Abstract
Background
The early identification of patients at risk of clinical deterioration is of interest considering the timeline of COVID-19 after the onset of symptoms.
Objective
The aim of our study was to evaluate the usefulness of testing serum IL-6 and other serological and clinical biomarkers, to predict a short-term negative clinical course of patients with noncritical COVID-19.
Methods
A total of 208 patients with noncritical COVID-19 pneumonia at admission were consecutively enrolled. Clinical and laboratory findings obtained on admission were analyzed by using survival analysis and stepwise logistic regression for variable selection. Three-day worsening as outcome in a logistic model to generate a prognostic score was used.
Results
Clinical worsening occurred in 63 patients (16 = died; 39 = transferred to intensive care unit; 8 worsening of respiratory failure). Forty-five of them worsened within 3 days after admission. The risk of clinical worsening was progressively enhanced along with increasing quartiles of IL-6 levels. Multivariate analysis showed that IL-6 (P = .005), C-reactive protein (CRP) (P = .003), and SaO2/FiO2 (P = .014) were the best predictors for clinical deterioration in the first 3 days after admission. The combined score yielded an area under the curve = 0.88 (95% confidence interval: 0.83-0.93). A nomogram predicting the probability of 3-day worsening was generated. The score also showed good performance for 7-day and 14- or 21-day worsening and in predicting death occurring during all the follow-up.
Conclusions
Combining IL-6, CRP, and SaO2/FiO2 in a score may help clinicians to identify on admission those patients with COVID-19 who are at high risk for a further 3-day clinical deterioration.
Key words: COVID-19, IL-6, SARS-CoV-2, C-reactive protein, Risk factors
Abbreviations used: AACI, Age-adjusted Charlson index; ARDS, Acute distress respiratory syndrome; AUC, Area under the curve; CI, Confidence interval; COVID-19, Coronavirus disease; CRP, C-reactive protein; CRS, Cytokine release syndrome; ICU, Intensive care unit; LDH, Lactate dehydrogenase; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; SD, Standard deviation; SF ratio, SaO2/FiO2
What is already known about this topic? Several clinical and laboratory factors have been reported to be associated with disease severity and death in patients with COVID-19. The time between hospital admission and clinical deterioration may be very short.
What does the article add to our knowledge? We showed that elevated serum IL-6 levels at admission correlate with clinical worsening in COVID-19. We identified a 3-variable score (IL-6, C-reactive protein [CRP], SaO2/FiO2) able to predict further clinical deterioration of patients with moderate-to-severe COVID-19 early in the course of admission.
How does the study impact current management guidelines? IL-6, CRP, and SaO2/FiO2 ratio, combined in our proposed score, could help clinicians to identify on admission those patients with COVID-19 who are at high risk for a further 3-day clinical deterioration.
In December 2019, a novel coronavirus disease (COVID-19) was reported in China caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The number of infected patients has increased rapidly worldwide, and at the end of January 2020, the first 2 cases in Italy were recorded. More than 140,000 cases have been confirmed in Italy, so far, with a high case fatality rate. The majority of patients with COVID-19 experience a mild influenza-like illness. However, a significant proportion develop pneumonia that may evolve in acute distress respiratory syndrome (ARDS) in nearly 15% to 25% of cases.1 Old age and chronic comorbidieties such as hypertension, diabetes mellitus, renal, and heart diseases have been associated with the negative clinical outcome of COVID-19 cases.2 , 3
The immune response to SARS-CoV-2 is key for control and resolution of infection. Specifically, a rapid innate immune response is the first line of defense against viruses, but dysregulated and excessive immune responses may cause immunopathology.4 , 5 In fact, inflammatory cytokines (IL-6, TNF-α, IL-10, IL-2, IL-7, CXCL10, CCL2, CCL3) are higher in plasma of severe cases (“cytokine storm”) and are associated with pulmonary inflammation and extensive lung tissue damage in patients with COVID-19.5 In other coronavirus infections (SARS-CoV), it has been shown that type I IFNs are critical for initiation of the response and viral clearance, whereas delayed production of type I IFNs is associated with a severe clinical disease, as they are also involved in activation and recruitment of inflammatory monocytes in target tissues such as lung.6
Laboratory hallmarks in patients with COVID-19, according to the current literature, are decreased white blood cell counts, lymphocytopenia, high level of neutrophil count, increase of D-dimer, and C-reactive protein (CRP) particularly in more severe cases.7
IL-6 is a proinflammatory multifunctional cytokine released by several cell types during SARS-CoV-2 infection, including endothelial cells. In a cross-sectional study where receiver operating characteristic (ROC) analysis was applied, IL-6 serum levels above the value of 24.3 pg/mL were associated with severe pneumonia in patients with COVID-19.8 Notably, it has been shown that both in human and mice, IL-6 downmodulates the cytotoxic activity of natural killer cells, by decreasing the release of perforin and granzyme B involved in the lysis of infected cells.9
The aim of our study was to evaluate the usefulness of testing levels of serum IL-6, alone and together with other serological and clinical biomarkers, to predict who is at risk for a short-term negative clinical course of COVID-19 among patients admitted to a noncritical hospital setting.
