Abstract
Drosophila melanogaster harbors a simple gut microbial community, or microbiome, that regulates several facets of its physiology. As a result, the host employs multiple mechanisms of maintaining control over its microbiome in an effort to promote overall organismal homeostasis. Perturbations to the balance between microbiome and host can result in states of instability or disease, making maintenance of microbial homeostasis a fundamental physiologic aspect of D. melanogaster biology. While the interactions between microbes and their hosts can be direct, particularly in the context of immunity and gut renewal, effects resulting from indirect interactions, such as those between microbiota members, can be equally as important. This review highlights the major ways in which D. melanogaster regulates microbial homeostasis, the consequences of disruptions to homeostasis, and the different mechanisms by which the microbiome interacts with its host.
Keywords: Drosophila-microbe interactions, gut immunity, microbe-microbe interactions
Introduction
Drosophila melanogaster is established as a powerhouse model for host-microbe interactions owing to the tractability of both the host and the microbial community. A large array of traditional and multi-omic techniques are available in D. melanogaster to aid investigations into physiologic consequences of microbial interactions in the gut. Use of the fly model has identified core aspects of host physiology that are influenced or regulated by the microbiome, increasing our understanding of how the microbiota can tip the balance between states of homeostasis and instability, or disease. In this review, we highlight how an understanding of the interactions of D. melanogaster with its microbiome will reveal relationships between homeostasis and disease.
The microbiome is a malleable organ that modulates host physiology
D. melanogaster typically harbors simple bacterial communities containing species primarily from the Lactobacillus and Acetobacter genera [1,2]. The nature of the microbial community-including factors shaping the microbiome, its diversity and abundance, its roles in host physiology, and the ecological consideration of these associations- has been extensively reviewed elsewhere [3,4]. Some of the more well- studied contributions of the fly microbiome to physiology include impacts on innate immunity, intestinal homeostasis, reproduction, nutrition, and metabolism (Figure 1) [5–22]. These studies have revealed the fly microbiome to be, in some ways, an additional organ that interacts with other host systems to influence overall biology. Like other organs, the microbiome is a distinct entity that mediates host processes to promote overall organismal homeostasis. What is unique about considering the microbiome as an organ is its fluidity or malleability, as while it can have powerful influences over physiology, it is also subject to being influenced or controlled itself by the host environment and host biology.
