Abstract
Several studies have indicated that dietary silicon (Si) is beneficial for bone homeostasis and skeletal health. Furthermore, Si-containing bioactive glass biomaterials have positive effects on bone regeneration when used for repair of bone defects. Si has been demonstrated to stimulate osteoblast differentiation and bone mineralisation in vitro. However, the mechanisms underlying these effects of Si are not well understood. The aim of the present study was to investigate the effects of soluble Si on osteogenic differentiation and connexin 43 (CX43) gap junction communication in cultured pluripotent cells from human dental follicles (hDFC). Neutral Red uptake assay demonstrated that 25 μg/ml of Si significantly stimulated hDFC cell proliferation. Dosages of Si above 100 μg/ml decreased cell proliferation. Alizarin Red staining showed that osteogenic induction medium (OIM) by itself and in combination with Si (25 μg/ml) significantly increased mineralisation in hDFC cultures, although Si alone had no such effect. The expression of osteoblast-related markers in hDFC was analysed with RT-qPCR. OSX, RUNX2, BMP2, ALP, OCN, BSP and CX43 genes were expressed in hDFC cultured for 1, 7, 14 and 21 days. Expression levels of BMP-2 and BSP were significantly upregulated by OIM and Si (25 μg/ml) and were also induced by Si alone. Notably, the expression levels of OCN and CX43 on Day 21 were significantly increased only in the Si group. Flow cytometric measurements revealed that Si (50 μg/ml) significantly increased CX43 protein expression and gap junction communication in hDFC. Next-generation sequencing (NGS) and bioinformatics processing were used for the identification of differentially regulated genes and pathways. The influence of OIM over the cell differentiation profile was more prominent than the influence of Si alone. However, Si in combination with OIM increased the magnitude of expression (up or down) of the differentially regulated genes. The gene for cartilage oligomeric matrix protein (COMP) was the most significantly upregulated. Genes for the regulator of G protein signalling 4 (RGS4), regulator of G protein signalling 2 (RGS2), and matrix metalloproteinases (MMPs) 1, 8, and 10 were also strongly upregulated. Our findings reveal that soluble Si stimulates Cx43 gap junction communication in hDFC and induces gene expression patterns associated with osteogenic differentiation. Taken together, the results support the conclusion that Si is beneficial for bone health.
Subject terms: Preclinical research, Biomaterials - cells
Introduction
Silicon (Si) is a trace element that is suggested to be essential for the normal development of connective tissues and the skeleton1–3. Dietary Si is mainly found in plant-based foods, such as rice, whole-grain cereals (including cereal products such as beer), vegetables, fruit, and mineral water. Absorption of Si depends on its chemical form, with the soluble orthosilicic acid (OSA, Si(OH)4) being the most bioavailable. The absorption of Si in the intestine is followed by a rapid increase in the serum levels of Si, and Si that is not retained by the body is excreted in the urine within 4–8 h4. Early studies of Si depletion in chicks and rats reported severely impaired growth and abnormal development of the skeletal and connective tissues5,6. Although more recent studies have failed to repeat these dramatic effects of Si deprivation in animals, there is strong evidence that Si is positively correlated with bone metabolism and homeostasis. Interestingly, young rats that were fed a Si-depleted diet showed increased longitudinal growth due to inhibition of growth plate closure and differences in calcium to phosphorus ratio7. In the Framingham offspring cohort, strong positive correlations between dietary Si intake and hip bone mineral density was demonstrated. The effect was most pronounced in pre-menopausal women and less so in men and absent in post-menopausal women. This indicated a relationship between oestrogen status and the effects of Si on bone health, which was confirmed in the Aberdeen Prospective Osteoporosis Screening Study (APOSS)8,9. The role of soluble Si in bone formation has been investigated at the cellular level, and Si was demonstrated to stimulate osteoblast differentiation, extracellular matrix synthesis, and increased bone mineralisation in vitro3,10–13. In line with these findings, it has been demonstrated that biomaterials that contain Si, e.g., bioactive glasses such as 45S5 Bioglass, stimulate osteoblast differentiation, as evidenced by increased expression of the osteoblastic markers osteocalcin and alkaline phosphatase and an increase in new bone formation14–16. However, the mechanisms underlying the effect of Si on osteogenic differentiation and bone formation are not well understood.
The dental follicle (DF) is an ectomesenchymal tissue sac that surrounds the un-erupted tooth. It appears to regulate osteoclastogenesis and osteogenesis needed for the eruption process17–19. Pluripotent mesenchymal stem cells have earlier been demonstrated to be present in the DFs of human wisdom teeth20 and in murine DFs21. Previous studies have demonstrated that stimulated DF cells (DFC) can form mineralised nodules in vitro, and that DFC express bone-related proteins, such as alkaline phosphatase (ALP), bone sialoprotein (BSP) and osteocalcin (OCN)20,22–24. Human DFC (hDFC) are easily accessible and do not entail the same ethical concerns and controversies as embryonic stem cells. The ability of hDFC to differentiate into different cell types makes them good candidates to study osteoinductive materials, as well as an alternative source of stem cells for tissue regeneration20.
The aim of the present in vitro study was to clarify the effects of soluble Si on osteogenic differentiation and bone formation using hDFC. We investigated the effects of Si on gene expression and bone nodule formation (matrix mineralisation) in hDFC compared to osteogenic induction media (OIM). We used next-generation sequencing (NGS) and bioinformatics processing to determine the transcriptomic profiles of hDFC that were cultured in the absence or presence of OIM and Si, alone or in combination. Furthermore, the effects of Si on Connexin 43 (CX43) expression and gap junction communication (GJC) in hDFC were assessed, since Cx43-mediated GJC is crucial for osteoblast differentiation and bone formation25–27.
Patients and Methods
Ethics
All experiments and methods were performed in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Regional Ethics Board at the University of Gothenburg (Dnr. 898–13) and by the National Data Inspection Board. Informed consent was obtained from the patients and their parents. The methods described below have been reproduced in part from Uribe et al.28
Establishment of cell cultures of hDFC
hDFC cultures were established from dental follicles obtained from four different patients with impacted canines who were referred for orthodontic reasons. The patients ranged in age from 12 to 16 years. After being rinsed in Minimum Essential Medium α (α-MEM) (Gibco, Life Technologies, USA), the dental follicle tissues were minced using a sterilised scalpel, and placed in culture plates with α-MEM that was supplemented with 10% (v/v) foetal bovine serum (FBS), 2 mM Glutamax, and an antibiotic-antimycotic solution (all from Gibco, Life Technologies) at 37 °C in humidified atmosphere consisting of 5% CO2. After 48 hours of incubation, non-adherent cells were removed by changing the growth medium. The adherent cells were cultured and maintained with medium change every 3 days. Passages 3–5 were used for all the experiments.