Methods
Study population
This was an observational retrospective study including 208 patients (M/F: 135/73) aged 21 to 94 years (mean, 65.7 ± 15 years) with laboratory-confirmed SARS-CoV-2 infection by positive reverse transcriptase polymerase chain reaction on nasopharyngeal swab, according to World Health Organization interim guidelines10 and admitted to Careggi University Hospital (Florence, Italy), from March 7, 2020, to March 30, 2020. All patients were followed until death or discharge, except for 4 patients who were still admitted and were improving after transfer from intensive care unit (ICU). The median length-of-stay in the infectious disease unit was 6 days (range, 1-20 days), and the median overall length-of-stay was 8 days (1-51 days). All 208 patients, at admission, displayed radiologic evidence of pneumonia and were classified as moderate (n = 91, 43.8%) or severe (n = 117, 56.2%), according to the Guidelines for Diagnosis and Management of COVID-19 (7th edition) issued by the National Health Commission of China.11 None of them displayed need of ICU ventilatory assistance at the time of hospital admission. For our model, we have used the pulse oximetric saturation SaO2/FiO2 (SF ratio) as reliable noninvasive surrogate for PaO2/FiO2.12, 13, 14 The procedures followed in the study were approved by the Local Ethical Committee (protocol 16859).
Data collection
We reviewed clinical records and laboratory findings (white blood cell count, neutrophils, eosinophils, lymphocytes, monocytes, lactate dehydrogenase [LDH], D-dimer, ferritin, fibrinogen, CRP, liver, and kidney function test) of patients with objectively confirmed SARS-CoV2 pneumonia. Normal range of laboratory examinations were defined according to the regional hospital laboratory practice. Chest imaging was also performed for all inpatients. Serum IL-6, TNF-α, IL-1β, and IL-10 were measured by using a commercially available ELISA test (Invitrogen, Thermo Fisher Scientific, Waltham, Mass). Clinical data about comorbidities were obtained and incorporated in the age-adjusted Charlson index (AACI).15 One of the following outcomes was selected for the definition of clinical worsening: (1) death during stay in admittance ward; (2) transfer to ICU due to severe worsening of respiratory failure requiring invasive support measures to correct hypoxiemia; and (3) worsening of respiratory failure but not requiring invasive ventilatory support. Clinical and demographic characteristics of patients are summarized in Table I .
Table I.
Demographic and clinical characteristics of study population
| All (n = 208) | 3-day clinical course |
||
|---|---|---|---|
| Worsened (n = 63) | Stable/improved (n = 145) | ||
| Gender (F/M) | 74/134 | 17/46 | 57/88 |
| Age (median ± SD) | 66 ± 15 | 72 ± 13 | 63 ± 15 |
| Smoking | |||
| Current | 11 | 4 | 7 |
| Former | 63 | 25 | 38 |
| Nonsmoker | 134 | 34 | 100 |
| Hypertension (%) | 98 (47.1) | 37 (58.7) | 61 (42.1) |
| Diabetes (%) | 40 (19.2) | 17 (27) | 23 (15.9) |
| Obesity (%) | 25 (16.2) | 7 (19) | 18 (15) |
| Chronic lung disease (%) | 21 (10.1) | 11 (17.5) | 10 (6.9) |
| Chronic heart failure (%) | 19 (9.1) | 7 (11.1) | 12 (8.3) |
| Chronic renal failure | |||
| Mild-moderate (%) | 8 (3.8) | 3 (4.8) | 5 (3.4) |
| Severe (%) | 6 (2.9) | 3 (4.8) | 3 (2.1) |
| Solid tumors∗ (%) | 15 (7.2) | 6 (9.5) | 9 (6.2) |
| Hematologic malignancy∗ (%) | 6 (2.9) | 2 (3.2) | 4 (2.7) |
| Charlson comorbidity index† (mean ± SD) | 3.43 ± 2.59 | 4.64 ± 2.96 | 2.91 ± 2.23 |
| IL-6 serum levels (pg/mL, mean ± SD) | 27 ± 40.9 | 53.6 ± 63.8 | 15.7 ± 15.6 |
| C-reactive protein | 85.4 ± 73 | 127 ± 92.6 | 67.7 ± 54.2 |
| SaO2/FiO2 ratio | 335 ± 131 | 243 ± 123 | 375 ± 114 |
Body mass index >30.
SD, Standard deviation.
Diagnosed in the last 5 years.
Age-adjusted.
Statistical analysis
We used survival analysis to investigate the prognostic value of basal IL-6, both as a linear term and as quartiles in a Cox model. We computed Harrell's C as an overall measure of discrimination for all candidate covariates as linear continuous variables.
