Abstract
Purpose of review
To summarize the most recent scientific progress in transfusion medicine genomics and discuss its role within the broad genomic precision medicine model, with a focus on the unique computational and bioinformatic aspects of this emergent field.
Recent findings
Recent publications continue to validate the feasibility of using next generation sequencing (NGS) for blood group prediction with 3 distinct approaches: exome sequencing, whole genome sequencing, and PCR-based targeted NGS methods. The reported correlation of NGS with serologic and alternative genotyping methods ranges from 92 to 99%. NGS has demonstrated improved detection of weak antigens, structural changes, copy number variations, novel genomic variants, and microchimerism. Addition of a transfusion medicine interpretation to any clinically-sequenced genome is proposed as a strategy to enhance the cost-effectiveness of precision genomic medicine. Interpretation of NGS in the blood group antigen context requires not only advanced immunohematology knowledge, but also specialized software and hardware resources, and a bioinformatics-trained workforce.
Summary
Blood transfusions are a common inpatient procedure, making blood group genomics a promising facet of precision medicine research. Further efforts are needed to embrace transfusion bioinformatic challenges and evaluate its clinical utility.
Keywords: genomics, next generation sequencing, blood group, bioinformatics
INTRODUCTION
Much like every other discipline in medicine and biology, transfusion medicine was transformed by the introduction of genotyping technologies – the next revolutionary blood grouping technique since Karl Landsteiner described ABO hemagglutination in 1900 [1]. Genotyping has become a valuable part of the blood bank laboratory toolkit, both as a complement and as an alternative to conventional serology. Its advantages in the clinical setting are widely documented, including blood group determination in recently-transfused patients, detection of rare blood antigens for which commercial serology is unavailable, blood typing in patients receiving monoclonal therapies that interfere with serology, determination of paternal zygosity, noninvasive fetal blood typing, and streamlining complex antibody workups [2*].
Established blood group genotyping platforms, such as probe arrays, tend to be targeted, address a restricted number of variants, and offer a limited throughput [3]. Since 2011 a growing number of publications have explored the application of Next Generation Sequencing (NGS) for prediction of red blood cell and platelet antigens [4**, 5*]. Broadly, NGS encompasses a group of sequencing platforms that vary in terms of underlying chemistry, read length, and error rate, but that uniformly offer a high-throughput and massively parallel approach. Since NGS precision is lower than the Sanger sequencing gold standard, experimental repetition (reported ‘read depth’) is one of the critical quality parameters when analyzing the results (Figure 1).
Figure 1. Schematic representation of NGS read depth.
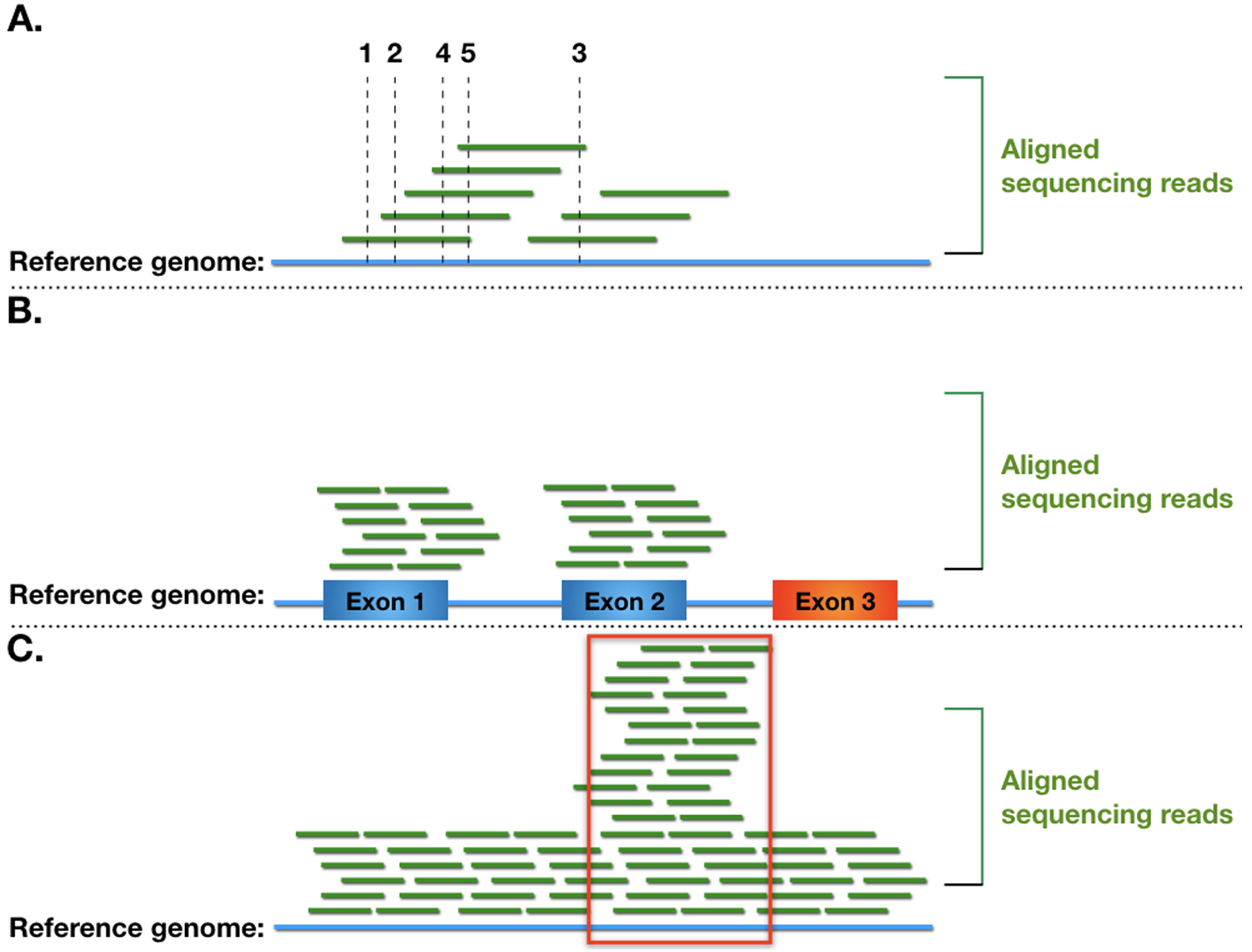
