Abstract
Objectives
Malaria, caused by Plasmodium infection, remains a major global health problem. Monocytes are integral to the immune response, yet their transcriptional and functional responses in primary Plasmodium falciparum infection and in clinical malaria are poorly understood.
Methods
The transcriptional and functional profiles of monocytes were examined in controlled human malaria infection with P. falciparum blood stages and in children and adults with acute malaria. Monocyte gene expression and functional phenotypes were examined by RNA sequencing and flow cytometry at peak infection and compared to pre‐infection or at convalescence in acute malaria.
Results
In subpatent primary infection, the monocyte transcriptional profile was dominated by an interferon (IFN) molecular signature. Pathways enriched included type I IFN signalling, innate immune response and cytokine‐mediated signalling. Monocytes increased TNF and IL‐12 production upon in vitro toll‐like receptor stimulation and increased IL‐10 production upon in vitro parasite restimulation. Longitudinal phenotypic analyses revealed sustained significant changes in the composition of monocytes following infection, with increased CD14+CD16− and decreased CD14−CD16+ subsets. In acute malaria, monocyte CD64/FcγRI expression was significantly increased in children and adults, while HLA‐DR remained stable. Although children and adults showed a similar pattern of differentially expressed genes, the number and magnitude of gene expression change were greater in children.
Conclusions
Monocyte activation during subpatent malaria is driven by an IFN molecular signature with robust activation of genes enriched in pathogen detection, phagocytosis, antimicrobial activity and antigen presentation. The greater magnitude of transcriptional changes in children with acute malaria suggests monocyte phenotypes may change with age or exposure.
Keywords: CHMI, interferon, malaria, monocytes, Plasmodium falciparum, RNA sequencing
We investigate the transcriptional, functional and phenotypic changes in monocytes during an experimental blood‐stage P. falciparum infection in malaria naive healthy subjects and during an episode of clinical malaria in children and adults residing in a malaria‐endemic region. We specifically isolated monocytes for transcriptome analysis by RNA sequencing, to determine monocyte gene expression during subpatent and clinical malaria. Monocyte activation during subpatent malaria is driven by an inflammatory molecular signature and a greater magnitude of transcriptional changes in children with acute malaria suggest monocyte phenotypes may change with age or exposure.
Introduction
Malaria remains an important global disease, with an estimated 228 million cases and 405 000 deaths in 2018, the majority caused by Plasmodium falciparum. 1 Upon transmission, Plasmodium parasites firstly infect hepatocytes to mature and multiply before release into the bloodstream and immediate infection of red blood cells (RBCs). Blood‐stage infection is characterised by cycles of asexual replication leading to RBC rupture and periodic malaria symptoms. Despite recent gains in reducing the burden of malaria, progress has stagnated in the last 3–5 years, with morbidity rising in several highly endemic countries. 1 In areas of unstable malaria transmission, morbidity and mortality affect adults as well as children.
Innate and adaptive immune responses mediate both tolerogenic and antiparasitic protective mechanisms that can result in reduced clinical symptoms in future reinfections, 2 yet acquisition of clinical immunity is complex and incompletely understood. Monocytes are integral to innate immune responses and may modulate adaptive immune responses during malaria. 3 , 4 An increased understanding of monocyte activation and function in subpatent Plasmodium infection may aid the development of strategies to enhance protective responses to improve the morbidity and mortality of malaria and assist in vaccine development.
Monocytes express a broad range of pattern recognition receptors, including toll‐like receptors (TLR), 5 as well as receptors for the detection and phagocytosis of opsonised or non‐opsonised parasites or infected RBC. 6 , 7 , 8 Besides parasite detection and phagocytosis, monocytes are major producers of predominantly inflammatory cytokines in malaria, including TNF and IL‐12; these assist immune control of Plasmodium infection, 9 , 10 but also contribute to the clinical symptoms of malaria. 11 , 12
Three distinct subsets of monocytes are identified by differential expression of the LPS receptor (CD14) and FcγRIII (CD16), and are defined as classical (CD14+CD16−), intermediate (CD14+CD16+) and non‐classical (CD14−CD16+) monocytes. 13 These subsets have differing capacities to mediate innate and adaptive immune responses. 13 , 14 Classical monocytes mount a highly inflammatory response following exposure to bacterial TLR ligands, 14 while non‐classical monocytes respond to viral antigens 15 and intermediate monocytes are the most proficient at antibody‐mediated phagocytosis of P. falciparum. 8 , 16 Plasmodium infection can change the composition of circulating monocytes. In children with asymptomatic 17 or uncomplicated malaria, 16 and in malaria in pregnancy, 18 CD16+ intermediate/non‐classical monocytes increase proportionally in the peripheral blood. However, there is a lack of information regarding prospective changes to monocyte composition during primary blood‐stage infection and acute malaria in non‐immune adults. Recent whole‐blood transcriptional analysis suggests that monocytes may have an anti‐inflammatory role in antimalarial immunity. 19 Genetically susceptible Mossi groups in Mali lack transcriptionally active monocytes while, relatively resistant Fulani populations have transcriptionally active monocytes (but not T cells, B cells or NK cells) during P. falciparum infection 20 , highlighting the importance of monocytes in protective immune responses.
Here, we investigate the transcriptional, functional and phenotypic changes in monocytes during an experimental blood‐stage P. falciparum infection in malaria‐naive healthy subjects and during an episode of clinical malaria in children and adults residing in a malaria‐endemic region. We specifically isolated monocytes for transcriptome analysis by RNA sequencing, to determine monocyte gene expression during subpatent and clinical malaria. In primary infection, we focus on blood‐stage only infection, allowing us to examine how P. falciparum infection affects monocyte responses independent of the liver stage. Furthermore, our study design allows us to do paired analyses, as each patient's baseline or convalescence sample served as their own control.
Results
Monocyte gene expression during primary P. falciparum infection
To determine the immune pathways and genes activated early during a primary P. falciparum blood‐stage infection, we performed a paired transcriptome analysis of isolated monocytes via RNA sequencing. Blood monocytes were isolated from five volunteers at baseline immediately prior to intravenous infection with P. falciparum blood stages and on day 8 post‐infection allowing each individual's baseline sample to act as their own control (Figure 1a and b). On day 8 post‐infection, when the infection was still subpatent, 509 differentially expressed genes (DEGs, FDR < 0.05) were identified, of which 71% (360) were upregulated (Figure 1c, Supplementary table 1).
Figure 1.
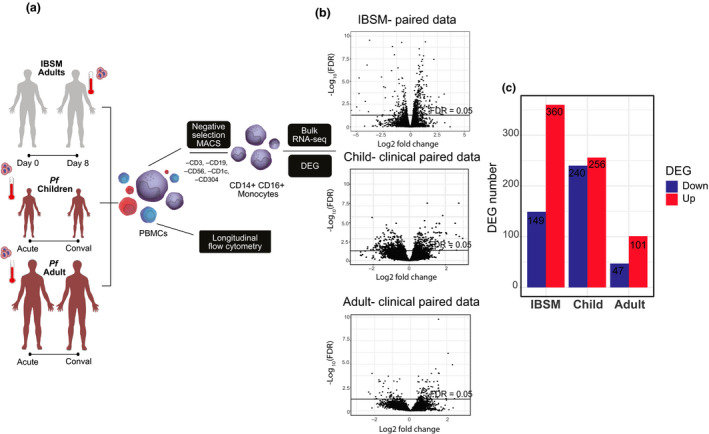
Cohort and RNA sequencing workflow. (a) Schematic of IBSM clinical trial cohort (grey) and acute clinical malaria trial cohorts with children (dark red) and adults (dark red). Monocytes were isolated by negative selection MACS separation and bulk‐RNAseq performed. Alongside, RNAseq monocyte number and activation were assessed by flow cytometry. (b) Total gene expression changes for IBSM, child and adult clinical cohorts, line indicates FDR < 0.05. (c) Total differentially expressed gene (DEG) numbers for the three cohorts, red indicates increased expression and blue indicates reduced expression. conval, convalescence (day 28); DEGs, differentially expressed genes; FC, fold change; FDR, false discovery rate; IBSM, induced blood‐stage malaria; PBMCs, peripheral blood mononuclear cells; Pf, P. falciparum.
