Summary
Decades of artificial selection have significantly improved performance and efficiency of animal production systems. However, little is known about the microevolution of genomes due to intensive breeding. Using whole-genome sequencing, we document dynamic changes of chicken genomes under divergent selection on adiposity over 19 generations. Directional selection reduced within-line but increased between-line genomic differences. We observed that artificial selection tended to result in recruitment of preexisting variations of genes related to adipose tissue growth. In addition, novel mutations contributed to divergence of phenotypes under selection but contributed significantly less than preexisting genomic variants. Integration of 15 generations genome sequencing, genome-wide association study, and multi-omics data further identified that genes involved in signaling pathways important to adipogenesis, such as autophagy and lysosome (URI1, MBL2), neural system (CHAT), and endocrine (PCSK1) pathways, were under strong selection. Our study provides insights into the microevolutionary dynamics of domestic animal genomes under artificial selection.
Subject Areas: Biological Sciences, Evolutionary Biology, Genetics, Genomics
Graphical Abstract
Highlights
-
•
Directional selection reduces within-line but increases between-line genomic difference
-
•
Artificial selection tends to recruit preexisting variations of genes for fatness
-
•
Novel mutations contribute to the divergence of fatness under artificial selection
-
•
Genes involved in signaling pathways important to adipogenesis are under selection
Biological Sciences; Evolutionary Biology; Genetics; Genomics
Introduction
Observance of domesticated animals provided Darwin insights that contributed to the conceiving of the theory of evolution (Darwin, 1859, Darwin, 1868). Modern comparative genomics further contributes to our understanding of the molecular dynamics underlying evolution, character displacement, adaptation, and formation of species (e.g., Darwin's Finches) (Lamichhaney et al., 2015, Lamichhaney et al., 2016, Lamichhaney et al., 2018). Microevolution, in particular, is concerned about the changes within or among populations or within species over a relatively short time interval (Hendry and Kinnison, 2001), which not only can be used to investigate basic evolutionary questions such as evolutionary patterns, convergence, and determinism of different traits but also can identify the impacts of artificial or natural selection (Hendry and Kinnison, 2001).
After only a few decades, intensive selection and breeding has resulted in unprecedented efficiency and performance of animal production systems (Falconer and Mackay, 1996, Hill and Kirkpatrick, 2010, Hill, 2014, Hill, 2016). However, the underlying molecular mechanisms contributing to the increased efficacy of animal production systems remain largely unknown. Recently, experimental evolution coupled with genome sequencing has been adopted as a powerful approach for evolutionary studies (Barrick and Lenski, 2013, Long et al., 2015, Schlötterer et al., 2015, Good et al., 2017, Lillie et al., 2018). However, the majority of evolutionary studies implementing genome sequencing have focused on lower organisms (e.g., bacteria, yeast, and fruit fly) and have been retrospective or used single time point to characterize dynamic changes in allele frequencies. Retrospective or single point in time studies are not able to fully distinguish between adaptive “driver” and non-adaptive “passenger” genomic variants. For domestic animals, a number of selective sweeps were identified in chickens (Rubin et al., 2010), pigs (Rubin et al., 2012), dogs (Axelsson et al., 2013), or rabbits (Carneiro et al., 2014), and recent functional annotation of genomic elements (Andersson et al., 2015, Clark et al., 2017) further reveal the molecular clues underlying phenotypic diversification (Naval-Sanchez et al., 2018) and convergent selection (Alberto et al., 2018). An understanding of genome dynamics, within a microevolutionary framework, might improve our understanding of the genetic basis for complex traits (Siegel, 2014, NCD Risk Factor Collaboration, 2016, Fuchsberger et al., 2016, Marouli et al., 2017).