Figure 1.
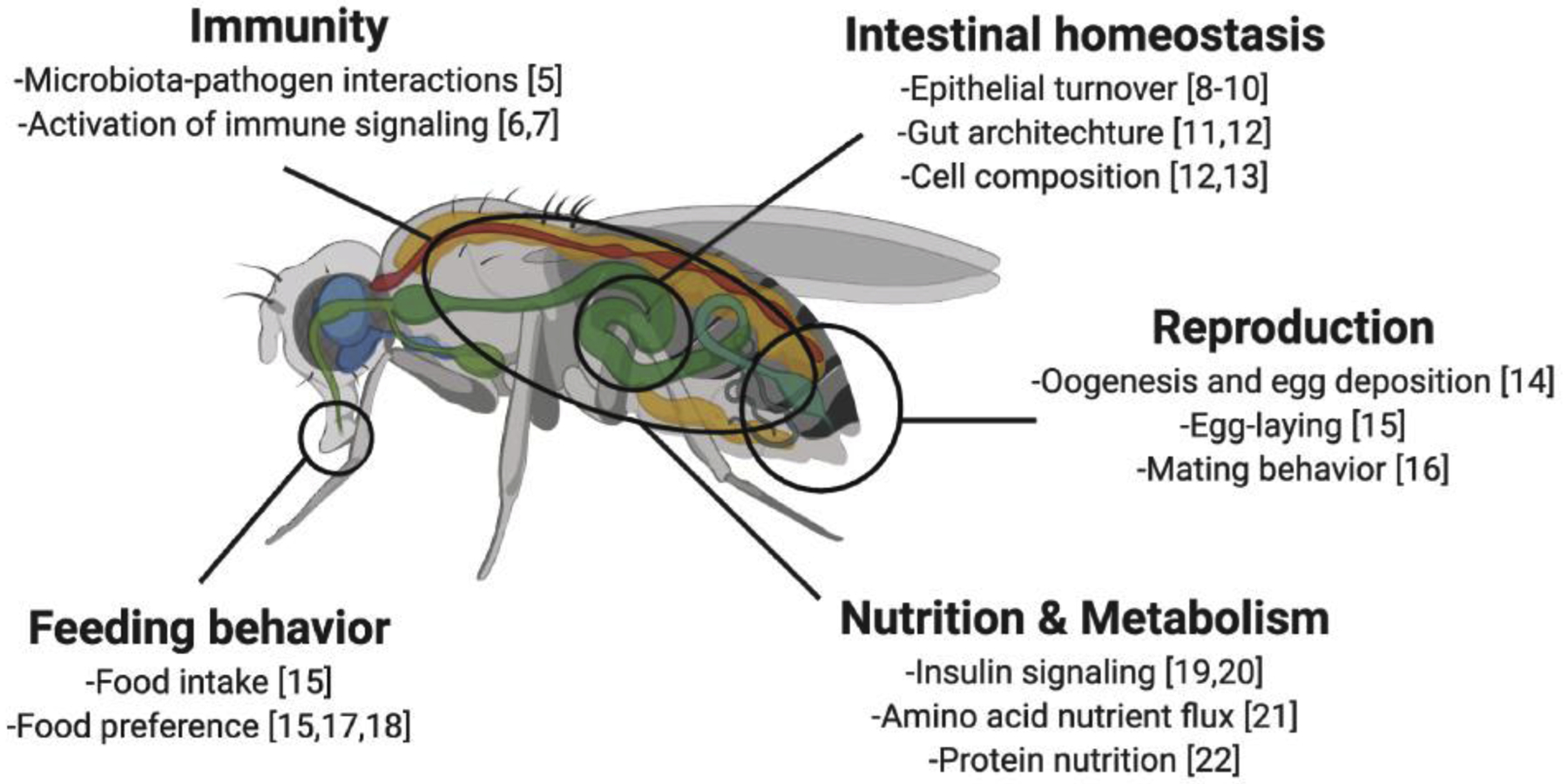
The D. melanogaster microbiome influences nearly all aspects of host physiology.
Host immunity as a mechanism of microbiome control
As an important component of host physiology, the microbiome contributes to establishment of the basal expression of many physiological functions. In turn, there is feedback of a number of these pathways on the microbiome, and their ability to maintain the microbiome (location, density, diversity) is critical for balancing host responses and functions. The host immune response is an important regulator of the microbiome of eukaryotic organisms and can be considered the most critical way by which a host communicates with its microbiome and vice versa.
The D. melanogaster immune repertoire includes a humoral response, characterized by the release of antimicrobial peptides (AMPs) and reactive oxygen and chlorine species, and a cellular response, which involves phagocytic cells and an invertebrate-specific melanization response [23,24]. These immune pathways are induced in response to both beneficial and pathogenic microbes. One of the initial, and most rapidly induced, host responses is the production of reactive oxygen and chlorine species (ROS and RCS, respectively). Lactate derived from Lactobacilli activates the Nox pathway in gut cells to release reactive oxygen species (ROS) as a mechanism to reduce overgrowth of the microbiota [7,25], while pathogenic bacteria stimulate ROS and RCS production via the dual oxidase system (Duox; Figure 2) [26]. These different mechanisms of microbial control via ROS allow the host to mount variable responses to control gut homeostasis, but the distinction between pathogen and microbiota is not absolute. Lactobacillus brevis, a common microbiota member, was found to release uracil and stimulate ROS production via Duox, indicating the importance of specific microbial members in immune-mediated regulation of homeostasis [26]. Less is known about the role of RCS, which is also regulated by Duox, in the D. melanogaster gut. However, a recent study has shown that RCS can limit colonization by an invader (a human-derived E. coli probiotic) to the fly gut [RM Derke et al., https://www.biorxiv.org/content/10.1101/690669v2].
Figure 2.