Preparation of Silica solutions
The Si-containing medium used in the experimental setup was prepared using the method of Sripanyakorn et al.29, with minor alterations28. A stock solution (350 μg/ml) was prepared by adding 0.1 ml of concentrated sodium silicate (Sigma-Aldrich) to 49.9 ml of α-MEM. The pH was adjusted to 7.0–7.2 using HCl before 10% FBS (v/v) (Gibco Invitrogen) was added. Solutions with final concentrations of 0–250 μg/ml Si were prepared by diluting the stock solution with α-MEM + 10% FBS.
Cell proliferation assay
Neutral Red (NR) uptake assays were used to estimate cell proliferation, as previously described by Repetto et al.30. hDFC were cultured for 48 h in 96-well plates with 100 μl growth medium (α-MEM and 10% FBS) per well and in the absence or presence of 0–250 μg/ml Si. NR dye (Merck, Dannstadt, Germany) was dissolved in PBS (4 mg/ml), diluted 1:100 in α-MEM, and incubated for 24 h at 37 °C to obtain the NR incubation medium (4 μg/ml). The NR incubation medium was centrifuged at 200 × g for 5 min prior to usage and added 100 μl/well. After 2 h of incubation at 37 °C, the cells were washed with PBS (150 μl/well), and the NR destaining solution (150 μl/well; 10 ml H2O, 10 ml EtOH 99.5%, and 0.2 ml glacial acetic acid) was added to release NR from the lysosomes in the cells. After 10 min, the absorbance of the solubilised dye was quantified at 540 nm in a spectrophotometer multi-plate reader (Multiskan FC Microplate Photometer; Fisher Scientific). Protocol validated previously by Uribe et al.28. The experiment was repeated three times with four replicates per treatment.
Osteogenic induction medium
To determine osteogenic differentiation capabilities, hDFC were grown in 24-well plates to confluency. Thereafter, the hDFC were cultured for 1, 7, 14 and 21 days with osteogenic induction medium (OIM) that contained α-MEM, 10% FBS, 50 mg/ml L-ascorbic acid 2-phosphate sesquimagnesium salt and 10 mM β-glycerophosphate disodium salt hydrate, with or without Si (25 μg/ml). Untreated cells cultured in α-MEM were used as controls.
Alizarin red staining
Alizarin Red S (ARS) staining of hDFC cultures was used to study the effects of Si on bone mineralisation. ARS is a dye that binds selectively to calcium salts and is widely used for calcium mineral histochemistry. As previously described by Saugspier et al.31, monolayers of hDFC cultured for 1, 7, 14, 21 and 28 days with OIM ± Si were washed twice with PBS, fixed with ice-cold 70% ethanol for 30 min, and stained for 30 min with 40 mM ARS (Sigma Aldrich, St. Louis, MO, USA), at pH 4.2 at room temperature with rotation on an orbital shaker. The samples were then rinsed five times with water to reduce non-specific ARS staining and allowed to air-dry. Stained cells were photographed and this was followed by a quantitative extraction method adapted from the protocol described by Stanford et al.32. Briefly, ARS was released from the cell matrix by incubation of the monolayers with 10% (w/v) cetylpyridinium chloride (CPC) in 10 mM sodium phosphate (pH 7.0) for 1 h (1 ml/well). The ARS extracts were then removed and 100-μl aliquots were transferred in to a 96-well plate prior to reading the absorbance at 562 nm in a multi-plate reader (Multiskan FC Microplate Photometer; Fisher Scientific). Control values were obtained from control samples without cells. The experiment was repeated three times with four replicates per treatment.
Real-time quantitative polymerase chain reaction (RT-qPCR)
The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were followed to ensure the relevant experimental conditions and assay characteristics33. RNA extraction was carried out using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, cell monolayers were rinsed with PBS and lysis buffer was added to each well. RNA was bound to the silica membrane using ethanol. The column was rinsed and washed with the TR2 and TR3 buffers, and DNase I treatment was performed to remove any contaminating DNA. The RNA was eluted, and then stored at −20 °C. The RNA concentrations were quantified with the Qubit Fluorometer (Invitrogen, Burlington, ON, Canada).
All the reverse-transcription steps were performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) with 1 μg of total RNA. Two microliters of Universal RNA Spike (TATAA Biocenter, Gothenburg, Sweden) were added to each sample to allow quality control throughout the entire RT-qPCR experimental workflow as previously described in Uribe et al.28.
To select the most stable reference genes for normalization, a panel of 12 reference genes was screened in 5 samples from every group and time-point. The expression profiles of the screened reference genes were evaluated using the geNorm34 and Normfinder35 programs. The HPRT1 and GUSB genes showed the most stable expression among the samples and were therefore selected as the reference genes for the subsequent analyses. The measured Cq value and the shape of the amplification curve revealed no inhibition in the presence of RNA spiking in the control assays.
The primers used in the RT-qPCR were purchased from Bio-Rad Laboratories (Table 1). The analysis of the target genes and the two selected reference genes was performed in a 10-μl reaction volume (10 ng of cDNA per reaction) in duplicate on a CFX 96 Real-Time System (Bio-Rad Laboratories) using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories). An inter-plate calibrator (TATAA Biocenter) was added to each plate to compensate for the variation between runs. The quantities of the target genes were normalised using the geometric mean of the Cq values of the selected reference genes. Gene expression was quantified according to the comparative threshold cycle method Δ − ΔCq and 90% PCR efficiency36.
Table 1.
