Abstract
There is an unmet need for treatments for diseases associated with aging. The anti-aging, life-extending and cognition-enhancing protein Klotho is neuroprotective due to its anti-inflammatory, anti-oxidative and pro-myelinating effects. In addition, Klotho is also a tumor suppressor and has beneficial roles in multiple organs. Klotho is downregulated as part of the aging process. Thus, upregulating Klotho in the brain may lead to novel therapeutics to people suffering or at risk for neurodegenerative diseases such as Alzheimer’s, Parkinson’s and amyotrophic lateral sclerosis, and demyelinating diseases such as multiple sclerosis. We attempted to upregulate Klotho for its beneficial effects in the brain and elsewhere. Here we describe a method to specifically activate Klotho gene expression. To accomplish this task, we designed zinc finger proteins (ZFPs) targeting within −300 bps of the human Klotho promoter. We designed the ZPF constructs either de novo from modular building blocks, or modified sequences from the natural endogenous Egr1 transcription factor backbone structure. Egr1 is known to upregulate Klotho expression. We tested the transcriptional activation effects of these ZFPs in a dual luciferase coincidence reporter system under the control of 4kb promoter of human Klotho in stable HEK293 cells and in HK-2 cells that express Klotho protein endogenously. We found that the best ZFPs are the de novo designed ones targeting −250 bps of Klotho promoter and one of the Egr1 binding sites. We further enhanced Klotho’s activation by using p65-Rta transcriptional activation domains in addition to VP64. These upregulation approaches could be useful for studying Klotho’s protective effects and designing Klotho boosting therapeutics for future in vivo experiments.
Keywords: Aging, Alzheimer’s disease, kidney disease, cancer, neurodegeneration, transcription factor
Introduction
Klotho.
The anti-aging protein Klotho was named after the goddess who spins the thread of life (1). Klotho knockout mice have an accelerated aging phenotype that recapitulates many of the features observed in aged humans (1). Conversely, lifespan is extended by ~30% and increased resistance to oxidative stress is observed in Klotho overexpressing mice (2). The single copy gene Klotho, is a type-I transmembrane protein which is mainly expressed in the kidney, brain and reproductive organs (3). Klotho is also shed by proteolytic cleavage resulting in a soluble form that circulates in serum and CSF (4–6). A third form of Klotho, found mainly in the brain, results from differential mRNA splicing and is secreted from the cell into the blood and CSF (7). The transmembrane isoform of Klotho serves as a co-receptor for FGF23 signaling, while the soluble Klotho has pleiotropic actions throughout the body (for extensive reviews see (8,9)).
Klotho in the brain.
We pioneered the exploration Klotho’s role in the brain. Pivotal studies from our lab discovered diminished Klotho expression in the brain of aged healthy monkeys, mice and rats (10). Impaired cognition is observed in Klotho-deficient mice (11), while enhanced Klotho expression reduces cognitive deficits in a mouse model of AD (12) and improves cognitive functions in young (13) and old mice (14). Recent work on Klotho’s functions in the CNS of mouse models of Parkinson’s (PD) and ALS, performed by us and by others, demonstrated that Klotho can combat processes associated with neurodegeneration through diverse mechanisms (15–18).
Together, these findings suggest that increasing Klotho levels in the brain would have a beneficial effect to prevent cognitive impairment associated with normal aging and neurodegenerative diseases and also prevent motor deficits in PD and ALS. There are several approaches to regulate Klotho expression, one being regulation of gene transcription.
The Klotho promoter and its transcriptional regulation.
The Klotho promoter and first exon lay within a CpG island and indicates a possible mechanism of pre-transcriptional epigenetic silencing as a potential candidate for age-related downregulation (19). The Klotho promoter is methylated with age (19), in several types of cancer (20–22), renal fibrosis (23) and Duchenne muscular dystrophy (DMD) (24). Several transcription factors (TFs) were reported as transcriptional regulators of Klotho, including Vitamin D receptor (VDR) (25,26), Egr1 (early growth response protein 1) (27) and peroxisome proliferator activation receptor γ (PPARγ) (28).
Zinc Finger Proteins (ZFPs).
ZFPs belong to the class of DNA binding proteins. They bind to DNA through a finger-shaped fold, which is stabilized by zinc ions coordinated to a combination of cysteine and histidine residues. Advancements in design and engineering of ZFPs have led to genome targeting by fusing them to other functional domains for several therapeutic purposes (29–31). A great deal of progress has been made to develop the modular building blocks that recognize specific three nucleotide sequence (triplet) of DNA sequence (32). These modular building blocks can be fused together to create proteins that can bind to a specific DNA sequence. Combined with effector domains that result in transcriptional activators or repressors, these artificial engineered ZFPs can have transcriptional control of any specific gene of interest (29,30).
The Cys2-His2 zinc finger motif was first identified in the DNA and RNA binding transcription factor TFIIIA (33). It provides the ideal structural scaffold on which a sequence-specific ZFP might be constructed. Each ZFP motif consists of 30 amino acids with a simple ββα fold stabilized by hydrophobic interactions and the chelation of a single zinc ion. Presentation of the α-helix of this motif into the major groove of DNA allows for sequence-specific base contacts. Each zinc finger motif typically recognizes a DNA triplet, simple covalent tandem repeats of the zinc finger motif allow for the recognition of longer asymmetric sequences of DNA by this motif. The recognition specificity is due to the amino acids located at positions −1, +1, +2, +3, +4. +5 and +6 relative to the start of α- helix (34). Change in these amino acids can change its specificity. The remaining amino acids of the 30 amino acids can remain unchanged, and constitute the backbone of ZFP. Each zinc finger motif typically binds a triplet, a complete recognition of all possible triplets requires the characterization of 64 motifs. Computational tools are available to predict and design ZFP specific to certain target sequences. For example, Zinc Finger Tools (32) employs a non-redundant set of 49 helices to target as many DNA triplets. This set includes zinc fingers that recognize 16 GNN, 15 ANN, 15 CNN and 3 TNN triplets (32).
In this study, we aimed to design a method to significantly upregulate Klotho by targeting its promoter sequence within 300 bp upstream of the translation start site. We designed the ZPF constructs either de novo from modular building blocks or modified sequences from the natural Egr1 transcription factor backbone structure. We tested the transcriptional activation effects of these ZFPs in HEK cells expressing the dual luciferase reporter linked to the 4kb human Klotho promoter and in HK-2 human kidney cells. We found the best ZFPs and further enhanced the activation effects of Klotho expression by using various transcriptional activation domains.
Materials and Methods
Plasmid construction.
To make mutations in the Egr1 binding sites in the Klotho promoter, the mutation was introduced by amplifying DNA with ClonAmp HiFi polymerase and self-ligating of the PCR product using In Fusion kit (Clontech).
ZFP1 and ZFP52 cDNA were commercially synthesized (Genewiz) and cloned into pcDNA3.1TOPO (Invitrogen). ZFP1-Egr1-site1-sense, ZFP1-Egr1-site2, ZFP3-Egr1-site3-antisense, and ZFP4-Egr1-site1-antisense cDNA fragments (BamHI-NheI) were synthesized (Genewiz) and cloned into ZFP52/pcDNA3.1TOPO vector digested with BamHI and NheI. To construct ZFP3-VPR (V64, p65 and Rta activation domains), and ZFP52-VPR, the VPR insert and the vector containing either ZFP3 or ZFP52 were amplified with ClonAmp HiFi polymerase according to the manufacturer’s protocol.
The insert and vector bands (100 ng each) were ligated together using In Fusion kit (Clontech) to generate the final ZFP3-VPR or ZFP52-VPR constructs.
To construct Egr1-Insert2 and Egr1-Insert3 construct, the additional 3 zinc finger motif insertions were synthesized (Genewiz) and cloned into Egr1 by ClonAmp HiFi polymerase and In Fusion kit (Clontech).
To clone the Egr1-Insert2-RR (repressor removed) and Egr1-Insert3-RR constructs, the repressor domain was removed by amplifying DNA with ClonAmp HiFi polymerase followed by self-ligation of the PCR product using In Fusion kit (Clontech).
To clone the inducible ZFP3_VPR and ZFP52_VPR, the ZFP3_VPR and ZFP52_VPR cDNA inserts and the inducible vector (Lenti-iCas9-neo, Addgene #85400) were amplified using the following primers using Clontech HiFi according to the manufacturer’s protocol:
5’- GACGATGACGATAAGGCCCAGGCGGCCCTGGAGCCC-3’ (forward primer for ZFP3_VPR);
5’- GCTGAAGTTGGTGGCATGGTGATGGTGATGATGACCGGTAC-3’ (reverse primer for both ZFP3_VPR and ZFP52_VPR);
5’- gacgatgacgataagGCCCAAGCTGCCTTAGAACCCGGCG-3’ (forward primer for ZFP52_VPR);
5’- GCCACCAACTTCAGCCTGCTGAAG-3’ (forward primer for inducible vector);
5’- CTTATCGTCATCGTCTTTGTAATCCATGG-3’ (reverse primer for inducible vector).