Read depth refers to the number of aligned NGS reads that overlap with a given genomic coordinate. (A) Broken lines indicate the read depth at different genomic positions for a simulated low-depth NGS alignment. Low read depths are associated with lower nucleotide call precision and may miss heterozygous polymorphic sites. (B) Simulation of low read depth in an exome sequencing NGS dataset. Boxes represent predicted exons, a low read depth exon is colored in red. Possible interpretations include: genomic deletion, loss of sequences in the initial quality trimming process, or failure of exome capture strategy design. (C) Higher read depth in a select genomic region (marked by the red rectangle). This may represent misalignment of short reads that in reality derive from a homologous gene; in this case a real heterozygous polymorphism may manifest as significantly less than 50% of the nucleotide calls. Other important NGS quality parameters, not shown in the figure, include mapping quality and fraction of alternate allele.
NGS technological advances have led to a drastic decline in the time and cost to sequence an entire human genome [6], bringing about many large-scale global sequencing projects (1000 Genomes, ExAC, UK10, TopMed), with more ambitious projects brewing in the near future [7–11]. Thus, thousands of human exome and whole genome sequences have flooded public and private databases, exponentially escalating our knowledge about worldwide human genetic variations, often at a quicker pace than our ability to interpret them thoroughly. Indeed, NGS platforms converge in the generation of large amounts of bioinformatic data and the need for sophisticated computational systems and analytic tools to interpret them. In fact, genomics has taken a place as one of the 4 ‘Big Data’ sciences, next to astronomy, Twitter and youTube [12]. Genomics has grown thanks to, and parallel with, computational and software innovations, bringing new informatics challenges to the health care industry at an unprecedented speed. Bioinformatics is emerging as the main bottleneck in clinical genomics and warrants careful attention by stakeholders [13**]. The goal of this review is to summarize the most recent scientific progress in transfusion medicine genomics and discuss its role within the broad genomic precision medicine model, with a focus on the unique computational and bioinformatic aspects of this emergent field.
RECENT PROGRESS IN TRANSFUSION MEDICINE GENOMICS
Transfusion literature on NGS applications has continued to expand since this journal’s last review [5*]. Three main NGS strategies continue to be explored (Table 1): analysis of whole-genome sequencing (WGS) datasets [14*, 15], exome sequencing (ES) [16*], and, most commonly, a PCR-based targeted approach [17*,18, 19*,20**]. Recent WGS and ES investigations were based on analysis of previously-generated datasets, while targeted NGS was pursued with both ion semiconductor chemistry [18, 19*, 21*] and sequencing by synthesis with fluorescent reversible terminators [17*, 20**]. Amplification strategies varied, including long range PCR for the RHD and RHAG genes [21*]. Wu et al. amplified and sequenced the entire ABO gene and flanking regions, describing optimization parameters for the challenging GC-rich ABO exon 1 [20**]. Orzinska et al. employed a primer design software to target 9 blood group antigens and 5 human platelet antigens, reporting failure of RhC and MN detection, and the need to adjust primers for these regions [18]. In addition, Boccoz et al. describe a protocol to sequence 15 blood group variants in 95 patients in a single run, where read depth correlated with the extremes of amplicon sizes and with lower precision in genotype predictions [17*].