Bio-Rad SYBR Green primers used for the RT-qPCR analyses.
| Gene identification | Abbreviation | Unique Assay ID |
|---|---|---|
| Target gene encoding: | ||
| Sp7 transcription factor | OSX | qHsaCED0003759 |
| Runt-related transcription factor 2 | RUNX2 | qHsaCED0044067 |
| Bone morphogenetic protein 2 | BMP2 | qHsaCID0015400 |
| Alkaline phosphatase | ALP | qHsaCID0010031 |
| Gap junction protein, alpha 1, 43 kDa | CX43 | qHsaCID0012977 |
| Bone gamma-carboxyglutamate protein (BGLAP) | OCN | qHsaCED0038437 |
| Integrin-binding sialoprotein | BSP | qHsaCED0002933 |
| Reference gene encoding: | ||
| β-Glucuronidase | GUSB | qHsaCID0011706 |
| Hypoxanthine phosphoribosyltransferase 1 | HPRT1 | qHsaCID0016375 |
CX 43 protein expression and Functional analysis of gap junctions
The methods described below have been reproduced in part from Uribe et al.28 To quantify the expression of the CX43 protein, hDFC were prepared as a cell suspension and stained for analysis with flow cytometry. Briefly, hDFC were cultured with or without Si (50 µg/ml) for 48 h. Cells were detached with ice-cold EDTA with the aid of a cell scraper. Aliquots of 106 cells in 100 µl of α-MEM were stained and labelled with a human connexin43/GJA APC-conjugated antibody (monoclonal mouse IgG2A Clone 578618, R&D systems) for 20 min at 4 °C. Thereafter, the cells were washed twice with PBS, and finally suspended in 500 µl of PBS. An isotype b antibody (Mouse IgG2A Allophycocyanin conjugated, R&D systems) with a different spectrum of detection than the antibody of interest was used as a control.
Intercellular communication via gap junctions was quantified using the fluorescent dye transfer parachute technique described by Czyz et al.37. Briefly, hDFC were cultured for 3 days in 12-well plates in α-MEM medium that contained 10% FBS, in the absence or presence of 25 μg/ml Si. For the donor cells, a cell suspension from a parallel culture was prepared with a staining solution using two dyes: the membrane permeable green dye calcein-AM (1 μg; Molecular Probes Inc., Eugene OR, USA); and the lipophilic red dye Dil (1 mM; Molecular Probes), used at final concentrations of 2 μM and 10 mM, respectively, for 30 min at 37 °C. The stained cells were washed with PBS and centrifuged three times (200 × g for 5 min), and thereafter re-suspended in 1 ml PBS with 2% FBS. Then, 2% of the double-stained donor cells were added to the unstained recipient cells at a ratio of 1:50 (donor:recipient) and incubated at 37 °C in 5% CO2 for 1, 2 and 3 h. Carbenoxolone (CBX) was added as an inhibitor of GJC, and used as a negative control. A parallel plate was placed on ice before the donor cells were added, to allow blocking of GJC, and used as a negative control. The non-fluorescent dye calcein-AM is hydrolysed by intracellular esterases into the fluorescent calcein, and can, thereafter, only pass from the donor to recipient cells through functional gap junction channels. Second-, third-and higher-order cells will acquire the dye depending on the degree of coupling. GJC is quantified by flow cytometry. The flow cytometer detected Dil-stained cells by FL3 and calcein-AM-stained cells by FL1. Donor cells were detected as cells with high intensities for both FL1 and FL3, while the recipient cells were distinguished by their increased intensity of FL1. Flow cytometry analysis has a high sensitivity and provides an appropriate resolution that allows the detection of small changes in gap-junctional communication by examining a large number of cells in a single experiment. The gating strategy for the parachute analyses is based on location of donor cells and recipient cells incubated on ice. The gated area should include as few donor cells as possible, but include the recipient cells with increased color uptake. This strategy is based on the original study by Czyz et al.35. By gating the populations, the recipient cells could be identified and quantified.
Next-generation sequencing
Whole-transcriptome RNA sequencing using next-generation sequencing (NGS-RNAseq) technologies and bioinformatics was used to compare the gene expression levels in hDFC cultured in the absence or presence of OIM and Si. RNA was collected from hDFC that had been cultured for 1, 7, 14 and 21 days with OIM with or without Si (25 μg/ml). Untreated cells cultured in α-MEM were used as controls. RNA extraction was carried out with the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions, and the samples were frozen at −80 °C before sending for library preparation.
The quality control (Trimmomatic), alignment (TopHat), and differential expression analysis (Cuffdiff) were performed at GATC (https://www.gatc-biotech.com). The resulting data were further analysed using R (ver. 3.4.2,38). Graphical visualisations, such as heat maps and functional profiles, were created using the pheatmap (ver. 1.0.8) and goProfiles (ver. 1.40.0).
Statistical analysis
The gene expression data were analysed using PRISM 7 (GraphPad Software Inc., San Diego, CA, USA). Analysis of variance (ANOVA) was performed to test for significant differences in gene expression and absorbance percentages between controls and treated groups. Statistical significance was adjudged for p-values ≤0.05. Each experiment was repeated three times with four biological replicates per treatments, unless otherwise stated in legends to figures.
Results
Cell viability and proliferation
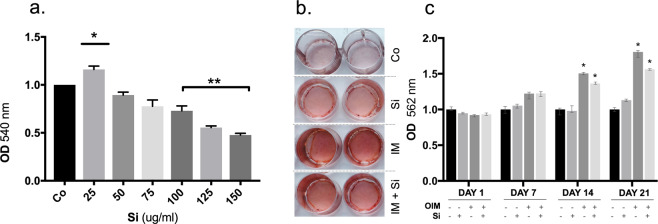
hDFC were cultured with different concentrations of Si for 48 h, Cell viability and proliferation was anlysed using the neutral red (NR) assay. A dose-dependent change in NR uptake was demonstrated (Fig. 1a). 25 μg/ml Si caused a significant (p = 0.001) stimulatory effect on cell proliferation. No significant effects on of Si was seen at concentrations >25 and up to 75 μg/ml, as compared to the control cells. A significant decrease in the number of viable hDFC was seen when cells were cultured with Si ≥100 μg/ml (p < 0.0001) (Fig. 1a)
Figure 1.
Effects of Si and OIM on proliferation and mineralization in cultures of hDFC. (a) Neutral Red uptake by hDFC cultured for 48 h with different concentrations of Si. Data are normalised to control, where the control (Co) absorbance values are set to 1. Values presented are mean ± SEM (n = 3). (b) Representative images of nodule mineralisation in hDFC cultured for 21 days in the absence or presence of Si (25 µg/ml) and OIM, and then stained with Alizarin Red. (c) Quantification of calcium deposition after 21 days. OIM has a significant effect on the mineralisation process after 21 days of culture. A significant effect of calcium deposition is observed in the presence of Si (25 µg/ml) plus OIM after 14 days and 21 days, as compared to the control cells. Data are normalised to control, where the control absorbance values are set to 1. Values shown are the averages of three different experiments (n = 15). *p ≤ 0.001.