A 3-day worsening as a response variable in a logistic model to generate a prognostic score was used because most patients worsened within 3 days after admission. We used stepwise logistic regression for variable selection, with a forward procedure and an entry probability of 0.1.
Model-based discrimination was assessed using the area under the curve (AUC), and calibration was assessed using the Hosmer and Lemeshow goodness-of-fit test.
Finally, we generated a nomogram to calculate an overall score and the corresponding probability of 3-day worsening, with the main purpose of presenting graphically the impact of each predictive variable.
Continuous variables were summarized using mean and standard error and compared using Student's t-test. All tests were 2-sided, and a P value of <.05 was considered significant. All calculations were performed using Stata 16.1 software (StataCorp, College Station, TX).
Results
Serum IL-6 in patients with COVID-19 with moderate-to-severe disease
We first investigated IL-6 serum levels in all enrolled patients. Of 208 patients, 131 (62.9%) displayed IL-6 serum levels higher than the positive cutoff (10 pg/mL), with a mean value (± standard deviation [SD]) of 27 ± 40.9. No significant differences (P = .158) in IL-6 serum levels were noted between males (29.9 ± 45.6) and female (21.5 ± 29.9). It is important to note that baseline IL-6 was weakly but significantly associated with age (Spearman r = 0.24, P < .001).
In a small proportion of our cases, an increase of TNF-α and/or IL-10 was observed. Specifically, TNF-α was assessed in 61 patients (29.3%); 15 of 61 (19.7%) showed TNF-α levels higher than the positive cutoff (15 pg/mL) with a mean value (±SD) of 39.1 ± 26.6. Regarding IL-10, it was measured in 65 patients (31.2%) and it resulted in higher than the cutoff (10 pg/mL) in 21 patients (32.3%) with a mean value (±SD) of 35.7 ± 50.5. The small number of patients did not allow the involvement of these cytokines in the subsequent statistical analysis of the study. None of 65 patients displayed increased serum levels of IL-1β (data not shown).
Usefulness of IL-6 for predicting the clinical course of patients with COVID-19
Of 208 patients, 63 (30.3%) suffered from clinically significant worsening according to the criteria reported in the Methods section. In detail, 8 patients (12.7%) exhibited progressive respiratory failure, 39 (61.9%) were transferred to ICU, and 16 (25.4%) died in the admitting unit. Most patients (n = 45, 71.4%) started to experience such worsening within the first 3 days of admission. The remaining 141 patients (67.7%) were stable or improved during the period of clinical observation until the discharge. Four patients are still hospitalized at the moment of final analysis, and they are improving after transfer back from ICU.
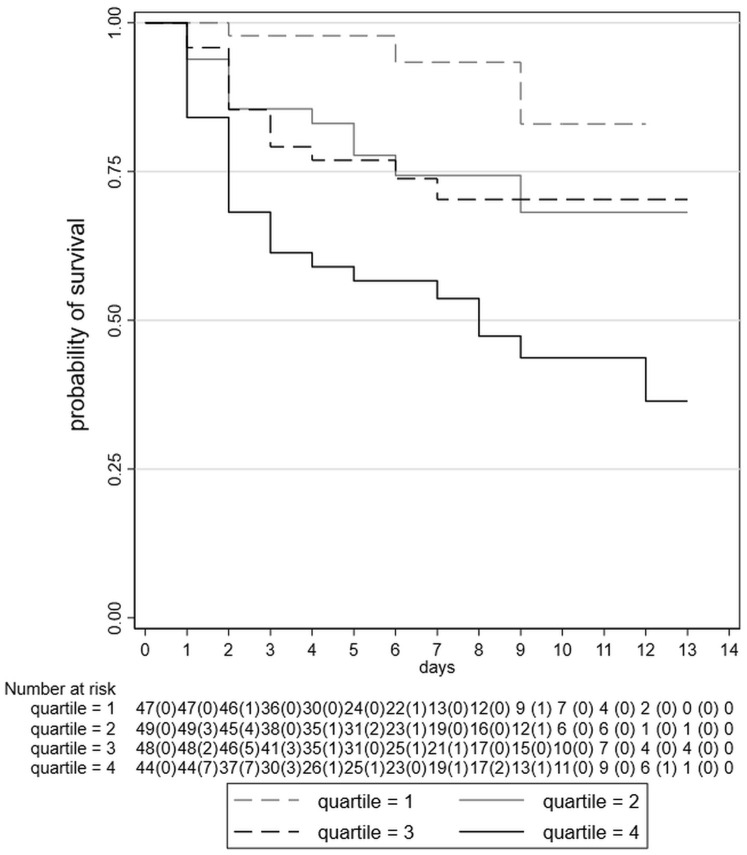
We investigated the predictive ability of basal IL-6 values as quartiles, which were formed at approximate cutoffs (<6.8, ≥6.8-<13.8, ≥13.8-<27.8, ≥27.8 pg/mL), reaching a maximum value of 250.7 pg/mL. Figure 1 shows Kaplan-Meier survival plots for IL-6 quartile subgroups. Using Cox regression analysis, we observed a progressive and statistically significant decreased risk for developing the negative clinical outcome (worsening) moving from the third quartile of IL-6 to the lowest one, in comparison with the highest one (Table II ) (P < .001). Harrell's C was 0.72 when IL-6 was used as a continuous covariate in the model. These data are suggestive of a moderate/satisfactory predictive performance of IL-6 serum levels at admission regarding the clinical course of COVID-19 disease.