Using IPA (Ingenuity Pathway Analysis), we identified DEG enrichment of canonical pathways, including IFN signalling, communication between innate and adaptive immune cells (including CD40), antigen presentation, neuroinflammation signalling (including CCL2, CD40, CXCR10), triggering receptor expressed on myeloid cells‐1 (TREM‐1) signalling, role of pattern recognition receptors in recognition of bacteria and viruses (including TLR1, TLR7, TLR8), activation of IRF by cytosolic pattern recognition receptors, phagosome formation [including FCGR1A (CD64)], FcγR‐mediated phagocytosis and production of reactive oxygen (RO) species [including CYBB, the gene for NADPH oxidase 2 (NOX2) and NCF1, another subunit of NADPH oxidase] (Figure 2a and c). STRING analysis revealed similar GO (Gene Ontology) biological pathways relating to innate immune response (including TLR1, TLR7, TLR8), cytokine‐mediated signalling, type I IFN signalling, regulation of immune system process, cell surface signalling (including FCGR1A, the gene for the high‐affinity Fc‐gamma receptor CD64; CD36, a receptor involved in non‐opsonised phagocytosis; and inflammatory chemotactic cytokines CXCL9, CXCL10), IFN‐γ‐mediated signalling, signal transduction, regulation of innate immune response [including CD40 and signal transduction (including chemokine CCL2 (MCP‐1) and its receptor CCR2)] (Figure 2c, Supplementary figure 1).
Figure 2.
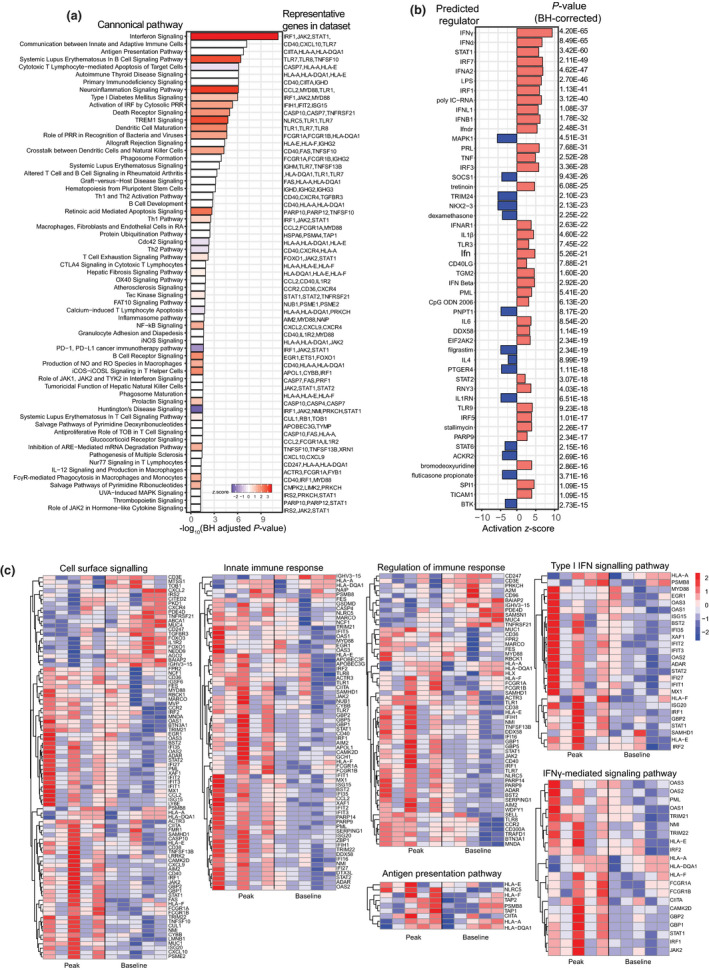
Transcriptional profile of monocytes after primary P. falciparum infection (IBSM cohort). (a) Canonical pathway analysis performed by IPA (Ingenuity Pathway Analysis), Benjamini–Hochberg‐adjusted significant pathways are shown, bar colour indicates activation z‐score (blue = reduced, white = 0 or no score and red = increased). (b) Predicted upstream regulators induced analysis performed using IPA, and red bars indicate activated, blue bars indicate inhibited, Benjamini–Hochberg‐adjusted P‐values. (c) Normalised gene expression of genes involved in enriched pathways represented in heat maps. Each column is one volunteer at ‘peak infection’ and ‘baseline’ (before infection). Red indicates increased gene expression, and blue indicates reduced gene expression. For all analyses, DEG input FDR < 0.01, n = 5 paired samples. DEGs, differentially expressed genes; FC, fold change; FDR, false discovery rate.
Using IPA, we further identified predicted upstream regulators of the DEGs (Figure 2b), with predicted upstream activation of IFN‐related genes (including IFN‐α, IFN‐γ, IRF7, IFNA2, IRF1, IFNL1, IFNB1, IRF3, IFNαR, STAT1), and other inflammatory cytokines (TNF, IL1B, IL6), pattern recognition receptors (TLR3 and TLR9), and TNF superfamily member CD40LG (CD40 ligand) (Figure 2b). Upstream regulators predicted as inhibited included mitogen‐activated protein kinase 1 (MAPK1), prostaglandin E2 receptor 4 (PTGER4), suppressor of cytokine signalling 1 (SOCS1) and interleukin 1 receptor antagonist (IL1RN), and tripartite‐motif protein 24 (TRIM24), the negative regulator of p53 stability. 21 Increased TP53 along with increased p53 gene expression in monocytes has recently been identified to attenuate malaria‐induced inflammation. 19
Across all identified pathways, the magnitude of response varied across participants (Figure 2c), consistent with the known heterogeneity of immune responses to primary Plasmodium infection in adults. 22 However, overall, the monocyte transcriptional profile during subpatent P. falciparum blood‐stage infection is dominated by an IFN‐driven signature accompanied by pathways relating to pathogen detection, phagocytosis, antimicrobial activity and antigen presentation.
Monocytes have an activated phenotype after a primary infection
TLR1, TLR7 and TLR8 signalling pathways were enriched in monocytes during primary P. falciparum infection (Figure 2). To assess whether these transcriptional changes resulted in functional changes to TLR responsiveness, monocyte cytokine production was measured following stimulation with TLR agonists and P. falciparum‐infected RBCs (Figure 3a). TLR responsiveness was assessed on total CD14+ monocytes because surface expression of CD16 (FcγRIII) is rapidly lost during short‐term culture and stimulation. 23 In response to TLR1/2 (PamCys2) or TLR4 (LPS) stimulation, monocytes produced a strong TNF and moderate IL‐12 response (Figure 3b), which increased at peak infection. In response to TLR7 (imiquimod) stimulation, monocytes had a modest TNF response, which significantly increased at peak infection (Figure 3b). TLR agonists induced low IL‐10 cytokine production which did not change following infection (Figure 3b). Increased expression of SLAMF7 on monocytes indicates activation specifically by TLR triggering. 24 Consistent with increased slamf7 gene expression (Figure 5c) and increased TLR responsiveness (Figure 3b), phenotypic expression of SLAMF7 was also increased at peak infection on classical monocytes (Figure 3c).
Figure 3.
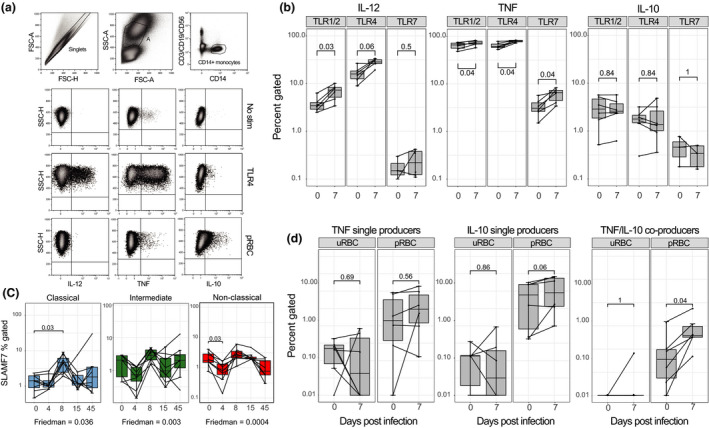
Monocytes increase cytokine production upon TLR stimulation during subpatent primary P. falciparum infection (IBSM cohort). (a) Representative gating of monocytes in whole‐blood intracellular cytokine assay. Monocytes were identified as lineage (CD3, CD19 and CD56)−, CD14+. Intracellular cytokine production by monocytes (IL‐12, TNF and IL‐10) in three conditions: no stimulation, TLR4 and pRBC stimulation. (b) Longitudinal monocyte responsiveness to TLR1/2 (Pam3CSK4/HKLM), TLR4 (LPS) or TLR7 (imiquimod) stimulation during IBSM, left plot; IL‐12 production, middle plot; TNF production, right plot; IL‐10 production. (c) Confirmation of SLAMF7 gene expression by flow cytometry on monocyte subsets: classical (blue), intermediate (green) and non‐classical (red), monocyte subset gating in Figure 4. The Friedman test was used to compare longitudinal data. (d) Monocyte cytokine production in response to uRBC or pRBC stimulation, left plot; TNF single producers, middle plot; IL‐10 single producers, right plot; TNF/IL‐10 co‐producers. Boxplot lower and upper hinges represent first and third quartiles with median line indicated across the box. Whisker lines correspond to highest and lowest values no further than 1.5 interquartile range from the hinges, whereas dot points beyond whisker lines are outliers. The Wilcoxon matched‐pairs sign‐rank test was used to compare paired data. Tests were two‐tailed and considered significant if P‐values < 0.05, n = 6 paired samples. FSC, forward scatter; pRBC, parasitised RBC; SSC, side scatter; TLR, toll‐like receptor; uRBC, uninfected RBC.