Mineralised nodule formation
Alizarin Red staining (ARS) was used to determine the formation of mineralised nodules in hDFC cultures exposed to osteogenic induction medium (OIM) and Si. OIM stimulated the formation of mineralisation nodule in cultured hDFC, as revealed by ARS staining. The effect of Si on ARS staining was small and Si did not have any additive effect when combined with OIM (Fig. 1b). When ARS release from the matrix was determined, a significant stimulatory effect of OIM (p = 0.0102) on the mineralisation process was seen after 14 and 21 days. There was no additive effect of Si (25 μg/ml) in combination with OIM (p = 0.0063). There was no significant effect of Si alone on ARS/nodule formation at any time-point (Fig. 1c).
Gene expression analyses
The expression levels of the osteoblast markers OSX, RUNX2, BMP2, ALP, OCN, and BSP and the gap junction gene CX43 was investigated with Quantitative RT-qPCR in hDFC cultured for 1, 7, 14 21 days in the presence or absence of OIM and Si (25 µg/ml) (Fig. 2). All the above-listed genes were expressed in control cultures of hDFC and also in the cells cultured in the presence of OIM and/or Si (25 µg/ml) for 21 days. The expression levels were low on Day 1, except for CX43 which was highly expressed in all groups at Day 1, however, no statistically significant differences were seen between groups. Significantly (p = 0.001) increased expression of BMP2 and OCN were demonstrated in hDFC exposed to Si or to Si plus OIM for 21 days, as compared to controls. A time-dependent significant increase in BSP levels (p < 0.0001) was seen in the OIM treated groups. CX43 was significant up-regulated after 21 days, but only in the Si-treated group (p = 0.0054), (Fig. 2). The expression of BMP2 was significantly higher in the Si group, as compared to the OIM group on Day 7 (p < 0.0001) and Day 21 (p = 0.0025). BSP was significantly higher expressed in the OIM + Si group than in the other groups on Day 7 (p = 0.001). BSP was significantly higher in the OIM + Si (p < 0.0001) and Si (p < 0.0001) groups on Day 14,, as compared to the control group. The expression level was highest with OIM alone (p < 0.0001 and this was also seen on Day 21. (Fig. 2).
Figure 2.
Effects of Si on mRNA levels of osteoblastic phenotypic markers expressed in hDFC. Expression levels of markers of osteoblastic differentiation in hDFC that were cultured for 1, 7, 14, and 21 days in the absence or presence of Si (25 µg/ml) and OIM. Presented are the relative values normalised to HPRT1 and to the control. Values shown are mean ± SEM (n = 3). *p ≤ 0.01.
Next-generation sequencing (NGS-RNAseq)
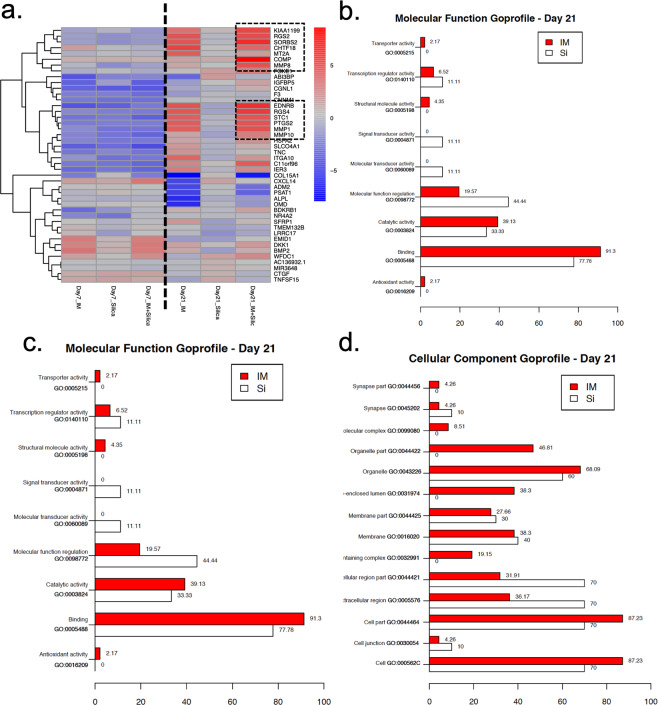
The heat map of expression data shown in Fig. 3a demonstrates the impacts of OIM, OIM + Si and Si alone on gene expression in hDFC. Regardless of the group examined, two main gene expression patterns were noted: (i) a pattern representative of cells cultured for 7 days (Fig. 3a, left); and (ii) a pattern representative of cells cultured for 21 days (Fig. 3a, right). hDFC that were cultured with OIM and OIM + Si clustered together, while those cultured with Si alone formed a separate branch. These results demonstrate the influence that OIM exerts on the cell differentiation profile, which is stronger than the influence of Si alone. However, Si in combination with OIM showed an augmented effect on the differentially regulated genes that were subjected to hierarchical cluster analysis, which is in line with the results previously reported for the functional test by matrix deposition and PCR quantification of osteoblast-related markers. The ten most highly up-regulated genes, as well as the three most down-regulated genes are listed in Table 2. Cartilage oligomeric matrix protein (COMP) was the most significantly up-regulated gene. Genes for the regulator of G protein signalling 4 (RGS4), regulator of G protein signalling 2 (RGS2), and matrix metalloproteinases (MMP) 1, 8, and 10 were also highly up-regulated (Table 2). Regulated genes were further clustered according to gene functions and according to their associated biological processes, annotated to gene ontology (GO) terms. The GO profiles for hDFC in the presence of Si or in OIM medium are listed in terms of molecular functions (Fig. 3b), biological processes (Fig. 3c), and cellular components (Fig. 3d). Most of the clustered genes were regulated by both Si and OIM. However, the molecular function gene-sets for signal transducer activity and molecular transducer activity were significantly differentially expressed only in hDFC cultured with Si alone.
Figure 3.
Impacts of Si on gene expression in hDFC during osteoblastic differentiation. (a) Heat map representation of medium-dependent effects on hDFC that were cultured for 7 and 21 days in growth medium that contained OIM, Si, or OIM + Si. Genes enclosed by a discontinuous line are highly expressed and regulated by the presence of Si during osteoblastic differentiation. (b, c, d) The gene ontology of hDFC after 21 days of differentiation regulated by OIM and Si.
Table 2.