Figure 1.
Kaplan-Meier survival curves for clinical worsening, as defined in the Methods section, by quartiles of baseline IL-6 serum levels. Number of patients at risk for each interval, with number of failures in parentheses, is shown.
Table II.
Hazard ratios of clinical worsening according to IL-6 quartiles
| HR values (95% CI), univariate | HR values (95% CI), multivariate | |
|---|---|---|
| IL-6 levels (pg/mL) | ||
| <6.8 | 0.17 (0.06-0.43) | 0.20 (0.08-0.51) |
| 6.8-13.8 | 0.34 (0.18-0.67) | 0.42 (0.21-0.85) |
| 13.8-27.8 | 0.48 (0.26-0.89) | 0.50 (0.27-0.92) |
| >27.8 | 1 | 1 |
| Age (10 y) | 1.35 (1.12-1.64) | 1.25 (1.02-1.53) |
| Sex (male) | 1.81 (1.03-3.18) | 1.68 (0.95-2.97) |
CI, Confidence interval; HR, hazard ratio.
The predictive value of IL-6 was maintained when age and sex were added to the Cox model (Table II; likelihood ratio test for the overall effect of IL-6 quartiles, P = .008), with age (P = .028) but not sex (P = .073) crossing the threshold of nominal statistical significance.
Relationship between IL-6 and other serological measures
Taking into account that IL-6 is essential for CRP synthesis in the liver and many other laboratory alterations are detectable in patients with COVID-19, we then analyzed the correlation of IL-6 serum levels with other parameters. A statistically, although moderately, significant (P < .05) Spearman r correlation with basal IL-6 levels was found for CRP (0.58), LDH (0.45), ferritin (0.45), SF ratio (0.30), neutrophil count (0.29), fibrinogen (0.26), and D-dimer (0.24). IL-6 concentration was negatively correlated with lymphocyte (−0.37), monocyte (−0.29), and eosinophil counts (−0.21) (P < .05) (Table E1, available in this article's Online Repository at www.jaci-inpractice.org).
Predictive performance of a combined score including IL-6, CRP, and SF ratio
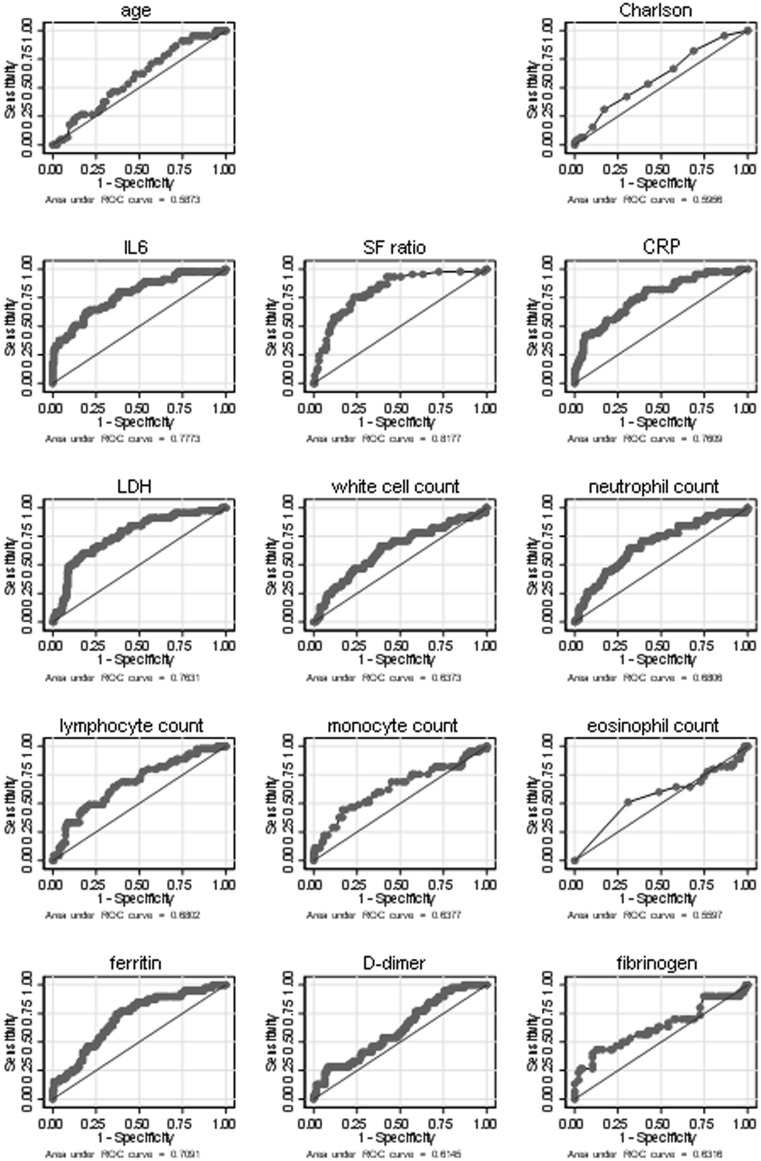
In logistic regression models, several variables were strongly associated with 3-day worsening (AUC > 0.70, P ≤ .001). The highest AUC was achieved by SF ratio (0.82), followed by IL-6 (0.77), CRP (0.76), LDH (0.76), and ferritin (0.70). A weaker but statistically significant association (AUC < 0.70, P < .05) was found for lymphocyte count (0.69), neutrophil count (0.68), fibrinogen (0.65), white cell count (0.64), and AACI (0.59). Figure E1 (available in this article's Online Repository at www.jaci-inpractice.org) presents additional information on the ROC curves and AUCs for all variables recorded in the study.
