Abstract
Human Survival Motor Neuron (SMN) genes code for SMN, an essential multifunctional protein. Complete loss of SMN is embryonic lethal, while low levels of SMN lead to spinal muscular atrophy (SMA), a major genetic disease of children and infants. Reduced levels of SMN are associated with the abnormal development of heart, lung, muscle, gastro-intestinal system and testis. The SMN loci have been shown to generate a vast repertoire of transcripts, including linear, back- and trans-spliced RNAs as well as antisense long noncoding RNAs. However, functions of the majority of these transcripts remain unknown. Here we review the nature of RNAs generated from the SMN loci and discuss their potential functions in cellular metabolism.
Keywords: spinal muscular atrophy, SMA; Survival Motor Neuron, SMN; backsplicing; circRNA; Alu elements; long noncoding RNA and microRNA
1. Introduction
Humans have two nearly identical copies of the Survival Motor Neuron (SMN) gene: SMN1 and SMN2 [1]. Both SMN genes code for SMN, a multifunctional protein implicated in snRNP assembly, pre-mRNA splicing, transcription, translation, stress granule formation, macromolecular trafficking and maintenance of cytoskeletal dynamics [2,3]. With the exception of plants, SMN or SMN-like proteins are expressed in all eukaryotic organisms [3]. While complete loss of SMN is embryonic lethal, deletions of or mutations in SMN1 causes spinal muscular atrophy (SMA), a leading genetic disease of children and infants [3–6]. SMN2 cannot compensate for the loss of SMN1 due to predominant skipping of exon 7 [7]. However, the presence of SMN2 provides a readymade target for SMA therapy through manipulation of SMN2 exon 7 splicing. A critical C-to-T mutation at the 6th position (C6U substitution in RNA) of exon 7 distinguishes SMN2 from SMN1 [7,8]. C6U substitution itself is sufficient to trigger exon 7 skipping [7]. Currently, there is a diversity of opinions with respect to the mechanism by which this nucleotide change (C6U substitution) exerts its negative effect on exon 7 splicing [9–12]. Studies spanning over two decades highlight the roles of multiple cis-elements and transacting factors in regulation of SMN exon 7 splicing [13–15].
Several critical cis-elements, including Intronic Splicing Silencer N1 (ISS-N1), located within intron 7 support a point of view that regulation of SMN2 exon 7 splicing is governed by an intron definition model [16,17]. Of a particular significance to SMA therapy was the discovery that a deletion of ISS-N1 or its blocking by an antisense oligonucleotide (ASO) fully restored SMN2 exon 7 inclusion [16]. In a major development, independent in vivo studies reported within a short span of three years (from 2011 to 2013) validated the unprecedented high therapeutic efficacies of ISS-N1-targeting ASOs [18–21]. Findings also underscored that the efficacy of ISS-N1-targeting ASOs was not dependent on the specific ASO chemistry [21]. Consequently, the ISS-N1-targeting ASO Nusinersen (Spinraza™) emerged as the first approved therapy for SMA [22–24]. Recently published reports on the outcomes of the clinical trials support the therapeutic efficacy of this drug [25–27]. Multiple cis-elements, including structural elements, overlap ISS-N1 [28–34]. Based on in vivo studies, two of these cis-elements appear to be good targets for SMA therapy as well [35,36]. Restoration of SMN levels via gene therapy is another approved approach for SMA treatment [37]. Several small compounds presently in clinical trials are likely to further expand the therapeutic choices for SMA patients [6,38–41].
Parallel to the advances in SMA therapy, there has been transformative progress in our understanding of tissue-specific pathologies caused by the reduced levels of SMN [6]. Animal models suggest that all cell/tissue types, including brain, spinal cord, muscle, heart, gastrointestinal system, liver, lung, pancreas, spinal cord and testis, are intrinsically affected by low SMN [5,42–55]. The spectrum of SMA is broad ranging from embryonic lethality to a nearly normal life expectancy [4,6]. The expression of SMN and SMA modifying factors determine the severity of the disease and the timing of its manifestation [56–65]. Based on a recent study that analyzed insurance claims, patients frequently reported initial pathology associated with peripheral tissues, including issues with male fertility, prior to diagnosis with mild SMA [66]. This is a major departure from the severe SMA that is believed to be caused primarily by the degeneration of motor neurons [5]. A study conducted in a mild SMA mouse model suggests that testis need high levels of SMN [52]. This requirement is met in part by a testis-specific splicing switch from skipping to inclusion of SMN2 exon 7 in adult animals [52]. Hence, it is not surprising that patients in the insurance claim study sought medical treatment for fertility-related issues before being diagnosed with mild SMA [66]. Another frequently reported concern for patients with mild SMA is cardiac rhythm disorder [66,67]. Based on the emerging evidence, it appears that in cases of mild form of the disease, the developmental defects of heart, male reproductive organ and other peripheral tissues may precede neurodegeneration. Timing of the clinical disease manifestations as a consequence of a defined SMN concentration in specific tissues remains a matter of intense investigations.
The loci of the SMN genes generate a diversity of transcripts, including multiple alternatively spliced RNAs [68–71], a large repertoire of circular RNAs (circRNAs) [72,73], and two antisense long noncoding RNAs (lncRNAs) [74,75]. Except for the antisense RNAs, most transcripts, including circRNAs, are produced from the same pre-mRNA. In other words, generation of one SMN mRNA/circRNA most likely comes at the expense of another mRNA/circRNA. Currently, it is not known if a disbalance in a diverse SMN transcript “pool” itself contributes towards the tissue-specific SMA pathology. In this review, we survey the overall nature of transcripts generated from the SMN loci and discuss their potential role in maintaining the transcriptome and proteome homeostasis.
2. SMN gene organization
Located within an Alu repeat-rich region of human chromosome 5, the SMN genes are annotated to have 9 exons, which are exons 1, 2A, 2B, 3, 4, 5, 6, 7 and 8 (Fig. 1, Supplementary Fig. S1) [76]. The translation initiation (AUG) and termination (UAA) codons for the full-length mRNA are located within exons 1 and 7, respectively (Fig. 1) [39]. The noncoding exon 8 serves as the 3′ untranslated region (3′UTR). Transcripts lacking exon 7 code for SMNΔ7, which is less stable and only partially functional [77–79]. The size of exon 1 varies depending on the transcription start site (described below). There is a 3′ splice site (3′ss) within exon 1 that is used for backsplicing during circRNA production (Supplementary Fig. S1) [72]. Intron 1 is the largest intron; it contains three recently identified human-specific exons [I1(NE1–98), I1(NE2–79) and I1(NE3–33)] found to be incorporated into circRNAs (Fig. 1, Supplementary Fig. S1) [72]. The second largest intron is intron 6; it contains an Alu-derived exon 6B that is included into both linear and circRNAs (Fig. 1, Supplementary Fig. S1) [71,72]. Four additional novel exons (exons 9, 10, 11 and 12) derived from the intergenic sequences are mostly incorporated into circRNAs (Fig. 1, Supplementary Fig. S1) [72,73]. The discovery of four novel intergenic exons downstream of exon 8 makes the human SMN genes much larger than originally established.
Figure 1. Organization of the SMN genes.
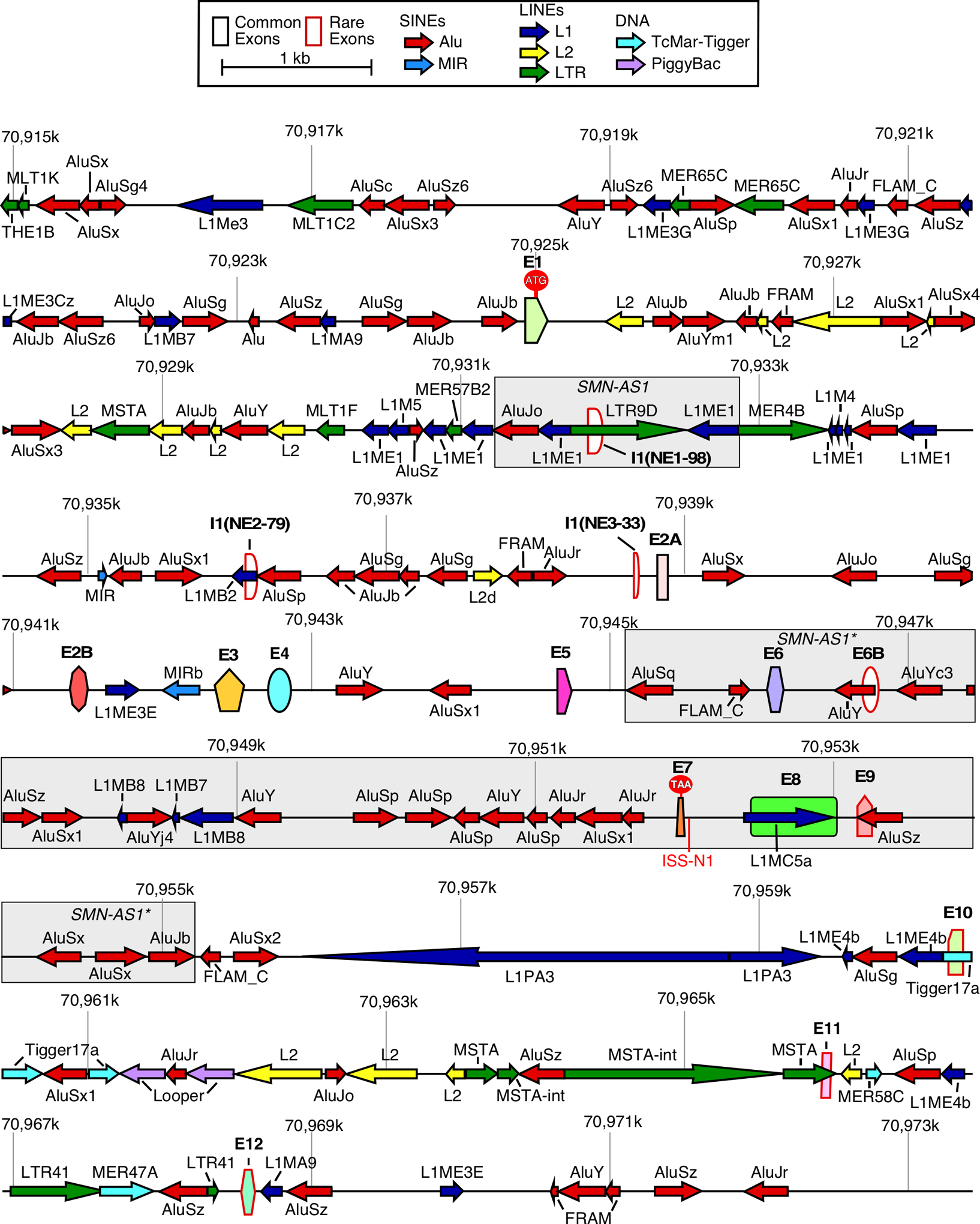
A scale depiction of the SMN1 gene (SMN2 has an identical overall structure) and flanking sequences are shown. Exons are depicted by colored shapes. Common exons (≥10% of total SMN RNA) are outlined in black, rare exons (<10% of total SMN RNA) are outlined in red. The start codon (ATG) located within exon 1 and the stop codon (TAA) located within exon 7 are indicated. Large grey boxes mark approximate locations of the antisense lncRNAs. Repeat sequences as identified by Repeatmasker are depicted by colored arrows. Arrow direction indicates the orientation of the repeat sequence. ISS-N1, a critical splicing regulatory sequence, is shown in red. Numbers indicate position within chromosome 5 of the GRCh38 human genome build.