Table 1.
Comparison of the 3 NGS strategies in transfusion genomics.
| Whole Genome Sequencing (WGS) | Exome Sequencing (ES) | Targeted NGS |
|---|---|---|
| Untargeted | Some commerical exome panels do not capture select blood group genes | Often involves initial PCR amplification |
| Largest dataset | Smaller data files than WGS | Smallest data file sizes per sequenced subject |
| More mature CNV algorithms | CNV algorithms are available, but more challenging | Read depth based CNV analysis feasable |
| Clinically-actionable secondary variants may be identified | Clinically-actionable secondary variants may be identified | Design may circumvent genes with secondary findings |
| Lowest throughput | Intermediate throughput | Highest throughput |
| Lower read depth typically achieved | Higher read depth than WGS | Read depths typically >1000 |
| Short reads may misalign to homologous regions | Short reads may misalign to homologous regions | Targeted PCR design coupled to customized alignment pipeline may avoid some misalignments |
| Detects variations in non-coding regions | Typically does not sequence noncoding regions | May detect non-coding genomic variants if targeted by design |
CNV = copy number variants.
Recent publications continue to confirm a high correlation (92–99%) between NGS predictions and alternative serology or genotyping methods. Furthermore, they contribute to the accumulated evidence of 4 distinct NGS advantages in the precision setting: improved detection of weak antigens (e.g. detection of ABO*AEL.02 and ABO*BEL.02 [20**]); superior ability to address structural and copy number variations (e.g. detection of a novel ABO large deletion [20**]); the ability to detect novel blood group variants [19*]; and detection of microchimerism [18, 20**]. Discrepancies with NGS continue to be reported; these are commonly ascribed to low read depth (Figure 1), mislabeling, or early PCR-induced error, but sometimes remain unresolved even after careful study.
Genomics was also applied in the recently reported discovery of a non-coding transcription regulatory element for the Xga antigen [14*, 22**, 23**] and to validate the RUNX1-dependent regulatory site for the P1 antigen [14*]. These publications illustrate the discovery power that can be unleashed by analyzing NGS datasets in correlation with known phenotype frequencies, and the ability of WGS to uncover functional erythroid regulatory elements in non-coding regions.
To date, transfusion genomics research has employed relatively short-read NGS platforms, which preclude phasing of heterozygous variants when these are farther than the read length. Nonetheless, Wu et al. report successful haplotype construction in 17 of 20 samples [20**], while Tounsi et al. focused on hemizygous samples to circumvent this problem [21*]. Longer read-length platforms, but with a higher error rate, are available and remain to be tested in transfusion settings. Sequencing of cDNA has been useful to detect blood group gene rearrangements, verify expressed isoforms, and phase variants after cloning. The value, if any, of transcript NGS sequencing (‘RNA-seq’) in immunohematology has not yet been explored. If pursued, read length remains an important consideration, and RNA-seq analysis algorithms, which currently focus on expression level rather than detection of variants or rearrangements, may need modification.
TRANSFUSION AS AN ACTIONABLE PART OF GENOMIC MEDICINE
The Human Genome Project was conceived as the ambitious foundation for a precise and personalized form of medicine, a goal that continues to be ardently supported by international research initiatives [7, 8, 10, 13**]. Genomic sequencing is predicted to become an integral part of future health care; the number of sequenced human genomes is expected to escalate rapidly [12], while evidence of the potential clinical benefit of NGS continues to accumulate [13**, 24*]. A recent study reports that 9.3% of 3315 patients with chronic kidney disease who underwent ES carried diagnostic variants, which often led to diagnostic reclassification or altered referral and clinical management decisions [25]. In addition, pathogenic or likely pathogenic germline variants were identified in 8% of 10,389 Cancer Genome Atlas samples, which rose to 22.9% in the Pediatric Cancer Genome Project subset [26].
The American College of Medical Genetics and Genomics has designated 59 actionable genes to be disclosed as secondary findings of ES or WGS [27]; 1 in 38 healthy Dutch individuals were reported to carry a likely pathogenic variant in one of the actionable dominant genes [28]. The most cited priorities for current and future precision medicine research include cancer genomics, prenatal testing, rare diseases, and polygenic risk scores for common disease prediction [13**, 29**]. Although transfusion medicine is not commonly recognized as a critical precision medicine focus, it is important to keep in mind that everyone has a blood type; therefore, every patient could potentially benefit from transfusion genomics. Indeed, red cell transfusions that matched for ABO blood type could be considered, historically, as the first example of precision medicine, even though agglutination methods, rather than nucleic acid sequencing methods were used to determine the most probable genotype. Analysis of the REDS-III recipient database revealed that 12.5% of inpatient encounters report a transfusion [30]. For these patients, in addition to those transfused as outpatients, a blood-type genomic interpretation could be beneficial, particularly in the setting of alloimmunization or extended antigen matching. Given that cancer is one of the primary diagnoses associated with transfusion [30], the cost-effectiveness of WGS or ES performed for oncologic indications could conceivably improve by adding a blood type interpretation. Transfusion medicine genomics could also aid in transplant donor selection and for identifying and recruiting rare blood donors [2*, 4**]. Blood type interpretation algorithms, however, entail special challenges and considerations, which are discussed below.