Figure E1.
ROC curves with area under the curve (AUC) values for prediction of clinical worsening within 3 days of admission for each study variable (age, IL-6, SF ratio, CRP, LDH, white blood cells, neutrophils, lymphocytes, monocytes, eosinophils, ferritin, D-dimer and fibrinogen, age-adjusted Charlson comorbidity index). CRP, C-reactive protein; LDH, lactate dehydrogenase; ROC, receiver operating characteristic; SF ratio, SaO2/FiO2.
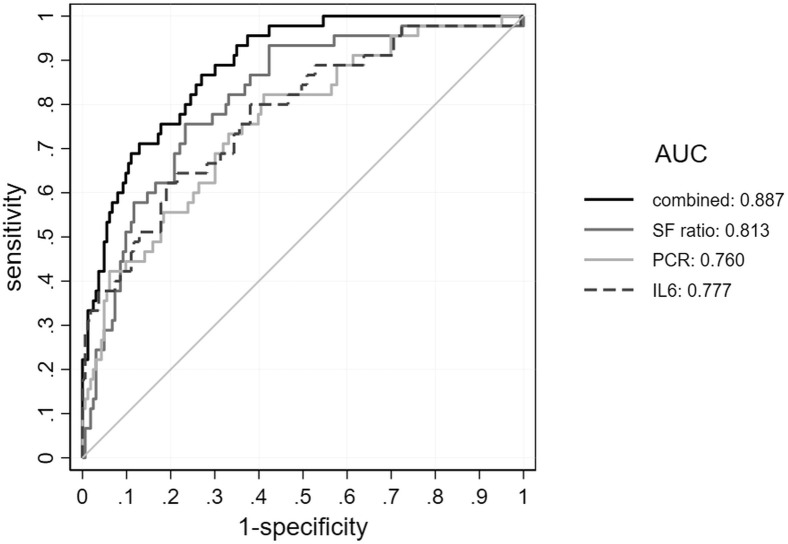
The stepwise logistic regression model retained IL-6, CRP, and SF ratio as predictors and yielded good discrimination (AUC = 0.879) and good calibration (Hosmer and Lemeshow goodness-of-fit test, P = .577). The 3-variable model (Logit(p) = −0.5541 + SF × −0.00774 + PCR × 0.00946 + IL-6 × 0.01717) was significantly better in predicting 3-day worsening than IL-6 (P = .005), CRP (P = .003), and SF ratio alone (P = .014), as shown with ROC curves in Figure 2 . The score computed to predict 3-day worsening also showed good performance for 7-day worsening (AUC = 0.86) and 14- or 21-day worsening (AUC = 0.83). The model also had a good performance in predicting death (n = 26) occurring during all the follow-up in the admitting unit or in ICU (AUC = 0.82). Fibrinogen and D-dimer were not included in the stepwise regression model due to incomplete data, but they showed a modest univariate predictive performance.
Figure 2.
ROC curves with area under the curve (AUC) values for prediction of clinical worsening within 3 days of admission for each predictor included in the model (SF ratio, IL-6, and CRP) and for the combined score. CRP, C-reactive protein; ROC, receiver operating characteristic; SF ratio, SaO2/FiO2.