Figure 5.
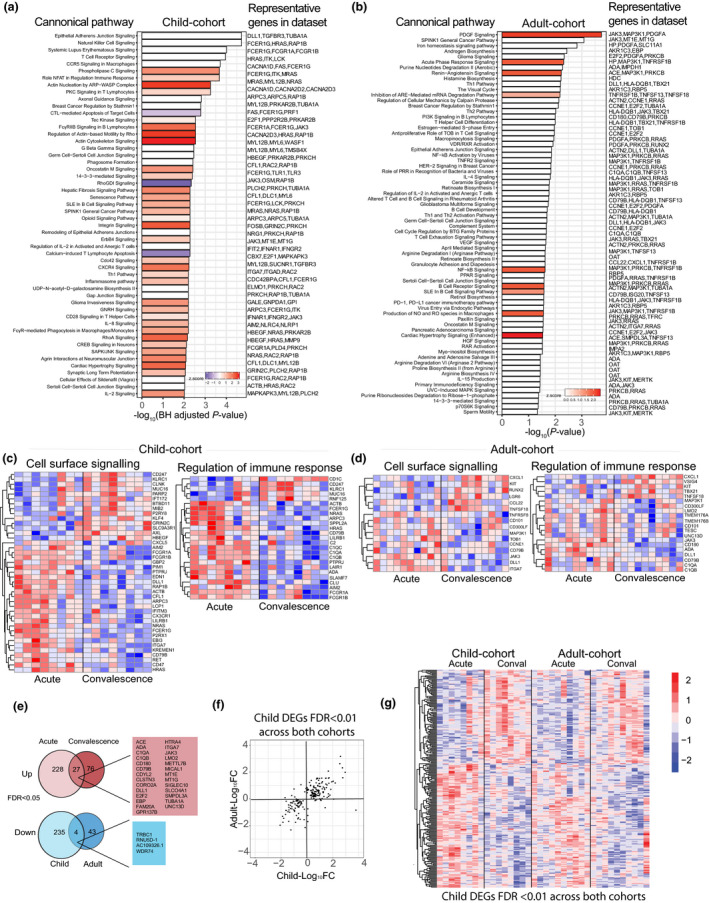
Transcriptional profile of monocytes in children and adults with acute clinical malaria (clinical malaria cohort). (a) Canonical pathway analysis performed by IPA, Benjamini–Hochberg‐adjusted significant pathways are shown, and bar colour indicates activation z‐score (blue = reduced, white = 0 or no score and red = increased). (b) Canonical pathway analysis performed by IPA, Fisher's exact test, significant pathways are shown, and bar colour indicates activation z‐score (blue = reduced, white = 0 or no score and red = increased). Normalised gene expression of genes involved in enriched pathways represented in heat maps. Each column is one patient at ‘acute’ infection and ‘convalescence’, 28 days after treatment. Red indicates increased gene expression, and blue indicates reduced gene expression. (c) Child cohort and (d) adult cohort. (e) Venn diagrams of DEGs in adult and child cohorts, red indicates increased gene expression and blue indicates reduced gene expression, FDR < 0.05. (f) Scatter plot showing fold change in DEGs across both adult and child cohorts, FDR < 0.01. (g) Heat map of normalised gene expression across child and adult acute malaria cohorts, FDR < 0.01, child cohort; n = 8 paired samples, adult cohort; n = 10 paired samples. conval, convalescence (day 28); DEGs, differentially expressed genes; FC, fold change; FDR, false discovery rate; Pf, P. falciparum.
Monocyte responsiveness to pRBC in vitro stimulation at baseline and at peak infection was also assessed. Prior to infection, pRBC stimulation resulted in low but detectable parasite‐specific TNF and IL‐10 production [P = 0.06 and 0.03 for pRBC versus uninfected RBC (uRBC), respectively; Supplementary figure 2]. At peak infection, IL‐10 production increased (P = 0.06), with a significant increase in TNF/IL‐10 co‐production in response to pRBC restimulation (P = 0.04) (Figure 3d). There was no IL‐12 production detected in response to pRBC or uRBC stimulation (data not shown).
Taken together, these data suggest that during a subpatent P. falciparum blood‐stage infection, monocytes responded to TLR stimulation with increased inflammatory cytokine production, while they co‐produced regulatory cytokines in response to restimulation with P. falciparum parasites, that is increased IL‐10 producers, suggesting that during subpatent P. falciparum malaria monocytes develop a regulatory capacity, potentially to minimise tissue damage following chronic activation. 25
Phenotypic changes to monocyte subsets persist after parasite clearance
To determine whether the transcriptional changes we identified translated to phenotypic changes in monocytes, we assessed monocyte subsets (Figure 4a) for cell surface marker expression, prior to, during and after infection by flow cytometry. Despite monocyte subset frequency normally being tightly regulated, 14 during a primary subpatent P. falciparum infection, classical (CD14+CD16−) monocyte frequency increased significantly on day 15 and remained significantly elevated at 45 days after infection (Figure 4b), whereas non‐classical (CD14dimCD16+) monocytes showed a corresponding decrease (Figure 4b). This increase in classical monocytes was also observed when classical monocytes were enumerated to all live PBMCs (Supplementary figure 3).
Figure 4.
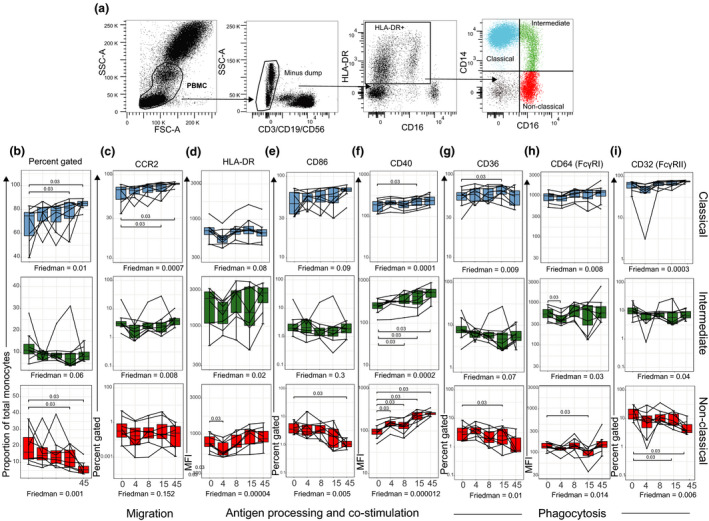
Differential activation of monocyte subsets during subpatent primary P. falciparum infection (IBSM cohort). (a) Fresh whole‐blood gating strategy for total monocyte subsets. Subsets were identified as lineage (CD3, CD19 and CD56)−, HLA‐DR+ and differential expression of CD14+ and CD16+. Classical (CD14+ CD16−), blue; intermediate (CD14+ CD16+), green; and non‐classical (CD14dimCD16+), red. (b) The proportion of monocyte subsets within the total monocyte population: classical (blue), intermediate (green) and non‐classical (red). Confirmation of genes differentially expressed in Figure 2 by flow cytometry, (c) CCR2 percent gated, (d) HLA‐DR MFI, (e) CD86 percent gated, (f) CD40 MFI, (g) CD36 percent gated, (h) CD64/FcyRI MFI and (i) CD32/FcyRII percent gated. Boxplot lower and upper hinges represent first and third quartiles with median line indicated across the box. Whisker lines correspond to highest and lowest values no further than 1.5 interquartile range from the hinges, whereas dot points beyond whisker lines are outliers. The Friedman test was used to compare longitudinal data. Tests were two‐tailed and considered significant if P‐values < 0.05, n = 8 paired samples. FSC, forward scatter; MFI, median fluorescence intensity; SSC, side scatter.
We further measured cell surface marker expression relating to activated pathways. CC‐chemokine receptor 2 (CCR2) gene expression was significantly increased in isolated monocytes at day 8 of blood‐stage infection. In accordance with the literature, 26 , 27 we observed CCR2 cell surface expression predominantly on classical monocytes, with a significant increase from day 15, which remained elevated at day 45 (Figure 4c). In line with the increased CD40 gene expression following P. falciparum infection, we observed increased CD40 cell surface expression on all three monocyte subsets, with expression also remaining significantly elevated 45 days after infection (Figure 4f). This increase was especially prominent on intermediate (CD14+CD16+) and non‐classical (CD14dimCD16+) monocytes. CD40 is a member of the TNF receptor family, and increased expression of CD40 on monocytes is reported to increase monocyte pro‐inflammatory cytokine production and expression of other co‐stimulatory receptors. 28 The increase in CD40 was not accompanied by an increase in co‐stimulatory molecule CD86 or HLA‐DR expression (Figure 4d and e).