In broiler chickens, substantial improvements have been made in the selection of strains with high rates of growth. However, rapid growth of broiler chickens is generally accompanied by rapid increase in abdominal fat, which has low commercial value and decreases feed efficiency (Demeure et al., 2013). To investigate genetic mechanisms of fat deposition, we constructed two broiler lines divergently selected for abdominal fat content (AFC) over 19 generations since 1996. Tackling the microevolutionary dynamics of chicken genomes under divergent selection for adiposity over the 19-year timeline might improve our understanding of the genetic basis for fat deposition. A similar experimental design has previously been used to investigate body weight of chickens resulting in the ultimate selection of the high-weight selected (HWS) and low-weight selected (LWS) Virginia lines (Pettersson et al., 2013). Adaptive allele dynamics in generation 53 of LWS and HWS lines were investigated and results indicated that existing genetic variations of the founder population made significant contributions to their selection and were as important as novel mutations (Pettersson et al., 2013). Furthermore, Dunnington et al. (2013) confirmed that pre-existing genetic variants and major novel mutations influenced body weight of generation 54 of LWS and HWS lines. However, the latter study has not explicitly explored the contributions by novel mutations using birds from an Advanced Intercross Line (AIL) bred from generation S40 parents from the HWS and LWS lines (Sheng et al., 2015). Competing theories related to the role of mutations in selection responses have also been proposed (Hill and Kirkpatrick, 2010, Mackay, 2010, Barrick and Lenski, 2013, Hill, 2014, Hill, 2016, Laland et al., 2014). Therefore, the contribution of novel mutations to phenotypes remain unanswered.
In the current study, we performed a large-scale 15 generations genome sequencing on chicken lines divergently selected for AFC for 19 generations, coupled with a sequencing-based genome-wide association study (GWAS) and multi-omics analyses to investigate the dynamic changes of chicken genome under artificial selection.
Results
Genome Dynamics during Microevolution
The birds used in this study were selected from two broiler lines divergently selected for AFC over 19 generations (one generation per year) since 1996. A total of 60 pooled DNA samples (60 = 15 generations×2 sexes×2 lines) were collected over 15 generations (G4–G18). Each pooled sample consisted of 8–35 male birds or 27 to 138 female birds collected between G4 and G18 (3,642 birds in total, Table S1). Samples were sequenced to an average depth of 30.53-fold, and ∼2.35 Tb (terabases) high-quality data (medium 95.53% bases ≥ Q20) were generated (Figure S1 and Table S2). Stringent variant calling produced millions of single-nucleotide polymorphisms (SNPs) and insertion/deletion (InDel) polymorphisms (Tables S3A and S3B). Both fat and lean lines had 12.32 million SNPs and approximately 1.60 million and 1.58 million InDel polymorphisms for the fat and lean lines, respectively.
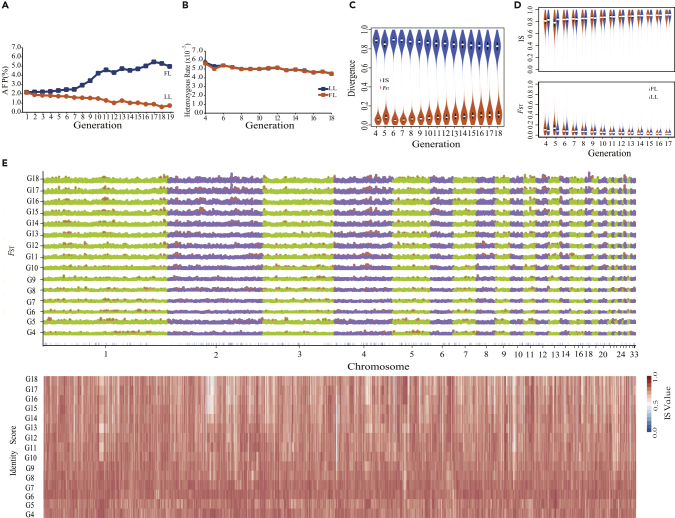
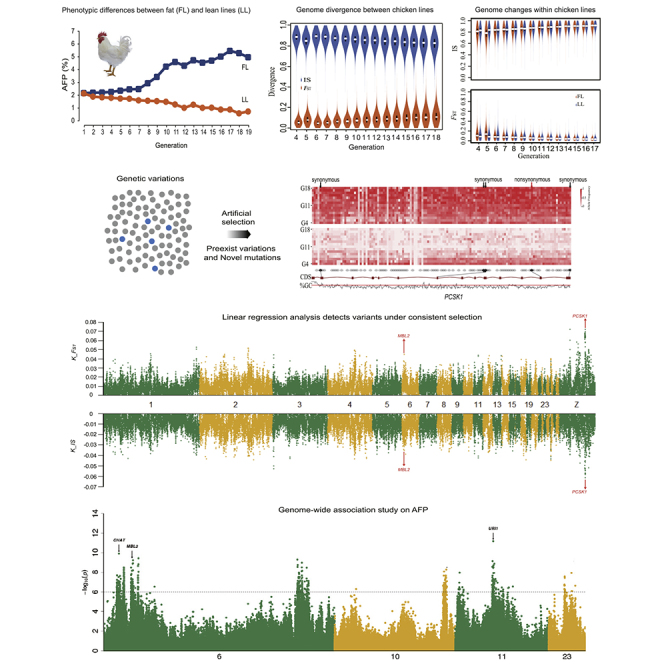