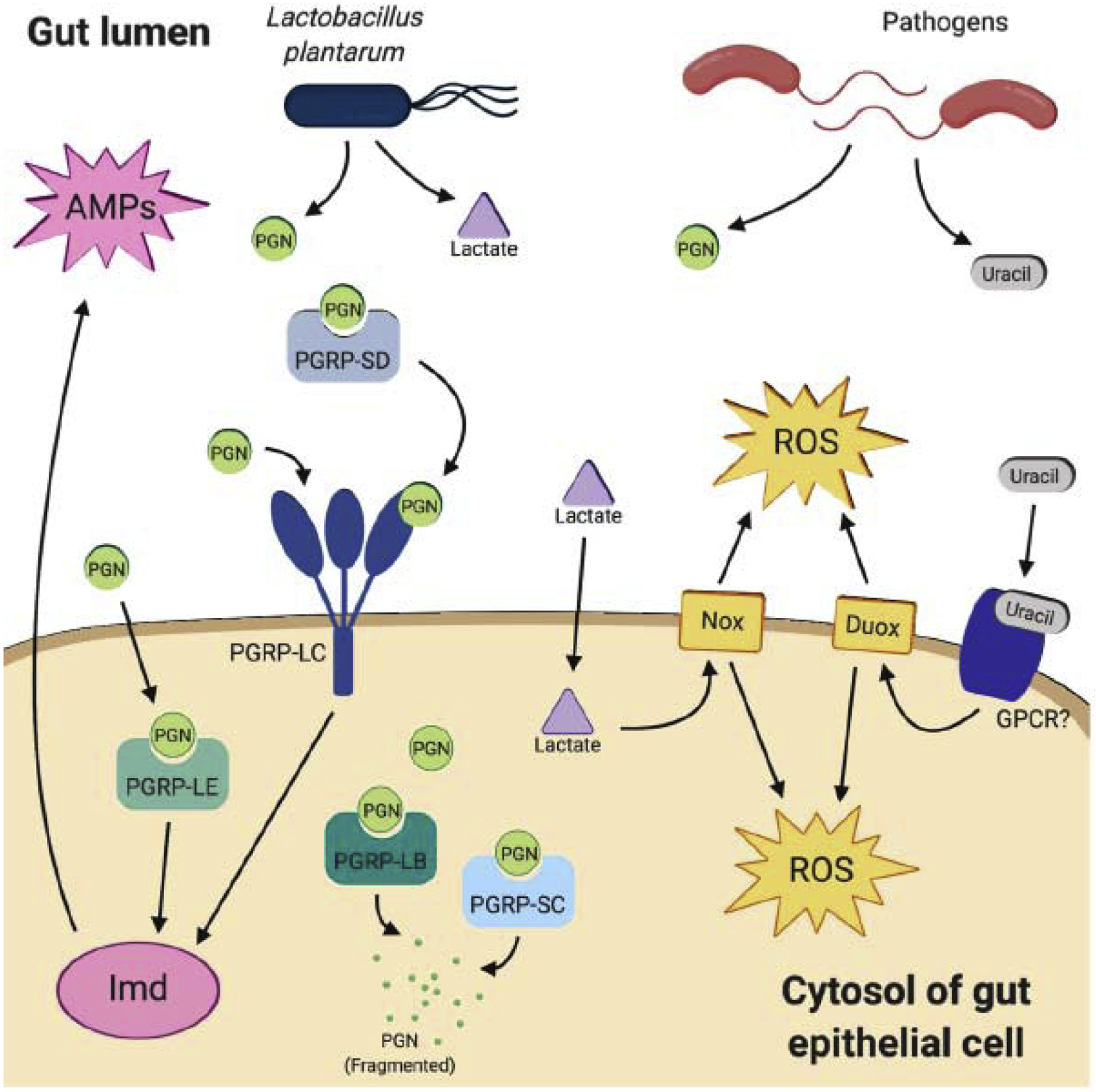
Gut bacteria modulate Imd activation, leading to control of both microbiome and pathogen load via AMPs and ROS. PGN = peptidoglycan; PGRP = peptidoglycan receptor protein; AMPs = antimicrobial peptides; ROS = reactive oxygen species; GPCR = G-protein coupled receptor
As in the oxidative burst response, the humoral immune response, characterized by the production of AMPs, is similarly stimulated by both pathogens and the microbiota. This is due to the nature of the two regulatory pathways, the immune-deficiency (Imd) and Toll, which are activated by pattern recognition receptors (PRRs) in response to conserved microbial associated molecular patterns (MAMPs). The immune-deficiency (Imd) signaling pathway is primarily induced by recognition of Gram-negative, or- DAP-type, peptidoglycan by peptidoglycan receptor proteins (PGRPs) present either in the membrane or cytosol of gut epithelial cells [27]. Many fly pathogens and microbiome members including Acetobacter spp., as well as some Lactobacilli (See Table 1) possess DAP-type peptidoglycan [28,29], whereas activation of the Toll pathway is initiated either by recognition of lysine (Lys)-type peptidoglycan by PGRP-SA or fungal ß-glucans by the misnamed Gram-negative binding proteins (GNBPs; not involved in sensing Gram-negative peptidoglycan) [30]. D. melanogaster PGRPs within the Imd signaling cascade known to recognize peptidoglycan derived from the microbiota include PGRP-LC, -LE, -SD, -LB, and -SC (Figure 2) [31,32]. Recently, PGRP-LB was shown to protect the host from deleterious effects caused by the dissemination of peptidoglycan into the hemolymph and other organ systems, demonstrating the delicate balance that exists between the immune system and microbes in the gut (Figure 2) [33].