The most highly up- and down-regulated genes in hDFC induced to undergo osteoblastic differentiation.
| Ensembl ID | Gene symbol | Gene name | Fold-change | p-value |
|---|---|---|---|---|
| Up-regulated genes | ||||
| ENSG00000105664 | COMP | cartilage oligomeric matrix protein | 8.58 | 0.013 |
| ENSG00000154556 | SORBS2 | sorbin and SH3 domain-containing 2 | 6.60 | 0.013 |
| ENSG00000136160 | EDNRB | endothelin receptor type B | 6.43 | 0.013 |
| ENSG00000117152 | RGS4 | regulator of G protein signalling 4 | 5.99 | 0.024 |
| ENSG00000103888 | KIAA1199 | Cell migration inducing hyaluronidase 1 | 5.50 | 0.013 |
| ENSG00000116741 | RGS2 | regulator of G protein signaling 2 | 5.23 | 0.013 |
| ENSG00000196611 | MMP1 | matrix metalloproteinase 1 | 5.14 | 0.013 |
| ENSG00000118113 | MMP8 | matrix metalloproteinase 8 | 5.02 | 0.031 |
| ENSG00000159167 | STC1 | stanniocalcin-1 | 4.93 | 0.013 |
| ENSG00000073756 | PTGS2 | prostaglandin-endoperoxide synthase 2 | 4.93 | 0.013 |
| ENSG00000178919 | FOXE1 | forkhead box E1 | 3.26 | 0.038 |
| ENSG00000166670 | MMP10 | matrix metalloproteinase10 | 3.04 | 0.013 |
| ENSG00000103175 | WFDC1 | WAP four-disulfide core domain 1 | 2.68 | 0.038 |
| ENSG00000137331 | IER3 | immediate early response 3 | 2.62 | 0.013 |
| Down-regulated genes | ||||
| ENSG00000128165 | ADM2 | adrenomedullin 2 | −3.08 | 0.013 |
| ENSG00000135069 | PSAT1 | phosphoserine aminotransferase 1 | −3.66 | 0.013 |
| ENSG00000204291 | COL15A1 | collagen type V alpha 1 chain | −6.89 | 0.013 |
Cx43 expression and Gap Junction communication
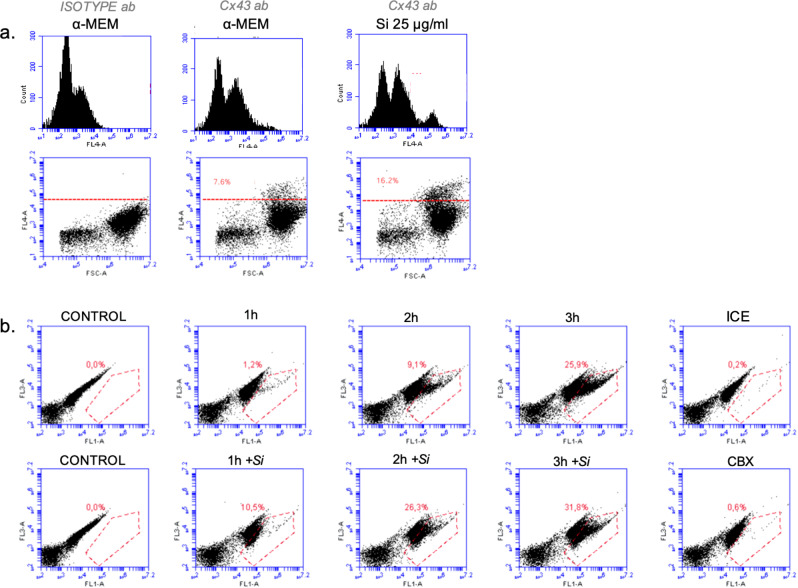
Flow cytometry quantification of the surface expression of CX43 protein revealed that a small fraction of hDFC express CX43 (7.6%) on their surface. An increase in the expression of CX43 (16.2%) was observed after exposure to Si (25 μg/ml) for 48 h (Fig. 4a). GJC in the hDFC was then analysed using the parachute dye-loading technique. Control cells were incubated at 37 °C in α-MEM and the results demonstrate an increased dye-uptake in the recipient cells, which indicate an active GJC communication (Fig. 4b). The presence of Si (25 μg/ml) in the culture medium progressively increased GJC after 1, 2, and 3 h of incubation, resulting in a significant increase in the numbers of stained-recipient hDFC compared to the corresponding controls without Si treatment (Fig. 4b). This outcome strongly suggests that Si increases the GJC function. The transfer of dye between the donor and recipient cells was limited when the cells were kept on ice, and dye transfer was totally blocked by the gap junction blocking compound CBX, demonstrating that the dye transfer is due to GJC activity.
Figure 4.
CX43 protein expression and functional analysis of gap junction communication in hDFC. (a) Expression of CX43 protein in hDFC cultured for 48 h with or without Si (25 µg/ml) presented as dot plots and histograms. Representative graphs and proportion of recipient cells are shown. (b) Flow cytometry of the coupling ratio in hDFC. Gating strategy for the percentage of recipient-positive cells after 1 h, 2 h and 3 h with or without Si (25 µg/ml). Donor cells are characterised by high intensities of green (FL1) and red (FL2) fluorescence; recipient cells are identified by colouration with the green transferred dye (FL1). Representative graphs and proportion of recipient cells are shown.
Discussion
Earlier studies have demonstrated that dietary Si is important for wound healing as well as for bone homeostasis1–3 confirming its important role in connective tissues. Si has been reported to be taken up from the plasma and incorporated into bovine and rat bones, although the amount of Si in human bone tissue is not yet established39,40. The mechanisms of action of soluble Si on bone remain unclear, although it has been demonstrated that Si enhances prolyl hydroxylase activity, stabilizes collagen cross-linking, and increases bone matrix mineralisation in vitro1,3,8,9,41. To date, it has not been established if the effects of Si are mainly extracellular. However, direct regulatory effects of Si on osteoblast activities have been demonstrated. Furthermore, we have recently reported that soluble Si inhibits osteoclast formation and bone resorption in vitro by interacting with both inter- and intra-cellular pathways42. Si transporters have been demonstrated in rice and in diatoms. The Lsi1 transporter in rice is a membrane protein with similarities to mammalian aquaporins43. Even if there is still no evidence that Si can be transported into bone cells, there are some indications that Si is transported into mammalian cells, as provided by the finding that Slc34a2, a sodium-phosphate co-transporter, acts as an active Si transport protein in rat kidney cells44 that appears to be involved in the reabsorption of Si from the pre-urine.