To clone the VPR domain shortening constructs, the following primers were used to remove the short segment to generate VPR single, double, triple and quadruple deletion constructs:
Short1_FWD:
5’-TCTCAGGCCTCTGCTCTGGCTCCAGCC-3’
Short1_RV:
5’-AGCAGAGGCCTGAGAACCAACTTTGCGTTTCTTTTTCGGAG-3’
Short2_FWD:
5’-CCACTGGATCCAGCGCCCGCAGTG-3’
Short2_RV:
5’-CGCTGGATCCAGTGGGCCCTCAAACACGTCACTAATAGC-3’
Short3_FWD:
5’-GGCACACTGTCTGAAGCTCTGCTGC-3’
Short3_RV:
5’-TTCAGACAGTGTGCCCACCTGAGGAGGGGCTGGAGCCAGAG-3’
Shor4_FWD:
5’-GATGAGCTGACAACCACACTTGAGTC-3’
Short4_RV:
5’-GGTTGTCAGCTCATCGGCCACAGGGATGCCCTGGTTCAGC-3’
The cDNAs and protein sequences of the constructs are listed in Supplemental materials.
Cell lines and transfections.
We used HEK293 and HK-2 cell lines for the studies. Cell culture and transfections were performed as described previously (35).
Luciferase reporter assay.
For measurement of firefly luciferase (FLuc) expression, the coincidence reporter vector under Klotho promoter was used and measured with a Luciferase kit (Promega) as described (35).
Klotho protein analysis.
ELISA assay for human Klotho was purchased from IBL and performed according to the manufacturer’s protocol.
Cell viability and cytotoxicity were calculated as described previously (16).
Results
Searching for potential Egr1 binding sites in the Klotho promoter region
Researchers examined the transcriptional regulation of the Klotho gene by epidermal growth factor (EGF) in human embryonic kidney cells (HEK293) (27). Serial deletion of promoter fragments, identified a proximal 45 bp (−90 to −45) region responsible for EGF-induced promoter activity that contains the Egr1 binding element (27). The researchers showed that over-expression of Egr1 increased Klotho gene promoter activity, while a point mutation in the Egr1 binding element abrogated promoter activation. We further expanded studies of Klotho promoter regulation by Egr1 in order to upregulate Klotho expression in the brain. We identified two additional binding sites for Egr1 in the Klotho promoter not previously described: in the −89 to −97 bp (sense orientation) and −158 to −166 bp (anti-sense orientation) regions upstream of the start codon (Figure 1).
Figure 1.
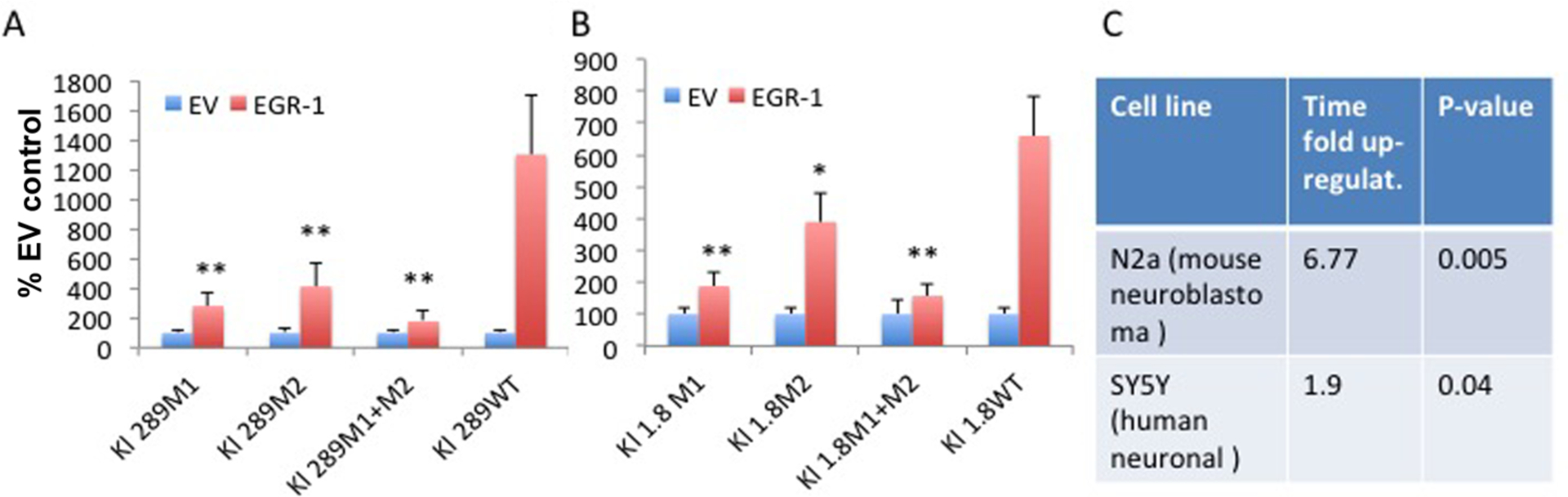
Mutations in the Egr-1 binding sites (M1 and M2) in the Klotho promoter fragments −289 (A) and −1800 bp from the translation start site ((+1) ATG). (B) Increased Klotho transcription in Egr-1 transfected cells. Mutating both sites prevents upregulation by Egr-1 almost completely. *, p<0.05. **, p<0.01 (transfected with empty vector (EV) or Egr-1; mutated vs. WT). (C) qPCR analysis of Klotho expression in 2 neuronal cell lines 48 hours after transfection with the Egr1 transcription factor.
First we cloned a mutation of each one of these sites (M1: GCGGGGGTG → GCTGTTTTG); (M2: CTGGGGGTGGGGGCA → CTAAGGACAAGGACA) into two vectors containing firefly luciferase reporters under the regulation of Klotho promoter fragments −289bp and Klotho promoter −1800bp, respectively, from the translation start site (+1ATG), in HEK293 cells. Activation of the promoter was reduced significantly, but not completely by either of the mutations. However, when we mutated both sites, the Egr1 effect on promoter activation was abrogated almost completely. Having such a convincing result obtained previously, and confirmed and expanded by us, we questioned whether we can upregulate Klotho expression in brain cells transfected with the Egr-1 transcription factor. We detected a significant increase in Klotho mRNA in 2 neuronal cell lines 48 hours after transfection, supporting the potential of this transcription factor to enhance Klotho expression in the brain through specific Klotho promoter binding sites.
Target site selection and ZFP design
As recently reported, targeting the proximal promoter region closer to the transcription start site results in the maximal level of gene activation using the CRISPR system (35,36), and because of the divergent transcription activities from active promoters around 250 bps upstream of transcription start site (37,38), we therefore selected ZFPs target sites within −300 bps of the Klotho promoter region (Figure 2). As mentioned above, we identified two additional binding sites for Egr1 in the Klotho promoter region in addition to the reported Egr1 binding site in the −51 to −59 bp (GCG GGG CGC, site 1): −89 to −97 bp (GCG GGG GTG, site 2) and −158 to −166 bp (CTG GGG GTG, anti-sense orientation, site 3) upstream of the start codon. To activate endogenous Klotho gene transcription using ZFPs, we searched in the human Klotho promoter within −300 bp region for the optimal ZFP binding sites using the ZFP design tool (32). There are other ZFP design tools available, however, the database we used is the original ZFP design tool, and we found it very effective. We found two target sites between −300 to −200 bp upstream, one is at −300 bp (name ZFP1) and another at around −249 bp (name ZFP52, 52 bp from −300) (Figure 2A). In addition, we chose the 3 Egr1 binding sites because we were able to consistently activate Klotho gene expression by overexpressing Egr1 transcription factor (Figure 2A). We used two design strategies to construct ZFPs. One is the de novo method to artificially design ZFPs, and another method is to design a ZFP based on the natural existing Egr1 protein (Figure 2B). The de novo design consists of the ZF domain, a nuclear localization sequence (NLS) and VP64 transcription activator domain together with a HA-tag for protein detection (Figure 2B). For the natural engineered method, we inserted 3 additional zinc fingers downstream of the original existing ZF domain to increase specificity. It is well established that to uniquely recognize only one target out of the entire human genome, at least 18 bps (418 ~ 69 billion) are required to ensure uniqueness in the genome (~3 billion total bps). We, therefore, designed ZF domains ranging from a minimum of 6 fingers to 10 fingers that recognize 18 to 30 bps of DNA target sequences. We designed 6 ZFPS by the de novo method: ZFP1, ZFP52, ZFP1-site1, ZFP2-site2, ZFP3-site3, and ZFP4-site1-AS (targeting Egr1 site1 at anti-sense orientation) (Figure 2A); and 2 ZFPs by natural method: Egr1-insert2 (targeting Egr1 binding site 2) and Egr1-insert3 (targeting Egr1 binding site 3) (Figure 2A). The ZFP ID name, binding position, binding sequence, and the zinc finger domain recognition amino acid sequences are listed in Tables 1 and 2. Structural studies have suggested that each finger contacts DNA in an antiparallel manner (33,34). In a typical 3 zinc fingers that recognize 9 bps DNA sequence, the 1st finger would recognize the 3’-most triplet, the 2nd finger recognizes the 2nd triplet, the 3rd finger recognizes the 5’−1st triplet. Table 2 lists the ZFPs and their seven specificity-determining residues in each finger.
Figure 2.