Table 1.
Gram stain results and peptidoglycan structure of common D. melanogaster microbiota members.
Interestingly, the majority of commonly reported members of the bacterial microbiome have cell walls containing DAP-type peptidoglycan. In addition, the nature of Toll activation, which involves complex serine protease cascades downstream of PRR binding, is not considered compatible with the normal pH and enzyme profile of the fly gut [31]. For these reasons, the Toll pathway is not regarded as a major immune response pathway responding to and controlling microbial load in the fly gut. At least two common Lactobacillus spp. in the fly, L. brevis and L. fructovorans, contain Lys-type peptidoglycan (Table 1), but interactions between these bacteria and Toll activation have not been extensively examined. Lys-type peptidoglycan-mediated Toll activation was recently shown to be modulated by nephrocytes, which filter hemolymph. Flies lacking nephrocytes experienced constitutive Toll activation due to microbiota-derived peptidoglycan in the hemolymph, identifying a novel contribution of Toll signaling in homeostatic regulation of the microbiome [34]. Additional recent work suggests a role for Toll in activating epithelial renewal in the gut in response to the microbiota, but further studies are necessary to better understand the role of Toll signaling in regulating gut homeostasis [ML Atilano & M Glittenberg et al., https://www.biorxiv.org/content/10.1101/248138v1].
Microbiome and the art of fly homeostatic maintenance
While the host immune response has an important role in negotiating interactions between a host and its microbiome, mechanisms that control or repair damage and maintain homeostasis have emerged as being equally critical for fly health. Perturbations that alter the homeostatic balance between the host and microbiome can lead to states of instability or disease.
ROS and RCS, in addition to regulating microbial load in the gut, damage gut cells that in turn stimulate epithelial renewal via EGFR and JAK-STAT signaling pathways, leading to gut repair [25,35]. These pathways are induced in response to both microbiota and pathogens, but the level of damage they cause, and thus, the level to which the pathways and renewal program are activated differs [8,12]. One consequence is that the damage inflicted by microbiota can in essence prime the gut environment, thereby contributing to maintenance of homeostasis by allowing a more rapid renewal response to pathogens [7,25,26].
Dysbiosis, a state of disrupted microbiome composition and/or abundance, is associated with many disease states across diverse eukaryotes. In flies, dysbiosis is associated with overactive immune stimulation [36,37], and has been linked to aging- related pathologies, such as increased gut permeability and mortality due to decreased intestinal stem cell turnover [8,9]. Because intestinal stem cell turnover is intimately linked to immune activation in the gut, the effects the microbiome exerts on the immune system has downstream impacts on intestinal integrity [38]. Two mechanisms by which dysbiotic microbiomes synergize with host immunity to cause gut instability are via overaccumulation of lactate and ROS. When increased due to the presence of L. plantarum, the microbial byproduct lactate causes gut acidification and epithelial dysplasia, resulting in ROS production and reduced fly lifespan-pathologies that are not observed in flies containing lactate-deficient L. plantarum [7]. Additionally confirming L. plantarum as a microbe with the potential to promote disease phenotypes, mono-association with L. plantarum was shown to result in an overall loss of gut barrier integrity, possibly due to overactivation of damaging ROS in the gut [10]. However, L. plantarum has also been associated with a reduction in dysbiosis. In flies deficient in the histone demethylase KDM5, which leads to an overactive Imd response and dysbiosis, L. plantarum supplementation was sufficient to restore normal activation of Imd [37], indicating that additional factors may be involved that modulate the impact of gut microbes on host health.