Si-containing biomaterials, e.g., bioactive glasses, promote bone regeneration when used to treat local bone defects14–16,45. The effects have been attributed to locally released ionic dissolution products (Si, Ca and P;46,47), although it is possible that physical and chemical properties and ions other than Si are also involved48. The concentrations of Si released from bioactive glass 45S5 into cell culture medium, that we previously determined with inductively coupled plasma optical emission spectrometry (ICP-OES), were used in the present study.42. We used hDFC from dental follicles of impacted teeth as the source of mesenchymal pluripotent cells, to investigate the process of osteogenic induction with soluble Si. Multipotent adult stem cells from dental tissues represent a potential source of mesenchymal stem cells for experimental studies, as well as for clinical treatments to repair local bone defects. It has previously been demonstrated that hDFC have a higher proliferation ability than mesenchymal stem cells derived from the pulp49. hDFC forms an alternative pool of cells that can easily be obtained from impacted teeth, since the follicle is usually otherwise discarded following surgical procedures.
A small proliferation was demonstrated when the hDFC were cultured in the presence of Si at 25 µg/ml. This concentration also had stimulatory effects on the differentiation markers in long-term experiments and therefore used in most of the subsequent experiments. The results also showed that Si at concentrations ≥100 μg/ml had inhibitory effects on cell proliferation. One reason for this is that Si at high concentrations (above 2 mM or 50–60 μg/ml) autocondenses to form small polymers50 that are less bioavailable but also interacts readily with cell membranes. Thus, the inhibitory effects of higher concentrations of Si seen in the present study could be due to the presence of polymeric forms of Si, which would likely have negative effects on cell growth.
OIM has earlier been demonstrated to promote osteoblast phenotypic differentiation, as evidenced by the increased expression of late osteoblastic markers, collagen accumulation, and ALP activity in osteoblast-like cells51. In line with these findings, exposure to an osteogenic differentiation environment, such as that with ascorbic acid and β-glycerol phosphate or growth factors (BMP-2 and TGFβ1), has previously been demonstrated to induce osteogenic differentiation of DFC24,31. Our present results confirm that OIM itself has osteogenic effects, as demonstrated by significant stimulation of matrix mineralisation and increased expression of the BSP gene. Although the increase in matrix mineralisation could not be demonstrated in hDFC cultured with Si alone, there was significant up-regulation of the expression of BMP2 and BSP in hDFC treated with only Si. In contrast, the expression of BMP2 was not increased with OIM alone, but in combination with Si. Our results suggests that although Si does not have the capacity by itself to support matrix mineralisation, it can stimulate osteogenic differentiation in hDFC. This role is supported by BMP2 which plays an important role in the regulation of osteogenic differentiation52. In line with this, increased expression of OCN was only demonstrated in the Si group. NGS-RNAseq analysis and bioinformatics processing were used to identify differentially regulated genes and pathways. The results demonstrate that effect of OIM over the differentiation profile was more significant than the effects of Si alone. However, Si in combination with OIM had an additive effect (increase) on the expression of the differentially regulated genes. Interestingly, COMP was the most significantly up-regulated gene. COMP expression had earlier been demonstrated in MG-63 cells and is suggested to be important for osteogenesis53. Our results are in line with those earlier reports of a significant up-regulation of both BMP2 and COMP during osteogenic differentiation of human dental follicle stem cells54. Our results further demonstrate that the genes for regulator of G protein signalling 4 (RGS4), regulator of G protein signalling 2 (RGS2), and matrix metalloproteinases (MMPs) 1, 8, and 10 are also highly up-regulated in hDFC undergoing osteogenic differentiation. The RGS proteins are important for the regulation of G protein-coupled receptors and they influence important signalling pathways in many tissues, including bone. RGS2 has been reported as being expressed in osteoblasts and induced by the cAMP-PKA pathway. The transcriptome analysis in our present study thus supports the notion that Si could have a regulatory role in the signalling pathways involved in osteogenic differentiation and bone formation.
GJC is an important signaling system which contributes to the differentiation of cells and tissues by transferring ions and second messengers between adjacent cells. It has earlier been demonstrated that CX43 and gap junction channels in bone are crucial for osteoblast differentiation, bone matrix formation, and the normal mineralization of the bone tissue55. Moreover, osteoblast dysfunction, craniofacial defects, and a delayed skeletal mineralization, is reported in CX43-null mice26,56. In addition, we, and others, have earlier demonstrated the importance of CX43 and GJC for osteoclast formation and for the bone resorbing activities of osteoclasts55,57,58. We have recently reported CX43 gene expression in human dental follicles59,60. A novel finding in the present paper is our results demonstrating significantly higher CX43 gene and protein expression and GJC in Si-treated hDFC. These findings suggest that signals that are dependent upon GJC are crucial during the differentiation process regulated by Si. We used a dye-transfer assay to evaluate the possible functional role of Si on GJC in hDFC. The GJC activity in hDFC was significantly stimulated by Si. This suggest that GJC could be a signaling pathway to mediate effects of Si in osteogenic differentiation. The results demonstrating that OIM has no effect on CX43 gene expression agree with Hashida et al.27. They reported an important role for CX43 GJC in BMP2-induced osteoblast differentiation, and furthermore, that CX43 GJC did not mediate the osteogenic effects by ascorbic acid. This is interesting since our results in the present study validate that Si, but not OIM, increase the expression of BMP2.
Even though Si by itself is not sufficient to initiate a mineralisation process, our results show that Si can stimulate factors and signals important for osteoblastic differentiation. This strongly suggests that dietary Si is beneficial for bone formation and skeletal health. Furthermore, it has been reported that Si-containing biomaterials stimulate bone regeneration when used for repair of bone defects. Gaining evidence of the mechanisms involved with Si interactions on osteogenic differentiation is necessary to better use Si-containing biomaterials in the clinical practice. Furthermore, our results point to an interesting opportunity to use undifferentiated mesenchymal cells obtained from human dental follicles for the validation of osteoinductive materials and for bone regeneration.
Supplementary information
Acknowledgements
This study was supported by grants from TUA Research Funding, The Sahlgrenska Academy at the University of Gothenburg, and Region Västra Götaland, Sweden. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.Open access funding provided by University of Gothenburg.
Author contributions
P.U. and M.R. did all methodology and wrote the main manuscript text; A.J. performed and prepared Figure 4; M.D. performed and prepared Figure 3; A.W., C.M., R.J. and J.P. help during silica-data analysis. All authors reviewed and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-66939-1.