TRANSFUSION GENOMICS FROM THE BLOOD COLLECTOR PERSPECTIVE
Using genomic sequencing technologies to predict blood group antigen phenotypes in blood donors is a different venture given that it targets an essentially healthy population. Targeted NGS approaches are particularly attractive to circumvent concerns about secondary findings, improve alignment precision compared to untargeted genome-wide short read sequencing methods, achieve the highest possible throughput, and reduce the data storage burden (Table 1). Further work is needed to evaluate the cost-effectiveness of NGS versus repeated serologic and targeted genotyping tests to provide antigen-negative blood for patients.
HARNESSING GENOMIC BIG DATA IN TRANFUSION
It is widely accepted that the current major bottleneck in genomics is the analysis of the large amounts of resulting data [13**]. The size of an NGS output depends on the strategy (WGS, ES, or targeted), read depth (Figure 1), specific sequencing platform, and type of file that is stored (raw, aligned, or annotated summary), and we can expect swift future growth along with sequencing and computational technology. In our experience, storage of trimmed, processed, and aligned 80x ES generated by reversive terminator chemistry occupied an average of 9.5GB per subject 5 years ago. In contrast, storage of recent 30x WGS raw, aligned, and annotated files reached upwards of 70GB per individual. If we consider that the average scientific manuscript is 1.5MB, the volume of NGS output data per patient would manually equate to reading 45 thousand research articles. Due to this volume of data, and to the complexity of algorithms that process and interpret it, computational support is fundamental for genomics to become a pragmatic clinical reality.
Software and analysis pipelines
A representative NGS analysis pipeline is depicted in Figure 2. Research software is available at each step from both commercial and open sources. Many pipeline variations exist, particularly across sequencing technologies, and software is continuously evolving through the release of updated versions or entirely novel algorithms. Three key analysis steps highlighted in Figure 2 are discussed below in the context of immunohematology.
Figure 2. Simplified, schematic diagram of an NGS bioinformatic pipeline.
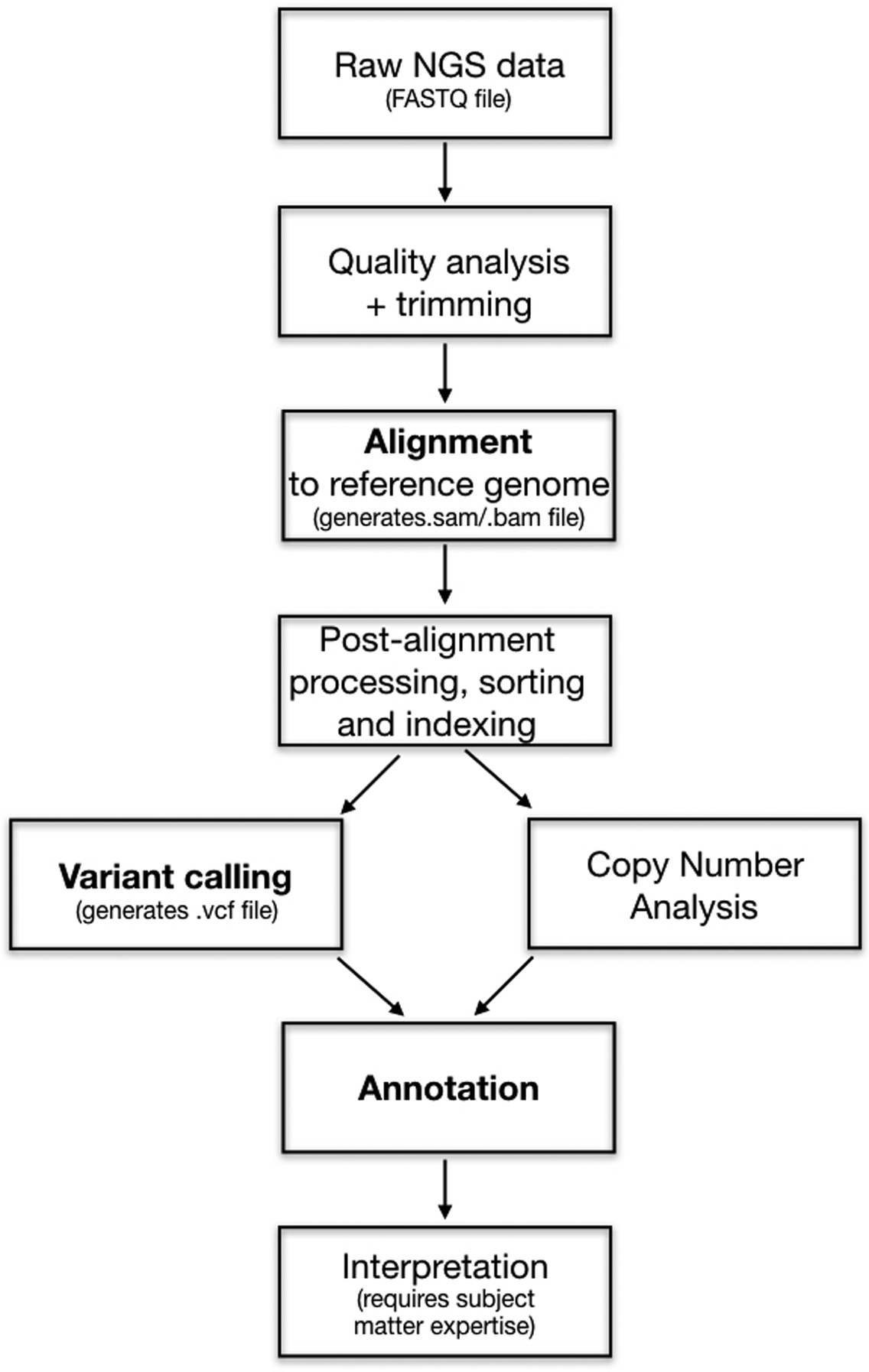
Processing steps in bold are discussed in the manuscript text. Pipelines for specific software may vary.
Abbreviations: FASTQ = text file that contains nucleotide sequence and quality strings; sam = sequence alignment map; bam = binary, smaller version of a sam file; vcf = variant call file.