Indirect contributions of the microbiome to homeostasis
The microbiome and microbiome products can directly affect host signaling pathways to both maintain and perturb homeostasis. Equally important are indirect effects of the microbiome, but we currently know far less about their contributions and underlying mechanisms. One such indirect effect is via microbe-microbe interactions, either between microbiota members or between microbiota and pathogens. Certain host responses have been attributed to mixed (but defined) communities that are not seen in hosts mono-colonized with individual members of the mixed community [39]. Recently, Sommer & Newell reported a mutualistic relationship between two D. melanogaster gut microbes, A. fabarum and L. brevis, in which the Acetobacter sp. utilized Lactobacillus fermentation products for its growth, which in turn impacted host metabolic status [40]. Another recent study showed that a specific consortium of Lactobacillus and Acetobacter species, when present in the gut, prevented epithelial renewal responses to Vibrio cholerae infection, indicating that interactions between microbiota members influenced the pathologic host response to V. cholerae infection [13]. Together, these studies demonstrate how species-specific interactions between microbes in the gut can modulate host physiology.
Interactions have also been reported between the microbiome and invading pathogens, either to the host’s benefit or peril. L. plantarum is protective of flies challenged with Pseudomonas aeruginosa or Serratia marcescens, but the mechanism behind this protection has not been fully explored [41]. In addition, certain strains of yeast, which are considered part of the D. melanogaster microbial community in nature, offer similar protection against the pathogen Aspergillus flavus when administered live (but not dead), but again, the underlying mechanisms are not well understood [42]. In contrast, Fast et al. identified a mechanism by which the fly microbiome increases Vibrio cholerae virulence. This deleterious effect of a single microbiome member was mediated by an as yet uncharacterized interaction between Acetobacter pasteurianus and the V. cholerae Type 6 Secretion System (T6SS), possibly through the Imd pathway [5]. Interestingly, the pathologies associated with A. pasteurianus-T6SS interactions were found to be independent of V. cholerae’s effects on host epithelial renewal as seen in [13], reinforcing that microbiome-mediated effects on physiology can be multi-faceted.
It is also possible that observed interactions between the microbiota and pathogens can be attributed to the role of the microbiome as barriers to pathogen colonization. While many members of the D. melanogaster microbiome are thought to colonize due to continuous re-inoculation from the food substrate, their presence in the gut may yet block pathogens from establishing themselves. This is suggested by work showing that prior colonization of microbiota members reduced subsequent invasion by strains of the same species (Lactobacillus plantarum). An L. plantarum strain isolated from wild-caught flies was best at colonizing the gut and less likely to be displaced by L. plantarum strains from lab-reared flies or humans [43]. In another study, an A. thailandicus isolated from wild flies also colonized more stably than other common microbiome members [44]. Together, these studies highlight that strain-level differences within the microbiome can affect host interactions. Future work investigating the importance of species- and strain-specific microbe-microbe interactions in host physiology will be of great interest to better understand the microbiome as a homeostatic regulatory organ.
Nutritional and metabolic contributions to gut homeostasis
While regulation of immunity and epithelial renewal pathways are perhaps the most well-studied aspects of microbiome-mediated gut homeostasis, the D. melanogaster microbiota additionally contributes to host nutrition and metabolic function. Gut bacteria activate insulin signaling, either via the TOR pathway or acetic acid metabolism, to promote larval growth [19,20,45]. Axenic flies are metabolically deficient compared to their conventionally reared counterparts [39,45], suggesting that the host utilizes the microbiome to promote normal metabolism and development. Indeed, both living and dead gut microbes rescue undernutrition phenotypes on protein-poor diets, indicating that the microbiome can be a direct nutritional source [22]. It is likely that microbiome-mediated impacts on nutrition and metabolism have downstream implications for immune function and gut renewal, and vice-versa, but these relationships require more attention in future work.