We also assessed expression of CD36 which mediates non‐opsonic phagocytosis of parasites 29 and CD64/FcγRI and CD32/FcγRII which are involved in opsonic phagocytosis. 8 CD36 was differentially expressed across monocyte subsets, with classical monocytes expressing high levels of CD36, followed by intermediate monocytes, while < 10% of non‐classical monocytes expressed CD36 (Figure 4g). On classical monocytes, CD36 expression increased further by day 15 post‐infection. Classical and intermediate monocytes predominantly expressed CD64/FcγRI (Figure 4h). CD32/FcγRII expression was increased on classical monocytes but decreased on non‐classical monocytes, with significant changes observed at day 45 post‐infection (Figure 4i).
Taken together, these data indicate that the transcriptional changes observed during subpatent primary P. falciparum infection (Figures 1 and 2) result in multiple sustained changes to the composition and phenotypes of monocytes, suggestive of changes to innate immune effector mechanisms, antigen capture and cell migration pathways which are maintained after parasite clearance.
Monocyte gene expression following naturally acquired acute uncomplicated malaria
Following the investigation of monocyte responses during a first P. falciparum blood‐stage infection in previously malaria‐naive adult volunteers, we next isolated monocytes from ten adults and eight children living in malaria‐endemic Papua who presented to the clinic with acute P. falciparum malaria and again 28 days after malaria treatment and successful parasite clearance (Figure 1a, Table 1). Lowland Papua is an area of perennial, unstable malaria transmission, where both children and adults are susceptible to symptomatic malaria because of incomplete development of immunity.
Table 1.
Characteristics of participants
| Malaria‐naive volunteers (IBSM) | Malaria‐exposed adults (acute malaria) | Malaria‐exposed children (acute malaria) | P‐value a | |
|---|---|---|---|---|
| Number | 19 | 15 | 8 | |
| Median age in years [IQR] | 23 [20–26] | 30 [20–38] | 11 [8–13] | |
| Male, number (%) | 17 (89) | 10 (66) | 3 (38) | |
| Median parasite density b (parasites per μL) [IQR] | 10 [4–19] | 2275 [788–5712] | 7377 [2965–13 644] | 0.03 |
IBSM, induced blood‐stage malaria; IQR, interquartile range.
Mann–Whitney U‐test compared malaria‐exposed adults and malaria‐exposed children at presentation to clinic.
Determined by PCR for malaria‐naive adults (IBSM) and by microscopy for all other groups.
We performed a paired analysis of transcriptional changes in isolated monocytes via RNA sequencing allowing each patient's convalescence sample to act as their own control. Overall, children had more differentially expressed genes at acute infection than adults (children = 496, adults = 148, FDR < 0.05, Figure 1b and c, Supplementary tables 2 and 3). In children with acute malaria, more IPA canonical pathways were predicted to be upregulated, including actin‐related pathways, integrin, CXCR4, IL‐8 signalling and Fcγ receptor‐mediated phagocytosis in macrophages and monocytes (Figure 5a), reflective of the larger number of DEGs identified. As seen in subpatent primary P. falciparum infection (Figure 2b), IPA‐predicted activated upstream regulators included IFN‐γ‐related gene products, including IFNG and STAT1 in children (Supplementary figure 4b); in adults, IL‐4, IL‐6, the corresponding signal transducers STAT6 and STAT3, and triggering receptor expressed on myeloid cells‐1 (TREM1) (Supplementary figure 5b), a pathway activated in subpatent primary infection (Figure 2a). In adults with acute malaria, IPA canonical pathway analysis identified upregulation of pathways related to platelet‐derived growth factor (PDGF) signalling, acute‐phase response signalling [including HP, the gene for haptoglobin; TNFRSF1B, the gene for TNF receptor 2 (TNFR2); and mitogen‐activated protein kinase kinase kinase 1 (MAP3K1), a molecule involved in TNFR signalling] and NF‐κB signalling [including MAP3K1, TNFRSF1B and PRKCB (protein kinase C beta type)]. OAT, the gene for ornithine aminotransferase, an enzyme involved in L‐proline metabolism from L‐ornithine, was also differentially expressed in adults, suggestive of arginine degradation (Figure 5c).
As seen in experimental infection, STRING analysis showed DEGs were enriched for immune response pathways and more specifically, pathways involved in leucocyte activation [including Notch receptor ligand delta‐like 1 (DLL1) in both adults and children, and SLAMF7 and FCER1G, the Fc receptor gamma chain (FcRγ) 30 in children], regulation of immune response, complement activation and cell surface receptor signalling [including FCGR1A, the gene for the high‐affinity Fc‐gamma receptor CD64, and FCGR1B (CD64b) in children] (Supplementary figures 4 and 5c,d). More pathways were enriched in children when compared to adults (Figure 5a and c, Supplementary figures 4a and 5a). Of the DEGs, only 27 upregulated DEGs were shared between children and adults (FDR < 0.05, Figure 5e), including C1QA and C1QB encoding C1q, the first component of the classical complement pathway and DLL1.
Overall, despite not sharing many significantly deferentially expressed genes at FDR < 0.05, of the genes significantly differentially expressed in children with acute malaria, the direction of the fold change of each individual gene was positively correlated between children and adults (Figure 5f and g), suggesting that the pattern of gene changes is independent of age, but the magnitude of changes may be reduced in adults (Figure 5g).
Intermediate monocytes increased in children with clinical malaria
In addition to transcriptional changes, we also assessed monocyte subset phenotype in children and adults with malaria and at convalescence (Figure 6a). The proportion of intermediate (CD14+CD16+) monocytes was significantly higher in children during acute infection than convalescence (P = 0.02; Figure 6b), while we observed no significant changes in monocyte subsets in adult patients. The increase in intermediate monocytes in children during clinical infection is consistent with previous findings in uncomplicated malaria in Kenyan children, 16 during asymptomatic infection, 17 and in pregnant woman. 18 There were no statistically significant changes in markers associated with antigen presentation (HLA‐DR, Figure 6c) or co‐stimulatory capacity (CD86, Supplementary figure 6). However, consistent with transcriptional upregulation of CD64/FcγRI genes FCGR1A and FCER1G (common FcRγ chain) in children (Figure 5b), CD64/FcγRI cell surface expression on monocyte subsets was increased in both children and adults during malaria (Figure 6d). Expression of the haemoglobin/haptoglobin scavenger receptor CD163 on classical monocytes was significantly increased in children, but not adults with acute malaria (Figure 6e). While the CXCR4 signalling pathway was upregulated in children with acute malaria (Supplementary figure 4a), we saw no significant change in CXCR4 expression on the cell surface (mobilises monocytes to bone marrow) across any of the monocyte subsets during acute malaria infection (Supplementary figure 6). One predicted activated upstream regulator in adults was IL‐4 (Supplementary figure 5b). IL‐4 can polarise monocytes towards alternative activation and was recently associated with nitric oxide insufficiency and disease severity. 31 However, no significant change in monocyte surface expression of the mannose receptor CD206, a marker of alternatively activated monocytes, 32 was observed in children or adults with uncomplicated acute malaria (Supplementary figure 6c).
Figure 6.
