-S | NS3 | 4587 | 1497 | C | T | 99.1 | Syn-A | 252-C | NS5 | 10606 | 3504 | C | G | 7.9 | R-G |
| 256-S | NS3 | 4587 | 1497 | C | T | 100.0 | Syn-A | 256-S | NS5 | 10606 | 3504 | C | G | 31.9 | R-G |
aSame sample ID number means same patient, C: CSF, S: serum.
bDENV-2 (Sri Lanka/Kandy/231-2017/MT180479) isolated from serum of non-encephalitis patient during 2017 dengue outbreak was used as reference strain.
cAlternative allele in DENV-2 isolates.
dSynonymous and nonsynonymous mutations.
e2K peptide, 17 amino acid peptide linking NS4A with NS4B of DENV.
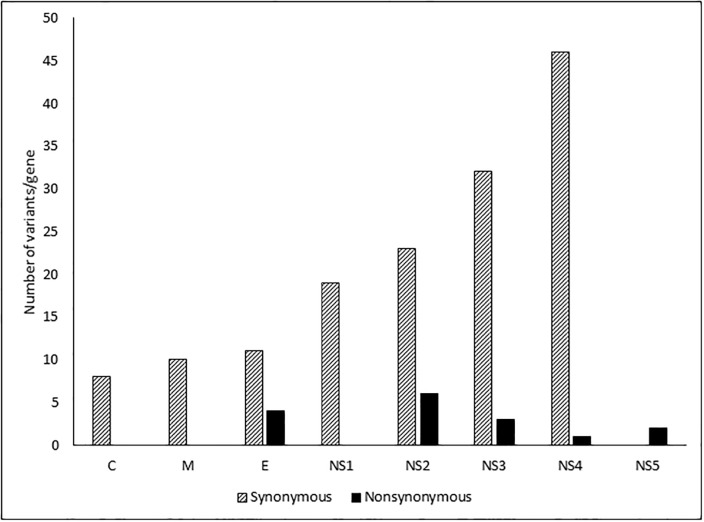
We analyzed the total number of synonymous and nonsynonymous mutations in each gene, independent of the frequency (Fig 2). The number of synonymous variants was higher than the number of nonsynonymous substitutions at each site. The greater variability was observed among the nonstructural genes than the structural genes. There was significant association between the mutations with DENV genes (p = <0.0001, chi-square test). Significantly higher proportion of nonsynonymous mutations was observed in the NS2 gene compared to other genes. There was no difference in the ratios of frequencies of non-synonymous to synonymous mutations (nS/S) between the structural and non-structural genes (p = 0.85, Mann Whitney U test) in (S1 Fig).
Fig 2. Number of positions with variant incidence > 1% per gene among the DENV-2 isolates.
Discussion
The 2017 dengue outbreak in Sri Lanka was the largest in the island’s history [13]. Out of 295 dengue-suspected patients, 5 patients (1.7%) who presented with neurological symptoms and confirmed to have dengue encephalitis were the subjects of this study. DENV infections associated with neurological manifestations have been reported in Thailand, India, Vietnam, and Cambodia at 1–20% [32–36], in Brazil at 21–47% [37, 38], and in Puerto Rico at 26% of patients [39]. Neurological manifestations caused by DENV have been reported in patients with ages ranging from 3 months to 60 years [6]. In the present study, 2 children and 3 adults were identified with DENV-associated neurological manifestations. CNS involvement occurs 4–30 days after the first dengue symptoms appear, with a median of 12 days after the onset of fever, and CNS symptoms were identified between days 3–10 after the onset of fever in our study [38].
The protein NS1 is present at high concentrations in the sera from dengue-infected patients during the early clinical phase of the disease and can be found between days 1–9 after the onset of fever in samples from primary or secondary dengue-infected patients [40]. The detection of NS1 in serum samples was positive in 4 of the 5 patients in this study, starting between 3–5 days after the onset of illness. The patient who was negative for dengue NS1 Ag was tested for the presence of this antigen on day 10 of the illness The detection of dengue IgM in CSF has been shown to have a high specificity (97%) for the diagnosis of neurologic dengue and might be associated with the neurovirulence of DENV and its ability to cause encephalitis [41]. Together, our results showed that all patients were DENV-IgM-positive, except for the fatal case. In a previous study, dengue RNA was found in the CSF from 7 out of 13 Brazilian patients, and the CSF viral load was lower than 1,000 copies/ml [37]. However in the present study, dengue viral genome detected in CSF samples from 3 patients showed high viral loads (> 1,000,000 copies/ml) and the remaining 2 patients had viral loads higher than 1,000 copies/ml.