Comparison of the two chicken lines demonstrated striking phenotypic differences in the continuous and directional selection of adiposity (p < 0.05) (Figures 1A and S2). In addition, we investigated how chicken genomes changed dynamically by testing the changes in heterozygosity of the lean line (LL) and fat line (FL) between G4 and G18, which decreased significantly from 0.57% and 0.56% of G4 to 0.44% and 0.45% of G18 in LL and FL, respectively (p = 0.04, paired t test). Rates of heterozygosity between FL and LL within each generation from G4 to G18 did not significantly diverge (p value = 0.39, paired t test) (Figure 1B). A significant reduction in rates of heterozygosity was detected via linear regression (p = 8.744 × 10−5 (FL) and 9.926 × 10−7 (LL), F-statistic, Figure S3). Comparison of allele frequency differences (△AF) demonstrated that the number of SNPs associated with population genetic differentiation (△AF ≥ 0.8) increased sharply from G9 onward and was closely correlated with the pattern of selection responses (Pearson's correlation coefficients: △AF and AFC were 0.91 (FL) and −0.95 (LL), Figure S4), indicating that selection could recruit genomic variants to drive the phenotypic improvement. Furthermore, we investigated patterns of dynamic changes in allele frequency differences, by clustering positively selected SNPs over generations, and identified five main groups. Allele frequencies of two groups (I and IV) increased consistently throughout all generations, whereas the other three groups (II, III, and V) initially increased and then decreased (Figure S5). Thus, divergent selection acted dynamically on different genomic regions/loci impacting phenotypes across generations.
Figure 1.
Genome Dynamics during the Microevolutionary Process
(A) Phenotypic differences between fat (FL) and lean (LL) lines. Chickens from the same base population were selected divergently for abdominal fat percentage (AFP) over 19 generations, while maintaining similar body weights at 7 weeks of age (Note S1).
(B) Genome heterozygosity decreased in both lines.
(C) Genome divergence between chicken lines indicated by increasing FST and decreasing IS values.
(D) Genomic changes within the fat and lean chicken lines, as indicated by increasing FST and decreasing IS values.
(E) Genomic regions under selection based on sequencing of pooled-DNA samples over 15 generations (G4–G18). The line below G4 presents genes significantly differentially expressed in abdominal fat tissue by RNA sequencing.
Genomic Regions under Positive Selection
Identity score (IS) and fixation index (FST) were calculated using a sliding-window approach (40-kb windows sliding in 20-kb steps). Between fat and lean lines, FST increased but IS decreased from G4 to G18 (Figure 1C), whereas the opposite trend was found within both fat line and lean line (Figure 1D). Divergent selection thus increased between-line but reduced within-line genomic differences.
To investigate if genomic regions were subject to directional selection for each generation, we analyzed overlapping genomic windows that had high IS and FST values (top 5%). We observed divergent changes between and within the fat and lean lines (Table S4, Figures 1D and S6), in 11,409 regions on chromosomes 1–8, accounting for 80% of the genome across the 15 generations (Figure 1E). Multiple peaks observed on chromosomes 1, 2, 13, 19, or 26 covered large regions of the genome (Figure 1E) and contained genes potentially under selection. Chicken-human genome co-linearity analysis of the positively selected regions identified a number of genes and one conserved lncRNA (Table S5). Identified candidate genes in the selected regions were enriched in pathways related to adipose tissue growth across generations, such as biosynthesis and metabolism of fatty acids and ribosome (Figure S7).