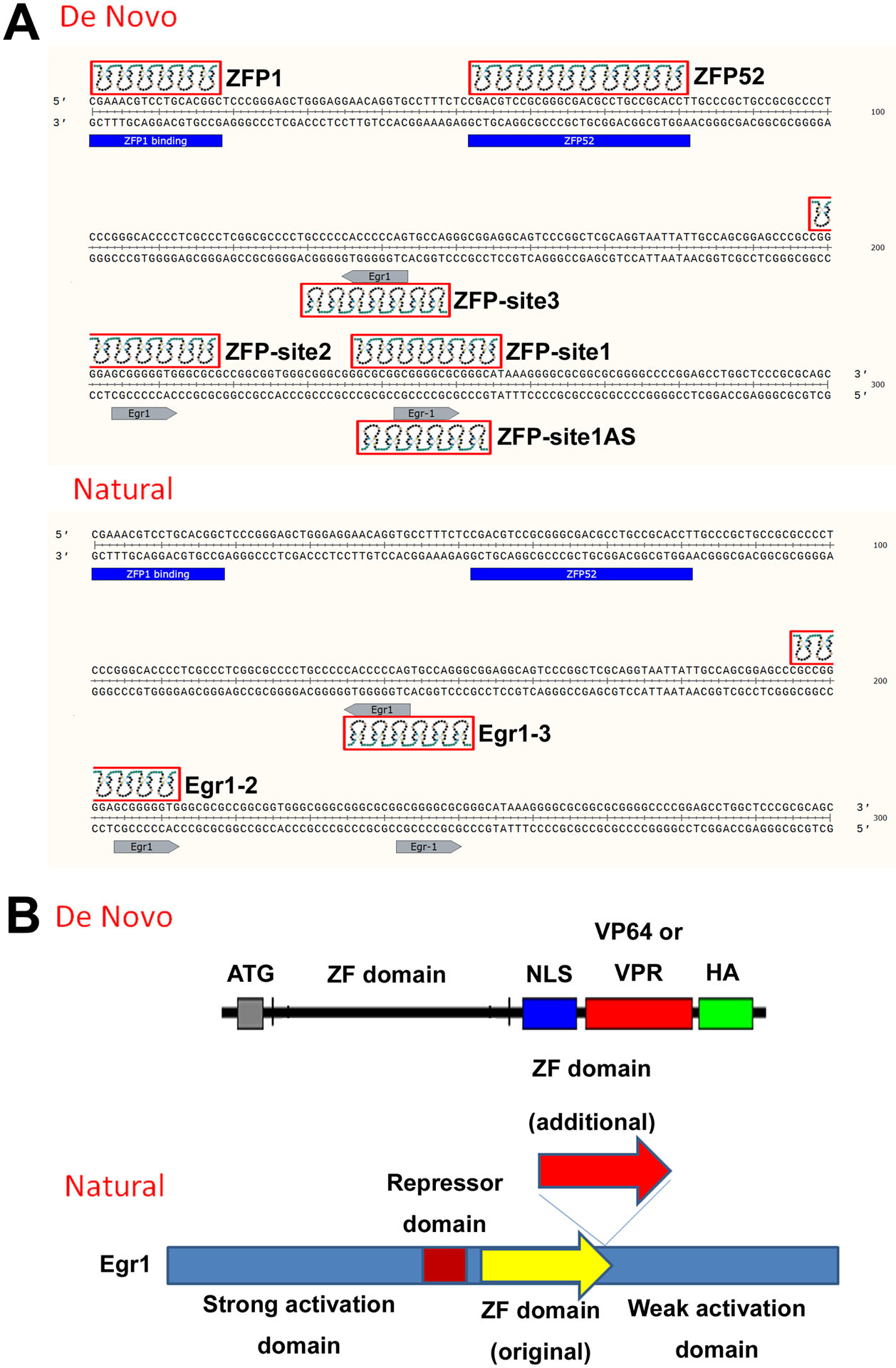
A) Diagram of ZFP target sites in the Klotho promoter region (−300 bps) of the four de novo ZFPs, and two natural-engineered ZFPs. ZFP ID name and target are indicated. B) Diagram of the de novo and natural engineered ZFPs. NLS: nucleus localization sequence. HA: HA epitope tag. Red arrow (additional) shows the location where three additional fingers were inserted to construct the ZFP into six fingers.
Table 1.
ZFP ID, binding position and binding sequences
| ZFP ID | Position | Sequence (5’−3’) |
|---|---|---|
| ZFP1 | −283 to −300 | CGA AAC GTC CTG CAC GGC |
| ZFP52 | −220 to −249 | CGA CGT CCG CGG GCG ACG CCT GCC GCA CCT |
| ZFP1_Egr1_site1 | −45 to −65 | GGC GCG GCG GGG CGC GGG CAT |
| ZFP2_Egr1_site2 | −83 to −103 | CGG GGA GCG GGG GTG GGC GCG |
| ZFP3_Egr1_site3 | −152 to −172 | CTG GCA CTG GGG GTG GGG GCA |
| ZFP4_Egr1_site1_AS | −47 to −64 | GCC CGC GCC CCG CCG CGC |
| Egr1_insert_site2 | −89 to −106 | CGC CGG GGA GCG GGG GTG |
| Egr1_insert_site3 | −149 to −166 | GCC CTG GCA CTG GGG GTG |
The position numbers are in reference to translation start codon (+1)ATG.
Egr1 binding site 1 (GCG GGG CGC), site 2 (GCG GGG GTG) and site 3 (CTG GGG GTG) are indicated in red.
Table 2.
ZFP ID and zinc finger domain sequences
| ZFP ID | ZF1 | ZF2 | ZF3 | ZF4 | ZF5 | ZF6 | ZF7 | ZF8 | ZF9 | ZF10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ZFP1 | DPGHLVR | SKKALTE | RNDALTE | DPGALVR | DSGNLRV | QSGHLTE | ||||
| ZFP52 | TKNSLTE | QSGDLRR | DCRDLAR | TKNSLTE | RTDTLRD | RSDDLVR | RSDKLTE | RNDTLTE | SRRTCRA | QSGHLTE |
| ZFP1_Egr1_site1_sense | TSGNLTE | RSDKLVR | HTGHLLE | RSDKLVR | RSDDLVR | RSDDLVR | DPGHLVR | |||
| ZFP2_Egr1_site2_sense | RSDDLVR | DPGHLVR | RSDELVR | RSDKLVR | RSDDLVR | QRAHLER | RSDKLTE | |||
| ZFP3_Egr1_site3_antisense | QSGDLRR | RSDKLVR | RSDELVR | RSDKLVR | RNDALTE | QSGDLRR | RNDALTE | |||
| ZFP4_Egr1_site1_antisense | HTGHLLE | RNDTLTE | RNDTLTE | DCRDLAR | HTGHLLE | DCRDLAR | ||||
| Egr1_insert_site2 | RSDELTR* | RSDHLTT* | RSDERKR* | QRAHLER | RSDKLTE | HTGHLLE | ||||
| Egr1_insert_site3 | RSDELTR* | RSDHLTT* | RSDERKR* | QSGDLRR | RNDALTE | DCRDLAR |
The seven specificity-determining residues from position −1 to +6 relative to the start of the α- helix were derived and characterized in the Barbas lab (32). Each set of zinc fingers are given in the amino acid one-letter code. It is important to remember that in reading the table, the first finger binds the 3′-most DNA triplet. For example, the 3’-most GGC triplet in the ZFP1 target in table 1 is recognized by ZF1 (DPGHLVR) and the 2nd 3’-most CAC triplet is recognized by ZF2 (SKKALTE), and so on. The ZF recognition domain ranges from minimum of 6 fingers to 10 fingers that recognize 18 to 30 bps of DNA target sequences.
ZF1, ZF2, and ZF3 are original ZFs from Egr1 TF. Egr1 binds to the consensus sequence: (http://crunch.unibas.ch/ENCODE_REPORTS/Myers_HudsonAlpha/BG_7_8/report_EGR1/EGR1..3.p2.html)
Evaluation of Klotho gene activation using a dual luciferase coincidence reporter system
We cloned the human Klotho promoter of about 4kb into a dual luciferase reporter system. This system stoichiometrically expresses two orthologous reporters, firefly luciferase (FLuc) and PEST-destabilized nanoLuc luciferase (NLuc) under the control of the Klotho promoter (35). To examine the efficiency of the ZFPs on Klotho gene activation, we transfected the ZFP expression plasmids into HEK293 cells stably expressing the FLuc NLuc coincidence reporter driven by the 4kb Klotho promoter. The results showed that ZFP3-site3, Egr1-insert2 and Egr1-insert3 significantly activated Klotho gene transcription, with the natural engineered constructs Egr1-insert2 and Egr1-insert3 performing better (4–6 fold increase) (Figure 3). In the 4kb Klotho promoter reporter system, the positive control transcription factor Egr1 for Klotho gene activation resulted in 3–4 fold increase of Klotho gene expression, compared to empty vector control (Figure 3).
Figure 3.