A number of studies have reported the presence of DENV RNA and the isolation of DENV from CSF samples taken from encephalitis patients. However, only a few studies, including one study that reported the complete genome sequence for DENV-4, two studies that reported partial C-PrM-E sequences for DENV-3, one study that reported the complete genome sequence for DENV-3 and two studies that reported partial E-NS1 sequences for DENV-2, have examined the genetic characteristics of neurotropic DENV. All of these DENV were isolated from CSF samples [8, 42–45]. To our knowledge, this is the first report to describe the complete genome and the molecular and virological characteristics of DENV-2 isolated from CSF samples from fatal and non-fatal dengue case-patients with neurological manifestations. Compared with other dengue serotypes, DENV-2 and DENV-3 appear to be more frequently associated with encephalitis and other neurological manifestations, regardless of whether it is primary or secondary infections [43, 45]. In a meta-analysis examining the severity of dengue during primary or secondary infection with different DENV serotypes, secondary infections with DENV-2 were reported to be associated with severe cases [46]. In our study, 4 out of 5 patients with SD was due to secondary infections. However, only one case was associated with a primary DENV-2 infection and this might be due to other viral and/ or host factors. The detection of DENV in the brain and CSF by PCR and virus isolation and the detection of NS1 and dengue IgM provided strong evidences that DENV has neurovirulent properties. The evidence of DENV in the CSF, as assessed by RT-PCR and ELISA (NS1/IgM) in this study, is consistent with a CNS infection.
A complete genomic phylogenetic analysis revealed that DENV-2 isolates from the serum and CSF of the same patients showed high similarity and appeared in the same branch. The full genomes of serum- and CSF-derived DENV-2 from the same patient shared 99.7% similarity, indicating the virus dissemination across the blood-brain barrier. Two novel mutations, S643N (E) and V1779I (NS3), which have not been reported previously, were identified in CSF and serum of patient 2. Furthermore, our study compared the genetic background of DENV-2 strains from the 5 dengue encephalitis patients in this study and of DENV-2 strain (MT180479) from non-encephalitis patient who was admitted to the hospital where the 5 patients were also admitted during the dengue outbreak. There were 16 amino acid changes found in neurotropic DENV-2 isolates compared with MT180479 strain. However, whether these amino acid changes are associated with the neurovirulence properties of these DENV-2 isolates remains unclear. Both viral mutations and the genetic or immunological status of the hosts may contribute to the development of dengue encephalitis in patients [47–49]. In addition, further studies including cell tropism studies using in vivo and in vitro experiment are necessary to elucidate the virulence of these isolates.
In conclusion, our study reported for the first time the isolation and the complete genome analysis of a new cosmopolitan genotype clade of neurotropic DENV-2 isolated from serum and CSF samples taken from the same patients during the unprecedented dengue outbreak in Sri Lanka, in 2017. Our findings suggested that pathogenic DENV-2 caused neurological complications, including one fatal case. We concluded that genetic variations were increased among the non-structural DENV-2 genes. In addition, dengue should be suspected in patients with neurological manifestations in dengue-endemic areas, regardless of whether the patient presents with classical dengue features.
Supporting information
(DOCX)
(TIF)
Acknowledgments
We thank the Director of Teaching Hospital Kandy and Dr. Corazon C. Buerano for providing support and advice. We are grateful for the support of the members of the Department of Virology, Institute of Tropical Medicine, Nagasaki University.
Data Availability
All relevant data are within the manuscript.
Funding Statement
This work was supported by grants from the Japan Initiative for Global Research Network on Infectious Diseases (JGRID) of the Japan Science and Technology Agency (JST) (Grant number JP19fm0108001), and Research Program on Emerging and Re-emerging Infectious Diseases of the Agency for Research and Development (AMED, 19fk0108035h1203).