3. Antisense transcripts of SMN
The SMN loci generate two antisense lncRNAs, SMN-AS1 and SMN-AS1* (Fig. 1) [74,75]. While transcription of SMN-AS1 begins and ends within a central region of intron 1 [74], transcription of SMN-AS1* begins in the intergenic region downstream of the annotated SMN gene and ends in intron 5 (Fig. 1) [75]. SMN-AS1* is ~10 kb long and shows size heterogeneity [75]. It is not known if the shorter variants of SMN-AS1* are generated by removal of introns from this large transcript or by use of alternative transcription initiation and/or termination sites. Since both SMN-AS1 and SMN-AS1* overlap Alu elements, they are likely to be associated with primate-specific functions of the SMN genes. Both antisense lncRNAs are proposed to regulate SMN expression in a transcription-dependent manner by recruiting the Polycomb Repressive Complex 2 (PRC2) to the SMN promoter region or SMN gene body [74,75]. PRC2 recruitment promotes di- and tri-methylation of Lysine 27 of histone H3 (H3K27) leading to repression of SMN transcription, and, as a consequence, a decrease in SMN protein production [74,75]. Therefore, SMN-AS1 and SMN-AS1* may serve as targets for SMA therapy. Indeed, the proof of principle that targeting of SMN-AS1 or SMN-AS1* may ameliorate SMA pathology has already been presented [74,75]. It will be of great consequence to know whether the antisense transcripts of SMN display a spatiotemporal pattern of expression (which appears to be the case for SMN-AS1) and how the expression of these antisense lncRNAs is regulated. It will be equally important to uncover the tissue-specific targets of these lncRNAs. Since the antisense lncRNAs are expressed from the SMN loci, they may anneal to the complementary regions within SMN pre-mRNAs. Consequently, these lncRNAs may have a direct role in SMN transcript processing steps, including pre-mRNA splicing and 3′-end formation.
4. Transcript diversity due to variability of the 5′UTR
The transcription start site (TSS) defines the length and sequence composition of the 5′UTR. While the portion of exon 1 coding for the first 27 amino acids is generally present in all linear SMN transcripts [39], early reports confirm the diversity of the 5′UTR due to usage of alternative TSSs upstream of the translation initiation codon (Fig. 2A) [80–82]. As per 5′ RACE (rapid amplification of cDNA ends) conducted using poly(A) RNA from adult human spinal cord, a TSS (TSS296) was determined to be located 296 bp upstream of the first ATG (Figs. 2A, 2B) [80]. However, the results of primer extension using total RNA isolated from HeLa cells showed TSS at 161 bp upstream of the first ATG (TSS161, Figs. 2A, 2B) [81]. Subsequent studies also captured tissue-specific and/or developmental stage-specific differences. For example, a primer extension approach using total RNA from human fetal tissue, adult cerebellum and human cell lines identified two more TSSs (TSS163 and TSS242, Figs. 2A, 2B), one at 163 bp and the other at 242 bp upstream of the first ATG codon, with the latter TSS being predominantly used during fetal development (Figs. 2A, 2B) [82].
Figure 2. Transcription initiation of the SMN genes.
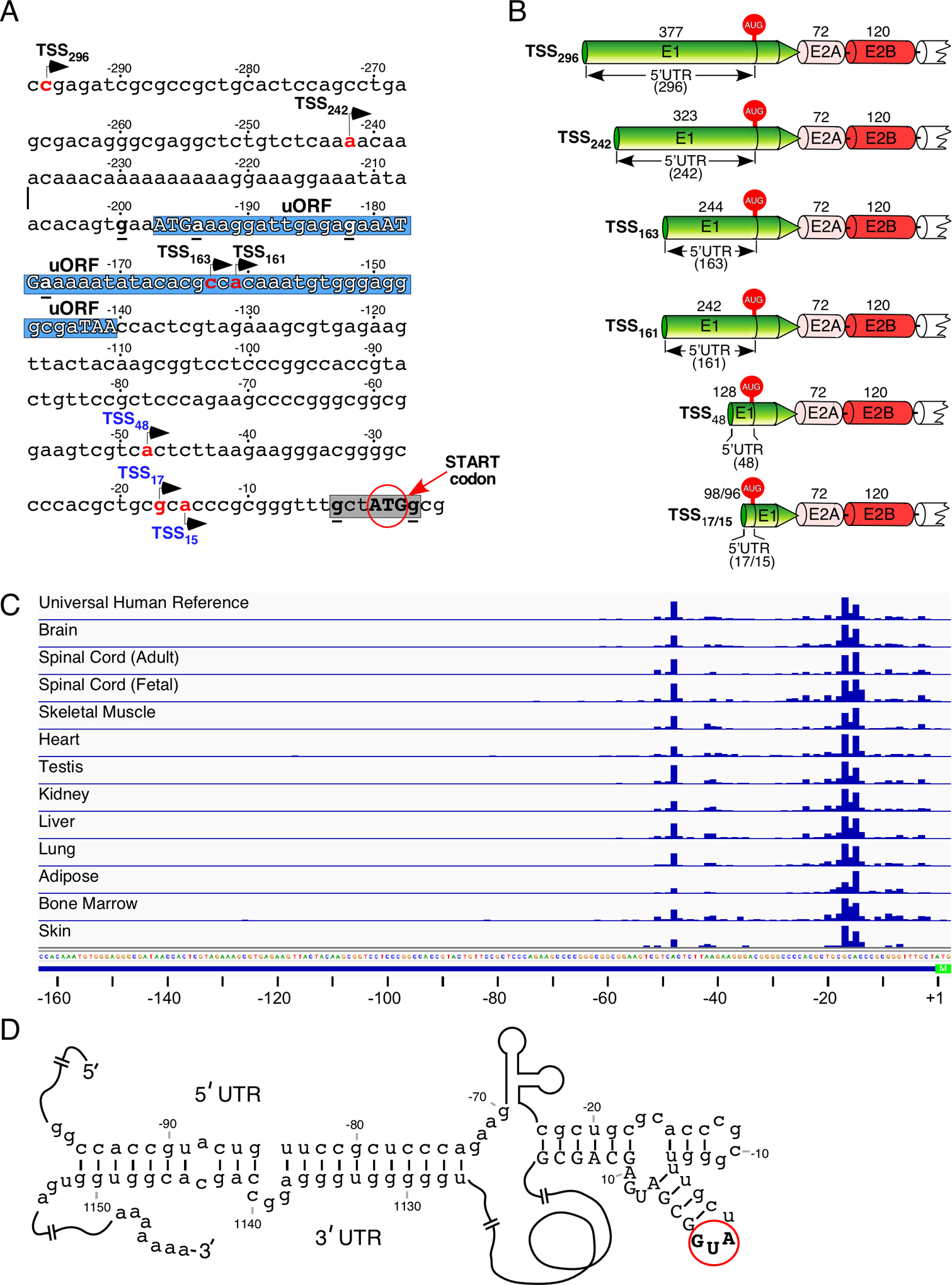
(A) The 297-nucleotide-long DNA sequence immediately upstream of the first ATG (start codon) for the main protein coding sequence. Three nucleotides immediately downstream of the start codon are included as well. The start codon itself is shown in bold capital letters and highlighted with a red circle and an arrow. Negative nucleotide numbering begins from the start codon. The seven most commonly used transcription start sites are indicated by arrows, with their names written in black or blue. Black color indicates sites identified using gene-specific approaches, blue indicates sites identified using CAGE (see panel C). The number in the name of each transcription start site corresponds to a position of the first transcribed nucleotide (highlighted in red) relative to the start codon. The core Kozak context of the first ATG codon is highlighted in grey and the consensus nucleotides are underlined and in bold. Hypothetical upstream open reading frames (uORFs) are shown on the blue background. Their start codons and in-frame stop codon are marked in capital letters; the consensus nucleotides of the Kozak sequence are in bold and underlined. Abbreviation: TSS, transcription start site; uORF, upstream open reading frame. (B) Diagrammatic representation of six SMN transcript isoforms with different 5′UTRs generated as the result of alternative transcription start site usage. Exons 1, 2A and 2B are shown in different colors, their sizes (in nucleotides) are indicated on top. Alternative 5′UTRs are marked and their sizes are given in the parentheses. The start codon (AUG) located within exon 1 is indicated. Abbreviations: UTR, untranslated region; E, exon. (C) TSS usage determined by CAGE sequencing. The relative TSS usage of the SMN genes is represented by blue bars. The bar height corresponds to read counts. Sample type is indicated at the left. Genomic sequence is given at the bottom. The annotated 5′ untranslated region (for TSS163) is shown as a blue line, the start codon is highlighted in green. Nucleotide positions relative to the start codon are indicated at the bottom. (D) Predicted secondary structure formed by base pairing between the 5′UTR and the 3′UTR of SMN mRNA. Numbering (negative and positive) begins from the start codon (AUG) shown in bold capital letters and highlighted with a red circle. Negative numbers represent sequences upstream of the AUG. Positive numbers represent sequences downstream of the AUG, with A in this codon counted as “1”. Positive numbering is given in the context of included exon 7.
Analyses of the publicly available FANTOM 5 (Functional Annotation of Mammalian cDNA) data using the Cap Analysis of Gene Expression (CAGE) method revealed a much shorter 5′UTR of SMN in both neuronal and non-neuronal tissues (Fig. 2C) [83,84]. In particular, two primary transcription start sites, TSS48 and TSS17/TSS15, mapped to 48 and 17/15 bp upstream of the first ATG codon, respectively (Fig. 2C). It should be noted that the region immediately upstream of TSS48 is extremely CG-rich with a potential for stable RNA structure formation, which might affect the results of CAGE (Fig. 2A). Notwithstanding, it appears that TSS48 and TSS17/TSS15 are used with nearly equal frequency, as indicated by relatively even number of reads corresponding to the above-mentioned sites, with the noticeable exception of adipose tissue, kidney and heart (Fig. 2C). In these three tissues, the proximal initiation of transcription is preferred. A much lower number of reads located at other sites may generate low levels of transcripts with the heterogeneous 5′UTR (Fig. 2C). However, it is likely that these low-abundance transcripts represent background noise with no real functional significance. Considering the median 5′UTR length of human mRNAs is ~218 nucleotides, the 5′UTR of SMN based on unbiased FANTOM 5 data appears to be very short [83].