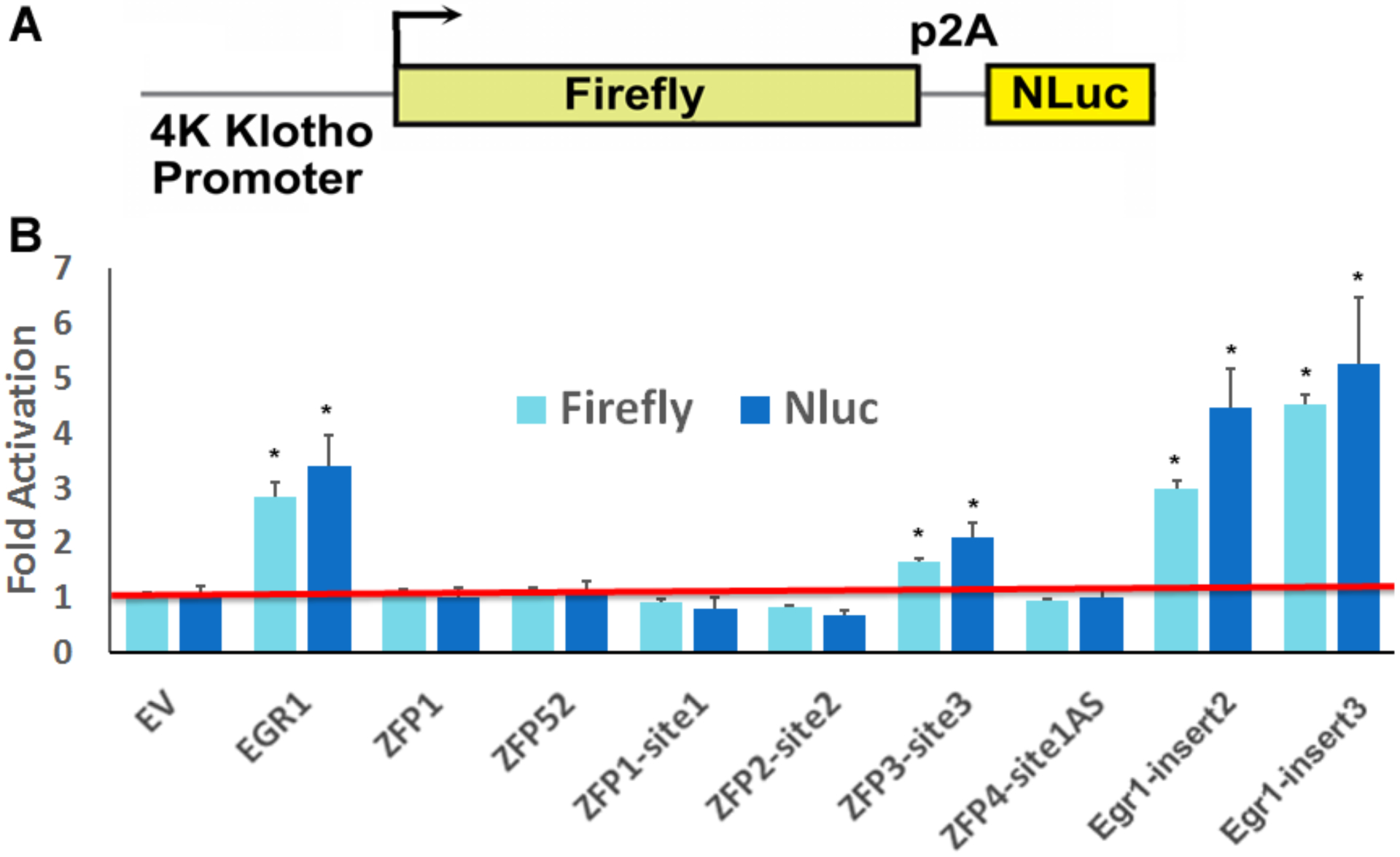
Klotho gene activation in a dual luciferase coincidence reporter system. (A) Schematic view of the Firefly, NLuc coincidence reporter system under control of Klotho 4kb promoter. The P2A ribosome skipping sequence is indicated. (B) Fold activation of Klotho gene expression by ZFPs was evaluated with the dual luciferase coincidence reporter system in stable HEK293 cells. Cells were analyzed by the dual luciferase assay 2 days after transfection with ZFPs. Negative control: empty vector (EV). Positive control: Egr1 transfected cells. Data are expressed as fold over negative control. Error bars show standard deviation among 6–8 replicates.
Evaluation of Klotho gene activation in HK-2 cells
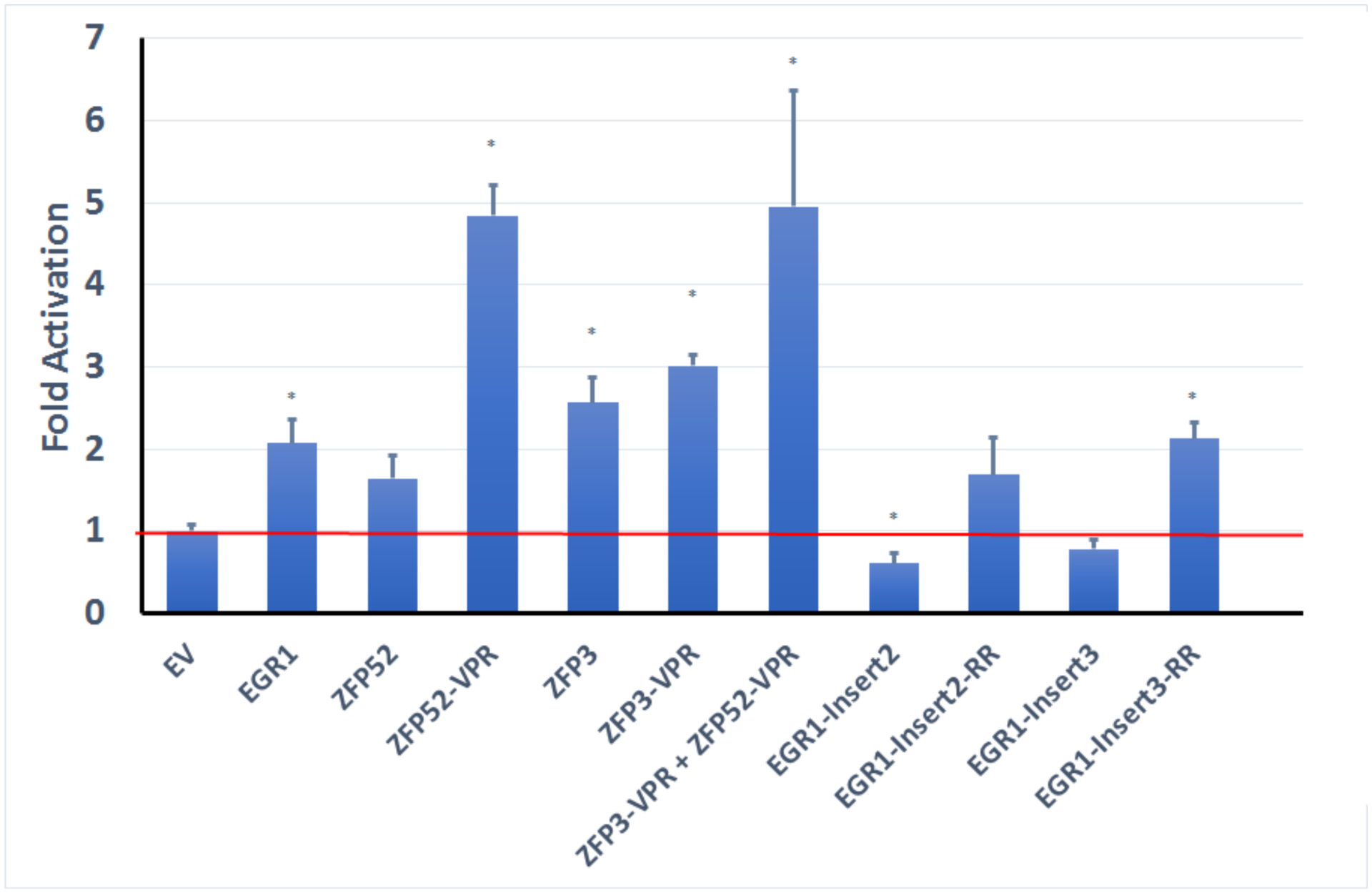
Klotho is mainly expressed in brain and kidney (3). Therefore, we sought to investigate whether the ZFPs can activate Klotho expression in cell lines from these organs that express Klotho endogenously. We transfected the ZFPs into HK-2 cells and examined Klotho protein levels and found that ZFP52 and ZFP3-site3 enhanced Klotho protein expression 2 and 3 fold, respectively (Figure 4). Klotho protein level was evaluated using the human ELISA kit. The ELISA kit from IBL is very sensitive and can measure human Klotho protein quantitatively with great accuracy. The Egr1 overexpression resulted in significant Klotho gene expression compared to empty vector control (Figure 4). However, the effects of Klotho activation by Egr1 were not as strong compared to the stable FLuc NLuc coincidence reporter HEK293 cells system.
Figure 4.
Klotho fold activation in HK-2 cells using ZFP constructs measured by Klotho ELISA kit. *, p<0.05. Results normalized to total protein measured by BCA.