References
- 1.Massad E, Coutinho FA. The cost of dengue control. Lancet. 2011;377(9778):1630–1. Epub 2011/05/07. 10.1016/S0140-6736(11)60470-4 . [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. Epub 2013/04/09. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: a continuing global threat. Nature reviews Microbiology. 2010;8(12 Suppl):S7–16. 10.1038/nrmicro2460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubler DJ. Dengue, Urbanization and Globalization: The Unholy Trinity of the 21(st) Century. Tropical medicine and health. 2011;39(4 Suppl):3–11. Epub 2012/04/14. 10.2149/tmh.2011-S05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. World Health Organization. Dengue Guidelines for Diagnosis, Treatment, Prevention, and Control. Geneva. 2009. [PubMed]
- 6.Li GH, Ning ZJ, Liu YM, Li XH. Neurological Manifestations of Dengue Infection. Frontiers in cellular and infection microbiology. 2017;7:449 Epub 2017/11/10. 10.3389/fcimb.2017.00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini L, Chakrabarty B, Pastel H, Israni A, Kumar A, Gulati S. Dengue fever triggering hemiconvulsion hemiplegia epilepsy in a child. Neurology India. 2017;65(3):636–8. Epub 2017/05/11. 10.4103/neuroindia.NI_1367_15 . [DOI] [PubMed] [Google Scholar]
- 8.Solomon T, Dung NM, Vaughn DW, Kneen R, Thao LT, Raengsakulrach B, et al. Neurological manifestations of dengue infection. Lancet. 2000;355(9209):1053–9. Epub 2000/04/01. 10.1016/S0140-6736(00)02036-5 . [DOI] [PubMed] [Google Scholar]
- 9.Carod-Artal FJ, Wichmann O, Farrar J, Gascon J. Neurological complications of dengue virus infection. The Lancet Neurology. 2013;12(9):906–19. Epub 2013/08/21. 10.1016/S1474-4422(13)70150-9 . [DOI] [PubMed] [Google Scholar]
- 10.Lum LC, Lam SK, Choy YS, George R, Harun F. Dengue encephalitis: a true entity? The American journal of tropical medicine and hygiene. 1996;54(3):256–9. Epub 1996/03/01. 10.4269/ajtmh.1996.54.256 . [DOI] [PubMed] [Google Scholar]
- 11.Soares CN, Cabral-Castro MJ, Peralta JM, Freitas MR, Puccioni-Sohler M. Meningitis determined by oligosymptomatic dengue virus type 3 infection: report of a case. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2010;14(2):e150–2. Epub 2009/06/09. 10.1016/j.ijid.2009.03.016 . [DOI] [PubMed] [Google Scholar]
- 12.Araujo FM, Brilhante RS, Cavalcanti LP, Rocha MF, Cordeiro RA, Perdigao AC, et al. Detection of the dengue non-structural 1 antigen in cerebral spinal fluid samples using a commercially available enzyme-linked immunosorbent assay. Journal of virological methods. 2011;177(1):128–31. Epub 2011/07/30. 10.1016/j.jviromet.2011.07.003 . [DOI] [PubMed] [Google Scholar]
- 13.Ali SKA, Taylor-Robinson AW, Adnan M ea. The unprecedented magnitude of the 2017 dengue outbreak in Sri Lanka provides lessons for future mosquito-borne infection control and prevention. Infection, Disease & Health. 2017;23(2):114–20. [DOI] [PubMed] [Google Scholar]
- 14.Bundo K, Igarashi A. Antibody-capture ELISA for detection of immunoglobulin M antibodies in sera from Japanese encephalitis and dengue hemorrhagic fever patients. Journal of virological methods. 1985;11(1):15–22. Epub 1985/05/01. 10.1016/0166-0934(85)90120-x . [DOI] [PubMed] [Google Scholar]
- 15.Ngwe Tun MM, Thant KZ, Inoue S, Kurosawa Y, Lwin YY, Lin S, et al. Serological characterization of dengue virus infections observed among dengue hemorrhagic fever/dengue shock syndrome cases in upper Myanmar. Journal of medical virology. 2013;85(7):1258–66. Epub 2013/04/19. 10.1002/jmv.23577 . [DOI] [PubMed] [Google Scholar]
- 16.Inoue S, Alonzo MT, Kurosawa Y, Mapua CA, Reyes JD, Dimaano EM, et al. Evaluation of a dengue IgG indirect enzyme-linked immunosorbent assay and a Japanese encephalitis IgG indirect enzyme-linked immunosorbent assay for diagnosis of secondary dengue virus infection. Vector borne and zoonotic diseases (Larchmont, NY). 2010;10(2):143–50. Epub 2009/10/31. 10.1089/vbz.2008.0153 . [DOI] [PubMed] [Google Scholar]