Conclusions
Investigations into interactions between microbes and D. melanogaster have revealed the microbiome to be an important, yet malleable aspect of fly physiology. As such, the host has mechanisms in place to maintain homeostasis of its resident community, some of which depend on direct host-microbe interactions, others of which are dependent on cross-talk between microbiota members themselves. Breakdown of these control mechanisms lead to loss of both host and microbiome homeostasis, resulting in states of instability that can be detrimental to fly health. As the microbiome is such a complex and pervasive entity with regard to physiology, future work exploring additional mechanisms of host microbial maintenance and influences of species-specific, and even strain-specific, interactions within the gut will be of great value in understanding the microbiome as a fundamental system.
Figure 3.
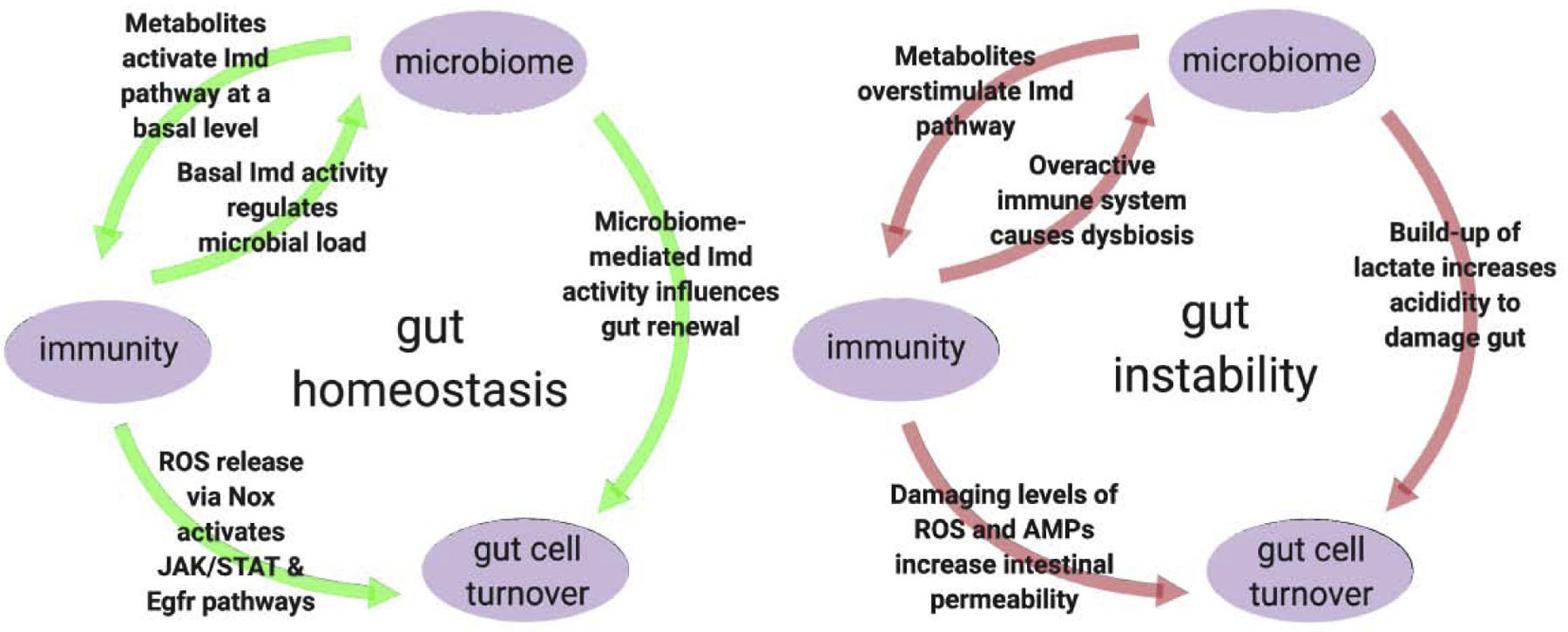
Gut homeostasis is regulated as a tripartite balance between immune activation, gut repair, and actions of the microbiome.
Highlights.