To further enhance Klotho gene expression, we improved the activation domain to include additional p65 and Rta domains in addition to the VP64 domain. The constructs ZFP52_VPR and ZFP3_VPR showed enhanced activation abilities that further increased Klotho gene expression to 5 and 3 folds, respectively (Figure 4). For the engineered Egr1 constructs, we sought to increase the activity by removing of the repressor domain (Egr1 repressor (39,40)) (Figure 2). We were able to enhance Klotho activation with the Egr1-insert2-RR (remove repressor) and Egr1-insert3-RR constructs, the effects were significant for Egr1-insert3-RR mutant, but were not as strong compared to the de novo constructs (Figure 4). We then combined the best two constructs to determine whether there are synergistic effects for activation, but we only detected a slight and not significant increase with the ZFP52_VPR and ZFP3_VPR constructs (Figure 4). These results showed that we were able to activate Klotho protein expression up to 5 folds with our ZFP constructs.
Evaluation of cross reactivities of the ZFP constructs in mouse cells
In order to check whether the ZFP constructs we designed can also work on the mouse Klotho promoter region to activate mouse Klotho gene expression, we transfected ZFP3 and ZFP52 constructs in a N2a knock-in line with NLuc inserted into 3’-UTR of the Klotho gene by CRISPR genomic editing technology. In this system, Klotho transcript and NLuc are expressed off the same Klotho promoter by the P2A sequence. Thus, we can monitor Klotho gene transcription using NLuc activity. We found that, as expected, ZFP52 constructs did not activate Klotho gene expression, however, to our surprise, both ZFP3 and ZFP3_VPR constructs were able to activated mouse Klotho gene expression (Figure 5A). The effects were also observed in mouse N2a cells using qPCR, where we were able to detect more than 390 fold increase in Klotho mRNA (Figure 5B and C). We were surprised by the results of cross reactivity by ZFP3 constructs. The alignment of the mouse and human Klotho promoter region showed that indeed, the similarity between human and mouse Klotho promoter sequence at the ZFP52 binding site is very low, however, they exhibit fairly high similarity between ZFP3 binding site with 16/24 identical bases (Figure 5D). The results suggest that ZFP3 constructs can work on both human and mouse systems to activate Klotho gene expression.
Figure 5.
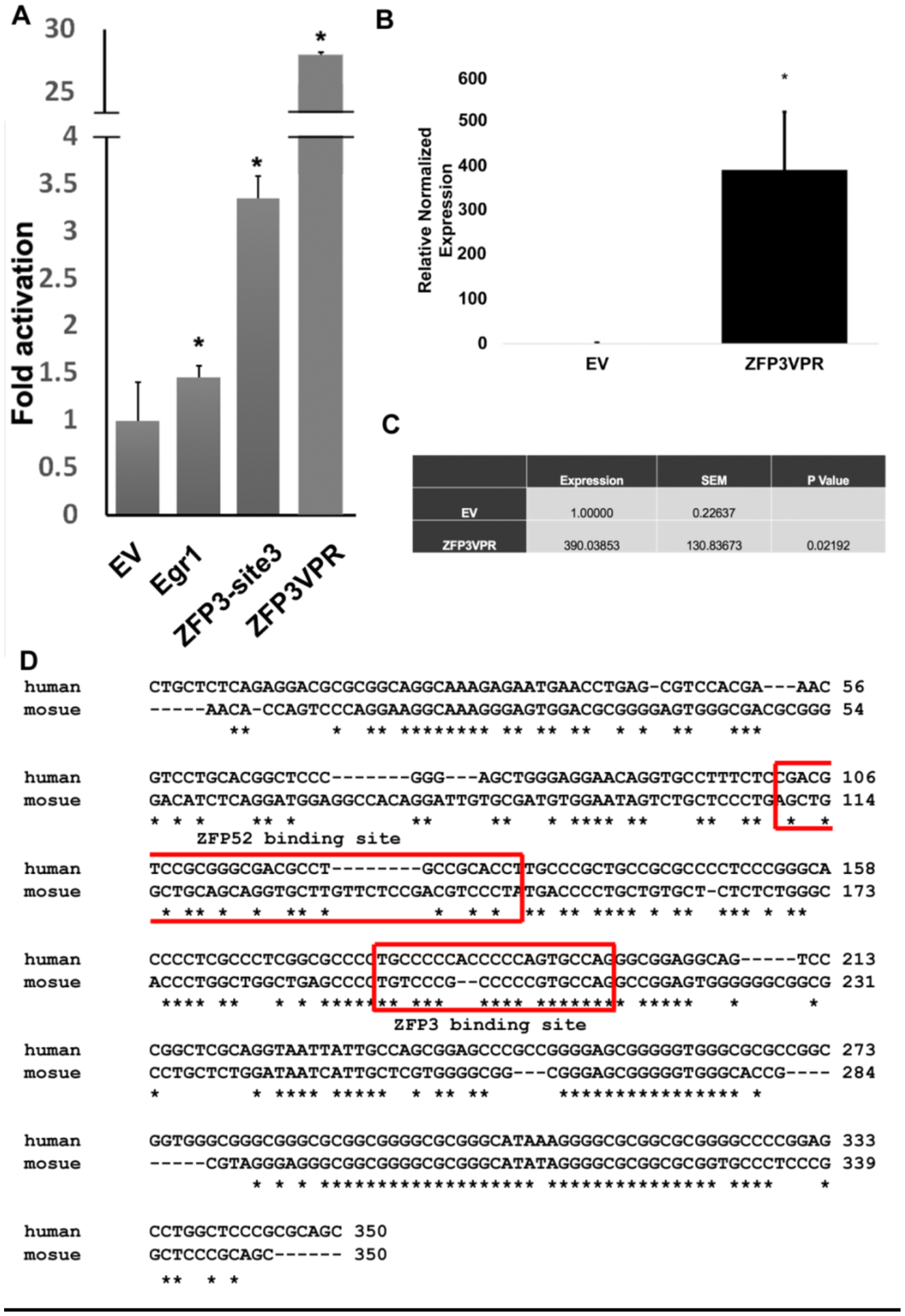
Klotho activation in mouse N2a cells using ZFP constructs. (A) Fold activation of Klotho gene expression in N2a Nluc knockin cells using ZFP constructs measured by Nanoluc assay. Negative control: empty vector (EV). Positive control: Egr1 transfected cells. Data are expressed as fold over negative control. *, p<0.05.(B, C) Klotho activation in mouse N2a cells using ZFP constructs measured by qPCR. D) Alignment of human and mouse Klotho promoter sequence (−350 to −1). The ZFP3 and ZFP52 binding sites are indicated.
Evaluation of the neuroprotective activity of the ZFP construct in HT-22 cells
To determine whether the ZFP construct that activates Klotho in mouse cells can protect against glutamate cytotoxicity, we employed the glutamate toxicity assay in the HT-22 hippocampal cell line as a model for neuronal death by oxidative damage. We found that transfection of ZFP3_VPR into HT-22 cells can significantly increase cell viability in the presence of either 3 mM or 4mM glutamate as measured by CellTiter-Glo (Figure 6). Overexpression of the extracellular domain of Klotho (KL980 construct) can also increase cell viability; however, the effects were not as significant as ZFP3_VPR construct (Figure 6). These results suggest that ZFP construct can work on a hippocampal cell line to protect against neurotoxicity caused by glutamate.
Figure 6.
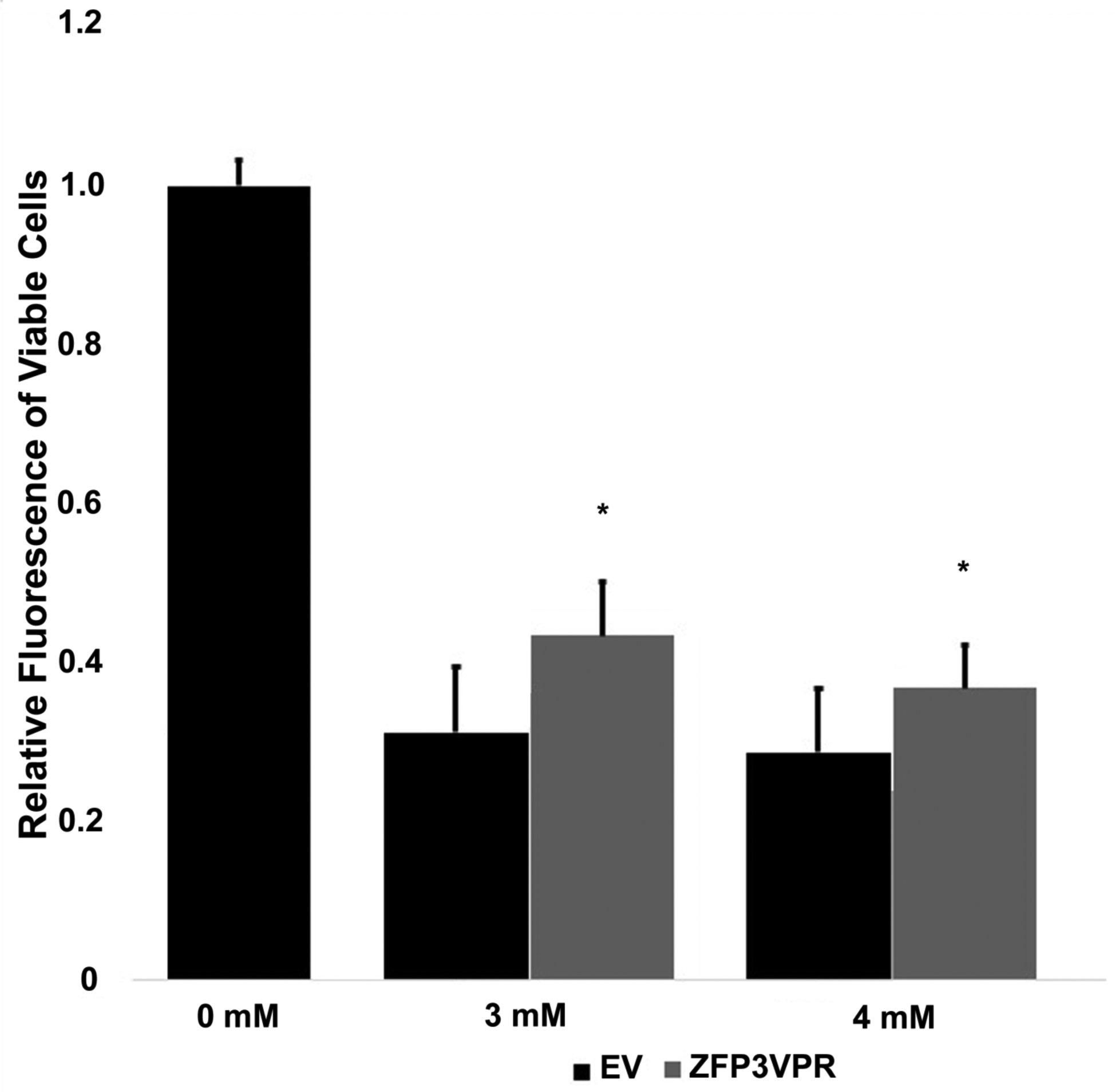
Evaluation of neuroprotective activity of the ZFP construct in HT-22 cells. Relative fluorescence of viable cells was measured in HT-22 cells transfected with ZFP3_VPR in the presence of either 3 or 4 mM of glutamate. *, p<0.05 compared to empty vector control (EV).