Translation of mRNAs with 5′UTRs shorter than 20 nucleotides happens to be suboptimal, although RNA secondary structures downstream of the ATG increase its recognition [85,86]. In case of SMN, the G-rich sequences upstream and downstream of the first ATG have potential to form G-quadruplex structures. The role of G-quadruplex structures has been implicated in the regulation of translation initiation [87,88]. Moreover, RNA helicases, such as DHX36 and DHX9, were shown to modulate mRNA translation through unfolding G-quadruplex structures formed within the upstream open reading frame (uORF) of 5′UTRs [87]. Incidentally, there is a hypothetical uORF in the long 5′UTRs of SMN (Fig. 2A). It will be of great interest to find out if this uORF is functional and can modulate SMN expression. It will be also important to know if RNA helicases regulate SMN translation through their effect on putative G-quadruplex and/or other structures located within the 5′UTR of SMN.
In an alternative scenario, GC-rich sequences within the 5′UTR of SMN could base pair with complementary GC-rich sequences within the 3′UTR of SMN, forming duplex structures (Fig. 2D). Such RNA structures have been suggested to play a role in regulation of translation through enhanced cooperativity (negative or positive) between proteins interacting with the 5′UTR and the 3′UTR [89]. While RNA-protein interactions within the 5′UTR of SMN have not been examined yet, multiple proteins have been shown to bind to the 3′UTR of SMN [90]. Future experiments will determine if the RNA structure between the two UTRs of the SMN transcript is indeed formed and whether it plays a role in regulation of SMN translation.
5. Transcript diversity due to alternative splicing
Alternative splicing is one of the greatest sources of SMN transcript diversity. While the eight coding exons (exons 1, 2A, 2B, 3, 4, 5, 6 and 7) of human SMN show strong homology with the analogous exons of mouse Smn [3], there are substantial differences between human and mouse exon 8 as well as intronic sequences. These differences could be significant for regulation of both forward splicing and backsplicing that generate mRNAs and circRNAs, respectively. With the exception of testis, SMN2 exon 7 is predominantly skipped in all studied organs and tissues [52]. Also, low levels of skipping of SMN exons 3 and 5 are observed in most tissues [52]. Multi-exon-skipping detection assay (MESDA), first reported in 2012, has been used to capture the relative abundance of the alternatively spliced SMN transcripts containing exons 1 through 8 [52,69–72,91]. MESDA has the unique ability to identify SMN splice isoforms lacking multiple exons in different combinations. For instance, co-skipping of exons 5 and exon 7 together with other exons has been detected (Fig. 3B) [69,70,91]. MESDA has been particularly useful in revealing novel SMN splice isoforms under the conditions of oxidative stress [69,70]. Additional novel variants of the alternatively spliced transcripts of SMN have been captured upon depletion of DHX9 or U1 snRNA [72,91]. Overall, it appears that under specific conditions, any of the internal exons of SMN (from exon 2A to exon 7) could be skipped. Since their length (in nucleotides) is divisible by three, skipping of the internal exons in any combination could potentially generate short SMN isoforms. Notably, skipping of exon 7 alone or with any other internal exon(s) creates a degradation signal at the C-terminus of the SMN protein [71,79]. This could be the reason why shorter SMN isoforms encoded by splice variants with co-skipped exon 7 cannot be detected. At the same time, shorter SMN isoforms encoded by transcripts that do harbor exon 7 but lack other exons are also not detectable. This could be due to several reasons, including low relative expression, low protein stability or poor antibody reactivity.
Figure 3. SMN pre-mRNA alternative splicing.
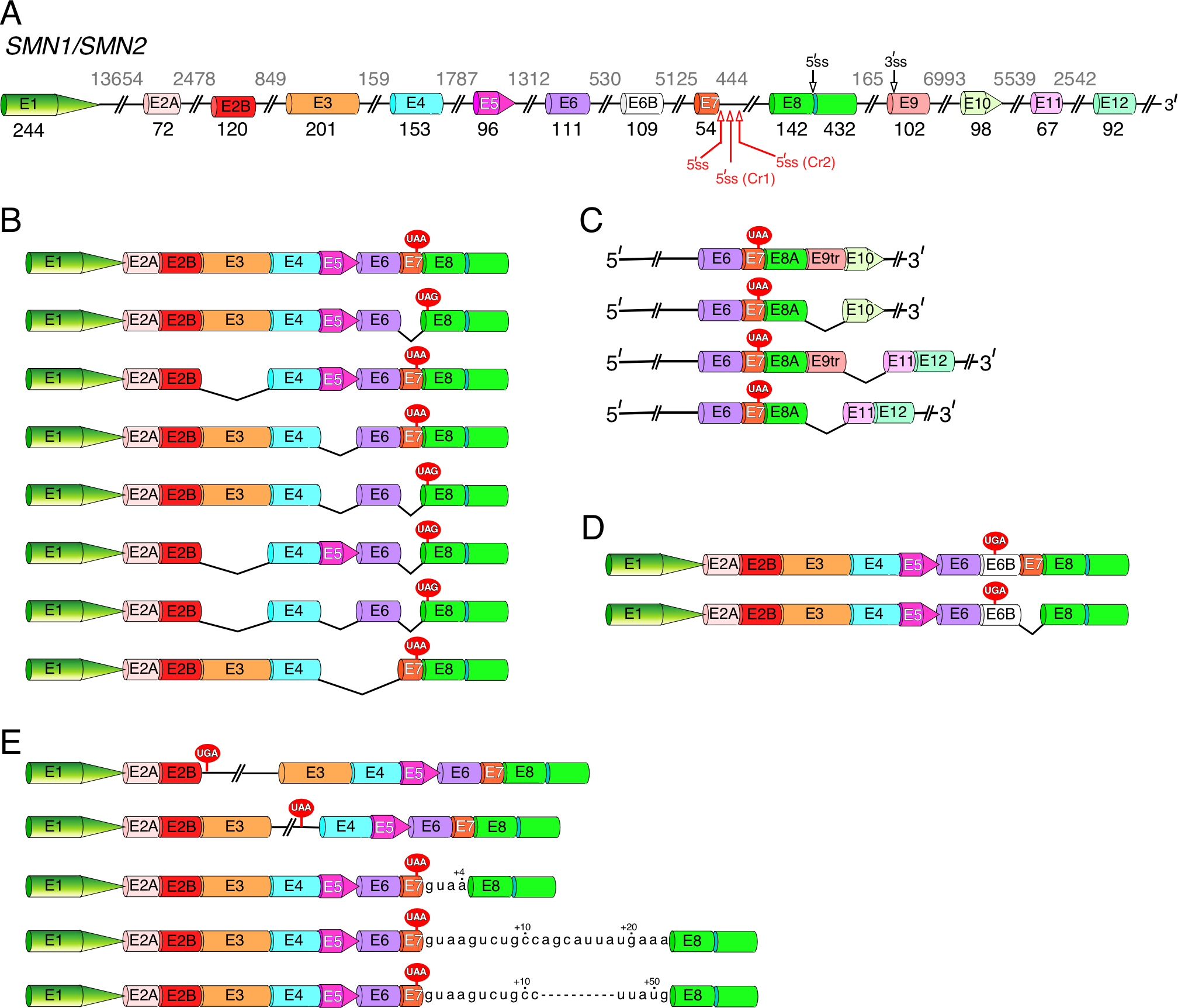
(A) Diagrammatic representation of SMN1/SMN2 pre-mRNA. Exons are shown as different colored shapes, introns, as broken lines. Exon sizes are indicated by numbers in black, intron sizes, by numbers in grey. Alternative and cryptic splice sites we identified are marked by black and red arrows, respectively. When applicable, the corresponding names of the cryptic splice site are given in parentheses. Abbreviation: E, exon; ss, splice sites. (B) Diagrammatic representation of the most common linear SMN splice isoforms generated by exon skipping. Skipped exons are indicated by black “V” shapes. The UAA and UAG stop codons located in exon 7 and exon 8, respectively, are shown. (C) Diagrammatic representation of linear SMN splice isoforms generated by inclusion of newly identified “intergenic” exons 9tr, 10, 11 and 12. Only the 3′-end exons are shown. Exon 9tr is a truncated version of exon 9 produced when the alternative 3′ss located in exon 9 (see (A)) is used. Exon 8A is a truncated version of exon 8 generated when the alternative 5′ss located in exon 8 (see (A)) is used. (D) Diagrammatic representation of linear SMN splice isoforms generated by inclusion of alternative exon 6B, together with or without exon 7. Inclusion of exon 6B will change the C-terminus of the SMN protein. (E) Diagrammatic representation of linear SMN splice isoforms with retained intronic sequences. Intronic sequences included due to activation of the cryptic 5′ss located in intron 7 are given. Usage of these cryptic sites increases the length of exon 7 by 4, 23 or 51 nucleotides.
Inclusion of novel exons derived from intron 1, intron 6 (exon 6B) as well as intergenic sequences (exon 9 through exon 12) further expand the diversity of SMN transcripts (Figs. 3C, 3D) [72]. The protein encoded by exon 6B-containing transcripts, termed SMN6B, possesses an altered C-terminus that enhances the stability of the protein compared to that of SMNΔ7 (Fig. 3, Supplementary Fig. S1) [71]. Furthermore, based on the shared domain structure, it is likely that SMN and SMN6B have overlapping functions. The possibility that SMN6B plays additional or special functions remains to be explored. In general, inclusion of Alu-derived exons, such as exon 6B, is suppressed by splicing factor hnRNP C [71]. Low abundance of exon 6B-containing linear transcripts could be also attributed to their degradation by nonsense-mediated decay (NMD) [71]. Inclusion of newly identified exons I1(NE1–98) and I1(NE3–33) derived from intron 1 introduces premature termination codons. Insertion of another intron 1-derived novel exon I1(NE2–79) causes a frameshift that creates a premature termination codon within exon 2A (Fig. 1, Supplementary Fig. S1) [72]. Hence, linear transcripts harboring these intron 1-derived exons would be likely degraded by NMD as well. The fact that linear transcripts containing these novel exons are generally not detected supports this hypothesis.