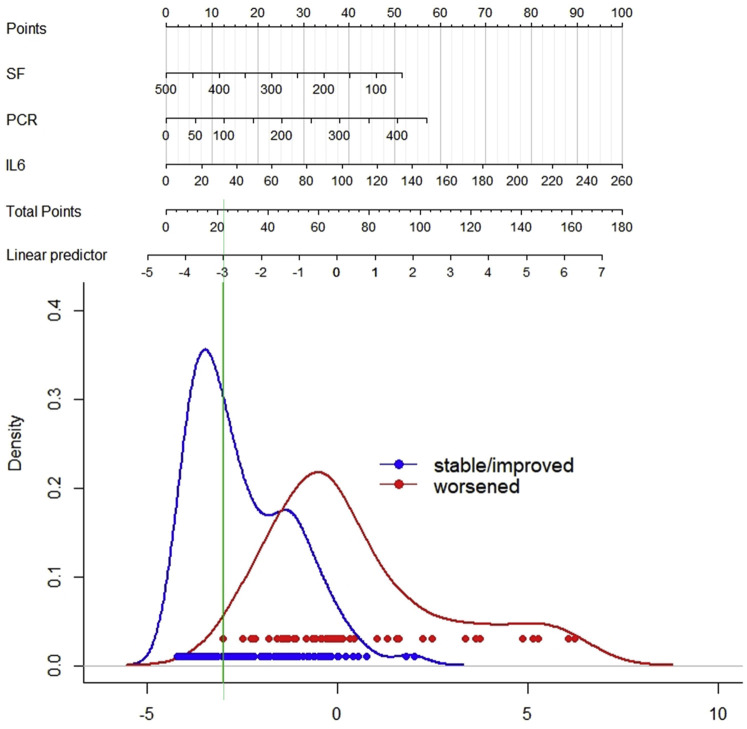
Figure 3 shows a nomogram predicting the probability of 3-day worsening. The best discrimination between stable or improved versus worsened patients was obtained at low scores, that is, when all 3 parameters were near normal. At less than 10% predicted probability of worsening, 3 of 109 patients worsened (negative predictive value = 97%, 95% confidence interval [CI]: 22%-99%), and 42 of 99 patients worsened above this level (positive predictive value = 42%, 95% CI: 33%-53%).
Figure 3.
Nomogram for predicting clinical deterioration of noncritical COVID-19 within 3 days on admission. To use the nomogram: for each predictor, determine the corresponding points by drawing a straight line up from the patient's value; sum the points obtained for each predictor; and locate the total sum on the upper point line. Identify the corresponding value in the linear predictor scale by drawing a straight line down. Values to the right of the green line predict 3-day worsening, and values to the left predict no-worsening. PCR, Polymerase chain reaction; SF ratio, SaO2/FiO2.
Discussion
Because of the rapid evolution of lung inflammatory processes leading to severe and potentially life-threatening forms of COVID-19, the early identification of predictive clinical signs and biochemical markers is still a medical unmet need in SARS-CoV-2-infected patients. In our study, regarding patients with moderate-to-severe COVID-19, elevated serum IL-6 levels, obtained at the time of ward admission, predicted high risk for further deterioration in clinical condition (worsening of respiratory failure during hospitalization, ICU admission, or death), occurring as early as the following 3 days. The study also showed that other variables correlate with the risk of worsening (age, CRP, LDH, SF ratio, D-dimer, leucocyte, and neutrophil counts). Among these, IL-6, CRP, and SF ratio were the best predictors and their combination improved the predictive power. Hence, we produced a nomogram allowing for a predictive score calculation with descriptive purposes regarding the relative contribution of each variable to prediction. As for any predictive score, further elaboration with external validation in large multicenter studies is necessary before it is used for clinical decision making. The best discrimination was achieved at low levels of predicted probability, and, if validated, the score we suggest may be used for triage of patients with COVID-19 to more appropriately allocate them into low- or medium-intensity care settings.
To our knowledge, this is the first study that combines clinical parameters (SF ratio) and serological biomarkers (IL-6 and CRP) in the prediction of clinical deterioration of patients with moderate-to-severe COVID-19.
It is important to note that our suggested combined score is predictive of disease evolution within the first 3 days after hospital admission. The early identification of patients at risk of clinical deterioration is of particular interest considering the timeline of COVID-19 cases after the onset of symptoms. In fact, the experience in Wuhan Hospital (China) showed a 3.5-day median time from first hospital admission to ICU admission.5
Our study has some limitations. First, patients were included at a single university-based institution, and as for any predictive model, an external validation in other clinical settings is also mandatory Second, the sample size is relatively small considering the commonly accepted rule-of-thumb that at least 15 events per covariate are needed in a regression model; future confirmations of our data in a larger case series of patients with COVID-19 are desirable. Because of the limited sample size, further studies may lead to finding different sets of predictors with similar or better performance. Third, our primary analysis was short term (3 days); however, this is related to the aim of our model regarding clinical decision making, which was triage of patients admitted for moderate-to-severe COVID, and our model did well also for long-term outcome and death. Fourth, partial data were available for variables of interest such as D-dimer, which on the other hand did not show a strong univariate predictive value. Nonetheless, we have shown that there is potential for a predictive model using objective biochemical and physiological measures to allocate patients with moderate-to-severe COVID to the appropriate care setting.