Improving VPR transcription activation domain (TAD)
The length of the VPR transcription activation domain is 57 kD. The size of the protein is an important limiting factor when considering protein production in the E. coli system. In order to improve the size of the TAD by searching for the “minimum requirement TAD”, we sought to remove or shorten an unnecessary amino acid “spacer” to achieve equal or better transcription activity of the VPR. We used the nine amino acids transcription activation “9aaTAD” prediction tools (https://www.med.muni.cz/9aaTAD/) to retain the predicted TAD motifs and remove unnecessary segments in the VPR. We predicted two segments for removal from original VPR sequence (Short 1 and Short 2 segments in Figure 7A), and the resulting constructs were VPR single deletion (VPR-SD) and VPR double deletion (VPR-DD) constructs (Figure 7A). We further predicted and removed two additional segments (Short 3 and Short 4) to produce VPR triple deletion (VPR-TD) and VPR quadruple deletion (VPR-QD) (Figure 7B). The detailed amino acid sequence of all the deletion constructs are listed in the “Sequence list”. We tested the ZFP3 deletion constructs in both mouse and human cell systems for activities since it worked in both systems, and ZFP52 only in human HK-2 cells. We found that in the case of ZFP3 construct, deleting the first two segments resulted in similar or slightly better transcription activities; however, further deletion of the Short 3 and 4 segments resulted in reduction of transcription activities (Figure 7C). In the case of the ZFP52 construct, deletion of the first 3 segments improved the transcription activities; however, further deletion of the Short 4 segment resulted in reduction of transcription activities (Figure 7D).
Figure 7.
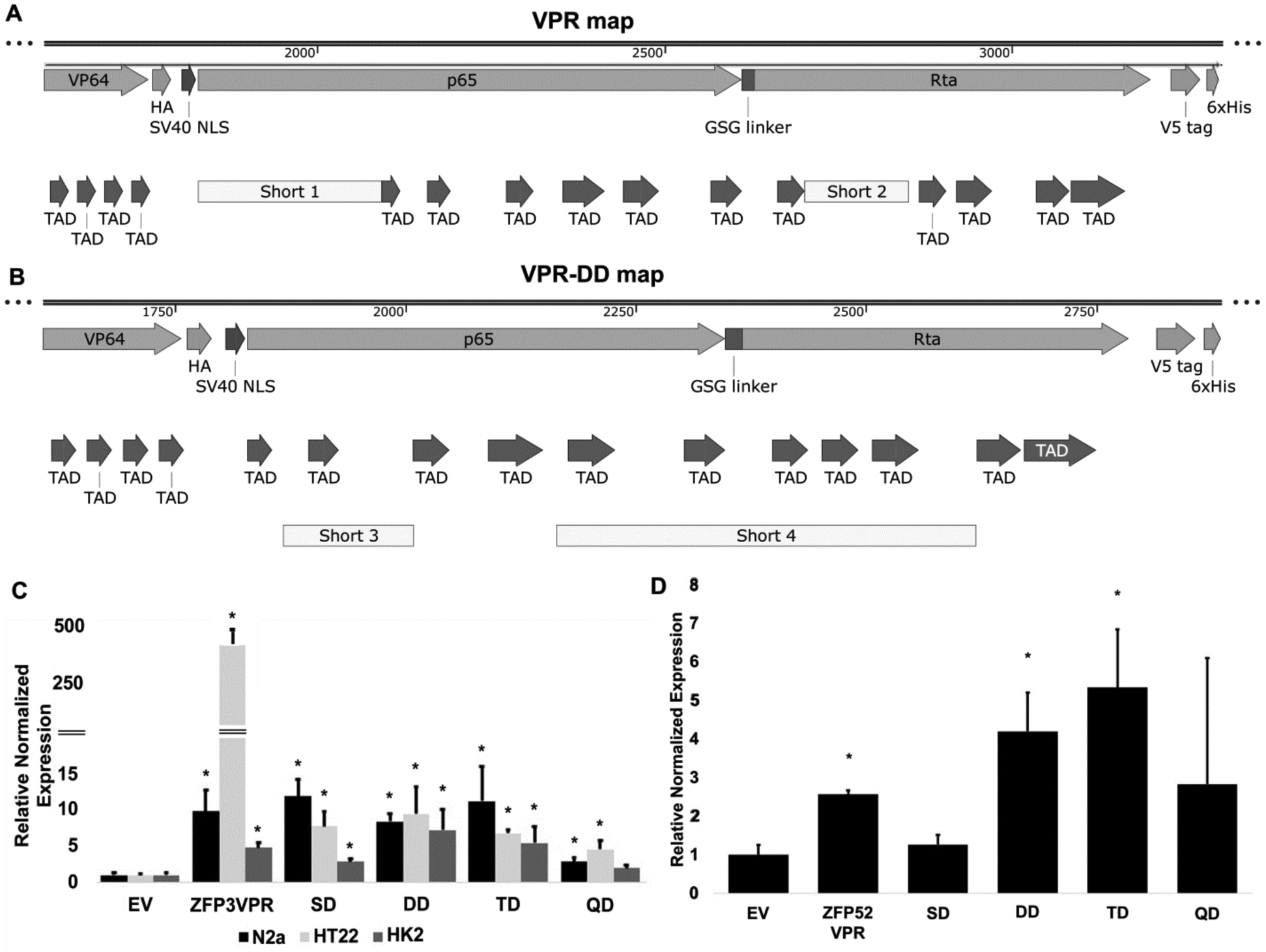
Evaluation of Klotho activation using VPR deletion constructs. Diagram of the VPR map (A) and VPR-DD map (B) showing VP64, p65, and Rta transcription activation domains. TAD: 9aaTAD motifs. The Short 1, 2, 3 and 4 segments removed are indicated. (C) Klotho activation using ZFP3_VPR deletion constructs in mouse (N2a and HT22) and human HK-2 cells measured by qPCR. *, p<0.05 compared to empty vector control (EV). (D) Klotho activation using ZFP52_VPR deletion constructs in human HK-2 cells measured by qPCR. *, p<0.05 compared to empty vector control (EV).
Discussion
ZFPs that recognize unique DNA sequences are a potentially powerful technology with multiple uses in drug discovery for the design of novel human therapeutics. Notably, a ZFP combined with a nuclease has been used for targeted DNA cleavage (31), and has resulted in more than 23 clinical trials (https://clinicaltrials.gov ZFN keyword search). Other applications of combining ZFP with different functional domains include a ZFP combined with a ligand binding domain for use in high throughput screening, a ZFP combined with a DNA methylation enzyme or histone deacetylase for DNA and chromatin modification, a ZFP combined with a transcriptional activator or repressor for gene regulation, and a ZFP combined with retroviral intergrase for targeted DNA integration (for a review see (41)).
The ultimate and perhaps most important application of ZFPs may be their use as therapeutics to control gene expression to prevent or cure diseases. It is especially an attractive alternative way to activate or repress genes where no conventional pharmaceutical drugs are available, since it is estimated that only 25–50% of the human genes implicated in disease will provide appropriate targets for small molecule drugs (42). The anti-aging and cognition-enhancing gene Klotho may be one of the ideal candidate genes to activate using the ZFP approach since all the existing drugs or chemicals that activate Klotho are either non-specific or the effects are not significant. Klotho is a multifunctional protein that has the potential to cure or prevent diseases such as neurological disorders including AD (12), ALS (18,43), MS (44), and several age-related diseases including cancer (45), chronic kidney disease (46) and cardiovascular disease (47).