Intron retention is another contributor to the diversity of SMN transcripts (Fig. 3E) [68,92]. For instance, intron 3 retention gives rise to a short protein, a-SMN, shown to play an important role in the development of the mammalian brain [68]. However, it is not known if other intron-retained SMN isoforms also code for proteins. Activation of the cryptic 5′ss (Cr1) located within intron 7 increases the size of exon 7 by 23 nucleotides and further expands the diversity of both linear and circular transcripts of SMN (Fig. 3E) [72,91]. Usage of yet another cryptic 5′ss located within exon 8 creates an exon we call exon 8A (Fig. 3A) [72,73]. This exon is often detected in circRNAs (Fig. 4) [72]. Considering the translation termination codon is located within exon 7, activation of Cr1 and/or inclusion of additional downstream exons does not alter the amino acid sequence of the SMN protein. However, activation of Cr1 and/or inclusion of any of the intergenic exons leads to changes in the 3′UTR. The significance of such changes has yet to be investigated.
Figure 4. Four types of SMN exonic circRNAs.
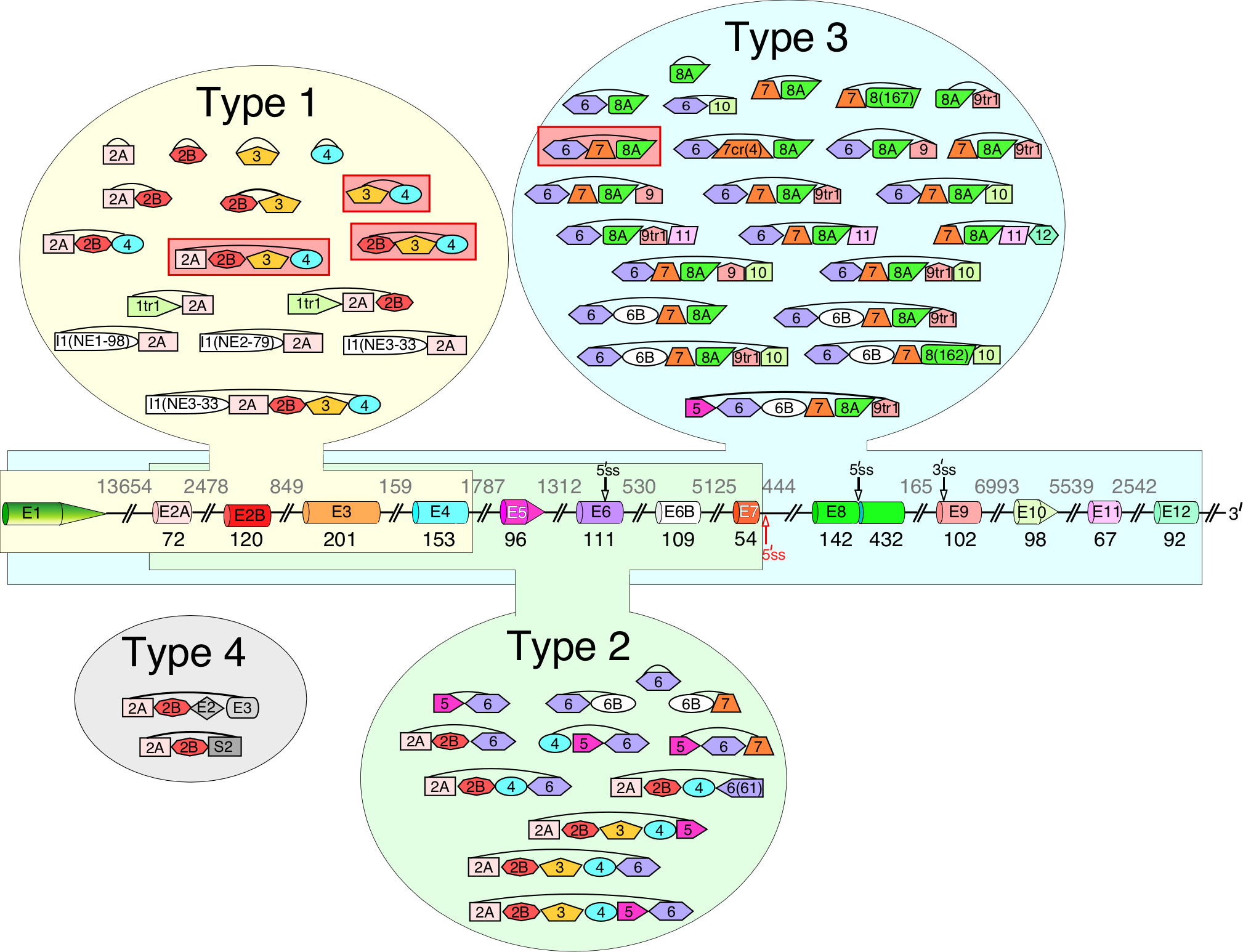
Diagrammatic representation of SMN pre-mRNA and circRNAs are given. The pre-mRNA diagram is labeled similarly as in Fig. 3A. Type 1 circRNAs and exons used to generate them are highlighted in yellow, type 2, in green and type 3, in blue. Type 4 circRNAs are highlighted in grey. Exons I1(NE1–98), I1(NE2–79) and I1(NE3–33) shown as white shapes are three novel exons derived from intron 1 sequences. The most abundant circRNAs are highlighted in red.
6. Circular SMN transcripts
The SMN genes generate a vast repertoire of circRNAs: ≥50 species of SMN circRNAs have been identified so far (Fig. 4) [72,73,76]. Generation of SMN circRNAs is generally attributed to backsplicing facilitated by RNA structures formed between inverted Alu repeats present within the transcribed region [72,93–95]. Depending on their exon content, SMN circRNAs are broadly categorized into four types (Fig. 4) [72]. Type 1 circRNAs contain early exons, namely exon 1 through exon 4, and represent the most highly-expressed circRNAs. Moderately expressed type 2 circRNAs contain middle exons (exon 5, 6 and 7) with or without early exons. Type 3 circRNAs contain at least one exon downstream of exon 7, including exon 8A and intergenic exons (Fig. 4) [72]. Type 3 represents the most diverse group of SMN circRNAs harboring different combinations of eight exons, namely exon 6, 6B, 7, 8A and four intergenic exons [72]. Interestingly, exon 6 is used most frequently for backsplicing, both in terms of the level of production (circRNA C6-7-8A is the most abundant type 3 circRNA) and the circRNA diversity. This observation suggests that Alu elements located in an upstream intron 5 might play a role in SMN backsplicing events. In fact, deletion experiments performed in a minigene engineered to produce circRNAs C6-7-8A and C6-7-8A-9 confirmed that removal of Alu elements from intron 5 resulted in a drastic decrease in backsplicing [73]. The next most used exon for backsplicing is exon 4: its 5′ss is used to generate three of the most abundant circRNAs, C2A-2B-3–4, C2B-3–4 and C3–4. Extensive usage of the 5′ss of exon 4 implies that one or more Alu elements in the vicinity of the 5′ss of intron 4 facilitate circRNA generation.
Type 4 circRNAs are formed by trans-splicing and in addition to SMN exons contain at least one exon from another Alu-rich gene [72]. The evidence that SMN transcripts are trans-spliced with transcripts of other genes was provided by two circRNAs, C2A-2B-SERF1A/B(E2) and C2A-2B-ERBIN(E2–) [72]. In both of these circRNAs, the 3′ and the 5′ss of SMN exons 2A and 2B were spliced with the 5′ and the 3′ss of exons of the transcripts of the neighboring genes, respectively. Most likely, the trans-splicing events are facilitated by pairing of the splice sites located on two different RNA molecules due to RNA structures formed by inverted Alu repeats. We speculate that trans-spliced linear molecules of SMN could also be formed by similar mechanisms. Future experiments will determine how frequently SMN transcripts undergo through trans-splicing events. One of the likely functions of trans-splicing is the cross-regulation of two genes through a single hybrid transcript, linear or circular. Similar to lncRNAs, a hybrid transcript could modulate transcription, pre-mRNA splicing, 3′-end processing and intra-cellular RNA trafficking. Mode of such cross-regulation could be very specific, rapid and independent of the protein encoded by the hybrid transcript.
Many circRNAs are conserved during evolution, suggesting their functional importance in cellular metabolism [96–98]. However, only a limited number of human SMN circRNAs, including C2A-2B-3–4, are expressed by mouse Smn [72]. Also, the repertoire of mouse Smn circRNA appears to be less diverse, likely due to the lower content of inverted repeat sequences. Further, type 4 circRNAs are not detected for mouse Smn [72]. At the same time, the longest circRNA comprised of eight exons was captured for mouse Smn but not for human SMN [72]. Generation of this long circRNA of mouse Smn is facilitated by pairing of a cryptic 5′ss located in exon 8 with the upstream 3′ss of exon 2A [72]. Lack of a similar circRNA in humans may suggest a competing mechanism that favors the usage of the 3′ss of exon 2A mostly for the generation of type 1 circRNAs. Based on the nature of SMN circRNAs and lncRNAs, an inference could be drawn that human SMN genes have recently acquired an additional layer of regulatory role in shaping the overall transcriptome diversity with significance to the tissue-specific functions.
We speculate that the production of different SMN circRNAs could be governed by different mechanisms and by combinatorial control by multiple factors. Some of the highly abundant SMN circRNAs were found to be negatively regulated by DHX9, an RNA helicase that unwinds RNA structures formed by inverted Alu repeats [72,73]. There also appears to be a correlation between exon skipping and the inclusion of the skipped exons into circRNAs. For instance, enhanced co-skipping of exons 3 and 4 upon depletion of DHX9 led to the upregulation of C3–4 [72]. Depletion of DHX9 is also linked to delayed intron removal in Alu-rich SMN introns, which could play a role in increased circularization [73]. A subset of SMN circRNAs was found to be positively regulated by the RNA-binding protein Sam68 that interacts with sequences close to the intronic Alu repeats [73]. Interestingly, Sam68 binding and Alu:Alu base pairing appear to be highly interrelated and cooperative, as DHX9 depletion increases Sam68 binding to SMN introns and deletion of intronic Alu elements and Sam68 binding sites cooperatively reduce production of C6-7-8A-9 from an SMN minigene construct [73]. Given the diversity of sequence and structural motifs that surround the splice sites involved in backsplicing, additional factors are likely to be implicated in the generation of SMN circRNAs.
CircRNAs could potentially produce polypeptides through an internal ribosome entry site (IRES)-mediated mechanism [99]. However, due to diverse structural features associated with IRESs, it is difficult to predict their presence. Assuming the presence of an IRES, an argument could be made in favor of the potential translation products from some of the SMN circRNAs. The highly expressed type 1 SMN circRNAs consist of coding exons that are divisible by three and contain an in-frame ATG in exon 4. Hence, the rolling circle translation of these circRNAs could produce a polypeptide containing numerous copies of the critical Tudor domain, and, in case of C2A-2B-3–4, an RNA-binding domain [100–103]. The most abundant type 2 circRNA is C5–6, which can produce rolling-circle translation products through two in-frame ATGs in the 3′ portion of exon 6. Similarly, type 3 circRNAs could be translated using ATGs in exon 6. However, due to presence of the stop codon in exon 7, these ATGs would produce short peptides containing the YG box that participates in SMN multimerization [104]. In addition, nearly all SMN circRNAs contain at least one out-of-frame ATG that could produce polypeptides of various lengths.