References
- 1.Jugdaohsingh R. Silicon and bone health. J. Nutr. Health Aging. 2007;11(2):99–110. [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen FH. Update on the possible nutritional importance of silicon. J. Trace Elem. Med. Biol. 2014;28(4):379–82.. doi: 10.1016/j.jtemb.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 3.Rodella LF, Bonazza V, Labanca M, Lonati C, Rezzani R. A review of the effects of dietary silicon intake on bone homeostasis and regeneration. J. Nutr. Health Aging. 2014;18(9):820–6. doi: 10.1007/s12603-014-0555-8. [DOI] [PubMed] [Google Scholar]
- 4.Reffitt DM, Jugdaohsingh R, Thompson RP, Powell JJ. Silicic acid: its gastrointestinal uptake and urinary excretion in man and effects on aluminium excretion. J. Inorg. Biochem. 1999;76(2):141–7. doi: 10.1016/s0162-0134(99)00126-9. [DOI] [PubMed] [Google Scholar]
- 5.Carlisle EM. Silicon as an essential trace element in animal nutrition. Ciba Found. Symp. 1986;121:123–39. doi: 10.1002/9780470513323.ch8. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz K, Milne DB. Growth-promoting effects of silicon in rats. Nature. 1972;239(5371):333–4. doi: 10.1038/239333a0. [DOI] [PubMed] [Google Scholar]
- 7.Jugdaohsingh R, et al. Increased longitudinal growth in rats on a silicon-depleted diet. Bone. 2008;43(3):596–606. doi: 10.1016/j.bone.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jugdaohsingh R, et al. Dietary silicon intake is positively associated with bone mineral density in men and premenopausal women of the Framingham Offspring cohort. J. Bone Min. Res. 2004;19(2):297–307. doi: 10.1359/JBMR.0301225. [DOI] [PubMed] [Google Scholar]
- 9.Macdonald HM, et al. Dietary silicon interacts with oestrogen to influence bone health: evidence from the Aberdeen Prospective Osteoporosis Screening Study. Bone. 2012;50(3):681–7. doi: 10.1016/j.bone.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz K. A bound form of silicon in glycosaminoglycans and polyuronides. Proc. Natl Acad. Sci. USA. 1973;70(5):1608–12. doi: 10.1073/pnas.70.5.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou S, Ireland D, Brooks RA, Rushton N, Best S. The effects of silicate ions on human osteoblast adhesion, proliferation, and differentiation. J. Biomed. Mater. Res. B Appl. Biomater. 2009;90(1):123–30. doi: 10.1002/jbm.b.31262. [DOI] [PubMed] [Google Scholar]
- 12.Kim EJ, Bu SY, Sung MK, Choi MK. Effects of silicon on osteoblast activity and bone mineralization of MC3T3-E1 cells. Biol. Trace Elem. Res. 2013;152(1):105–12. doi: 10.1007/s12011-012-9593-4. [DOI] [PubMed] [Google Scholar]
- 13.Reffitt DM, et al. Orthosilicic acid stimulates collagen type 1 synthesis and osteoblastic differentiation in human osteoblast-like cells in vitro. Bone. 2003;32(2):127–35. doi: 10.1016/s8756-3282(02)00950-x. [DOI] [PubMed] [Google Scholar]
- 14.Hench LL. The story of Bioglass. J. Mater. Sci. Mater Med. 2006;17(11):967–78. doi: 10.1007/s10856-006-0432-z. [DOI] [PubMed] [Google Scholar]
- 15.Jones JR. Review of bioactive glass: from Hench to hybrids. Acta Biomater. 2013;9(1):4457–86. doi: 10.1016/j.actbio.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 16.Tadjoedin ES, de Lange GL, Holzmann PJ, Kulper L, Burger EH. Histological observations on biopsies harvested following sinus floor elevation using a bioactive glass material of narrow size range. Clin. Oral. Implant. Res. 2000;11(4):334–44. doi: 10.1034/j.1600-0501.2000.011004334.x. [DOI] [PubMed] [Google Scholar]
- 17.Cahill DR, Marks SC., Jr. Tooth eruption: evidence for the central role of the dental follicle. J. Oral. Pathol. 1980;9(4):189–200. doi: 10.1111/j.1600-0714.1980.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 18.Wise GE, Yao S. Regional differences of expression of bone morphogenetic protein-2 and RANKL in the rat dental follicle. Eur. J. Oral. Sci. 2006;114(6):512–6. doi: 10.1111/j.1600-0722.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 19.Wise GE, Frazier-Bowers S, D’Souza RN. Cellular, molecular, and genetic determinants of tooth eruption. Crit. Rev. Oral. Biol. Med. 2002;13(4):323–34. doi: 10.1177/154411130201300403. [DOI] [PubMed] [Google Scholar]
- 20.Morsczeck C, et al. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24(2):155–65. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Luan X, Ito Y, Dangaria S, Diekwisch TG. Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cell development. 2006;15(4):595–608. doi: 10.1089/scd.2006.15.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malaval L, Modrowski D, Gupta AK, Aubin JE. Cellular expression of bone-related proteins during in vitro osteogenesis in rat bone marrow stromal cell cultures. J. Cell Physiol. 1994;158(3):555–72. doi: 10.1002/jcp.1041580322. [DOI] [PubMed] [Google Scholar]
- 23.Tsuchiya S, Ohshima S, Yamakoshi Y, Simmer JP, Honda MJ. Osteogenic differentiation capacity of porcine dental follicle progenitor cells. Connect. Tissue Res. 2010;51(3):197–207. doi: 10.3109/03008200903267542. [DOI] [PubMed] [Google Scholar]
- 24.Morsczeck C, et al. Gene expression profiles of dental follicle cells before and after osteogenic differentiation in vitro. Clin. Oral. Investig. 2009;13(4):383–91. doi: 10.1007/s00784-009-0260-x. [DOI] [PubMed] [Google Scholar]
- 25.Gupta A, et al. Communication of cAMP by connexin43 gap junctions regulates osteoblast signaling and gene expression. Cell Signal. 2016;28(8):1048–57. doi: 10.1016/j.cellsig.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecanda F, et al. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J. Cell Biol. 2000;151(4):931–44. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashida Y, et al. Communication-dependent mineralization of osteoblasts via gap junctions. Bone. 2014;61:19–26. doi: 10.1016/j.bone.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Uribe, P. Clinical and molecular studies on impacted canines and the regulatory functions and differentiation potential of the dental follicle. Kållered, Sweden: University of Gothenburg; (2017).