Influences of the microbiome on host physiology can be both direct and indirect
Hosts possess mechanisms to maintain homeostasis in response to the microbiome
Perturbations of homeostatic mechanisms can result in disease states in the host
Acknowledgements
This work was supported by the National Institutes of Health [R35GM128871] and the University of Connecticut.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: None
References
- 1.Wong CNA, Ng P, Douglas AE: Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol 2011, 13:1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A: Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 2011, 7:e1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broderick NA, Lemaitre B: Gut-associated microbes of Drosophila melanogaster. Gut Microbes 2012, 3:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas AE: The Drosophila model for microbiome research. Lab Animal 2018, 47:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fast D, Kostiuk B, Foley E, Pukatzki S: Commensal pathogen competition impacts host viability. Proc. Natl. Acad. Sci. U.S.A 2018, 115:7099–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]; * In this study, Fast et al. identify a microbiota-pathogen relationship between Acetobacter and Vibrio cholerae that is disadvantageous to the host, demonstrating that microbe-microbe interactions can have complex consequences for host biology.
- 6.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ: Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319:777–782. [DOI] [PubMed] [Google Scholar]
- 7.Iatsenko I, Boquete J-P, Lemaitre B: Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity 2018, 49:929–942. [DOI] [PubMed] [Google Scholar]; ** This study demonstrates the roles of PGRP-SD in maintaining homeostasis and identifies Lactobacillus-produced lactate as an important metabolite impacting physiology.
- 8.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B: Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & Development 2009, 23:2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, et al. : Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep 2015, 12:1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fast D, Duggal A, Foley E: Monoassociation with Lactobacillus plantarum disrupts intestinal homeostasis in adult Drosophila melanogaster. mBio 2018, 9:603. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Fast et al. demonstrate here that a common and widely studied D. melanogaster symbiont, Lactobacillus plantarum, when mono-associated with the fly, causes major disruptions to intestinal homeostasis leading to reduced longevity compared to germ-free flies. This study highlights the tight balance that must exist between microbes and host to maintain whole organism homeostasis.
- 11.Buchon N, Broderick NA, Kuraishi T, Lemaitre B: Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010, 8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broderick NA, Buchon N, Lemaitre B: Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 2014, 5:e01117–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fast D, Petkau K, Ferguson M, Shin M, Galenza A, Kostiuk B, Pukatzki S, Foley E: Vibrio cholerae-symbiont interactions inhibit intestinal repair in Drosophila. Cell Rep 2020, 30:1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]; * While Fast et al. (ref. 5) identified interactions between the V. cholerae type 6 secretion system and Acetobacter pasteurianus, this study further investigates mechanisms by which specific microbiomes affect epithelial renewal in response to pathogens. It identifies a consortium of bacterial species that, when mixed, inhibit epithelial repair during V. cholerae infection, cementing microbe-microbe interactions between microbiota members as significant factors impacting physiology.
- 14.Elgart M, Stern S, Salton O, Gnainsky Y, Heifetz Y, Soen Y: Impact of gut microbiota on the fly’s germ line. Nat Commun 2016, 7:11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitão-Gonçalves R, Carvalho-Santos Z, Francisco AP, Fioreze GT, Anjos M, Baltazar C, Elias AP, Itskov PM, Piper MDW, Ribeiro C: Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 2017, 15:e2000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E: Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A 2010, 107:20051–20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer CN, Trautman EP, Crawford JM, Stabb EV, Handelsman J, Broderick NA: Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife 2017, 6:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong AC-N, Wang Q-P, Morimoto J, Senior AM, Lihoreau M, Neely GG, Simpson SJ, Ponton F: Gut Microbiota Modifies Olfactory-Guided Microbial Preferences and Foraging Decisions in Drosophila. Curr. Biol 2017, 27:2397–2404. [DOI] [PubMed] [Google Scholar]
- 19.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F: Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011, 14:403–414. [DOI] [PubMed] [Google Scholar]
- 20.Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A, Yoon J-H, Ryu J-H, Lee W-J: Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011, 334:670–674. [DOI] [PubMed] [Google Scholar]
- 21.Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW: Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep 2015, 10:865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keebaugh ES, Yamada R, Obadia B, Ludington WB, Ja WW: Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 2018, 4:247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Keebaugh et al. contribute significant knowledge regarding the role of the microbiome as supplemental nutrition in nutrient-poor environments. This study shows that even heat-killed microbes rescue undernutrition phenotypes, suggesting that microbes could be an important source of protein independent of other metabolic contributions to physiology.