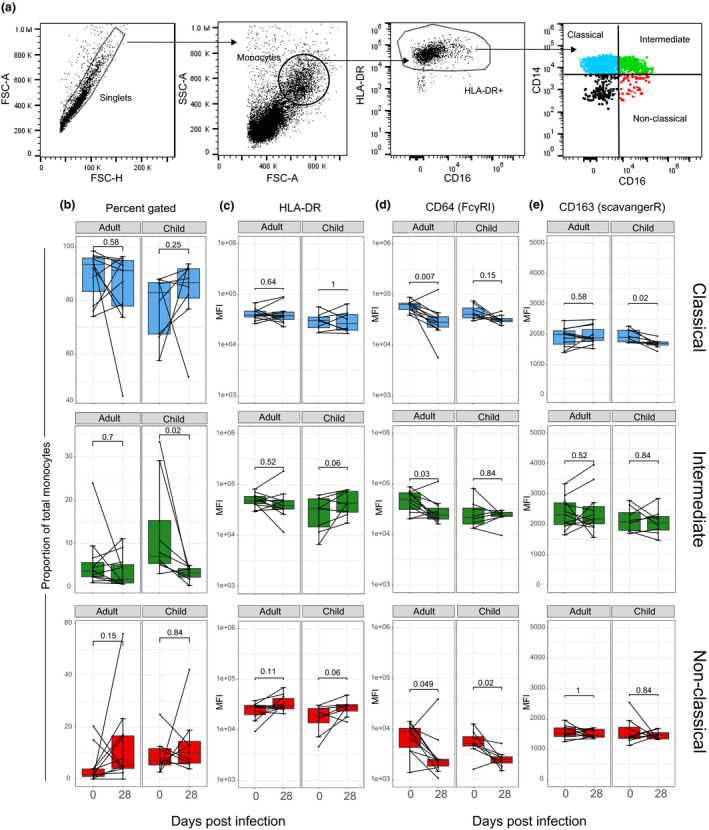
Increased intermediate monocytes in children with acute clinical malaria (clinical malaria cohort). (a) PBMC gating strategy for blood monocyte subsets. Monocyte subsets were identified by differential expression of CD14+ and CD16+. Classical (CD14+ CD16−), blue; intermediate (CD14+ CD16+), green; and non‐classical (CD14dimCD16+), red. (b) The proportion of monocyte subsets within the total monocyte population: classical (blue), intermediate (green) and non‐classical (red). Surface expression of (c) HLA‐DR MFI, (d) CD64/FcyRI MFI and (e) CD163 (scavenger receptor) MFI. Boxplot lower and upper hinges represent first and third quartiles with median line indicated across the box. Whisker lines correspond to highest and lowest values no further than 1.5 interquartile range from the hinges, whereas dot points beyond whisker lines are outliers. The Wilcoxon signed‐rank test was used to compare longitudinal data. Tests were two‐tailed and considered significant if P‐values < 0.05, child cohort; n = 8 paired, adult cohort; n = 11 paired samples. FSC, forward scatter; MFI, median fluorescence intensity; SSC, side scatter.
Discussion
Here, we evaluated transcriptional, phenotypic and functional monocyte responses in previously malaria‐naive volunteers undergoing their first P. falciparum blood‐stage infection. Our findings show that overall monocytes are transcriptionally, functionally and phenotypically activated during subpatent P. falciparum infection independent of Plasmodium liver stages. We identified robust activation of genes enriched in pathogen detection, phagocytosis, antimicrobial activity and antigen presentation. During primary infection, monocytes displayed heightened responsiveness to TLR stimulation and increased IL‐10 production upon re‐exposure to P. falciparum‐infected RBCs. Transcriptional activation resulted in compositional and phenotypical changes to monocyte subsets, suggesting altered migration, antigen presentation and phagocytosis function. In clinical malaria, the monocyte transcriptional response to infection appeared independent of age; despite more DEGs in children, the overall direction of transcriptional change was similar between adults and children. Together, these data provide an extensive map as to how human monocytes respond to P. falciparum infection and indicate monocytes have multiple roles in Plasmodium infection.
The monocyte response during subpatent primary P. falciparum was distinctively driven by a strong IFN molecular signature, including type I IFNs and gamma IFN, with the most activated pathways being interferon signalling and TREM1 signalling. The same two pathways were the most activated in a whole‐blood RNA sequencing analysis of healthy volunteers experiencing symptomatic malaria following controlled P. falciparum sporozoite infection. 33 This strong alignment with our findings in isolated monocytes suggests blood monocytes are the key responders during blood‐stage malaria despite their relatively small abundance amongst circulating white blood cells. Furthermore, we now report this response as activated at subpatent parasitaemia, independent of Plasmodium liver stages. We observed a strong inflammatory gene signature in isolated monocytes during subpatent infection, including increased CXCL10, CXCL9 and STAT1 gene expression. In contrast, the strong transcriptional response in monocytes we report here is absent in plasmacytoid dendritic cells, which have minimal gene expression changes within the same study cohorts. 34 These data extend previous research detailing PBMC or whole‐blood gene expression profiles in malaria patients 33 , 35 , 36 , 37 , 38 and are consistent with existing knowledge on the importance of monocytes in inflammatory responses during malaria. 19 , 20
In line with IFN‐γ having the highest activation score of the predicted upstream regulators, monocytes displayed a heightened responsiveness to TLR ligands during subpatent infection. This is in accordance with previous reports of PBMC hyper‐responsiveness to TLR ligation in febrile malaria. 39 , 40 In contrast, monocyte responsiveness to restimulation with parasite‐infected RBC is skewed towards a regulatory phenotype with increased IL‐10 production, similar to our previous reports for CD16+ dendritic cells (DCs). 23 The blunted inflammatory cytokine response to parasite restimulation may be explained by the enrichment of type I IFN signalling pathways, as IL‐6 production by monocytes is enhanced when type I IFN receptor is blocked. 41 During malaria, type I IFN is produced by multiple cell types including T cells, NK cells and monocytes. 41 While we did not evaluate monocyte cytokine production in children with uncomplicated clinical malaria, previous studies have demonstrated that monocytes isolated from children with uncomplicated malaria show increased IL‐12, IL‐6 and TNF cytokine production in response to TLR1/2 or TLR4 stimulation. 16 In contrast, a separate study showed that monocytes from children with severe malaria had an impaired ability to produce inflammatory cytokines (TNF and IL‐6) to in vitro TLR agonists such as LPS. 10 These differences in TLR responsiveness suggest that TLR signalling is influenced by parasite density and/or parasite exposure.
Besides IFNs, the CD40/CD40 ligand axis may contribute to monocyte activation following Plasmodium infection, with CD40LG being amongst the predicted activated upstream regulators. CD40 is considered a marker of monocyte activation, and CD40 ligation increases monocyte production of inflammatory cytokines 28 and prostaglandin E2. 42 CD40 may further contribute to monocyte interaction with activated endothelial cell and assist extravasation. 43 We demonstrated upregulation of its ligand CD40 at the protein level on all three monocyte subsets, with the greatest increase seen on non‐classical monocytes, a monocyte subset that proportionally declined in the circulation over time. Whether this decline is related to extravasation or sequestration remains to be determined.
Predicted inhibited upstream regulators of monocyte gene expression in subpatent primary infection included MAPK1, IL1RN and TRIM24, which overlapped with regulators predicted to be inhibited in malaria‐naive volunteers experiencing symptomatic malaria following P. falciparum sporozoite infection. 33 TRIM24 targets p53 degradation, 21 a molecule recently reported to play a role in clinical immunity, as p53 is upregulated at RNA and protein level in asymptomatic P. falciparum carriers during the malaria season in Mali. 19 The congruent overlap of upstream regulators between these studies suggests these predicted inhibited upstream regulators take effect early during blood‐stage infection, independent of P. falciparum liver stages, and warrants further investigation into the control (and timing) of upstream regulators.
Multiple lines of evidence suggest that during malaria, monocytes also have important roles in initiating adaptive responses. 8 , 44 , 45 Previous studies have shown that monocytes increase expression of B‐cell‐activating factor (BAFF) during experimental P. falciparum infection with sporozoites, 46 suggesting monocytes may contribute to B‐cell activation. 47 In this study, monocyte transcriptional profiling indicated a predominantly MHC class I antigen presentation pathway signature, suggesting monocytes may contribute to the activation of CD8 T cells that have recently been shown to kill Plasmodium‐infected reticulocytes 48 and parasitised erythroblasts in a murine malaria model. 49 Further, HLA‐DR expression on monocytes during infection was retained, consistent with our previous studies where monocytes retain their ability to upregulate HLA‐DR expression in response to TLR ligands or pRBC during subpatent primary P. falciparum infection. 50 In contrast, during both experimental subpatent infection and clinical malaria, HLA‐DR expression is reduced on classical CD1c+ DC, 50 , 51 which are typically considered superior antigen‐presenting cells. 52 Similarly, we have previously reported that clinical malaria drives impaired DC function and increased apoptosis. 51 Thus, together data support a role of monocytes as important antigen‐presenting cells, in contrast to DCs, for initiation of adaptive T‐ and B‐cell immune responses during malaria.
Plasmodium falciparum infection resulted in changes in monocyte composition and phenotype which was sustained for up to 1 month after a first infection. These results suggest that P. falciparum may drive ‘innate immune training’, 53 in which innate immune cells such as monocytes undergo specific changes in their chromatin profiles induced by pathogen stimulation. 53 Sustained changes seen here include an increased proportion of classical (CD14+CD16−) monocytes and reduced proportions of non‐classical (CD14−CD16+) monocytes in the periphery until at least day 45 following infection. In homeostasis, monocytes migrate from the bone marrow and can circulate in the periphery for approximately 1 day. 54 Subsequently, 90% of monocytes migrate to tissues and become macrophages, while 10% differentiate to intermediate monocytes. The elevated proportion of classical monocytes in the periphery following P. falciparum infection may be a consequence of a sustained increase in bone marrow production of monocytes. Consistent with this, we show increased transcription and cell surface expression of CCR2, which mediates monocyte emigration from the bone marrow. 26 Changes to the composition of monocytes occurred simultaneously with multiple phenotypic changes that were also maintained up to 45 days following infection. These sustained changes are consistent with monocyte training or imprinting. 53 , 55 Long‐term effects on monocytes post‐infection have been shown in both Q‐fever 56 and human Toxoplasma gondii infection. 57 These long‐term effects suggest these infections may influence the transcriptional programme of monocytes and potentially their myeloid progenitors. In support of this notion, heightened monocyte transcriptional responsiveness in Fulani populations infected with P. falciparum was suggested to be indicative of genomewide chromatin remodelling. 20 Future studies investigating parasite‐driven changes to the epigenetic landscape of monocytes following primary infection may be informative.