Alignment is the process by which NGS reads are lined up to a reference genome coordinate, along with important parameters, including mapping quality and nucleotide match information. Multiple alignment algorithms exist [13**, 31]; it remains to be determined if these differ in terms of suitability for blood group genes. The reference genome build must be clearly documented and remain consistent throughout the rest of the pipeline. Only a few published studies in transfusion [4**, 21*] have transitioned to the most recent GRCh38 reference build [32], but this recent release still has discrepancies with the blood group alleles considered historically as references by immunohematologists. Thus, blood group genomics needs to make the necessary adjustments in allele nomenclature and interpretation, participate in genome build releases and patches, or produce suitable reference sequences for transfusion. Efforts to produce the latter are underway [33]. Furthermore, short NGS sequences derived from genes that have homologous counterparts may misalign to the wrong coordinate. Documented examples include the RH and XG blood groups [14*, 34**,35,36].
Variant calling refers to the determination of which nucleotide calls are truly polymorphic in the sample, as opposed to sequencing errors, misalignment, or poor quality. Multiple algorithms exist, and their performance may differ when paired with different alignment software [37, 38]. As discussed previously, phasing of discrete variants within a single gene, or haplotype construction, may be necessary for accurate blood group antigen prediction. The proximity of these variants and the read length determine if phasing can be performed directly from NGS data; alternatively, known haplotype frequencies can be employed to infer phasing.
Annotation of a variant call file refers to the addition of notes or commentaries that aid in interpretation, such as dbSNP or ClinVar ids, known frequencies, regulatory site overlap, and functional consequence for predicted transcript and protein products.
The final blood group antigen interpretation derived from genomic sequencing undoubtedly requires immunohematology expertise. Software is available that automates the analysis pipeline starting from an aligned file [34**]. Unlike other clinical genomic applications, known and novel variants in transfusion medicine are interpreted on the basis of their potential antigenicity; the function of the corresponding protein product is often disconnected. For example, Kidd molecules carrying the Jka and the Jkb antigens have preserved function in terms of their ability to transport urea, yet they hold notable allosensitization potential. In this space, transfusion medicine could work synergistically with the promising field of cancer neoantigen vaccines [39*,40,41].
Data storage and computational equipment
The analysis algorithms described above require computational processing and data storage capabilities that exceed those traditionally available in blood banks. Local or cloud-based computer clusters [42*] allow for decreased processing time through parallelization – i.e. distribution of independent analytic threads among multiple processors. Graphical processing units have shown promise for genomics due to their powerful parallel processing architecture [43]. A final consideration is data storage and appropriate backup, replication, and recovery protocols.