- 17.WHO. World Health Organization. Dengue Guidelines for Diagnosis, Treatment, Prevention and Control. 2nd ed. Geneva1997. [PubMed]
- 18.Ngwe Tun MM, Thant KZ, Inoue S, Nabeshima T, Aoki K, Kyaw AK, et al. Detection of east/central/south African genotype of chikungunya virus in Myanmar, 2010. Emerging infectious diseases. 2014;20(8):1378–81. Epub 2014/07/26. 10.3201/eid2008.131431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngwe Tun MM, Kyaw AK, Makki N, Muthugala R, Nabeshima T, Inoue S, et al. Characterization of the 2013 dengue epidemic in Myanmar with dengue virus 1 as the dominant serotype. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;43:31–7. Epub 2016/05/08. 10.1016/j.meegid.2016.04.025 . [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. Journal of clinical microbiology. 1992;30(3):545–51. Epub 1992/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita K, Tanaka M, Igarashi A. Rapid identification of dengue virus serotypes by using polymerase chain reaction. Journal of clinical microbiology. 1991;29(10):2107–10. Epub 1991/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyaw AK, Ngwe Tun MM, Moi ML, Nabeshima T, Soe KT, Thwe SM, et al. Clinical, virological and epidemiological characterization of dengue outbreak in Myanmar, 2015. Epidemiology and infection. 2017;145(9):1886–97. Epub 2017/04/18. 10.1017/S0950268817000735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito M, Takasaki T, Yamada K, Nerome R, Tajima S, Kurane I. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. Journal of clinical microbiology. 2004;42(12):5935–7. Epub 2004/12/08. 10.1128/JCM.42.12.5935-5937.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–52. Epub 2011/05/17. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC bioinformatics. 2009;10:421 Epub 2009/12/17. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2010;26(5):589–95. Epub 2010/01/19. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic acids research. 2012;40(22):11189–201. Epub 2012/10/16. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22(3):568–76. Epub 2012/02/04. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30(4):772–80. Epub 2013/01/19. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59(3):307–21. Epub 2010/06/09. 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- 31.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods. 2012;9(8):772 Epub 2012/08/01. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thisyakorn U, Thisyakorn C, Limpitikul W, Nisalak A. Dengue infection with central nervous system manifestations. The Southeast Asian journal of tropical medicine and public health. 1999;30(3):504–6. Epub 2000/04/25. . [PubMed] [Google Scholar]
- 33.Chokephaibulkit K, Kankirawatana P, Apintanapong S, Pongthapisit V, Yoksan S, Kositanont U, et al. Viral etiologies of encephalitis in Thai children. The Pediatric infectious disease journal. 2001;20(2):216–8. Epub 2001/02/27. 10.1097/00006454-200102000-00020 . [DOI] [PubMed] [Google Scholar]
- 34.Kumar R, Tripathi S, Tambe JJ, Arora V, Srivastava A, Nag VL. Dengue encephalopathy in children in Northern India: clinical features and comparison with non dengue. Journal of the neurological sciences. 2008;269(1–2):41–8. Epub 2008/01/29. 10.1016/j.jns.2007.12.018 . [DOI] [PubMed] [Google Scholar]
- 35.Le VT, Phan TQ, Do QH, Nguyen BH, Lam QB, Bach V, et al. Viral etiology of encephalitis in children in southern Vietnam: results of a one-year prospective descriptive study. PLoS neglected tropical diseases. 2010;4(10):e854 Epub 2010/11/05. 10.1371/journal.pntd.0000854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srey VH, Sadones H, Ong S, Mam M, Yim C, Sor S, et al. Etiology of encephalitis syndrome among hospitalized children and adults in Takeo, Cambodia, 1999–2000. The American journal of tropical medicine and hygiene. 2002;66(2):200–7. Epub 2002/07/24. 10.4269/ajtmh.2002.66.200 . [DOI] [PubMed] [Google Scholar]