During clinical malaria, the monocyte transcriptional profile had both similarities (e.g. cell surface signalling and regulation of immune response) and differences (e.g. type I IFN signalling) to that seen during primary subpatent infection. Phagocytosis pathway was one pathway upregulated in both malaria cohorts and is a key function of monocytes in protective immunity. 58 In primary infection, both transcriptional pathways and cell surface expression of CD64/FcγRI and CD32/FcγRII on monocytes changed significantly and in clinical malaria CD64/FcγRI expression increased. These findings are in contrast with a previous study of Kenyan children with uncomplicated malaria, where monocyte phagocytosis of pRBC was impaired during both symptomatic infection and asymptomatic infection. 16 Differences between studies may be explained by differences in malaria transmission (perennial in lowland Papua; seasonal in Kenya), age of study participants or technical differences.
In acute clinical malaria, we show children have higher transcriptional activation of monocytes and more DEGs than adults. Despite greater transcriptional activation, the upstream regulators predicted for adults and children were similarly inflammatory, including IFN‐γ in children and IL‐6 and TREM1 (triggering receptor expressed on myeloid cells‐1) in adults. TREM1 is associated with amplifying the inflammatory response in sepsis 59 and cancer. 60 Moreover, the overall direction of transcriptional expression remained similar between children and adults, which suggests the magnitude of transcription could be influenced by parasite load, which was higher in children or that adults have an attenuated response as a consequence of prior infection, age and time to presentation, but otherwise similar responses to infection.
In summary, using transcriptional, functional and phenotypic analyses, we reveal monocyte function during primary subpatent infection as dominated by an IFN molecular signature, with increased proportion of activated classical monocytes. Alongside this inflammatory phenotype, monocytes increased IL‐10 production upon re‐exposure to parasite‐infected RBCs, suggesting a role in controlling inflammation upon multiple parasite exposure. Of note, malaria‐naive volunteers maintained phenotypic changes to monocytes up to 1 month post‐infection. Further studies are needed to understand whether these sustained changes are maintained upon repeat infection in previously malaria‐naive volunteers. Differences in transcriptional activation observed between adults and children with clinical malaria warrant further investigation to understand the biological basis driving these differences and the impact on immunity. Our data suggest that monocytes are active and integral immune cells assisting parasite elimination and controlling inflammation in malaria.
Methods
Experimental infection using induced blood‐stage malaria (IBSM)
Nineteen volunteers aged 18–31 years (median 23 [IQR 20–26] years, 89% male) consented to participate in a phase Ib clinical trial testing the efficacy of antimalarial drugs. All studies were registered with US NIH ClinicalTrails.gov (ACTRN12613000565741, ACTRN12613001040752, NCT02281344 and NCT03542149). Blood‐stage parasitaemia was initiated by inoculation of 1800 or 2800 parasitised RBCs (pRBC) as previously described. 50 In brief, healthy malaria‐naive individuals underwent induced blood‐stage malaria inoculation with 1800 or 2800 viable P. falciparum 3D7‐pRBCs, and peripheral parasitaemia was measured by qPCR as described previously. 61 Participants were treated with antimalarial drugs at day 7 or day 8 of infection, when parasitaemia reached median 10, 346 [IQR 4303–19, 681] parasites per mL (Table 1). Blood samples from 19 volunteers (across four independent studies) were collected prior to infection (day 0), at peak infection (day 7/8), post‐infection 15 and 45 days (end of study, EOS) after inoculation (in analyses, these time points are grouped as 0, 7/8, 15 and 45). Blood samples were taken at the same time each day before and during blood‐stage infection. Flow cytometry assays used fresh whole blood and were processed within 2 h of collection (Figure 1a).
Clinical malaria cohorts
Peripheral blood mononuclear cells (PBMCs) were collected from 15 adults aged 18–38 years (median 30 [IQR 20–38] years; 66% male) and eight children aged 5–13, (median 11 [IQR 8–13] years; 38% male) with acute uncomplicated P. falciparum malaria as part of artemisinin combination therapy efficacy studies conducted in southern Papua, Indonesia. 62 At presentation to the clinic, the median parasitaemia was 2275 [interquartile range IQR = 788–5712] parasites per µL in adults and 7377 [IQR = 2965–13, 644] parasites per µL in children. Gene expression, phenotype and activation of monocytes were assessed in cryopreserved PBMC samples collected prior to commencing treatment, and again 28 days after antimalarial drug treatment and parasite clearance (Figure 1a).
Ethics approval
IBSM was approved by the Human Research Ethics Committees of QIMR Berghofer Medical Research, NT Department of Health and Menzies School of Health Research. In the clinical malaria cohorts, written informed consent was obtained from participants, with the study approved by the Human Research Ethics Committees of the National Institute of Health Research and Development, Indonesian Ministry of Health (Jakarta, Indonesia), the NT Department of Health and Menzies School of Health Research.
Monocyte isolation and RNA sequencing
RNA sequencing was performed on paired samples collected prior to and at peak infection from five subjects experimentally infected with P. falciparum pRBC and on acute and convalescence samples from adults and children with acute clinical malaria. IBSM and clinical samples were selected based on the availability of PBMCs at paired time points. Isolation and RNA sequencing for IBSM and clinical samples were performed at separate times by different operators. Monocytes were isolated from PBMCs using the Pan Monocyte Isolation Kit (Miltenyi Biotec, Gladbach, Germany). In brief, monocytes were enriched by indirect magnetic labelling for the isolation of untouched monocytes (all subsets), from human PBMCs. Non‐monocytes, such as T cells, NK cells, B cells, dendritic cells and basophils, were indirectly magnetically labelled using a cocktail of biotin‐conjugated antibodies to anti‐biotin microbeads. Conjugated microbeads and untouched monocytes were washed over MAC isolation columns. Purity of isolated monocyte populations was checked using flow cytometry IBSM (baseline; median 90% [IQR = 84–92], peak infection; median 93% [IQR = 88–96]) and clinical cohorts (acute infection 96% [IQR = 87–98], convalescence 94% [IQR = 86–95]). The distribution of subsets in isolated monocytes was comparable to pre‐sorted populations (Supplementary figure 7). Isolated monocytes were resuspended in RNAprotect (Qiagen, Hilden, Germany) and stored immediately at −80 °C. RNA extraction and RNA sequencing of five paired IBSM study participants, eight paired children (clinical malaria) and ten paired adult (clinical malaria) samples were conducted by Macrogen© (Seoul, Korea) using the Illumina TruSeq Stranded mRNA LT Sample Kit and the HiSeq 2500 instrument. Transcriptome data were analysed using a modified version of an existing variant detection pipeline 63 consisting of software STAR aligner, 64 samtools, 65 HTSeq 66 and DESeq2. 67 Reads were first aligned to human reference genome GRCh37 using STAR with the gene model set to gencode v19 annotation and quantmode set to TranscriptomeSAM. The alignment files were sorted with samtools and the resultant reads input to HTSeq to generate raw read counts using the union overlap resolution mode. Read counts were input to DESeq2 and a paired analysis performed for all participants with acute infection and baseline or convalescence data.
Whole‐blood and PBMC monocyte analysis
Monocytes were characterised as lineage (CD3, CD56, CD19)‐negative, HLA‐DR+ and by differential expression of CD14 and CD16. In brief, 200 µL of whole blood or 1 million PBMCs were stained with surface antibodies, CD3 (HIT3a, SK7), CD14 (HCD14, M5E2), CD19 (HIB19, SJ25C1), CD56 (HCD56), HLA‐DR (L243), CD11c (B‐Ly6, Bu15), CD16 (3G8), CD86 (2331, IT2.2), CD319 (SLAMF7, 162.1), CD40 (5C3), CD32 (FUN‐2), CD64 (10.1), CCR2 (K036C2), CD206 (15‐2), CXCR4 (12G5) and CD163 (GHI/61), all antibodies were purchased from BD Biosciences (San Jose, CA, USA) or BioLegend (San Diego, CA, USA). For whole blood, RBCs were lysed with FACS lysing solution (BD Biosciences) and resuspended in 2% FCS/PBS to acquire.