Current bioinformatic challenges
Although promising, much work remains in the transfusion genomics informatics realm. Like all other blood bank processes, NGS analysis pipelines will require rigorous recordkeeping, validation, and standardization. Few published manuscripts in transfusion medicine provide a detailed description of the software versions employed, specific runtime parameters, and quality thresholds, but the practice of sharing these details and scripts is gaining traction [14*, 20**]. Sharing even basic working programming code is encouraged in an effort to enhance reproducibility and accelerate scientific progress [44, 45]. Genomic visualization and reporting tools for transfusion medicine, secure and standardized worldwide data sharing protocols, and systems that assure the confidentiality, authenticity, and integrity of genomic data are also essential. At any given time, processed genomic data should be accompanied by a description of its origin and the precise analytic processes that have been historically applied to it, also known as ‘data provenance’ [46]. Computational and software systems established for transfusion genomics must also retain plasticity to scale and grow with the constantly evolving sequencing technologies, hardware processing capabilities, data structures, and analysis algorithms. Processes should also assure the reusability of genomic data, so that previously produced genomic sequences can be re-interpreted as our knowledge of blood group variants and the regulation of erythroid gene expression continues to expand [24*].
OPPORTUNITIES AND FUTURE RESEARCH
NGS has demonstrated benefit as a complementary approach to resolve typing discrepancies, validate existing data, and detect reporting errors [4**, 5*]. Transfusion genomics can quickly grow through incorporation of tools designed for other specialties but will also need to develop resources of its own. While RH allele matching based on SNV (single nucleotide variant) genotype data has been used in alloimmunized patients [47**], genomic level blood group matching strategies have yet to be explored; these could range from strict allele matching to epitope-level compatibility. As transfusion medicine knowledge and its genomic algorithms mature, other predictive genomic features besides blood typing could be added, such as those addressing donor health, product storage, or a patient’s alloantibody ‘responder’ status. Long term outcome data will be needed to evaluate the true impact of genomics in terms of clinical decision support, transfusion safety, clinical outcomes, and appropriate allocation of the blood product inventory.
CONCLUSION
Transfusion genomics is a promising discipline that challenges us with a surge of a new type of data, calling for specialized information technology resources and a workforce with computational expertise. Given the vast volume of blood group genotype-phenotype correlations, transfusion should be considered as an additional facet of clinically-actionable knowledge that can be exploited when the clinical decision is made to sequence a patient, enhancing the cost-effectiveness of the precision medicine approach.
Key Points.
Recent research continues to validate the application of next generation sequencing for precise and comprehensive blood group antigen prediction.
A transfusion medicine interpretation represents an opportunity to increase the cost-effectiveness of a clinically sequenced genome.
Transfusion genomics is accompanied by unique bioinformatic challenges that call for specialized workforce training and focused research efforts.
Acknowledgements
The authors thank Harvey G. Klein, MD for critical review of this manuscript.
Financial support and sponsorship
Supported by the Intramural Research Program of the National Institutes of Health Clinical Center.
Footnotes
Publisher's Disclaimer: Statement of disclaimer:
Publisher's Disclaimer: The views expressed here do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the US Federal Government.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
- 1.Landsteiner K Zur Kenntnis der antifermentativen, lytischen und agglutinier- enden Wirkungen des Blutserums und der Lymphe. Zentralblatt fur Bakteriologie. 1900;27:357–62. [Google Scholar]
- *2.Westhoff CM. Blood group genotyping. Blood. 2019;133(17):1814–20. [DOI] [PubMed] [Google Scholar]; This is a thorough recent review on the application of blood group genotyping in various clinical contexts, including a brief discussion on NGS in transfusion.
- 3.Elkins MB, Davenport RD, Bluth MH. Molecular Pathology in Transfusion Medicine: New Concepts and Applications. Clin Lab Med. 2018;38(2):277–92. [DOI] [PubMed] [Google Scholar]
- **4.Hyland CA, Roulis EV, Schoeman EM. Developments beyond blood group serology in the genomics era. Br J Haematol. 2019;184(6):897–911. [DOI] [PubMed] [Google Scholar]; This manuscript reviews the genetic basis of blood groups and the published research on transfusion medicine genomics. The article provides blood group gene coordinates in the most recent human reference build GRCh38, and it discusses future applications of NGS in the blood bank.
- *5.Wheeler MM, Johnsen JM. The role of genomics in transfusion medicine. Curr Opin Hematol. 2018;25(6):509–15. [DOI] [PubMed] [Google Scholar]; This article is a preceding review on transfusion medicine genomics, on which the present manuscript is built.
- 6.NHGRI. The Cost of Sequencing a Human Genome. Available from: https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost.
- 7.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genomics England: 100,000 Genomes Project. Available from: https://www.genomicsengland.co.uk/.