Although older age and comorbidities were significantly associated with higher risks of the development of acute respiratory distress syndrome,5 an overall comorbidity measure such as the age-adjusted Charlson comorbidity index did not improve our predictive model.
The identification of patients at risk as early as possible would also allow more appropriate anti-inflammatory treatments before the full-blown ARDS phase, when cellular and molecular mechanisms are more difficult to control. Indeed, during severe coronavirus infections such as SARS and Middle East respiratory syndrome, pathology is frequently not directly due to the cytopathic activity of the virus, but instead related to a “cytokine storm.” Increased serum levels of proinflammatory cytokines and chemokines (IL-6, IL-1β, IL-12, CCL2, CXCL10) have been previously shown in patients with SARS with severe pulmonary involvement.16 Accordingly, in patients with severe and critical COVID-19, increased plasma levels of several cytokines and chemokines, including IL-6, have been recently reported.5 , 17 The increased ability to produce IL-6 under different clinical conditions seems to be dependent on single IL-6 gene polymorphisms.18 A dysregulation of the IL-6 production could be envisaged in COVID-19 subjects as responsible for the rapid evolution toward a more severe disease. For all these reasons, intercepting the initial phase of worsening during SARS-CoV-2 infection may be crucial for an early therapeutic intervention to modulate the exaggerated immune response.
IL-6 is a pleiotropic proinflammatory multifunctional cytokine, produced by a variety of cell types such as monocytes/macrophages, adipocytes, muscle cells, and hematopoietic cells.19 IL-6 produced during immune-mediated inflammatory processes is the main driver for CPR production by hepatocytes20; therefore, we might speculate a rise in serum IL-6 before the increase of CRP. Therefore, IL-6 serum levels could be considered as a very early biomarker in patients with COVID-19. This biological relationship between IL-6 and CRP synthesis may explain why both IL-6 and CRP maintain higher significant predictive power in comparison with the other parameters in our stepwise statistical analysis.
Thromboembolism is rapidly emerging as a key clinical concern in COVID-19 in addition to pneumonia. A massive endothelial dysfunction in COVID-19 is caused by a direct virus-induced damage, an IL-6-rich inflammatory microenvironment, and the development, at least in some patients, of procoagulant autoantibodies.21, 22, 23 Of note, endothelial cells are one of the most important sources of IL-6 during severe cytokine release syndrome (CRS).24 , 25 The initial coagulopathy of COVID-19 presents with prominent of D-dimer and fibrin/fibrinogen degradation products.26 In our study, we did not collect clinical and imaging data on thromboembolism, but we assessed the predictive value of D-dimer at admission and it was not selected as a key predictor in our model.
Although our study does not deal with therapeutic options in patients with COVID-19, the finding of specific pathogenic factors in individual subjects may support the choice of a treat-to-target strategy, as in the case of anti-IL-6R monoclonal antibody in patients with COVID-19.27 However, considering the multiple cytokines resulting from the activation of both innate and adaptive immune response during CRS, pending the results of ongoing clinical trials with various biotechnological drugs (anti-IL-1R, JAK inhibitors, BTK inhibitors, etc.), a therapeutic strategy able to simultaneously block the multiple mediators involved, such as steroids and antimalarial, may be considered.
In conclusion, taking the caveats we discussed into account, we have shown that early worsening of patients with moderate-to-severe COVID-19 can be predicted using a combination of simple clinical and biochemical parameters.
Acknowledgment
We thank Jane Griffith for English language revision.
Footnotes
No funding was received for this work.
Conflicts of interest: The authors declare that they have no relevant conflicts of interest.
Online Repository
Table E1.
Pairwise Spearman correlation between all study variables
| IL-6 | 1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 0.32 | 1 | |||||||||||
| SF ratio | −0.30 | −0.33 | 1 | ||||||||||
| CRP | 0.58 | 0.24 | −0.44 | 1 | |||||||||
| LDH | 0.45 | 0.23 | −0.51 | 0.48 | 1 | ||||||||
| WBC | 0.13 | 0.03 | −0.21 | 0.32 | 0.35 | 1 | |||||||
| Neu | 0.29 | 0.13 | −0.34 | 0.50 | 0.49 | 0.91 | 1 | ||||||
| Lymph | −0.37 | −0.31 | 0.31 | −0.36 | −0.23 | 0.26 | −0.06 | 1 | |||||
| Mono | −0.29 | −0.16 | 0.29 | −0.29 | −0.27 | 0.42 | 0.20 | 0.51 | 1 | ||||
| Eos | −0.21 | −0.19 | 0.38 | −0.12 | −0.10 | 0.29 | 0.17 | 0.34 | 0.38 | 1 | |||
| Ferritine | 0.45 | 0.18 | −0.54 | 0.49 | 0.53 | 0.12 | 0.27 | −0.24 | −0.29 | −0.30 | 1 | ||
| D-dimer | 0.24 | 0.33 | −0.31 | 0.28 | 0.44 | 0.24 | 0.36 | −0.27 | −0.18 | −0.09 | 0.28 | 1 | |
| Fibr | 0.26 | −0.18 | −0.18 | 0.49 | 0.19 | 0.13 | 0.19 | −0.08 | −0.11 | −0.10 | 0.26 | −0.11 | 1 |
| IL-6 | Age | SF ratio | CRP | LDH | WBC | Neu | Lymph | Mono | Eos | Ferritine | D-dimer | Fibr |
Correlations reaching statistical significance at P < .05 are in bold.