Interactions of proteins with circRNAs are suggested to play an important role in regulation of protein expression/function as well as pathophysiological processes [105]. A web-based tool using CLIP (UV Cross-Linking and Immunoprecipitation) tags has been employed to predict interaction of proteins with circRNAs [106]. Based on the CLIP tags, there are some potential interactions between SMN circRNAs and multifunctional RNA-binding proteins, including FUS, translation initiation factor 4A3 (EIF4A3) and LIN28. Considering CLIP tags do not distinguish interactions between linear or circular transcripts, any predicted protein-circRNA interaction based on CLIP tags would require further validation. Presence of FUS CLIP tags in exon 2B suggests interaction of FUS with C2A-2B-3–4, C2B-3–4 and several less abundant circRNAs. Of note, FUS has been implicated in circRNA generation through interaction with intronic seqeunces[107]. It remains to be seen if binding of FUS with exon 2B somehow facilitates the generation of exon 2B-containing circRNAs. Interestingly, FUS has been shown to interact with SMN [108], suggesting a possible cross-regulation between FUS, SMN, and SMN circRNAs. Presence of CLIP tag of EIF4A3 in exon 5 suggests its interaction with most type 2 circRNAs. Of note, the multifunctional EIF4A3 has been implicated in exon junction complex (EJC) formation [109], circRNA generation [110], nonsense-mediated mRNA decay (NMD) [111] and 18S rRNA biogenesis [112]. EIF4A3 is also associated with selenocysteine incorporation [113], an essential process in which SMN plays an important role [3]. It is likely that type 2 SMN circRNAs maintain a nucleocytoplasmic homeostasis of EIF4A3 by trapping a fraction of EIF4A3 in the cytoplasm. Based on the CLIP tag within exon 6, LIN28B is predicted to interact with types 2 and 3 circRNAs. The most characterized function of LIN28 proteins is the control of cell fate [114]. It has been shown that LIN28 proteins weakly stabilize thousands of mRNA targets, including its own mRNA [114]. Therefore, it is easy to envision that potential “sponging” of LIN28B by SMN circRNAs might have an effect on the stability of many mRNAs and the abundance of proteins they encode. Such perturbations in the transcriptome and the proteome in turn could affect cell cycle, development, differentiation and stem cell reprogramming.
7. Transcript diversity and microRNA sponging
MicroRNAs (miRNAs) are ~22-nt long RNAs associated with the translation inhibition and/or mRNA degradation [115]. The functions of miRNAs are mediated through an RNA-induced silencing complex (RISC) in which the Argonaute (AGO) proteins play an essential role [115,116]. Interactions of miRNAs with mRNAs are best captured by CLIP using antibodies against AGO followed by high throughput RNA sequencing termed CLIP-seq [116]. Analysis of the publicly available CLIP-seq data generated using HeLa cells reveals AGO interactions throughout the length of SMN transcripts (Fig. 5A) [116]. Based on the miRanda [117] and/or MirTarget [118] prediction algorithms, more than two dozen miRNAs might interact with SMN transcripts (Fig. 5B). Two prominent peaks, one close to the translation start site and the other located within the 3′UTR, alone harbor at least 10 potential miRNA target sites (Fig. 5A).
Figure 5. Predicted miRNA binding sites across SMN transcripts.
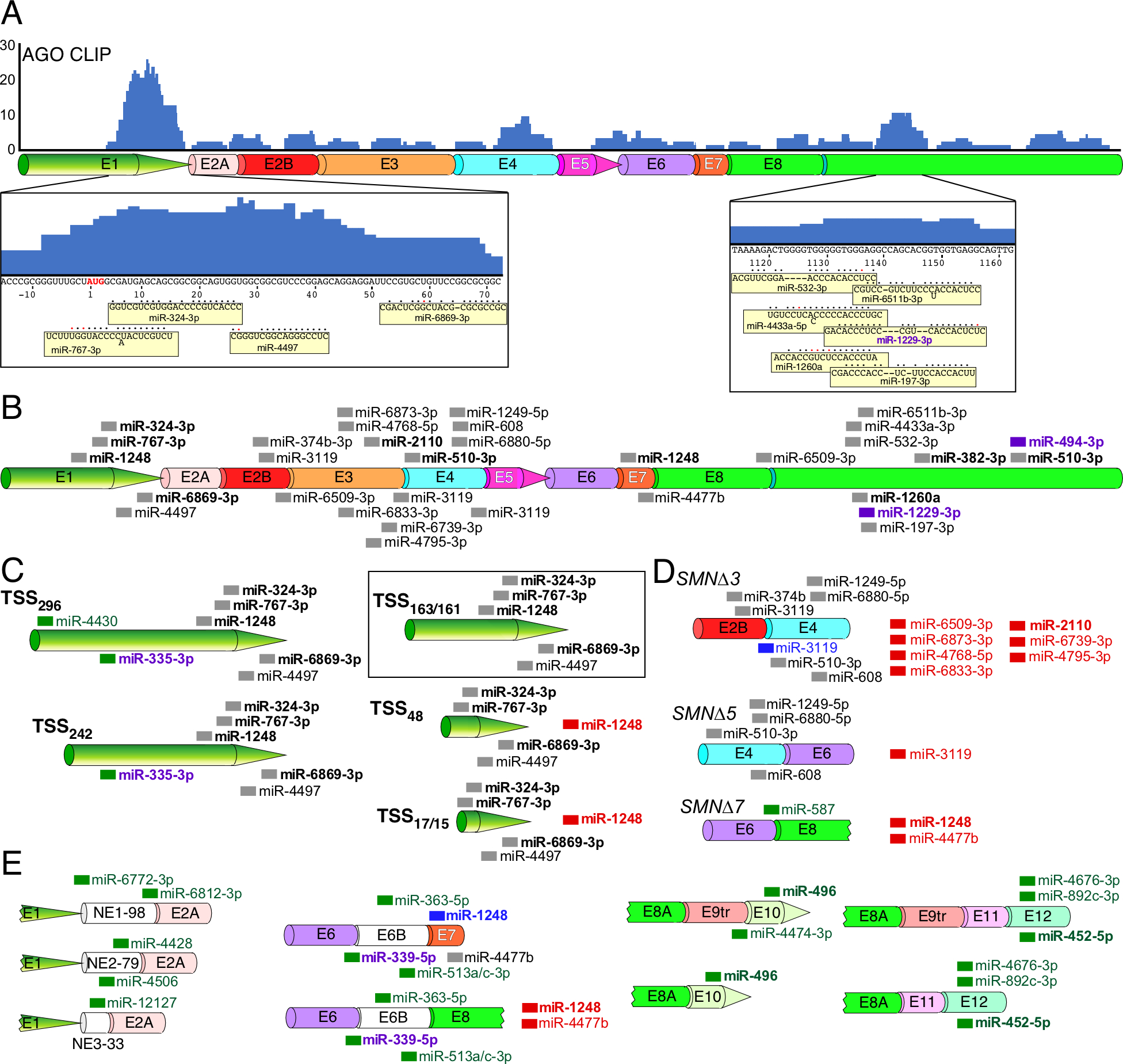
(A) AGO CLIP performed using HeLa cells reveals potential miRNA targets throughout the length of SMN mRNAs. Height of blue bars represents the number of overlapping reads at each position. Corresponding exons are shown diagrammatically below CLIP signal. Exon labeling and coloring are the same as in Fig. 3B. SMN sequences corresponding to two prominent peaks as well as miRNAs that target them are given at the bottom of panel A. The AUG start codon is shown in red letters. Nucleotide numbering is the same as in Fig. 2D. miRNAs are shown as yellow boxes. Their base pairing with the corresponding SMN targets is indicated with black circles; red circles mark wobble base pairing. Dashes indicate where base pairing between a miRNA and its SMN target is not continuous. “Out of line” letters in miRNA sequences indicate bulged nucleotides (miR-767–3p, miR-6511b-3p and miR-4433a-5p). (B) The canonical SMN mRNA transcript is shown. Exon labeling and coloring are the same as in Fig. 3B. miRNA target sites are marked with thick colored lines and miRNA names are given to the right of each site. miRNAs with names shown in bold correspond to miRNAs with known targets and/or functions. Purple color indicates that a miRNA is implicated in neurodegeneration. (C) Alternative TSS usage affects miRNA targeting profile. The name of each TSS is given at the left. The canonical transcript derived from Refseq annotation is boxed in black. miRNAs target sites unique to alternative (non-canonical) transcripts are labeled in green. miRNAs whose binding sites are lost in alternative transcripts are shown to the right side of the transcript and labeled in red. miRNAs with green lines and purple names indicate miRNAs unique to alternative transcripts that are also implicated in neurodegeneration. (D) Alternative splicing affects miRNA targeting profile. miRNAs shown in blue are predicted to have altered but preserved targets between two alternatively spliced junctions. Other labeling and coloring is the same as in (C) (E) Inclusion of rare exons results in novel miRNA target sites. Labeling and coloring is the same as in (C) and (D).
The differential usage of the TSS would alter the miRNA interaction profile with TSS242 and TSS296 adding 1 and 2 potential miRNA targets, respectively (Fig. 5C). The shortest isoforms, TSS48 and TSS17/15, result in a loss of the miR-1248 binding site (Fig. 5C). Alternative splicing also modifies the number of miRNA target sites present in a given SMN transcript. For instance, skipping of exon 3 would result in a loss of 7 potential miRNA targets as well as the shift of the miR-3119 target site outside of the seed sequence (Fig. 5D). Skipping of exon 5 would lead to the loss of a miR-3119 target site, while skipping of exon 7 would result into the loss of miR-1248 and miR-4477b target sites as well as the gain of the binding site for miR-587 (Fig. 5D). Inclusion of the rarely used SMN exons would potentially generate new miRNA target sites. For instance, inclusion of any of the 3 rare intron 1-derived exons would create at least one novel miRNA target site (Fig. 5E). Inclusion of exon 6B would generate target sites for miR-363–5p, miR-339–5p and miR-531a/c-3p (Fig. 5E). Exon 6B inclusion would also change the binding site for miR-1248 in the context of exon 7 being present (Fig. 5E). The intergenic rare exons also contain potential miRNA binding sites, in particular within exons 10 and 12 (Fig. 5E).