Identification of Genes under Selection
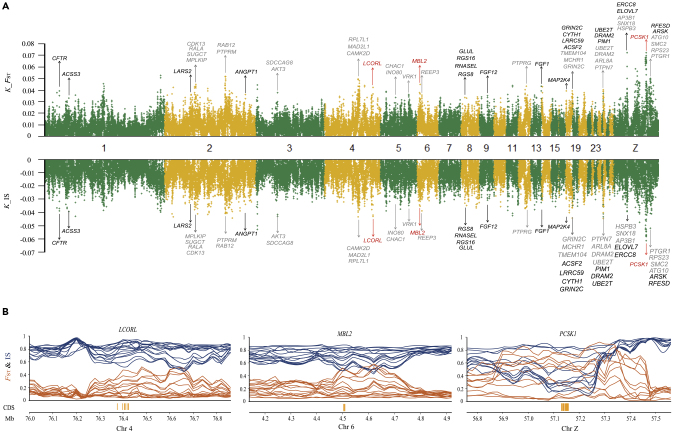
To distinguish genomic regions under selection from those impacted by genetic drift, a linear regression model was fitted to the 15 generations genome sequencing data (Transparent Methods) to identify genome regions subject to consistent selection over generations. Between-line comparison identified 1,068 genes (23.52-Mb region), and within-line analysis identified 897 annotated protein-coding genes (23.34 Mb) and 842 annotated protein-coding genes (22.80-Mb region) for the fat and lean lines, respectively (Figure 2A and Tables S6 and S7). Furthermore, the fixed divergent variations (delta allele frequency ≥0.95) were used to refine the gene list to 491 genes (17.32 Mb) between-line comparison and 288 (11.20 Mb) and 235 genes (9.52 Mb) in the fat and lean lines, respectively. Subsequent gene enrichment analysis of these identified genes identified enriched biological pathways important for adipose tissue growth and development, such as the p53 signaling pathway, cell cycle, cytokine-cytokine receptor interaction, MAPK (mitogen-activated protein kinase) signaling pathway, GnRH (gonadotropin releasing hormone) signaling pathway, cell adhesion molecules (CAMs), and steroid hormone biosynthesis (Table S8).
Figure 2.
Linear Regression Analysis Detects Variants under Consistent Selection
(A) Stringent linear regression analysis of time-series changes in allele frequencies. Variants detected to be under positive selection located in genes potentially important for AFP, such as PCSK1, LCORL, and MBL2 (this gene also identified by GWAS). Top and bottom panels, plots for genomic regions containing significant allele frequency changes detected between the fat and lean chicken lines, using FST and IS values calculated by comparing each line within the same generation. K_IS and K_FST showing regression coefficient of linear regression analysis for IS and FST values, Genes in black, in common pathways shared by male and female chickens; gray, sex-specific genes.
(B) Detailed regional plots for three genes across 15 generations under consistent and directional selection, LCORL, MBL2, and PCSK1 (IS, chartreuse; FST, purple). Vertical brown lines indicate coding sequence regions of target genes.
Furthermore, we analyzed SNPs with allele frequencies that continuously increased or decreased across generations in the fat and lean lines (Tables S9 and S10) and discovered that the genes related with these SNPs were enriched in neural system development, including cholinergic receptor muscarinic 2 (CHRM2), gamma-aminobutyric acid type A receptor subunit gamma1 (GABRG1), RasGEF domain family member 1A (RASGEF1A), and neuromedin U (NMU), implying the involvement of neural system in adipose tissue growth and development.
Interestingly, the genes involved in adipose tissue and neural system development were positively selected, such as ligand dependent nuclear receptor corepressor like (LCORL), proprotein convertase subtilisin/kexin type 1 (PCSK1), and mannose binding lectin 2 (MBL2) (Figures 2A and 2B). Previously, the PCSK1 gene was identified for its preferential selection in the breeding of broiler chickens (Zhang et al., 2012) and associations with abdominal fat weight and percentage in two populations of broiler chickens (Zhang et al., 2017). PCSK1 was highly expressed in neuroendocrine and intestinal tissues in broiler chickens (Zhang et al., 2017) and thus might contribute to the regulation of fat deposition via the neural and digestive systems. The MBL2 gene was associated with diabetes and affected mannan-binding lectin levels (Bijkerk et al., 2016). In addition, the LCORL gene was associated with average daily gain and body stature (Takasuga, 2016, Zhang et al., 2016).