In this study, we described a method to specifically and significantly upregulate Klotho by using ZFPs combined with transcriptional activation domains VP64, p65, and Rta. We also identified two additional Egr1 binding sites in the Klotho promoter region. We scanned the Klotho promoter for ZFP binding sites, selected target sites, designed and assembled ZFPs from an archive of pre-made modules and from natural engineered Klotho activating transcription factor Egr1, and we screened the ZFPs for efficacy in two cell-based assays. We found that two target sites are particularly efficient in activating Klotho gene expression: around −250 bps region and the 3rd Egr1 binding site located at −158 to −166 bps region of the Klotho promoter upstream of the start codon. Combination of multiple transcription activators increased Klotho activation abilities. Finally, using VPR as activation domain is better than using VP64 alone (Figure 4).
We found that the natural engineered Egr1 ZFP constructs worked better in the dual luciferase reporter system HEK cell line, however, the de novo designed ZFP constructs performed better in HK-2 cells that express Klotho endogenously. The difference in the two systems may be due to different cell environments in response to Egr1. In the HEK stable cell system, the Klotho promoter region is only 4 kb in length and the repressor regulatory elements present in the endogenous Klotho promoter are missing in this system. In contrast, the activation effect of Egr1 is not as strong in HK-2 cells which have the endogenous promoter containing repressors. We do not know whether these ZFPs can only activate cells that express Klotho endogenously or they can also increase Klotho expression in cells that normally do not express Klotho. When we evaluated the effects of our best two ZFPs, ZFP52_VPR and ZFP3_VPR constructs in Caco-2 cells, a continuous line of heterogeneous human epithelial colorectal adenocarcinoma cells, and examined the effects on Klotho expression by ELISA, we were not able to detect Klotho protein activation (data not shown). Based on our experiments, the ZFPs can activate Klotho transcription activities in cells that express Klotho endogenously such as in HK-2 cells, but not in an environment where the promoter is highly methylated such as in Caco-2 cells. In any case, the de novo designed ZFPs is a better approach because Egr1 is responsive to many stimulating factors such as growth factors, stress and insulin (48) and using the natural engineered Egr1 transcription factor backbone for Klotho activation may result in side effects due to unwanted stimuli. Whether ZFPs would affect other Egr1-regulated genes awaits further investigation.
We then evaluated the cross reactivates of the ZFP construct in mouse cells. We found that ZFP3 constructs can work on both human and mouse systems to activate Klotho gene expression. We plan to apply these ZFP constructs for in vivo studies in a mouse in the future.
In a functional assay, we then evaluated the neuroprotective effects of the ZFP construct in HT-22 cells. We found that the ZFP3_VPR construct was able to protect against neurotoxicity caused by glutamate in the hippocampal cell line HT-22. The implication of these results is that this construct could benefit patients with neurodegenerative diseases such as AD, MS, and ALS. It is likely that the ZFP constructs could operate in other diseases where Klotho overexpression is beneficial, such as kidney disease, cancer, and more.
Next, we then sought to improve the VPR transcription activation domain (TAD) in order to produce a shorter and better version of TAD. We found that we were able to remove 2 or 3 segments of spacer that do not contribute to transcriptional activities.
ZFP construct could be delivered with an inducible system using gene therapy for controlling the dose and timing of Klotho enhancement. In the future, we plan to apply these ZFP constructs for in vivo studies. The ZFP can be delivered by AAV or other viral based gene therapy methods. The advantage of using AAV-ZFP over AAV-Klotho are: 1) Safety: an extra copy of Klotho (inducible or not) is unfavorable because Klotho levels should be tightly controlled, and Klotho overexpression could have unwanted side effects. 2) AAV-ZFP method increases endogenous Klotho expression, under physiological conditions that produce both membrane and secreted isoforms of Klotho; whereas AAV-Klotho only delivers one isoform depending on which construct is delivered. When using ZFPs for in vivo studies, it might be useful to ensure that the amplitude and timing of Klotho activation can be regulated and controlled with a small molecule or other controllable means. One approach is to control ZFP expression using an inducible promoter such as the TET-ON system with doxycycline (48,49). Another example is to express the ZFP as inactive form in the cytosol, and then activate it as desired by a small molecule drug to induce nuclear translocation and gene activation. One approach has been to fuse the ZFP to a steroid hormone receptor ligand binding domain, and to control the nuclear translocation using drugs such as 4-hydroxytamoxifen or RU-486 (50). Another strategy is to use a rapamycin analogue to control the assembly of the ZFP and the transcription activation domain. In this case, the ZFP and the activation domains are each fused to the rapamycin binding domain, and the drug brings the two parts together to achieve gene activation (51). Another alternative approach is to use a blue light inducible system. It was demonstrated that blue light can induce heterodimerization of the proteins GIGANTEA (GI) and the light oxygen voltage (LOV) domain of FKF1 (52). The ZFP and the activation domains can be each fused to GI and LOV domains, and exposure to the blue light would bring the two parts together to achieve gene activation.
In addition to using the activation domain, we can combine ZFPs with a DNA methylation enzyme or a histone deacetylase for DNA and chromatin modification on the Klotho gene, since DNA and chromatin methylation are important factors for Klotho gene expression.
Once we show the feasibility of in vivo Klotho activation by ZFPs in wild type mice, this technique can be expanded into disease mouse models such as AD, ALS, MS, and other neurodegenerative diseases, as well as other age-related diseases including cancer, chronic kidney disease, and cardiovascular disease.
Supplementary Material
Acknowledgements
This work was supported by a small business innovation research (SBIR) grant 1R44 AG053084 from NIH-NIA to Klogene Therapeutics, Inc.
Funding: HHS | NIH | National Institute on Aging 1R44 AG053084 to CRA
List of abbreviations
- AD
Alzheimer’s disease
- AAV
Adenovirus-associated virus
- ALS
Amyotrophic lateral sclerosis
- DD
Double deletion
- DMD
Duchenne muscular dystrophy
- EGF
Epidermal growth factor
- Egr1
Early growth response protein 1
- FLuc
Firefly luciferase
- HK-2
Human kidney cell li
- LOV
Light oxygen voltage
- LPS
Lipopolysaccharide
- NLuc
Nano luciferase
- PD
Parkinson disease
- PPARγ
Peroxisome proliferator activation receptor γ
- QD
Quadruple deletion
- RR
Repressor removed
- SD
Single deletion
- TAD
Transcription activation domain
- TD
Triple deletion
- TET-ON
The positive inducible tetracycline ON
- VDR
Vitamin D receptor
- VPR
VP64, p65, and Rta domain
- ZFP
Zinc finger protein
Footnotes
Conflict of interest: All authors received salaries (full time or part time) from the company.