Although most sequences are shared between circRNAs and their linear mRNA counterparts, the backsplice junction is a feature unique to circRNAs. miRNA binding to the backsplice junction could render a distinct function of SMN circRNAs. SMN circRNAs with the highest expression level contain potential binding sites for miR-130b-5p, miR-6812–3p and miR-15b-3p (for exon 4-exon 2A backsplice junction), as well as miR-4774–5p and miR-221–5p (for exon 6-exon 5 backsplice junction). miR-92a-2–5p can potentially bind SMN with the seed sequence targeting the 5′-most region of exon 6; therefore, it could target all of the backsplice junctions for type 3 circRNAs with varying efficiency depending on the sequence of the downstream exon.
With direct significance to SMA, some of the miRNAs predicted to target SMN are associated with neuronal differentiation and neurodegeneration. Specifically, miR-324–3p, miR-2110 and miR-496 that interact with exons 1, 3, and 10, respectively, are known to regulate neuronal differentiation (Figs. 5B, 5E) [119–121]. Of note, exon 3 is also present in several abundantly expressed SMN circRNAs [72]. Hence, it is likely that in case of miR-2110 neuronal differentiation could be modulated through circRNA-miRNA-mRNA nexus. MiR-335–3p that targets the longer 5′UTR of SMN is produced from the same pre-miRNA as miR-335–5p, which has been found to be reduced in SMA iPSC-derived motor neurons (Fig. 5C) [122]. It will be interesting to see if low levels of miR-335 in SMA motor neurons correlate with the levels of SMN transcripts. MiR-494–3p, with a predicted binding site within the 3′UTR of SMN, is known to play a pro-neurodegenerative role in Parkinson’s disease (Fig. 5B) [123]. An additional two miRNAs, miR-339–5p and miR-1229–3p, predicted to target the exon 6/6B junction and the 3′UTR of SMN, respectively, are associated with Alzheimer’s disease (Fig. 5E) [124,125]. The multifunctional miR-1248 that modifies interferon and calcium signaling pathways by targeting RIG-1 and ITPR3 mRNAs, respectively [126], has two putative bindings sites within SMN mRNA, one located within the 5′UTR and the other within exon 7 (Fig. 5B). Alternative usage of the TSS and exon 7 skipping are likely to affect the interaction of miR-1248 with SMN mRNA. Notably, miR-510–3p with a potential binding site within the 3′UTR of SMN has been linked to non-obstructive azoospermia (Fig. 5B) [127]. It remains to be seen if there is any correlation between male infertility observed in mild SMA and the interactions between miR-510–3p and SMN transcripts, including circRNAs. Several miRNAs predicted to target SMN transcripts, including miR-510–3p, miR-494–3p, miR-496, miR-452–5p, miR-767–3p, miR-6869–3p, miR-335–3p, miR-1260a and miR-382–3p, are linked to cancer [128–134]. Future studies will determine if SMN transcripts, linear and circular, regulate the levels of the oncogenic proteins by sponging one or more of the above-mentioned miRNAs. It has been also suggested that SMN is directly or indirectly involved in the biogenesis of specific miRNAs [135]. We hypothesize that some of these miRNAs in turn could regulate SMN levels by targeting SMN transcripts. Finally, it will be interesting to see if noncoding SMN transcripts, including circRNAs, play a specific role in maintaining SMN levels through sequestration of a subset of miRNAs in a tissue-specific manner.
8. Conclusions
Knowledge of transcript diversity is essential to our understanding of overall gene function, which transcends the restricted point of view of gene function defined solely by the coding sequence. This could be particularly important for housekeeping genes involved in multiple and often diverse functions. SMN serves as a model human gene for which various complementary methods have been employed to identify/capture the diversity of transcripts it generates. Multiple types of RNAs, including sense, antisense, linear, circular and trans-spliced transcripts are generated from the SMN loci. While the rate of expression of various SMN transcripts cannot be accurately predicted (due to their different stability), the total sum of the low-abundance transcripts, including circRNAs, appear to be substantial. One of the potential functions of circRNA transcripts is the crosstalk with other genome outputs, particularly miRNAs and proteins. Sequestration of miRNAs and proteins has broad implications for cellular metabolism, considering that a single miRNA or a protein holds capability to regulate multiple genes. Incidentally, SMN itself is an RNA-binding protein with preference for structured RNA molecules [100,101,103]. Accordingly, the presence of highly-structured regions within the most abundant SMN circRNAs, particularly the ones containing exon 3, makes them potential targets for the interaction with the SMN protein. Beyond SMN stability, RNA-SMN interactions could be important for nucleation of large macro-molecular complex formation. Another likely function of the noncoding SMN transcripts could be regulation of transcription within the nucleus. In the cytosol, noncoding SMN transcripts could play an important role in macro-molecular trafficking. Although not yet examined, it is also possible that a fraction of noncoding SMN transcripts (circRNAs) could in fact code for small regulatory peptides with functions similar to those of cytokines. Future studies will determine if the diversity of transcripts generated by human SMN plays an important and even a tissue-specific role during development and maintenance.
Furthermore, the diversity of transcripts generated from the SMN loci opens up novel therapeutic and diagnostic avenues. As a potential new therapy, at least part of the noncoding SMN transcripts could be redirected to make full-length transcripts capable of producing functional SMN. Relative abundance of noncoding SMN transcripts, particularly highly stable circRNAs in biological fluids, including blood, saliva and urine may provide further insights into the status of the disease severity as well as the therapeutic efficacy. Now that we know of the existence of a large repertoire of transcripts generated from the SMN loci, the stage is set for uncovering their functions.
Supplementary Material
Acknowledgements
Authors acknowledge members of the Singh lab for critical reading of the manuscript and for valuable discussions and suggestions. While authors have attempted to include most contributions on SMN transcript diversity, they regret not being able to include several related references due to the lack of space.
Funding: This work was supported by grants from the National Institutes of Health (R01 NS055925).
Footnotes
Disclosures and competing interests: ISS-N1 target (US patent # 7,838,657) mentioned in this review was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including RNS, NNS and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target (US patent # 7,838,657) to IONIS Pharmaceuticals/Biogen, which is marketing Spinraza™ (Nusinersen), the FDA-approved drug, based on ISS-N1 target. RNS and NNS are founders of RNACorrect, Inc., an Iowa-based small business engaged in research and development.
References
- [1].Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Identification and characterization of a spinal muscular atrophy-determining gene, Cell 80 (1995) 155–165. [DOI] [PubMed] [Google Scholar]
- [2].Singh NN, Androphy EJ, Singh RN, The regulation and regulatory activities of alternative splicing of the SMN gene, Crit. Rev. Eukar. Gene 14 (2004) 271–285. [DOI] [PubMed] [Google Scholar]
- [3].Singh RN, Howell MD, Ottesen EW, Singh NN, Diverse role of survival motor neuron protein, Biochim. Biophys. Acta Gene Regul. Mech 1860 (2017) 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harding BN, Kariya S, Monani UR, Chung WK, Benton M, Yum SW, Tennekoon G, Finkel RS, Spectrum of neuropathophysiology in spinal muscular atrophy type I, J. Neuropathol. Exp. Neurol 74 (2015) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ahmad S, Bhatia K, Kannan A, Gangwani L, Molecular Mechanisms of Neurodegeneration in Spinal Muscular Atrophy, J. Exp. Neurosci 10 (2016) 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N, Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next, Annu. Rev. Genomics Hum. Genet 2020. January 31. doi: 10.1146/annurev-genom-102319-103602. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- [7].Lorson CL, Hahnen E, Androphy EJ, Wirth B, A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy, Proc. Natl. Acad. Sci. U.S.A 11 (1999) 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD, A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2, Hum. Mol. Genet 8 (1999) 1177–1183. [DOI] [PubMed] [Google Scholar]
- [9].Cartegni L, Krainer AR, Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1, Nat. Genet 30 (2002) 377–384. [DOI] [PubMed] [Google Scholar]
- [10].Kashima T, Manley JL, A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy, Nat. Genet 34 (2003) 460–463. [DOI] [PubMed] [Google Scholar]
- [11].Singh NN, Androphy EJ, Singh RN, An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy, Biochem. Biophys. Res. Commun 315 (2004) 381–388. [DOI] [PubMed] [Google Scholar]
- [12].Singh NN, Androphy EJ, Singh RN, In vivo selection reveals features of combinatorial control that defines a critical exon in the spinal muscular atrophy genes, RNA 10 (2004) 1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Singh RN, Evolving concepts on human SMN pre-mRNA splicing, RNA Biol 4 (2007) 7–10. [DOI] [PubMed] [Google Scholar]
- [14].Singh NN, Howell MD, Singh RN, Transcription and splicing regulation of spinal muscular atrophy genes, in: Sumner CJ, Paushkin S, Ko C-P (Eds.), Spinal Muscular Atrophy: Disease Mechanisms and Therapy, Academic Press, London, (2017) 75–97. [Google Scholar]
- [15].Singh RN, Singh NN, Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes, Adv. Neurobiol 20 (2018) 31–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh NK, Singh NN, Androphy EJ, Singh RN, Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron, Mol. Cell. Biol 26 (2006) 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Singh NN, Singh RN, Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model, RNA Biol 8 (2011) 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR, Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model, Nature 478 (2011) 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AH, A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse, Hum. Mol. Genet 21 (2012) 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou H, Janghra N, Mitrpant C, Dickinson RL, Anthony K, Price L, Eperon IC, Wilton SD, Morgan J, Muntoni F, A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice, Hum. Gene Ther 24 (2013) 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sivanesan S, Howell MD, DiDonato CJ, Singh RN, Antisense oligonucleotide mediated therapy of spinal muscular atrophy, Transl. Neurosci 4 (2013) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ottesen EW, ISS-N1 makes the First FDA-approved Drug for Spinal Muscular Atrophy, Transl. Neurosci 8 (2017) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh NN, Howell MD, Androphy EJ, Singh RN, How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy, Gene Ther 24 (2017) 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bennett CF, Krainer AR, Cleveland DW, Antisense Oligonucleotide Therapies for Neurodegenerative Diseases, Annu. Rev. Neurosci 42 (2019) 385–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashit M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM, Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study, Lancet 388 (2016) 3017–3026. [DOI] [PubMed] [Google Scholar]