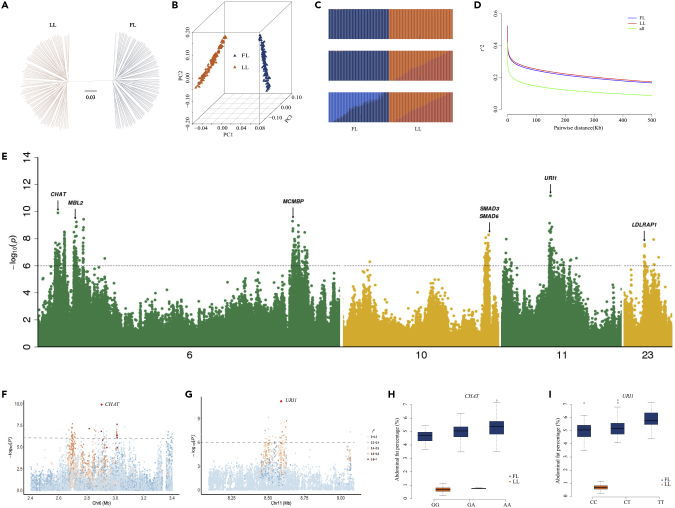
Next, we performed GWAS by sequencing 330 male birds from the G19 population (160 and 170 from the fat and lean lines, respectively) to an average coverage of 5.3-fold (Table S11) and generated ∼2.19 Tb of sequences (Figure S1). Overall, 7.07 million high-quality SNPs were obtained and were mainly located in the intergenic and intronic regions (Figure S8 and Table S12). All birds could be separated clearly into two clades (Figures 3A–3C), and similar linkage disequilibrium (LD) decay rates were observed in chickens from both fat and lean lines (Figure 3D). Furthermore, 226 significant quantitative trait loci (QTL) regions were detected (p < 10−6) for abdominal fat percentage (AFP) using a mixed linear model (Methods) (Figure 3E and Table S13). These regions contained 254 genes, of which 49 genes were common to those identified via the 15 generations genome sequencing selective sweep analyses (both within- and between-lines) (Table S14). Three genes, URI1 (URI1 prefoldin like chaperone), CHAT (choline O-acetyltransferase), and MBL2 were located in the most significant regions (p < 10−8) (Figures 3E–3I). URI1 was located in lysosomal lumen and crucial for autophagy (Haas, 2015). In addition, URI1 could integrate the nutrient surpluses signal to hepatic inflammation and induce neutrophil infiltration into white adipose tissue, resulting in insulin resistance and release of fatty acids into liver (Haas, 2015). Furthermore, URI1 might function through the autophagy pathway in the hepatic tissue and plays a role in adipogenesis via a tissue cross-talk fashion (Gomes et al., 2016). Moreover, CHAT and MBL2 were common genes identified by both GWAS and 15 generations genome sequencing (Table S14). As mentioned previously, CHAT knockout mice reduced the body weight via regulation of food intake in the dorsomedial hypothalamus (Jeong et al., 2017). In addition, CHAT might regulate the activity of the enteric nervous system (Fu et al., 2014, Stenkamp-Strahm et al., 2015) and is aberrantly expressed in the brain of obese rats (Goodman and Soliman, 1991). MBL2 is associated with diabetes as it affects levels of mannose-binding lectin (Bijkerk et al., 2016), which function via recognition and clearance of apoptotic adipocytes by macrophages infiltrated into adipose tissue (Stienstra et al., 2014). Thus, by combining our 15 generations genome sequencing and GWAS results, we observed that genes involved in autophagy (URI1, MBL2) or neural system (CHAT) pathways could contribute to the striking differences in adipose tissue growth and development between the fat and lean lines.
Figure 3.
A Sequencing-Based Genome-Wide Association Study on AFP
(A) Rooted neighbor-joining phylogenetic tree using the neighbor-joining method. The reliability of each branch was evaluated by bootstrapping with 1,000 replicates.
(B) Principal component analysis. The fractions of the variance explained were 20.98%, 1.42%, and 1.39% for eigenvectors 1, 2, and 3, respectively (p < 0.05, Tracy-Widom test).
(C) Genetic structure of fat and lean lines. The length of each colored segment represents the proportion of the individual's genome from K = 4 ancestral populations.
(D) LD decay determined by squared correlations of allele frequencies (r2) against distances between polymorphic sites in fat and lean lines.
(E) Manhattan plot for abdominal fat percentage (AFP). The dotted line indicates the significantly associated threshold of P < 10-6.
(F and G) Detailed overview of the associated regions containing CHAT (left) and URI1 (right), respectively. Plots show the association of SNPs with abdominal fat percentage (AFP) according to their chromosomal position. Different colors represent the linkage disequilibrium (R2) between the top SNP and all remaining SNPs.