References
- 1.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, and Nabeshima YI (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, and Kuro-o M (2005) Suppression of aging in mice by the hormone Klotho. Science 309, 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda H, Chikuda H, Suga T, Kawaguchi H, and Kuro-o M (2005) Regulation of multiple ageing-like phenotypes by inducible klotho gene expression in klotho mutant mice. Mech Ageing Dev 126, 1274–1283 [DOI] [PubMed] [Google Scholar]
- 4.Bloch L, Sineshchekova O, Reichenbach D, Reiss K, Saftig P, Kuro-o M, and Kaether C (2009) Klotho is a substrate for alpha-, beta- and gamma-secretase. FEBS Lett 583, 3221–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CD, Podvin S, Gillespie E, Leeman SE, and Abraham CR (2007) Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci U S A 104, 19796–19801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, and Nabeshima Y (1998) Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun 242, 626–630 [DOI] [PubMed] [Google Scholar]
- 7.Masso A, Sanchez A, Gimenez-Llort L, Lizcano JM, Canete M, Garcia B, Torres-Lista V, Puig M, Bosch A, and Chillon M (2015) Secreted and Transmembrane alphaKlotho Isoforms Have Different Spatio-Temporal Profiles in the Brain during Aging and Alzheimer’s Disease Progression. PloS one 10, e0143623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuro-o M (2012) Klotho in health and disease. Current opinion in nephrology and hypertension 21, 362–368 [DOI] [PubMed] [Google Scholar]
- 9.Abraham CR, Mullen PC, Tucker-Zhou T, Chen CD, and Zeldich E (2016) Klotho Is a Neuroprotective and Cognition-Enhancing Protein. Vitam Horm 101, 215–238 [DOI] [PubMed] [Google Scholar]
- 10.Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, and Abraham CR (2008) Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia 56, 106–117 [DOI] [PubMed] [Google Scholar]
- 11.Nagai T, Yamada K, Kim HC, Kim YS, Noda Y, Imura A, Nabeshima Y, and Nabeshima T (2003) Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. Faseb J 17, 50–52 [DOI] [PubMed] [Google Scholar]
- 12.Dubal DB, Zhu L, Sanchez PE, Worden K, Broestl L, Johnson E, Ho K, Yu GQ, Kim D, Betourne A, Kuro OM, Masliah E, Abraham CR, and Mucke L (2015) Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 2358–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubal DB, Yokoyama JS, Zhu L, Broestl L, Worden K, Wang D, Sturm VE, Kim D, Klein E, Yu GQ, Ho K, Eilertson KE, Yu L, Kuro-o M, De Jager PL, Coppola G, Small GW, Bennett DA, Kramer JH, Abraham CR, Miller BL, and Mucke L (2014) Life extension factor klotho enhances cognition. Cell reports 7, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masso A, Sanchez A, Bosch A, Gimenez-Llort L, and Chillon M (2018) Secreted alphaKlotho isoform protects against age-dependent memory deficits. Mol Psychiatry 9, 1937–1947 [DOI] [PubMed] [Google Scholar]
- 15.Cheng MF, Chen LJ, Niu HS, Yang TT, Lin KC, and Cheng JT (2015) Signals mediating Klotho-induced neuroprotection in hippocampal neuronal cells. Acta Neurobiol Exp (Wars) 75, 60–71 [PubMed] [Google Scholar]
- 16.Zeldich E, Chen CD, Colvin TA, Bove-Fenderson EA, Liang J, Tucker Zhou TB, Harris DA, and Abraham CR (2014) The neuroprotective effect of Klotho is mediated via regulation of members of the redox system. J Biol Chem 289, 24700–24715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brobey RK, German D, Sonsalla PK, Gurnani P, Pastor J, Hsieh CC, Papaconstantinou J, Foster PP, Kuro-o M, and Rosenblatt KP (2015) Klotho Protects Dopaminergic Neuron Oxidant-Induced Degeneration by Modulating ASK1 and p38 MAPK Signaling Pathways. PloS one 10, e0139914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeldich E, Chen CD, Boden E, Howat B, Nasse JS, Zeldich D, Lambert AG, Yuste A, Cherry JD, Mathias RM, Ma Q, Lau NC, McKee AC, Hatzipetros T, and Abraham CR (2019) Klotho Is Neuroprotective in the Superoxide Dismutase (SOD1(G93A)) Mouse Model of ALS. Journal of molecular neuroscience : MN 69, 264–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King GD, Rosene DL, and Abraham CR (2012) Promoter methylation and age-related downregulation of Klotho in rhesus monkey. Age (Dordr) 34, 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perveez M, Ajaz M, and Afroze D (2015) Promoter hypermethylation of KLOTHO; an anti-senescence related gene in colorectal cancer patients of Kashmir valley. Mol Biol Res Commun 4, 217–224 [PMC free article] [PubMed] [Google Scholar]
- 21.Qu Y, Dang S, and Hou P (2013) Gene methylation in gastric cancer. Clin Chim Acta 424, 53–65 [DOI] [PubMed] [Google Scholar]
- 22.Rubinek T, Shulman M, Israeli S, Bose S, Avraham A, Zundelevich A, Evron E, Gal-Yam EN, Kaufman B, and Wolf I (2012) Epigenetic silencing of the tumor suppressor klotho in human breast cancer. Breast Cancer Res Treat 133, 649–657 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Liu L, Lin W, Yin S, Duan A, Liu Z, and Cao W (2017) Rhein reverses Klotho repression via promoter demethylation and protects against kidney and bone injuries in mice with chronic kidney disease. Kidney Int 91, 144–156 [DOI] [PubMed] [Google Scholar]
- 24.Wehling-Henricks M, Li Z, Lindsey C, Wang Y, Welc SS, Ramos JN, Khanlou N, Kuro OM, and Tidball JG (2016) Klotho gene silencing promotes pathology in the mdx mouse model of Duchenne muscular dystrophy. Hum. Mol. Genet 25, 2465–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haussler MR, Whitfield GK, Kaneko I, Forster R, Saini R, Hsieh JC, Haussler CA, and Jurutka PW (2012) The role of vitamin D in the FGF23, klotho, and phosphate bone-kidney endocrine axis. Rev. Endocr. Metab. Disord 13, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haussler MR, Whitfield GK, Haussler CA, Sabir MS, Khan Z, Sandoval R, and Jurutka PW (2016) 1,25-Dihydroxyvitamin D and Klotho: A Tale of Two Renal Hormones Coming of Age. Vitam Horm 100, 165–230 [DOI] [PubMed] [Google Scholar]
- 27.Choi BH, Kim CG, Lim Y, Lee YH, and Shin SY (2010) Transcriptional activation of the human Klotho gene by epidermal growth factor in HEK293 cells; role of Egr-1. Gene 450, 121–127 [DOI] [PubMed] [Google Scholar]
- 28.Yamagishi T, Saito Y, Nakamura T, Takeda S, Kanai H, Sumino H, Kuro-o M, Nabeshima Y, Kurabayashi M, and Nagai R (2001) Troglitazone improves endothelial function and augments renal klotho mRNA expression in Otsuka Long-Evans Tokushima Fatty (OLETF) rats with multiple atherogenic risk factors. Hypertens Res 24, 705–709 [DOI] [PubMed] [Google Scholar]
- 29.Beerli RR, and Barbas CF 3rd. (2002) Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol 20, 135–141 [DOI] [PubMed] [Google Scholar]
- 30.Blancafort P, Segal DJ, and Barbas CF 3rd. (2004) Designing transcription factor architectures for drug discovery. Molecular pharmacology 66, 1361–1371 [DOI] [PubMed] [Google Scholar]
- 31.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, and Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33, 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandell JG, and Barbas CF 3rd. (2006) Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 34, W516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavletich NP, and Pabo CO (1991) Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252, 809–817 [DOI] [PubMed] [Google Scholar]
- 34.Elrod-Erickson M, Rould MA, Nekludova L, and Pabo CO (1996) Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure 4, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 35.Chen CD, Zeldich E, Li Y, Yuste A, and Abraham CR (2018) Activation of the Anti-Aging and Cognition-Enhancing Gene Klotho by CRISPR-dCas9 Transcriptional Effector Complex. Journal of molecular neuroscience : MN 64, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, and Zhang F (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, and Sharp PA (2008) Divergent transcription from active promoters. Science 322, 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core LJ, Waterfall JJ, and Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gashler AL, Swaminathan S, and Sukhatme VP (1993) A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol 13, 4556–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo MW, Matheny C, and Milbrandt J (1993) Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Mol Cell Biol 13, 6858–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamieson AC, Miller JC, and Pabo CO (2003) Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov 2, 361–368 [DOI] [PubMed] [Google Scholar]
- 42.Hopkins AL, and Groom CR (2002) The druggable genome. Nat Rev Drug Discov 1, 727–730 [DOI] [PubMed] [Google Scholar]
- 43.Anamizu Y, Kawaguchi H, Seichi A, Yamaguchi S, Kawakami E, Kanda N, Matsubara S, Kuro-o M, Nabeshima Y, Nakamura K, and Oyanagi K (2005) Klotho insufficiency causes decrease of ribosomal RNA gene transcription activity, cytoplasmic RNA and rough ER in the spinal anterior horn cells. Acta neuropathologica 109, 457–466 [DOI] [PubMed] [Google Scholar]
- 44.Zeldich E (2015) The Anti-Aging Protein Klotho Enhances Remyelination Following Cuprizone-Induced Demyelination. 57, 185–196 [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki Y, Imura A, Urakawa I, Shimada T, Murakami J, Aono Y, Hasegawa H, Yamashita T, Nakatani K, Saito Y, Okamoto N, Kurumatani N, Namba N, Kitaoka T, Ozono K, Sakai T, Hataya H, Ichikawa S, Imel EA, Econs MJ, and Nabeshima Y (2010) Establishment of sandwich ELISA for soluble alpha-Klotho measurement: Age-dependent change of soluble alpha-Klotho levels in healthy subjects. Biochem. Biophys. Res. Commun 398, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seibert E, Radler D, Ulrich C, Hanika S, Fiedler R, and Girndt M (2017) Serum klotho levels in acute kidney injury. Clin Nephrol 87 (2017), 173–179 [DOI] [PubMed] [Google Scholar]
- 47.Marcais C, Maucort-Boulch D, Drai J, Dantony E, Carlier MC, Blond E, Genet L, Kuentz F, Lataillade D, Legrand E, Moreau-Gaudry X, Jean G, Fouque D, and Project A (2017) Circulating Klotho Associates With Cardiovascular Morbidity and Mortality During Hemodialysis. J. Clin. Endocrinol. Metab 102, 3154–3161 [DOI] [PubMed] [Google Scholar]
- 48.Das AT, Tenenbaum L, and Berkhout B (2016) Tet-On Systems For Doxycycline-inducible Gene Expression. Curr Gene Ther 16, 156–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chtarto A, Yang X, Bockstael O, Melas C, Blum D, Lehtonen E, Abeloos L, Jaspar JM, Levivier M, Brotchi J, Velu T, and Tenenbaum L (2007) Controlled delivery of glial cell line-derived neurotrophic factor by a single tetracycline-inducible AAV vector. Exp Neurol 204, 387–399 [DOI] [PubMed] [Google Scholar]
- 50.Beerli RR, Schopfer U, Dreier B, and Barbas CF 3rd. (2000) Chemically regulated zinc finger transcription factors. J Biol Chem 275, 32617–32627 [DOI] [PubMed] [Google Scholar]
- 51.Pollock R, Giel M, Linher K, and Clackson T (2002) Regulation of endogenous gene expression with a small-molecule dimerizer. Nat Biotechnol 20, 729–733 [DOI] [PubMed] [Google Scholar]
- 52.Polstein LR, and Gersbach CA (2014) Light-inducible gene regulation with engineered zinc finger proteins. Methods Mol Biol 1148, 89–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