- 23.Buchon N, Silverman N, Cherry S: Immunity in Drosophila melanogaster — from microbial recognition to whole-organism physiology. Nature Reviews Immunology 2014, 14:796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broderick NA: Friend, foe or food? Recognition and the role of antimicrobial peptides in gut immunity and Drosophila-microbe interactions. Philos. Trans. R. Soc. Lond., B, Biol. Sci 2016, 371:20150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. : Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013, 32:3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K-A, Kim S-H, Kim E-K, Ha E-M, You H, Kim B, Kim M-J, Kwon Y, Ryu J-H, Lee W-J: Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 2013, 153:797–811. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Yano T, Aggarwal K, Lim J-H, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh B-H, et al. : PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol 2006, 7:715–723. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer KH, Kandler O: Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 1972, 36:407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espaillat A, Forsmo O, Biari El K, Björk R, Lemaitre B, Trygg J, Cañada FJ, de Pedro MA, Cava F: Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. J. Am. Chem. Soc 2016, 138:9193–9204. [DOI] [PubMed] [Google Scholar]
- 30.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J: The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002, 416:640–644. [DOI] [PubMed] [Google Scholar]
- 31.Buchon N, Broderick NA, Lemaitre B: Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol 2013, 11:615–626. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Su F, Li Q, Zhang J, Li Y, Tang T, Hu Q, Yu X-Q: Pattern recognition receptors in Drosophila immune responses. Dev. Comp. Immunol 2020, 102:103468. [DOI] [PubMed] [Google Scholar]
- 33.Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, Royet J: Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe 2018, 23:215–228. [DOI] [PubMed] [Google Scholar]; ** Charroux et al. demonstrate a mechanism of host control of the microbiota in cytosolic PGRP-LB, which prevents dissemination of bacterial peptidoglycan into hemolymph and other surrounding tissues. The degradation of peptidoglycan by PGRP-LB was found to regulate immune signaling, creating a fine-tuned feedback loop to maintain homeostatic immune activation.
- 34.Troha K, Nagy P, Pivovar A, Lazzaro BP, Hartley PS, Buchon N: Nephrocytes remove microbiota-derived peptidoglycan from systemic circulation to maintain immune homeostasis. Immunity 2019, 51:625–637. [DOI] [PubMed] [Google Scholar]; ** This study characterizes a novel mechanism by which the Toll pathway regulates gut homeostasis via interactions with the microbiota. Troha et al. show that the hemolymph filtration cells called nephrocytes are responsible for degrading bacterial peptidoglycan to reduce chronic Toll activation.
- 35.Morin-Poulard I, Vincent A, Crozatier M: The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT 2013, 2:e25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paredes JC, Welchman DP, Poidevin M, Lemaitre B: Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 2011, 35:770–779. [DOI] [PubMed] [Google Scholar]
- 37.Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, Mao J-H, Secombe J, Dan Z, Chen J-H, et al. : Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe 2019, 25:537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petkau K, Ferguson M, Guntermann S, Foley E: Constitutive immune activity promotes tumorigenesis in Drosophila intestinal progenitor cells. Cell Rep 2017, 20:1784–1793. [DOI] [PubMed] [Google Scholar]
- 39.Newell PD, Douglas AE: Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol 2014, 80:788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommer AJ, Newell PD: Metabolic basis for mutualism between gut bacteria and its impact on their host Drosophila melanogaster. Appl. Environ. Microbiol 2018, 85:e01882–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study found that gnotobiotic flies containing L. brevis and A. fabarum contained lower triglyceride levels compared to mono-colonized flies, suggesting that cross-feeding of metabolites between microbiome members is an important mechanism by which gut bacteria contribute to metabolic homeostasis.
- 41.Blum JE, Fischer CN, Miles J, Handelsman J: Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 2013, 4:e00860–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez-Camejo LA, Garcia-Alicea M, Maldonado-Morales G, Bayman P: Probiotics may protect Drosophila from infection by Aspergillus flavus. IJPSR 2017, 8:1624–1632. [Google Scholar]
- 43.Obadia B, Güvener ZT, Zhang V, Ceja-Navarro JA, Brodie EL, Ja WW, Ludington WB: Probabilistic invasion underlies natural gut microbiome stability. Curr. Biol 2017, 27:1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pais IS, Valente RS, Sporniak M, Teixeira L: Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018, 16:e2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, Watnick PI: The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 2014, 16:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]