(H and I) Box plots for abdominal fat percentage (AFP), based on SNP genotypes of CHAT (left) and URI1 (right), respectively. In the box plots, the center line denotes the median, box limits are the upper and lower quartiles, and whiskers mark the range of the data. FL and LL indicate fat line and lean line, respectively.
Function and Fate of Novel Mutations
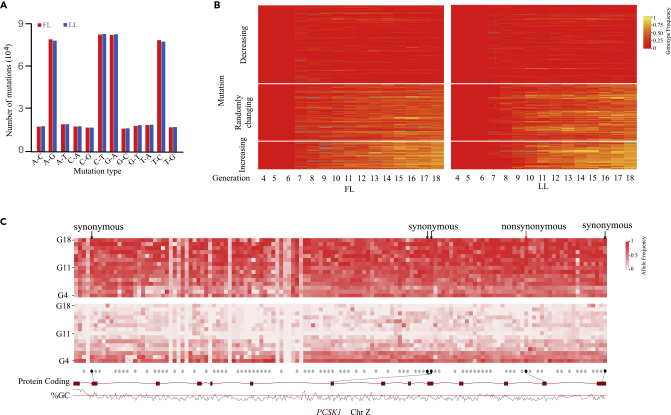
To investigate if novel mutations contribute to the improvement of complex traits and thus if they are favored by positive selection, we first characterized the occurrence of novel mutations in the two chicken lines. Novel mutations (0.28% of all identified SNPs) were detected by comparing each allele at all identified SNP sites over all generations (Figures S9A, S9B and Tables S15, S16, S17, and S18) (Methods). Novel mutations contained nucleotide transitions (69.34%) and nucleotide transversions (30.66%), suggesting that A-G or C-T mutations were common (Figure 4A). Of the identified novel mutations (total of 941,430), 4,590 mutations resulted in amino acid changes (Figure S10), of which the most frequent (7%) amino acid change was between alanine (Ala) and threonine (Thr).
Figure 4.
Mutations Detected to be Positively Selected
(A) Major types of mutation were nucleotide transitions (A-G, or C-T). The fat line had more mutations than the lean line.
(B) Heatmap showing mutations divided into three groups, beneficial, neutral, and deleterious.
(C) PCSK1 gene mutations. Novel mutations existed in PCSK1 were under positive selection in the two chicken lines. Allele frequencies of one non-synonymous mutation in exon 13 (Ala627Val) (red arrow) show divergent trends in the two lines.
We further investigated the function and fate of the identified novel mutations. Allele frequencies of most novel mutations of G18 were relatively high (Figure S11). Based on the changing patterns of allele frequencies, mutations were classified into three groups: increasing, randomly changing, and decreasing over time for the fat and lean lines, respectively (Figure 4B). Alleles related with low-abdominal-fat content increased in the lean line and decreased in the fat line over the generations. In contrast, alleles associated with high AFC increased in fat line and decreased in lean line, whereas alleles that have no effect on AFC changed randomly. Therefore, SNPs with allele frequencies that increased or decreased contributed positively or negatively to changes in phenotypes. GWAS analysis further demonstrated that novel mutations contributed 0.53% to the phenotype variance, whereas preexisting variations contributed 99.47%. Very few novel mutations (∼5% of all identified SNPs) were under positive selection, as demonstrated via linear regression model. The PCSK1 gene was consistently selected for and is closely related to fat deposition in broiler chickens as mentioned above, and one novel mutation (C to T) in exon 13 causing the change of amino acid (alanine to valine) in a conserved domain was discovered (Figure 4C). The T allele frequency of this novel mutation was similar in the lean line (0.75) and fat line (0.71) in G4; however, after several generations of artificial selection on AFC, TT was fixed (allele frequency = 1.00) in fat line and CC was fixed in lean line. Apart from the SNPs consistently and positively selected in PCSK1 gene (Zhang et al., 2017), this amino-acid-change mutation was also positively selected and contributed to phenotypic differences between the two chicken lines (Figures 2B and 4C). Results of our study suggested that, after 19 generations of artificial selection, not only standing genetic variants but also novel mutations contributed to the genetic modulation of complex phenotypes and provided empirical evidence about the role of mutations in microevolution.