- 29.Sripanyakorn S, et al. Moderate ingestion of alcohol is associated with acute ethanol-induced suppression of circulating CTX in a PTH-independent fashion. J. Bone Min. Res. 2009;24(8):1380–8. doi: 10.1359/JBMR.090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Repetto G, del Peso A, Zurita JL. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008;3(7):1125–31. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- 31.Saugspier M, et al. The differentiation and gene expression profile of human dental follicle cells. Stem Cell Dev. 2010;19(5):707–17. doi: 10.1089/scd.2010.0027. [DOI] [PubMed] [Google Scholar]
- 32.Stanford CM, Jacobson PA, Eanes ED, Lembke LA, Midura RJ. Rapidly forming apatitic mineral in an osteoblastic cell line (UMR 106-01 BSP) J. Biol. Chem. 1995;270(16):9420–8. doi: 10.1074/jbc.270.16.9420. [DOI] [PubMed] [Google Scholar]
- 33.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 34.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czyz J, Irmer U, Schulz G, Mindermann A, Hulser DF. Gap-junctional coupling measured by flow cytometry. Exp. Cell Res. 2000;255(1):40–6. doi: 10.1006/excr.1999.4760. [DOI] [PubMed] [Google Scholar]
- 38.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing2005.
- 39.Mladenovic Z, et al. In vitro study of the biological interface of Bio-Oss: implications of the experimental setup. Clin. Oral. Implant. Res. 2013;24(3):329–35. doi: 10.1111/j.1600-0501.2011.02334.x. [DOI] [PubMed] [Google Scholar]
- 40.Jugdaohsingh R, Pedro LD, Watson A, Powell JJ. Silicon and boron differ in their localization and loading in bone. Bone Rep. 2015;1:9–15. doi: 10.1016/j.bonr.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seaborn CD, Nielsen FH. Silicon deprivation decreases collagen formation in wounds and bone, and ornithine transaminase enzyme activity in liver. Biol. Trace Elem. Res. 2002;89(3):251–61. doi: 10.1385/bter:89:3:251. [DOI] [PubMed] [Google Scholar]
- 42.Mladenovic Z, et al. Soluble silica inhibits osteoclast formation and bone resorption in vitro. Acta Biomater. 2014;10(1):406–18. doi: 10.1016/j.actbio.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 43.Marron AO, et al. A family of diatom-like silicon transporters in the siliceous loricate choanoflagellates. Proc. Biol. Sci. 2013;280(1756):20122543. doi: 10.1098/rspb.2012.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratcliffe S, et al. Identification of a mammalian silicon transporter. Am. J. Physiol. Cell Physiol. 2017;312(5):C550–C61. doi: 10.1152/ajpcell.00219.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hench LL, Xynos ID, Polak JM. Bioactive glasses for in situ tissue regeneration. J. Biomater. Sci. Polym. Ed. 2004;15(4):543–62. doi: 10.1163/156856204323005352. [DOI] [PubMed] [Google Scholar]
- 46.Xynos ID, Edgar AJ, Buttery LD, Hench LL, Polak JM. Ionic products of bioactive glass dissolution increase proliferation of human osteoblasts and induce insulin-like growth factor II mRNA expression and protein synthesis. Biochem. Biophys. Res. Commun. 2000;276(2):461–5. doi: 10.1006/bbrc.2000.3503. [DOI] [PubMed] [Google Scholar]
- 47.Tsigkou O, Jones JR, Polak JM, Stevens MM. Differentiation of fetal osteoblasts and formation of mineralized bone nodules by 45S5 Bioglass conditioned medium in the absence of osteogenic supplements. Biomaterials. 2009;30(21):3542–50. doi: 10.1016/j.biomaterials.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 48.Bohner M. Silicon-substituted calcium phosphates - a critical view. Biomaterials. 2009;30(32):6403–6. doi: 10.1016/j.biomaterials.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Shoi K, Aoki K, Ohya K, Takagi Y, Shimokawa H. Characterization of pulp and follicle stem cells from impacted supernumerary maxillary incisors. Pediatr. Dent. 2014;36(3):79–84. [PubMed] [Google Scholar]
- 50.Iler, R. K. The chemistry of silica: solubility, polymerization, colloid and surface properties, and biochemistry: Wiley (1979).
- 51.Langenbach F, Handschel J. Effects of dexamethasone, ascorbic acid and beta-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013;4(5):117. doi: 10.1186/scrt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viale-Bouroncle S, Felthaus O, Schmalz G, Reichert TE, Morsczeck C. Transcription factors for dental stem cell differentiation. Int. J. Oral. Maxillofac. Implants. 2013;28(6):e478–86. doi: 10.11607/jomi.te28. [DOI] [PubMed] [Google Scholar]
- 53.Di Cesare PE, et al. Expression of cartilage oligomeric matrix protein (COMP) by embryonic and adult osteoblasts. J. Orthop. Res. 2000;18(5):713–20. doi: 10.1002/jor.1100180506. [DOI] [PubMed] [Google Scholar]
- 54.Vollkommer T, et al. Genome-wide gene expression profiles of dental follicle stem cells. Acta Odontol. Scand. 2015;73(2):93–100. doi: 10.3109/00016357.2014.956143. [DOI] [PubMed] [Google Scholar]
- 55.Zappitelli T, Aubin JE. The “connexin” between bone cells and skeletal functions. J. Cell Biochem. 2014;115(10):1646–58.. doi: 10.1002/jcb.24836. [DOI] [PubMed] [Google Scholar]
- 56.Civitelli R. Cell-cell communication in the osteoblast/osteocyte lineage. Arch. Biochem. Biophys. 2008;473(2):188–92. doi: 10.1016/j.abb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ransjo M, Sahli J, Lie A. Expression of connexin 43 mRNA in microisolated murine osteoclasts and regulation of bone resorption in vitro by gap junction inhibitors. Biochem. Biophys. Res. Commun. 2003;303(4):1179–85. doi: 10.1016/s0006-291x(03)00502-3. [DOI] [PubMed] [Google Scholar]
- 58.Matemba SF, Lie A, Ransjo M. Regulation of osteoclastogenesis by gap junction communication. J. Cell Biochem. 2006;99(2):528–37.. doi: 10.1002/jcb.20866. [DOI] [PubMed] [Google Scholar]
- 59.Uribe P, et al. Study on site-specific expression of bone formation and resorption factors in human dental follicles. Eur. J. Oral. Sci. 2018;126(6):439–48. doi: 10.1111/eos.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uribe, P., Larsson, L., Westerlund, A. & Ransjo, M. Gene expression profiles in dental follicles from patients with impacted canines. Odontology. (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.