- 9.NIH. All of Us Research Program. Available from: https://allofus.nih.gov/.
- 10.Adachi T, Kawamura K, Furusawa Y, et al. Japan’s initiative on rare and undiagnosed diseases (IRUD): towards an end to the diagnostic odyssey. Eur J Hum Genet. 2017;25(9):1025–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He KY, Ge D, He MM. Big Data Analytics for Genomic Medicine. Int J Mol Sci. 2017;18(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens ZD, Lee SY, Faghri F, et al. Big Data: Astronomical or Genomical? PLoS Biol. 2015;13(7):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Suwinski P, Ong C, Ling MHT, et al. Advancing Personalized Medicine Through the Application of Whole Exome Sequencing and Big Data Analytics. Front Genet. 2019;10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article is a recent, comprehensive review on exome sequencing as a tool for precision medicine and the bioinformatic challenges associated with big data in genomics.
- *14.Lane WJ, Aguad M, Smeland-Wagman R, et al. A whole genome approach for discovering the genetic basis of blood group antigens: independent confirmation for P1 and Xg(a). Transfusion. 2019;59(3):908–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript describes the use of genomics, in relation to known phenotype frequencies and serologic data, to validate transcription regulatory sites for the A4GALT and XG blood group genes. The article illustrates the difficulty in sequencing the XG gene in male participants due to misalignment of the NGS reads with a homologous region in the Y chromosome. Code employed for this research is publicly-available.
- 15.Montemayor-Garcia C, Karagianni P, Stiles DA, et al. Genomic coordinates and continental distribution of 120 blood group variants reported by the 1000 Genomes Project. Transfusion. 2018;58(11):2693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Schoeman EM, Roulis EV, Perry MA, et al. Comprehensive blood group antigen profile predictions for Western Desert Indigenous Australians from whole exome sequence data. Transfusion. 2019;59(2):768–78. [DOI] [PubMed] [Google Scholar]; This article illustrates the analysis of previously-generated exome sequencing data in the context of immunohematology, including a detailed diagram of the bioinformatic process. This research illustrates the identification of novel blood group variants, and the limitations of available exome sequencing assays that were not designed to target some blood group genes.
- *17.Boccoz SA, Fouret J, Roche M, et al. Massively parallel and multiplex blood group genotyping using next-generation-sequencing. Clin Biochem. 2018;60:71–6. [DOI] [PubMed] [Google Scholar]; This manuscript describes a high-throughput targeted NGS typing strategy for 15 blood group single nucleotide variants. A relationship between amplicon size and read depth is described.
- 18.Orzinska A, Guz K, Mikula M, et al. Prediction of fetal blood group and platelet antigens from maternal plasma using next-generation sequencing. Transfusion. 2019;59(3):1102–7. [DOI] [PubMed] [Google Scholar]
- *19.Wen J, Verhagen O, Jia S, et al. A variant RhAG protein encoded by the RHAG*572A allele causes serological weak D expression while maintaining normal RhCE phenotypes. Transfusion. 2019;59(1):405–11. [DOI] [PubMed] [Google Scholar]; This article describes the use of NGS as one of many assays for the discovery of an RHAG variant that results in normal RHAG and normal RHCE expression, but weak RHD. Data generated by this research corrected a misclasified RHAG allele and illustrates the limitations of the public available algorithms to predict damaging novel genomic changes.
- **20.Wu PC, Lin YH, Tsai LF, et al. ABO genotyping with next-generation sequencing to resolve heterogeneity in donors with serology discrepancies. Transfusion. 2018;58(9):2232–42. [DOI] [PubMed] [Google Scholar]; This articles uses NGS to resolve cases with ABO serologic discrepancies. The manuscript includes detailed documentation of bioinformatic processes and experimental optimization parameters, and it illustrates the potential for NGS to detect weak subtypes, novel variants, and microchimerism.
- *21.Tounsi WA, Madgett TE, Avent ND. Complete RHD next-generation sequencing: establishment of reference RHD alleles. Blood Adv. 2018;2(20):2713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes a long-range PCR targeted NGS strategy to establish reference RHD alleles. This manuscript is notable for using the most recent reference genome (GRCh38) for NGS read alignment. Phasing was avoided by focusing on hemizygous samples.
- **22.Moller M, Lee YQ, Vidovic K, et al. Disruption of a GATA1-binding motif upstream of XG/PBDX abolishes Xg(a) expression and resolves the Xg blood group system. Blood. 2018;132(3):334–8. [DOI] [PubMed] [Google Scholar]; One of two recent studies that employs genomics to eluciate the transcription regulatory site upstream of the XG gene, based on the correlation of known phenotype frequencies with polymorphic genomic sites in the 1000 Genomes Project database.