Discussion
Molecular mechanisms underlying the genetics and evolution of complex trait are determined by the etiology of the specific trait (Darwin, 1859, Darwin, 1868), affected by both genetic and environmental factors (Falconer and Mackay, 1996, Hendry and Kinnison, 2001). However, our understanding of genetic heterogeneity of populations and the complex development of traits require improved genetic models and experimental designs. The divergently selected chicken lines for abdominal fat content over 19 generations demonstrated striking differences in adiposity between fat and lean lines and provided insights into the growth and development of adipose tissues. In the current study, integration of 15 generations genome sequencing, a sequencing-based GWAS and comprehensive genomics approaches enabled us to reveal the dynamic processes of microevolution of chicken genomes under selection for adiposity.
Overall, our results demonstrate that, during breeding, phenotypic selection initiates accumulation and combination of preexisting genomic variations that contribute to phenotypic changes. Modern intensive molecular breeding of animals likely benefits from the same underlying principle (Rubin et al., 2010, Rubin et al., 2012, Axelsson et al., 2013, Carneiro et al., 2014, Lillie et al., 2018). Moreover, in addition to preexisting genomic variations that influence complex phenotypes, novel mutations also contribute to phenotypic changes, as demonstrated in the present study and previous reports (Dunnington et al., 2013, Pettersson et al., 2013, Tenaillon et al., 2016). These results might explain why fat line, which had greater numbers of mutations, did not reach a selection plateau after 19 generations of continuous selection. In both animal and plant breeding practices, it is well known that even as intensive selection continues over a long time, genetic variation is not depleted, and genetic progress is still possible (Hill and Kirkpatrick, 2010, Hill, 2014, Hill, 2016). As for the lean line, the slower rate of selection observed might be due to the fact that adipose tissue is essential to the proper functioning of the endocrine and physiological systems (Kershaw and Flier, 2004).
Chicken can be used as a model for human obesity (Ji et al., 2014, Shipp et al., 2016), and by integrating different methods (15 generations genome sequencing, GWAS, functional genomics), we identified a common set of important molecular circuits to adipogenesis (Figure S12), e.g., neural system (CHAT, PCSK1), autophagy, and lysosome (URI1, MBL2). Similar to human obesity, which is under the control of complex neural, gastrointestinal, endocrinal, and metabolic networks (De Vadder et al., 2014, Morton et al., 2016, Perry et al., 2016), chicken adiposity is also subject to complex control by genetic and epigenetic factors (Figure S12). As such, adipose functional genomics only (Figures S13 and S14) was insufficient to fully validate the positively selected genes identified by genome sequencing. For example, URI1 and PCSK1 were not differentially expressed in abdominal fat tissues between the two chicken lines (Table S19).
Results of the 15 generations genome sequencing and integrative omics analyses provide new insights into the genetic architecture of adiposity in vertebrates and contribute to advances in animal breeding and the understanding of human obesity and related metabolic diseases.
Limitations of the Study
The chickens used in this study were selected for AFC over 19 years, which can be considered long term for vertebrates but short for invertebrates such as Drosophila. The two chicken lines will continue to be bred and selected based on AFC, and more research will be carried out.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hui Li (lihui@neau.edu.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession numbers for the genome re-sequencing data reported in this paper are NCBI SRA: PRJNA353057 and PRJNA354990. Other data that support the findings of this study are available from the corresponding authors on request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank the Genome Sequencing Technology Platform at Novogene for performing the sequencing and suppling computer resources. The project was supported by the National 863 Project of China (No. 2013AA102501), the National Natural Science Foundation of China (No. 31472088), and the China Agricultural Research System (No. CARS-41).
Author Contributions
Hui Li, Z.D., and H.Z. conceived the study. Hui Li, H.Z., Q.W., L.L., Y.W., S.W., Y.L., Z.C., P.L., Zhipeng Wang, H.Y., and Z.D. collected the experimental material. H.Z., Z.D., J.Z., Hao Liang, L.L., and Xun Zhou performed the experiment. S.T., Q.L., H.Z., and Z.D. led the bioinformatics analysis, with contributions from J.M., Z.D., Hui Li, H.Z., Q.L., and S.T., and N.W. wrote the paper, with input from Zhiquan Wang, Xuming Zhou, S.J.L, and Y.D. Hui Li, Z.D., S.T., and R.L. coordinated the project. All authors approved the manuscript before submission.
Declaration of Interests
The authors declare no conflicts of interest.