Monocyte stimulation and intracellular cytokine staining (ICS)
Monocyte cytokine production was assessed in 1 mL of fresh whole blood unstimulated or stimulated with TLR agonists; TLR1: Pam3CSK4 100 ng per mL/TLR2: HKLM 108 cells per mL, TLR4: Escherichia coli K12 LPS 200 ng mL−1 or TLR7: imiquimod 2.5 µg mL−1 (Sigma‐Aldrich, St Louis, MO, USA), pRBC or unparasitised RBC (uRBC) prepared as previously described 50 at 5 × 106/mL. Protein transport inhibitor (Brefeldin A, GolgiPlug, BD Biosciences) was added after 2 h at 37 °C, 5% CO2. At 6 h, cells were stained to identify monocytes: lineage [CD3 (HIT3a), CD19 (HIB19), CD56 (HCD56)]− and CD14+ (M5E2), and RBCs were lysed with FACS lysing solution (BD Biosciences), washed with 2% FCS/PBS, cells permeabilised with 1× Perm/Wash™ (BD Biosciences) and stained with intracellular anti‐TNF‐α (MAB11), IL‐12/IL‐23p40 (C11.5), IL‐10 (JES3‐9D7) or IgG1 isotype controls (BioLegend). To determine the stimulant‐specific response, spontaneous cytokine production was subtracted from responses to pRBC, uRBC or TLR agonists.
FACS data were acquired using a FACSCanto™ II (BD Biosciences), Gallios™ (Beckman Coulter, Brea, CA, USA) or Fortessa 5 laser (BD Biosciences) and data analysed using Kaluza® 1.3 (Beckman Coulter) or FlowJo version 10.6 (BD Biosciences).
Statistics
Statistical analyses and graph generation performed using R studio and GraphPad Prism. The Friedman multiple comparisons test was used to compare longitudinal data in the IBSM cohort. For post hoc statistics, a pairwise sign test with the Bonferroni correction for multiple comparisons was performed. For clinical cohorts, the Wilcoxon matched‐pairs signed‐rank test was used as paired data. Tests were two‐tailed and considered significant if P‐values < 0.05. For pathway enrichment, the online tool STRING version 11 68 was used to identify GO biological pathways, and for all analysis, an interaction of high confidence was employed (score ≥ 0.7). Ingenuity Pathway Analysis (IPA) (Qiagen, Hilden, Germany) was used for canonical pathway enrichment and predicted upstream regulator analysis using DEGs with a false detection rate (FDR) < 0.05 unless stated otherwise, with no fold‐change cut‐off.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Jessica R Loughland: Formal analysis; Investigation; Visualization; Writing‐original draft; Writing‐review & editing. Tonia Woodberry: Conceptualization; Funding acquisition; Investigation; Methodology; Supervision; Writing‐review & editing. Matt Field: Data curation; Formal analysis; Methodology; Software; Writing‐review & editing. Dean W Andrew: Methodology; Writing‐review & editing. Arya SheelaNair: Methodology; Writing‐review & editing. Nicholas L Dooley: Methodology; Writing‐review & editing. Kim A Piera: Methodology; Project administration; Writing‐review & editing. Fiona H Amante: Project administration; Writing‐review & editing. Enny Kenangalem: Project administration; Supervision; Writing‐review & editing. Ric N Price: Funding acquisition; Project administration; Resources; Writing‐review & editing. Christian R Engwerda: Funding acquisition; Project administration; Resources; Writing‐review & editing. Nicholas M Anstey: Conceptualization; Funding acquisition; Project administration; Resources; Writing‐review & editing. James S McCarthy: Funding acquisition; Project administration; Resources; Writing‐review & editing. Michelle J Boyle: Funding acquisition; Investigation; Resources; Supervision; Writing‐review & editing. Gabriela Minigo: Conceptualization; Investigation; Methodology; Supervision; Writing‐review & editing.
Supporting information
Supplementary figures 1‐7
Supplementary tables 1‐3
Acknowledgments
We thank the Q‐pharm staff who conducted the human blood‐stage infection studies and in particular Dr Suzanne Elliot, Nannette Douglas and Gem Mackenroth for supporting the research. We thank Dr Paul Griffin for advice and technical assistance. We thank all staff with the Papuan Community Health and Development Foundation, and all teams involved in the Timika clinical malaria studies. We thank Dr Derek Sarovich for advice on study design and Fabian de Labastida Rivera for laboratory support. We thank the Australian Red Cross Blood Service for their support. We thank the volunteers who participated in all clinical trials and Medicine for Malaria Venture for funding these studies. This work was funded by the National Health and Medical Research Council (NHMRC, Program Grant 1132975 to NMA, JSM, CRE; Program Grants 1037304 and 1092789; Project Grant 1045156; and Senior Principal Research Fellowship 1042072 to NMA, Practitioner Fellowship 1135955 to JSM, Senior Research Fellowship 1154265 to CRE, Project Grant 1125656 and Career Development Award 141632 to MJB, and MF is funded by APP5121190). RP is a Wellcome Trust Senior Fellow in Clinical Science (200909) and Channel 7 Children's Research Foundation (Grant 151016). The clinical trials that enabled sample collection were funded by the Medicines for Malaria Venture. The Timika Malaria Research Program is supported by the Australian Department of Foreign Affairs. The views expressed in this publication are those of the authors and do not reflect the views of the NHMRC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. WHO . World Malaria Report. Geneva: World Health Organisation, 2019. [Google Scholar]
- 2. Rodriguez‐Barraquer I, Arinaitwe E, Jagannathan P et al Quantification of anti‐parasite and anti‐disease immunity to malaria as a function of age and exposure. Elife 2018; 7: e35832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rogerson S, Ortega‐Pajares A. The rough guide to monocytes in malaria infection. Front Immunol 2018; 9: 2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dobbs KR, Crabtree JN, Dent AE. Innate immunity to malaria—the role of monocytes. Immunol Rev 2020; 293: 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Mahony DS, Pham U, Iyer R, Hawn TR, Liles WC. Differential constitutive and cytokine‐modulated expression of human Toll‐like receptors in primary neutrophils, monocytes, and macrophages. Int J Med Sci 2008; 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baccarella A, Fontana MF, Chen EC, Kim CC. Toll‐like receptor 7 mediates early innate immune responses to malaria. Infect Immun 2013; 81: 4431–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gazzinelli RT, Kalantari P, Fitzgerald KA, Golenbock DT. Innate sensing of malaria parasites. Nat Rev Immunol 2014; 14: 744–757. [DOI] [PubMed] [Google Scholar]
- 8. Zhou J, Feng G, Beeson J et al CD14hi CD16+ monocytes phagocytose antibody‐opsonised Plasmodium falciparum infected erythrocytes more efficiently than other monocyte subsets, and require CD16 and complement to do so. BMC Med 2015; 13: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eriksson E, Sampaio N, Schofield L. Toll‐like receptors and malaria‐sensing and susceptibility. J Trop Dis 2013; 2: 126. [Google Scholar]
- 10. Mandala WL, Msefula CL, Gondwe E et al Monocyte activation and cytokine production in Malawian children presenting with P. falciparum malaria. Parasite Immunol 2016; 38: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chua CLL, Brown G, Hamilton JA, Rogerson S, Boeuf P. Monocytes and macrophages in malaria: protection or pathology? Trends Parasitol 2013; 29: 26–34. [DOI] [PubMed] [Google Scholar]
- 12. McGuire W, D'Alessandro U, Stephens S et al Levels of tumour necrosis factor and soluble TNF receptors during malaria fever episodes in the community. Trans R Soc Trop Med Hyg 1998; 92: 50–53. [DOI] [PubMed] [Google Scholar]
- 13. Ziegler‐Heitbrock L, Ancuta P, Crowe S et al Nomenclature of monocytes and dendritic cells in blood. Blood 2010; 116: e74–e80. [DOI] [PubMed] [Google Scholar]
- 14. Boyette LB, Macedo C, Hadi K et al Phenotype, function, and differentiation potential of human monocyte subsets. PLoS One 2017; 12: e0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cros J, Cagnard N, Woollard K et al Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 2010; 33: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobbs KR, Embury P, Vulule J et al Monocyte dysregulation and systemic inflammation during pediatric falciparum malaria. JCI Insight 2017; 2: e95352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fontana MF, Baccarella A, Craft JF et al A novel model of asymptomatic Plasmodium parasitemia that recapitulates elements of the human immune response to chronic infection. PLoS One 2016; 11: e0162132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaworowski A, Kamwendo DD, Ellery P et al CD16+ monocyte subset preferentially harbors HIV‐1 and is expanded in pregnant Malawian women with Plasmodium falciparum malaria and HIV‐1 infection. J Infect Dis 2007; 196: 38–42. [DOI] [PubMed] [Google Scholar]