- **23.Yeh CC, Chang CJ, Twu YC, et al. The molecular genetic background leading to the formation of the human erythroid-specific Xg(a)/CD99 blood groups. Blood Adv. 2018;2(15):1854–64. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript describes the work of a second research group that employed targeted NGS of a large region of the X chromosome to elucidate the regulatory site for the XG gene.
- *24.Alfares A, Aloraini T, Subaie LA, et al. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet Med. 2018;20(11):1328–33. [DOI] [PubMed] [Google Scholar]; This manuscript illustrates that the diagnostic yield of exome sequencing can improve just by reanalyzing the same data at a later time, in the context of the newly-described genotype to phenotype correlations.
- 25.Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic Utility of Exome Sequencing for Kidney Disease. N Engl J Med. 2019;380(2):142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang KL, Mashl RJ, Wu Y, et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell. 2018;173(2):355–70 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–55. [DOI] [PubMed] [Google Scholar]
- 28.Haer-Wigman L, van der Schoot V, Feenstra I, et al. 1 in 38 individuals at risk of a dominant medically actionable disease. Eur J Hum Genet. 2019;27(2):325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Shendure J, Findlay GM, Snyder MW. Genomic Medicine-Progress, Pitfalls, and Promise. Cell. 2019;177(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a recent and thorough review on the history, current progress, and expected future of genomic medicine, with a detailed discussion of historical milestones in the field and its main clinical applications.
- 30.Karafin MS, Bruhn R, Westlake M, et al. Demographic and epidemiologic characterization of transfusion recipients from four US regions: evidence from the REDS-III recipient database. Transfusion. 2017;57(12):2903–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Kawam A, Khatri S, Datta A. A Survey of Software and Hardware Approaches to Performing Read Alignment in Next Generation Sequencing. IEEE/ACM Trans Comput Biol Bioinform. 2017;14(6):1202–13. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Dai Y, Yu H, et al. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics. 2017;109(2):83–90. [DOI] [PubMed] [Google Scholar]
- 33.MacArthur JA, Morales J, Tully RE, et al. Locus Reference Genomic: reference sequences for the reporting of clinically relevant sequence variants. Nucleic Acids Res. 2014;42(Database issue):D873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Lane WJ, Westhoff CM, Gleadall NS, et al. Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study. Lancet Haematol. 2018;5(6):e241–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a software to automate the interpretation of whole genome sequences into a predicted exteded red blood cell and platelet antigen phenotype, and its validation in a total of 310 research participants.
- 35.Chou ST, Flanagan JM, Vege S, et al. Whole-exome sequencing for RH genotyping and alloimmunization risk in children with sickle cell anemia. Blood Adv. 2017;1(18):1414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoeman EM, Lopez GH, McGowan EC, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion. 2017;57(4):1078–88. [DOI] [PubMed] [Google Scholar]
- 37.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Xu C A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput Struct Biotechnol J. 2018;16:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript provides a comprehensive review of various variant calling strategies and their performance metrics, to aid researchers in the selection of the most appropriate pipeline for their work.
- *39.Ajina R, Zamalin D, Weiner LM. Functional genomics: paving the way for more successful cancer immunotherapy. Brief Funct Genomics. 2019;18(2):86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]; A recent comprehensive review on cancer immunotherapy that describes the genomics workflow for identifying tumor neoantigens for immunotherapy targeting.
- 40.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–6. [DOI] [PubMed] [Google Scholar]
- *42.Langmead B, Nellore A. Cloud computing for genomic data analysis and collaboration. Nat Rev Genet. 2018;19(4):208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review explains the advatanges of cloud-based computer clusters for analysis of large NGS datasets, emphasizing the cloud’s infrastructure flexibility and its inherent aptitude to enhance analytic reproducibility and scientific collaborations.
- 43.Yang A, Troup M, Ho JWK. Scalability and Validation of Big Data Bioinformatics Software. Comput Struct Biotechnol J. 2017;15:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng RD. Reproducible research in computational science. Science. 2011;334(6060):1226–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes N Publish your computer code: it is good enough. Nature. 2010;467(7317):753. [DOI] [PubMed] [Google Scholar]
- 46.Kanwal S, Khan FZ, Lonie A, Sinnott RO. Investigating reproducibility and tracking provenance - A genomic workflow case study. BMC Bioinformatics. 2017;18(1):337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47.Chou ST, Evans P, Vege S, et al. RH genotype matching for transfusion support in sickle cell disease. Blood. 2018;132(11):1198–207. [DOI] [PubMed] [Google Scholar]; This manuscript documents similar variant RH allele frequencies in a cohort of sickle cell disease patients and African American donors, and it documents the feasability of RH genotype blood product matching rather than matching at the serologic level.