- [26].Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, Topaloglu H, Tulinius M, Montes J, Glanzman AM, Bishop K, Zhong ZJ, Gheuens S, Bennett CF, Schneider E, Farwell W, De Vivo DC DC; ENDEAR Study Group, Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy, N. Engl. J. Med 377 (2017) 1723–1732. [DOI] [PubMed] [Google Scholar]
- [27].Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, Shieh PB, Tulinius M, Mazzone ES, Montes J, Bishop KM, Yang Q, Foster R, Gheuens S, Bennett CF, Farwell W, Schneider E, De Vivo DC, Finkel RS; CHERISH Study Group, Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy, N. Engl. J. Med 378 (2018) 625–635. [DOI] [PubMed] [Google Scholar]
- [28].Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN, A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy, RNA Biol 6 (2009) 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Singh NN, Hollinger K, Bhattacharya D, Singh RN RN, An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing, RNA 16 (2010) 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN, An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy, Nucleic Acids Res 41 (2013) 8144–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singh NN, Lee BM, Singh RN, Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions, Ann. N.Y. Acad. Sci 1341 (2015) 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Singh NN, Lee BM, DiDonato CJ, Singh RN, Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy, Future Med. Chem 7 (2015) 1793–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Singh NN, Singh RN, How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene, Biochim. Biophys. Acta Gene Regul. Mech. 1862 (2019) (11–12), pii: S1874–9399(18)30510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Singh RN, Singh NN, A novel role of U1 snRNP: Splice site selection from a distance, Biochim. Biophys. Acta Gene Regul. Mech 1862 (2019) 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kiel JM, Seo J, Howell MD, Hsu WH, Singh RN, DiDonato CJ, A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy, Mol. Ther. Nucleic Acids 3 (2014) e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Howell MD, Ottesen EW, Singh NN, Anderson RL, Singh RN, Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide, Mol. Ther 25 (2017) 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Al-Zaidy SA, Mendell JR, From Clinical Trials to Clinical Practice: Practical Considerations for Gene Replacement Therapy in SMA Type 1, Pediatr Neurol pii: S0887–8994 (2019) 31163–31169. [DOI] [PubMed] [Google Scholar]
- [38].Seo J, Howell MD, Singh NN, Singh RN, Spinal muscular atrophy: An update on therapeutic progress, Biochim. Biophys. Acta 1832 (2013) 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Howell MD, Singh NN, Singh RN, Advances in therapeutic development for spinal muscular atrophy, Future Med. Chem 6 (2014) 1081–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shorrock HK, Gillingwater TH, Groen EJN, Overview of Current Drugs and Molecules in Development for Spinal Muscular Atrophy Therapy, Drugs 78 (2018), 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Singh RN, More is needed to complement the available therapies of spinal muscular atrophy, Future Med. Chem (2019) doi: 10.4155/fmc-2019-0239. [DOI] [PubMed] [Google Scholar]
- [42].Michaud M, Arnoux T, Bielli S, Durand E, Rotrou Y, Jablonka S, Robert F, Giraudon-Paoli M, Riessland M, Mattei MG, Andriambeloson E, Wirth B, Sendtner M, Gallego J, Pruss RM, Bordet T, Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy, Neurobiol. Dis 38 (2010) 125–135. [DOI] [PubMed] [Google Scholar]
- [43].Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, Tronche F, Vadrot J, Moreau MH, Kemeny F, Melki J, Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload, Am. J. Pathol 165 (2004) 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shanmugarajan S, Tsuruga E, Swoboda KJ, Maria BL, Ries WL, Reddy SV, Bone loss in survival motor neuron (Smn(−/−) SMN2) genetic mouse model of spinal muscular atrophy, J. Pathol 219 (2009) 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, L Lorson C, Cardiac defects contribute to the pathology of spinal muscular atrophy models, Hum. Mol. Genet 19 (2010) 4059–4071. [DOI] [PubMed] [Google Scholar]
- [46].Heier CR, Satta R, Lutz C, DiDonato CJ, Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice, Hum. Mol. Genet 19 (2010) 3906–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bowerman M, Swoboda KJ, Michalski JP, Wang GS, Reeks C, Beauvais A, Murphy K, Woulfe J, Screaton RA, Scott FW, Kothary R, Glucose metabolism and pancreatic defects in spinal muscular atrophy, Ann. Neurol 72 (2012) 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schreml J, Riessland M, Paterno M, Garbes L, Roßbach K, Ackermann B, Krämer J, Somers E, Parson SH, Heller R, Berkessel A, Sterner-Kock A, Wirth B, Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585, Eur. J. Hum. Genet 21 (2013) 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gombash SE, Cowley CJ, Fitzgerald JA, Iyer CC, Fried D, McGovern VL, Williams KC, Burghes AHM, Christofi FL, Gulbransen BD, Foust KD, SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice, Hum. Mol. Genet 24 (2015) 3847–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Szunyogova E, Zhou H, Maxwell GK, Powis RA, Francesco M, Gillingwater TH, Parson SH SH, Survival Motor Neuron (SMN) protein is required for normal mouse liver development, Sci. Rep 6 (2016) 34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thomson AK, Somers E, Powis RA, Shorrock HK, Murphy K, Swoboda KJ, Gillingwater TH, Parson SH, Survival of motor neurone protein is required for normal postnatal development of the spleen, J. Anat 230 (2017) 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN, Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy, Sci. Rep 6 (2016) 20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nash LA, Burns JK, Chardon JW, Kothary R, Parks RJ, Spinal Muscular Atrophy: More than a Disease of Motor Neurons? Curr. Mol. Med 16 (2016) 779–792. [DOI] [PubMed] [Google Scholar]
- [54].Nery FC, Siranosian JJ, Rosales I, Deguise MO, Sharma A, Muhtaseb AW, Nwe P, Johnstone AJ, Zhang R, Fatouraei M, Huemer N, Alves CRR, Kothary R, Swoboda KJ, Impaired kidney structure and function in spinal muscular atrophy, Neurol. Genet 5 (2019) e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim JK, Jha NN, Feng Z, Faleiro MR, Chiriboga CA, Wei-Lapierre L, Dirksen RT, Ko CP, Monani UR, Muscle-specific SMN reduction reveals motor neuron-independent disease in spinal muscular atrophy models, J. Clin. Invest 2020. February 10 pii: 131989. doi: 10.1172/JCI131989. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J, Correlation between severity and SMN protein level in spinal muscular atrophy, Nat. Genet 16 (1997) 265–269. [DOI] [PubMed] [Google Scholar]
- [57].Wirth B, Brichta L, Schrank B, Lochmüller H, Blick S, Baasner A, Heller R, Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number, Hum. Genet 119 (2006) 422–428. [DOI] [PubMed] [Google Scholar]
- [58].Oprea GE, Kröber S, McWhorter ML, Rossoll W, Müller S, Krawczak M, Bassell GJ, Beattie CE, Wirth B, Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy, Science 320 (2008) 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Amara A, Adala L, Ben Charfeddine I, Mamaï O, Milli A, Lazreg TB, H’mida D, Salem N, Booughammura L, Saad A, Gribaa M, Correlation of SMN2, NAIP, p44, H4F5 and Occludin genes copy number with spinal muscular atrophy phenotype in Tunisian patients, Eur. J. Paediatr. Neurol 16 (2012) 167–174. [DOI] [PubMed] [Google Scholar]
- [60].Bosch-Marcé M, Wee CD, Martinez TL, Lipkes CE, Choe DW, Kong L, Van Meerbeke JP, Musarò A, Sumner CJ, Increased IGF-1 in muscle modulates the phenotype of severe SMA mice, Hum. Mol. Genet 20 (2011) 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Powis RA, Karyka E, Boyd P, Côme J, Jones RA, Zheng Y, Szunyogova E, Groen EJ, Hunter G, Thomson D, Wishart TM, Becker CG, Parson SH, Martinat C, Azzouz M, Gillingwater TH, Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy, JCI Insight 1 (2016) e87908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Howell MD, Ottesen EW, Singh NN, Anderson RL, Seo J, Sivanesan S, Whitley EM, Singh RN, TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy, Sci. Rep 7 (2017), 7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Custer SK, Astroski JW, Li HX, Androphy EJ, Interaction between alpha-COP and SMN ameliorates disease phenotype in a mouse model of spinal muscular atrophy, Biochem. Biophys. Res. Commun 514 (2019), 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kannan A, Jiang X, He L, Ahmad S, Gangwani L, ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy, Brain 143 (2020) 69–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Simon CM, Van Alstyne M, Lotti F, Bianchetti E, Tisdale S, Watterson DM, Mentis GZ, Pellizzoni L, Stasimon Contributes to the Loss of Sensory Synapses and Motor Neuron Death in a Mouse Model of Spinal Muscular Atrophy, Cell Rep 29 (2019), 3885–3901.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lipnick SL, Agniel DM, Aggarwal R, Makhortova NR, Finlayson SG, Brocato A, Palmer N, Darras BT, Kohane I, Rubin LL, Systemic nature of spinal muscular atrophy revealed by studying insurance claims, PLoS One 14 (2019) e0213680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wijngaarde CA, Blank AC, Stam M, Wadman RI, van den Berg LH, van der Pol WL, Cardiac pathology in spinal muscular atrophy: a systematic review, Orphanet. J. Rare Dis 12 (2017) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G, Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Singh NN, Seo J, Rahn SJ, Singh RN, A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes, PLoS One 7 (2012) e49595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN, Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene, PLoS One 11 (2016) e0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN, A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein, Sci. Rep 6 (2016) 30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ottesen EW, Luo D, Seo J, Singh NN, Singh RN, Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs, Nucleic Acids Res 47 (2019) 2884–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pagliarini V, Jolly A, Bielli P, Di Rosa V, De la Grange P, Sette C, Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization, Nucleic Acids Res 48 (2019) 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].d’Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ, The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy, Neuron 93 (2017) 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J 2nd, Schwartz B, Gordo S, Kasper A, Okamoto TR, Johansson HE, Mandefro B, Sareen D, Bialek P, Chau BN, Bhat B, Bullough D, Barsoum J, Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E1509–E1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ottesen EW, Seo J, Singh NN, Singh RN, A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements, Front. Microbiol 8 (2017) 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S,Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J, Refined characterization of the expression and stability of the SMN gene products, Am. J. Pathol 171 (2007) 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH, Regulation of SMN protein stability, Mol. Cell. Biol 29 (2009) 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cho S, Dreyfuss G, A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity, Genes Dev 24 (2010) 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Monani UR, McPherson JD, Burghes AH, Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT), Biochim. Biophys. Acta 1445 (1999) 330–336. [DOI] [PubMed] [Google Scholar]
- [81].Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J, The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements, Am. J. Hum. Genet 64 (1999) 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR, The SMN genes are subject to transcriptional regulation during cellular differentiation, Gene 279 (2001) 109–117. [DOI] [PubMed] [Google Scholar]