- 19. Tran TM, Guha R, Portugal S et al A molecular signature in blood reveals a role for p53 in regulating malaria‐induced inflammation. Immunity 2019; 51: 750–765.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quin JE, Bujila I, Chérif M et al Major transcriptional changes observed in the Fulani, an ethnic group less susceptible to malaria. Elife 2017; 6: e29156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allton K, Jain AK, Herz H‐M et al Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci USA 2009; 106: 11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yap XZ, McCall MB, Sauerwein RW. Fast and fierce versus slow and smooth: heterogeneity in immune responses to Plasmodium in the controlled human malaria infection model. Immunol Rev 2020; 293: 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loughland JR, Woodberry T, Boyle MJ et al Plasmodium falciparum activates CD16+ dendritic cells to produce tumor necrosis factor and interleukin‐10 in subpatent malaria. J Infect Dis 2018; 219: 660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim JR, Horton NC, Mathew SO, Mathew PA. CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes. Inflamm Res 2013; 62: 765–772. [DOI] [PubMed] [Google Scholar]
- 25. Guilliams M, Movahedi K, Bosschaerts T et al IL‐10 dampens TNF/inducible nitric oxide synthase‐producing dendritic cell‐mediated pathogenicity during parasitic infection. J Immunol 2009; 182: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 26. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 2011; 11: 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003; 19: 71–82. [DOI] [PubMed] [Google Scholar]
- 28. van Kooten C, Banchereau J. CD40‐CD40 ligand. J Leukoc Biol 2000; 67: 2–17. [DOI] [PubMed] [Google Scholar]
- 29. McGilvray ID, Serghides L, Kapus A, Rotstein OD, Kain KC. Nonopsonic monocyte/macrophage phagocytosis of Plasmodium falciparum–parasitized erythrocytes: a role for CD36 in malarial clearance. Blood 2000; 96: 3231–3240. [PubMed] [Google Scholar]
- 30. Brandsma AM, Hogarth PM, Nimmerjahn F, Leusen JH. Clarifying the confusion between cytokine and Fc receptor “common gamma chain”. Immunity 2016; 45: 225–226. [DOI] [PubMed] [Google Scholar]
- 31. Weinberg JB, Volkheimer AD, Rubach MP et al Monocyte polarization in children with falciparum malaria: relationship to nitric oxide insufficiency and disease severity. Sci Rep 2016; 6: 29151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem 2012; 287: 21816–21825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tran TM, Jones MB, Ongoiba A et al Transcriptomic evidence for modulation of host inflammatory responses during febrile Plasmodium falciparum malaria. Sci Rep 2016; 6: 31291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loughland JR, Minigo G, Sarovich DS et al Plasmacytoid dendritic cells appear inactive during sub‐microscopic Plasmodium falciparum blood‐stage infection, yet retain their ability to respond to TLR stimulation. Sci Rep 2017; 7: 2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ockenhouse CF, Hu W‐C, Kester KE et al Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun 2006; 74: 5561–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffiths MJ, Shafi MJ, Popper SJ et al Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis 2005; 191: 1599–1611. [DOI] [PubMed] [Google Scholar]
- 37. Lee HJ, Georgiadou A, Walther M et al Integrated pathogen load and dual transcriptome analysis of systemic host‐pathogen interactions in severe malaria. Sci Transl Med 2018; 10: eaar3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thiam A, Sanka M, Diallo RN et al Gene expression profiling in blood from cerebral malaria patients and mild malaria patients living in Senegal. BMC Med Genomics 2019; 12: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCall MB, Netea MG, Hermsen CC et al Plasmodium falciparum infection causes proinflammatory priming of human TLR responses. J Immunol 2007; 179: 162–171. [DOI] [PubMed] [Google Scholar]
- 40. Franklin BS, Parroche P, Ataíde MA et al Malaria primes the innate immune response due to interferon‐γ induced enhancement of toll‐like receptor expression and function. Proc Natl Acad Sci USA 2009; 106: 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Oca MM, Kumar R, de Labastida Rivera F et al Type I interferons regulate immune responses in humans with blood‐stage Plasmodium falciparum infection. Cell Rep 2016; 17: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inoue Y, Otsuka T, Niiro H et al Novel regulatory mechanisms of CD40‐induced prostanoid synthesis by IL‐4 and IL‐10 in human monocytes. J Immunol 2004; 172: 2147–2154. [DOI] [PubMed] [Google Scholar]
- 43. Wagner AH, Güldenzoph B, Lienenlüke B, Hecker M. CD154/CD40‐mediated expression of CD154 in endothelial cells: consequences for endothelial cell–monocyte interaction. Arterioscler Thromb Vasc Biol 2004; 24: 715–720. [DOI] [PubMed] [Google Scholar]
- 44. Chan JA, Stanisic DI, Duffy MF et al Patterns of protective associations differ for antibodies to P. falciparum‐infected erythrocytes and merozoites in immunity against malaria in children. Eur J Immunol 2017; 47: 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osier FH, Feng G, Boyle MJ et al Opsonic phagocytosis of Plasmodium falciparum merozoites: mechanism in human immunity and a correlate of protection against malaria. BMC Med 2014; 12: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teirlinck AC, Roestenberg M, Bijker EM et al Plasmodium falciparum infection of human volunteers activates monocytes and CD16+ dendritic cells and induces upregulation of CD16 and CD1c expression. Infect Immun 2015; 83: 3732–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scholzen A, Teirlinck AC, Bijker EM et al BAFF and BAFF receptor levels correlate with B cell subset activation and redistribution in controlled human malaria infection. J Immunol 2014; 192: 3719–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Junqueira C, Barbosa CR, Costa PA et al Cytotoxic CD8+ T cells recognize and kill Plasmodium vivax–infected reticulocytes. Nat Med 2018; 24: 1330–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Imai T, Ishida H, Suzue K et al Cytotoxic activities of CD8+ T cells collaborate with macrophages to protect against blood‐stage murine malaria. Elife 2015; 4: e04232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loughland JR, Minigo G, Burel J et al Profoundly reduced CD1c+ myeloid dendritic cell HLA‐DR and CD86 expression and increased TNF production in experimental human blood‐stage malaria infection. Infect Immun 2016; 84: 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pinzon‐Charry A, Woodberry T, Kienzle V et al Apoptosis and dysfunction of blood dendritic cells in patients with falciparum and vivax malaria. J Exp Med 2013; 210: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hart DNJ. Dendritic cells: unique leukocyte populations which control the primary immune response. Blood 1997; 90: 3245–3287. [PubMed] [Google Scholar]
- 53. Netea MG, Joosten LA, Latz E et al Trained immunity: a program of innate immune memory in health and disease. Science 2016; 352: aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patel AA, Zhang Y, Fullerton JN et al The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017; 214: 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schrum JE, Crabtree JN, Dobbs KR et al Cutting edge: Plasmodium falciparum induces trained innate immunity. J Immunol 2018; 200: 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raijmakers RP, Stenos J, Keijmel SP et al Long‐lasting transcriptional changes in circulating monocytes of acute Q fever patients. Open Forum Infect Dis 2019; 6: ofz296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ehmen HG, Lüder CG. Long‐term impact of Toxoplasma gondii infection on human monocytes. Front Cell Infect Microbiol 2019; 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Götz A, San Tang M, Ty MC et al Atypical activation of dendritic cells by Plasmodium falciparum . Proc Natl Acad Sci USA 2017; 114: E10568–E10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dimopoulou I, Pelekanou A, Mavrou I et al Early serum levels of soluble triggering receptor expressed on myeloid cells–1 in septic patients: correlation with monocyte gene expression. J Crit Care 2012; 27: 294–300. [DOI] [PubMed] [Google Scholar]
- 60. Tammaro A, Derive M, Gibot S et al TREM‐1 and its potential ligands in non‐infectious diseases: from biology to clinical perspectives. Pharmacol Ther 2017; 177: 81–95. [DOI] [PubMed] [Google Scholar]
- 61. Rockett RJ, Tozer SJ, Peatey C et al A real‐time, quantitative PCR method using hydrolysis probes for the monitoring of Plasmodium falciparum load in experimentally infected human volunteers. Malar J 2011; 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ratcliff A, Siswantoro H, Kenangalem E et al Two fixed‐dose artemisinin combinations for drug‐resistant falciparum and vivax malaria in Papua, Indonesia: an open‐label randomised comparison. Lancet 2007; 369: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Field MA, Cho V, Andrews TD, Goodnow CC. Reliably detecting clinically important variants requires both combined variant calls and optimized filtering strategies. PLoS One 2015; 10: e0143199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dobin A, Davis CA, Schlesinger F et al STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li H, Handsaker B, Wysoker A et al The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics 2014; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 2014; 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Szklarczyk D, Gable AL, Lyon D et al STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res 2018; 47: D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures 1‐7
Supplementary tables 1‐3