CRP, C-reactive protein; Eos, eosinophils; Fibr, fibrinogen; LDH, lactate dehydrogenase; Lymph, lymphocytes; Mono, monocytes; Neu, neutrophils; SF ratio, SaO2/FiO2; WBC, white blood cells.
References
- 1.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M. Comorbidity and its impact on 1590 patients with Covid-19 in China: a national analysis. Eur Respir J. 2020;26:2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q. Prevalence of comorbidieties in Coronavirus disease 2019 patients: a systematic review and meta-analysis. Inf J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan W., Ni Z., Hu Y., Ling W., Ou C., He J. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Li T., Han M., Li X., Wu D., Xu Y. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cifaldi L., Prencipe G., Caiello I., Bracaglia C., Locatelli F., De Benedetti F. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 2015;67:3037–3046. doi: 10.1002/art.39295. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases WHO—Interim Guide 2020. https://www.who.int/publications/i/item/10665-331501 Available from: Accessed March 4, 2020.
- 11.National Health Commission and National Administration of Traditional Chinese Medicine on the People's Republic of China Protocols for diagnosis and treatment of COVID-19 (7th Trial version) [in Chinese]. March 29, 2020. https://www.chinadaily.com.cn/pdf/2020/1.Clinical.Protocols.for.the.Diagnosis.and.Treatment.of.COVID-19.V7.pdf Available from: Accessed March 4, 2020.
- 12.Rice T.W., Wheeler A.P., Bernard G.R., Hayden D.L., Schoenfeld D.A., Ware L.B. Comparison of the SpO2/FiO2 ratio and the PaO2/FiO2 ratio in patients with acute lung injury or ARDS. Chest. 2007;132:410–417. doi: 10.1378/chest.07-0617. [DOI] [PubMed] [Google Scholar]
- 13.Chen W.-L., Lin W.-T., Kung S.-C., Lai C.-C., Chao C.-M. The value of oxygenation saturation index in predicting the outcomes of patients with acute respiratory distress syndrome. J Clin Med. 2018;7:205–213. doi: 10.3390/jcm7080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DesPrez K., McNeil J.B., Wang C., Bastarache J.A., Shaver C.M., Ware L.B. Oxygenation saturation index predicts clinical outcomes in ARDS. Chest. 2017;152:1151–1158. doi: 10.1016/j.chest.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlson M., Szatrowski T.P., Peeterson J., Gold J. Validation of a combined comorbidity index. J Clin Epidemiol. 1994;47:1245–1251. doi: 10.1016/0895-4356(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 16.Wong C.K., Lam C.W.K., Wu A.K., Ip W.K., Lee N.L., Chan I.H. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y. Detectable serum SARS-CoV-2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. [published online ahead of print April 17, 2020]. Clin Infect Dis. [DOI] [PMC free article] [PubMed]
- 18.Lorente L., Martin M.M., Perez-cejas A., Barrios Y., Solé-Violán J., Ferreres J. Association between IL-6 promoter polymorphism (-174G/C), serum IL-6 levels and mortality in severe septic patients. Int J Mol Sci. 2016;17:1861–1871. doi: 10.3390/ijms17111861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dempke W.C.M. Interleukin-6: a masterplayer in the cytokine network. Oncology. 2020;98:131–137. doi: 10.1159/000505099. [DOI] [PubMed] [Google Scholar]
- 20.Honsawek S., Deepaisarnsakul B., Tanavalee A., Sakdinakiattikoon M., Ngarmukos S., Preativatanyou K. Relationship of serum IL-6, C-reactive protein, erythrocyte sedimentation rate, and knee skin temperature after total knee arthroplasty: a prospective study. Int Orthop. 2011;35:31–35. doi: 10.1007/s00264-010-0973-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W. Coagulopathy and antiphospholipid antibodies in patients with Covid-19. N Engl J Med. 2020;382:e38. doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay K.A., Hanafi L.-A., Li D., Gust J., Liles W.C., Wurfel M.M. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor–modified T-cell therapy. Blood. 2017;130:2295–2306. doi: 10.1182/blood-2017-06-793141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Han W. Biomarkers of cytokine release syndrome and neurotoxicity related to CAR-T cell therapy. Biomark Res. 2018;6:4–14. doi: 10.1186/s40364-018-0116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J Med Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]