- [83].Noguchi S, Arakawa T, Fukuda S, Furuno M, Hasegawa A, Hori F, Ishikawa-Kato S, Kaida K, Kaiho A, Kanamori-Katayama M, Kawashima T, Kojima M, Kubosaki A, Manabe RI, Murata M, Nagao-Sato S, Nakazato K, Ninomiya N, Nishiyori-Sueki H, Noma S et al. , Data Descriptor: FANTOM5 CAGE profiles of human and mouse samples, Scientific Data 4 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Takahashi H, Kato S, Murata M, Carninci P. CAGE (Cap Analysis of Gene Expression): A Protocol for the Detection of Promoter and Transcriptional Networks In Gene Regulatory Networks: Methods and Protocols, Deplancke B, Gheldof N (eds) pp 181–200. Totowa: Humana Press Inc., 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kozak M, Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes, Proc. Natl. Acad. Sci. U. S. A 87 (1990), 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kozak M, Structural features in eukaryotic mRNAs that modulate the initiation of translation, J. Biol. Chem 266 (1991) 19867–19870. [PubMed] [Google Scholar]
- [87].Murat P, Marsico G, Herdy B, Ghanbarian AT, Portella G, Balasubramanian S, RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs, Genome Biol 19 (2018), 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jodoin R, Carrier JC, Rivard N, Bisaillon M, Perreault JP, G-quadruplex located in the 5’UTR of the BAG-1 mRNA affects both its cap-dependent and cap-independent translation through global secondary structure maintenance, Nucleic Acids Res 47 (2019), 10247–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lin YH, Bundschuh R, RNA structure generates natural cooperativity between single-stranded RNA binding proteins targeting 5’ and 3’UTRs, Nucleic Acids Res 43 (2015), 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Workman E, Veith A, Battle DJ, U1A regulates 3’ processing of the survival motor neuron mRNA, J. Biol. Chem 2014. February 7;289(6):3703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Singh NN, Del Rio-Malewski JB, Luo D, Ottesen EW, Howell MD, Singh RN, Activation of a cryptic 5’ splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene, Nucleic Acids Res 45 (2017) 12214–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Harahap NIF, Niba ETE, Ar Rochmah M, Wijaya YOS, Saito T, Saito K, Awano H, Morioka I, Iijima K, Lai PS, Matsuo M, Nishio H, Shinohara M, Intron-retained transcripts of the spinal muscular atrophy genes, SMN1 and SMN2, Brain Dev 40 (2018) 670–677. [DOI] [PubMed] [Google Scholar]
- [93].Baralle FE, Singh RN, Stamm S, RNA structure and splicing regulation, Biochim. Biophys. Acta Gene Regul. Mech 1862 (2019) 194448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Welden JR, Stamm S, Pre-mRNA structures forming circular RNAs, Biochim. Biophys. Acta Gene Regul. Mech 1862 (2019) 194410. [DOI] [PubMed] [Google Scholar]
- [95].Pervouchine DD, Circular exonic RNAs: When RNA structure meets topology, Biochim. Biophys. Acta Gene Regul. Mech 1862 (2019) 194384. [DOI] [PubMed] [Google Scholar]
- [96].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, Circular RNAs are abundant, conserved, and associated with ALU repeats, RNA 19 (2013) 141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown, Salzman, Circular RNA is expressed across the eukaryotic tree of life, PLoS One 9 (2014) e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C, Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes, Brief Bioinform 18 (2017) 984–992. [DOI] [PubMed] [Google Scholar]
- [99].Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S, Translation of CircRNAs, Mol. Cell 66 (2017) 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S, The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution, Hum. Mol. Genet 8 (1999) 775–782. [DOI] [PubMed] [Google Scholar]
- [101].Lorson CL, Androphy EJ, The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding, Hum. Mol. Genet 7 (1998) 1269–1275. [DOI] [PubMed] [Google Scholar]
- [102].Selenko P, Sprangers R, Stier G, Bühler D, Fischer U,Sattler M, SMNtudor domain structure and its interactionwith the Smproteins, Nat. Struct. Biol 8 (2001) 27–31. [DOI] [PubMed] [Google Scholar]
- [103].Ottesen EW, Singh NN, Luo D, Singh RN, High-affinity RNA targets of the Survival Motor Neuron protein reveal diverse preferences for sequence and structural motifs, Nucleic Acids Res 46 (2018) 10983–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Martin R, Gupta K, Ninan NS, Perry K, Van Duyne GD, The Survival Motor Neuron Protein Forms Soluble Glycine Zipper Oligomers, Structure 20 (2012) 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Huang A, Zheng H, Wu Z, Chen M, Huang Y, Circular RNA-protein interactions: functions, mechanisms, and identification, Theranostics 10 (2020) 3503–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Dudekulay DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M, CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs, RNA Biol 13 (2016) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Errichelli L, Modigliani SD, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, Lu L, Caffarelli E, Shneider NA, Morlando M, Bozzoni I, FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons, Nat. Commun 8 (2017) 14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yamazaki T, Chen S, Yu Y, Yan BA, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, Sharma A, Chandran S, Sullivan G, Nishimura AL, Shaw CE, Gygi SP, Shneider NA, Maniatis T, Reed R, FUS-SMN Protein Interactions Link the Motor Neuron Diseases ALS and SMA, Cell Rep 2 (2012) 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chan CC, Dostie J, Diem MD, Feng WQ, Mann M, Rappsilber J, Dreyfuss G, eIF4A3 is a novel component of the exon junction complex, RNA 10 (2004) 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang RJ, Zhang S, Chen XY, Li N, Li JW, Jia RC, Pan YQ, Liang HQ, EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis, Mol. Cancer 17 (2018) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Isken O, Maquat LE, The multiple lives of NMD factors: balancing roles in gene and genome regulation, Nat. Rev. Genet 9 (2008) 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Alexandrov A, Colognori D, Steitz JA, Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex, Genes Dev 25 (2011) 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Budiman ME, Bubenik JL, Miniard AC, Middleton LM, Gerber CA, Cash A, Driscoll DM, Eukaryotic Initiation Factor 4a3 Is a Selenium-Regulated RNA-Binding Protein that Selectively Inhibits Selenocysteine Incorporation, Mol. Cell 35 (2009) 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Hafner M, Max KEA, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T, Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition, RNA 19 (2013) 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Esteller M, Non-coding RNAs in human disease, Nat. Rev. Genet 12 (2011) 861–874. [DOI] [PubMed] [Google Scholar]
- [116].Zhang K, Zhang XR, Cai ZQ, Zhou J, Cao R, Zhao Y, Chen ZG, Wang DH, Ruan W, Zhao Q, Liu GQ, Xue YC, Qin Y, Zhou B, Wu LG, Nilsen T, Zhou Y, Fu XD, A novel class of microRNA-recognition elements that function only within open reading frames, Nat. Struct. Mol. Biol 25 (2018) 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS, MicroRNA targets in Drosophila, Genome Biol 5 (2004) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Liu WJ, Wang XW, Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data, Genome Biol 20 (2019) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zhao ZZ, Partridge V, Sousares M, Shelton SD, Holland CL, Pertsemlidis A, Du LQ, microRNA-2110 functions as an oncosuppressor in neuroblastoma by directly targeting Tsukushi, Plos One 13 (2018) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, Uhrberg M, Wernet P, Brustle O, MicroRNA-Based Promotion of Human Neuronal Differentiation and Subtype Specification, Plos One 8 (2013) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Rago L, Beattie R, Taylor V, Winter J, miR379–410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin, EMBO J 33 (2014) 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Murdocca M, Ciafre SA, Spitalieri P, Talarico RV, Sanchez M, Novelli G, Sangiuolo F, SMA Human iPSC-Derived Motor Neurons Show Perturbed Differentiation and Reduced miR-335–5p Expression, Int. J. Mol. Sci 17 (2016) 1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Xiong R, Wang ZQ, Zhao ZB, Li H, Chen W, Zhang B, Wang LL, Wu L, Li W, Ding JQ, Chen SD, MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration, Neurobiology of Aging, 35 (2014) 705–714. [DOI] [PubMed] [Google Scholar]
- [124].Long JM, Ray B, Lahiri DK, MicroRNA-339–5p Down-regulates Protein Expression of beta-Site Amyloid Precursor Protein-Cleaving Enzyme 1 (BACE1) in Human Primary Brain Cultures and Is Reduced in Brain Tissue Specimens of Alzheimer Disease Subjects, J. Biol. Chem 289 (2014) 5184–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ghanbari M, Ikram MA, de Looper HWJ, Hofman A, Erkeland SJ, Franco OH, Dehghan A, Genome-wide identification of microRNA-related variants associated with risk of Alzheimer’s disease, Sci. Rep 6 (2016) 28386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jang SI, Tandon M, Teos L, Zheng CY, Warner BM, Alevizos I, Dual function of miR-1248 links interferon induction and calcium signaling defects in Sjogren’s syndrome, EBioMedicine 48 (2019) 526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ji J, Qin YF, Zhou R, Zang RJ, Huang ZY, Zhang Y, Chen MJ, Wu W, Song L, Ling XF, Shen HB, Hu ZB, Xia YK, Lu CC, Wang XR, X chromosome-wide identification of SNVs in microRNA genes and non-obstructive azoospermia risk in han chinese population, Oncotarget 7 (2016) 49122–49129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yu XT, Zhang XC, Bi T, Ding YF, Zhao JY, Wang C, Jia TT, Han D, Guo G, Wang B, Jiang JY, Cui SY, MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma, Tumor Biol 34 (2013) 3501–3508. [DOI] [PubMed] [Google Scholar]
- [129].Zhan MN, Yu XT, Tang J, Zhou CX, Wang CL, Yin QQ, Gong XF, He M, He JR, Chen GQ, Zhao Q, MicroRNA-494 inhibits breast cancer progression by directly targeting PAK1, Cell Death Dis 8 (2017) e2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Alqurashi N, Hashimi SM, Alowaidi F, Ivanovski S, Farag A, Wei MQ, miR-496, miR-1185, miR-654, miR-3183 and miR-495 are downregulated in colorectal cancer cells and have putative roles in the mTOR pathway, Oncol. Lett 18 (2019) 1657–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zheng ZY, Liu JM, Yang Z, Wu LM, Xie HY, Jiang CZ, Lin BY, Chen TC, Xing CY, Liu ZK, Song PH, Yin SY, Zheng SS, Zhou L, MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting sox7 involving wnt/beta-catenin signaling pathway, Oncotarget 7 (2016) 28000–28012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Shu MF, Zheng XK, Wu SH, Lu HM, Leng TD, Zhu WB, Zhou YH, Ou YQ, Lin X, Lin Y, Xu D, Zhou YX, Yan GM, Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells, Mol. Cancer, 10 (2011) 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Khan FH, Pandian V, Ramraj S, Aravindan S, Herman TS, Aravindan N, Reorganization of metastamiRs in the evolution of metastatic aggressive neuroblastoma cells, BMC Genomics 16 (2015) 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wang JJ, Chen CF, Yan X, Wang P, The role of miR-382–5p in glioma cell proliferation, migration and invasion, Onco Targets Ther 12 (2019) 4993–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Magri F, Vanoli F, Corti S, miRNA in spinal muscular atrophy pathogenesis and therapy, J. Cell. Mol. Med 22 (2018) 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