- 37.Domingues RB, Kuster GW, Onuki-Castro FL, Souza VA, Levi JE, Pannuti CS. Involvement of the central nervous system in patients with dengue virus infection. Journal of the neurological sciences. 2008;267(1–2):36–40. Epub 2007/10/26. 10.1016/j.jns.2007.09.040 . [DOI] [PubMed] [Google Scholar]
- 38.Soares CN, Cabral-Castro MJ, Peralta JM, de Freitas MR, Zalis M, Puccioni-Sohler M. Review of the etiologies of viral meningitis and encephalitis in a dengue endemic region. Journal of the neurological sciences. 2011;303(1–2):75–9. Epub 2011/02/05. 10.1016/j.jns.2011.01.012 . [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Rivera EJ, Vorndam V, Rigau-Perez JG. Use of an enhanced surveillance system for encephalitis and aseptic meningitis for the detection of neurologic manifestations of dengue in Puerto Rico, 2003. Puerto Rico health sciences journal. 2009;28(2):114–20. Epub 2009/06/18. . [PubMed] [Google Scholar]
- 40.Wang SM, Sekaran SD. Early diagnosis of Dengue infection using a commercial Dengue Duo rapid test kit for the detection of NS1, IGM, and IGG. The American journal of tropical medicine and hygiene. 2010;83(3):690–5. Epub 2010/09/03. 10.4269/ajtmh.2010.10-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soares CN, Faria LC, Peralta JM, de Freitas MR, Puccioni-Sohler M. Dengue infection: neurological manifestations and cerebrospinal fluid (CSF) analysis. Journal of the neurological sciences. 2006;249(1):19–24. Epub 2006/07/28. 10.1016/j.jns.2006.05.068 . [DOI] [PubMed] [Google Scholar]
- 42.Hapuarachchi HC, Oh HM, Thein TL, Pok KY, Lai YL, Tan LK, et al. Clinico-genetic characterisation of an encephalitic Dengue virus 4 associated with multi-organ involvement. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2013;57(1):91–4. Epub 2013/02/19. 10.1016/j.jcv.2012.12.021 . [DOI] [PubMed] [Google Scholar]
- 43.Phu Ly MH, Takamatsu Y, Nabeshima T, Pham Hoai LL, Pham Thi H, Dang Thi D, et al. Isolation of dengue serotype 3 virus from the cerebrospinal fluid of an encephalitis patient in Hai Phong, Vietnam in 2013. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2015;70:93–6. Epub 2015/08/26. 10.1016/j.jcv.2015.07.295 . [DOI] [PubMed] [Google Scholar]
- 44.Oliveira DB, Machado G, Almeida GM, Ferreira PC, Bonjardim CA, Trindade Gde S, et al. Infection of the central nervous system with dengue virus 3 genotype I causing neurological manifestations in Brazil. Revista da Sociedade Brasileira de Medicina Tropical. 2016;49(1):125–9. Epub 2016/05/11. 10.1590/0037-8682-0208-2015 . [DOI] [PubMed] [Google Scholar]
- 45.Dhenni R, Karyanti MR, Putri ND, Yohan B, Yudhaputri FA, Ma’roef CN, et al. Isolation and complete genome analysis of neurotropic dengue virus serotype 3 from the cerebrospinal fluid of an encephalitis patient. PLoS neglected tropical diseases. 2018;12(1):e0006198 Epub 2018/01/13. 10.1371/journal.pntd.0006198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soo KM, Khalid B, Ching SM, Chee HY. Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections. PloS one. 2016;11(5):e0154760 Epub 2016/05/24. 10.1371/journal.pone.0154760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Araujo F, Nogueira R, Araujo Mde S, Perdigao A, Cavalcanti L, Brilhante R, et al. Dengue in patients with central nervous system manifestations, Brazil. Emerging infectious diseases. 2012;18(4):677–9. Epub 2012/04/04. 10.3201/eid1804.111552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clinical microbiology reviews. 2009;22(4):564–81. Epub 2009/10/14. 10.1128/CMR.00035-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misra UK, Kalita J, Mani VE, Chauhan PS, Kumar P. Central nervous system and muscle involvement in dengue patients: A study from a tertiary care center. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2015;72:146–51. Epub 2015/10/30. 10.1016/j.jcv.2015.08.021 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
Data Availability Statement
All relevant data are within the manuscript.