Published: June 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101193.
Contributor Information
Shilin Tian, Email: tianshilin@novogene.com.
Zhiqiang Du, Email: zhqdu@neau.edu.cn.
Hui Li, Email: lihui@neau.edu.cn.
Supplemental Information
FL, fat line; LL, lean line
References
- Alberto F.J., Boyer F., Orozco-terWengel P., Streeter I., Servin B., de Villemereuil P., Benjelloun B., Librado P., Biscarini F., Colli L. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018;9:813. doi: 10.1038/s41467-018-03206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L., Archibald A.L., Bottema C.D., Brauning R., Burgess S.C., Burt D.W., Casas E., Cheng H.H., Clarke L., Couldrey C. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015;16:57. doi: 10.1186/s13059-015-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Ratnakumar A., Arendt M.L., Maqbool K., Webster M.T., Perloski M., Liberg O., Arnemo J.M., Hedhammar A., Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- Barrick J.E., Lenski R.E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 2013;14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijkerk R., van der Pol P., Khairoun M., van Gijlswijk-Jansen D.J., Lievers E., de Vries A.P., de Koning E.J., de Fijter H.W., Roelen D.L., Vossen R.H. Simultaneous pancreas-kidney transplantation in patients with type 1 diabetes reverses elevated MBL levels in association with MBL2 genotype and VEGF expression. Diabetologia. 2016;59:853–858. doi: 10.1007/s00125-015-3858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M., Rubin C.J., Di Palma F., Albert F.W., Alföldi J., Martinez Barrio A., Pielberg G., Rafati N., Sayyab S., Turner-Maier J. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E.L., Bush S.J., McCulloch M.E.B., Farquhar I.L., Young R., Lefevre L., Pridans C., Tsang H.G., Wu C., Afrasiabi C. A high resolution atlas of gene expression in the domestic sheep (Ovis aries) PLoS Genet. 2017;13:e1006997. doi: 10.1371/journal.pgen.1006997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. John Murray; 1859. On the Origins of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life. [Google Scholar]
- Darwin C. John Murray; 1868. The Variation of Animals and Plants under Domestication. [PMC free article] [PubMed] [Google Scholar]
- De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A., Bäckhed F., Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Demeure O., Duclos M.J., Bacciu N., Le Mignon G., Filangi O., Pitel F., Boland A., Lagarrigue S., Cogburn L.A., Simon J. Genome-wide interval mapping using SNPs identifies new QTL for growth, body composition and several physiological variables in an F2 intercross between fat and lean chicken lines. Genet. Sel. Evol. 2013;45:36. doi: 10.1186/1297-9686-45-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnington E.A., Honaker C.F., McGilliard M.L., Siegel P.B. Phenotypic responses of chickens to long-term, bidirectional selection for juvenile body weight--historical perspective. Poult. Sci. 2013;92:1724–1734. doi: 10.3382/ps.2013-03069. [DOI] [PubMed] [Google Scholar]
- Falconer D.S., Mackay T.F.C. Prentice Hall; 1996. Introduction to Quantitative Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.Y., Li Z., Zhang N., Yu H.T., Wang S.R., Liu J.R. Effects of gastrointestinal motility on obesity. Nutr. Metab. (Lond) 2014;11:3. doi: 10.1186/1743-7075-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.L., Teijeiro A., Burén S., Tummala K.S., Yilmaz M., Waisman A., Theurillat J.P., Perna C., Djouder N. Metabolic inflammation-associated IL-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2016;30:161–175. doi: 10.1016/j.ccell.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Good B.H., McDonald M.J., Barrick J.E., Lenski R.E., Desai M.M. The dynamics of molecular evolution over 60,000 generations. Nature. 2017;551:45–50. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C.B., Soliman K.F. Altered brain cholinergic enzymes activity in the genetically obese rat. Experientia. 1991;47:833–835. doi: 10.1007/BF01922466. [DOI] [PubMed] [Google Scholar]
- Haas G. ETH Zürich; 2015. URI1c Is Located in the Lysosomal Lumen and Crucial for Autophagy. Ph.D. Thesis. [Google Scholar]
- Hendry A.P., Kinnison M.T. An introduction to microevolution: rate, pattern, process. Genetica. 2001;112-113:1–8. [PubMed] [Google Scholar]
- Hill W.G. Applications of population genetics to animal breeding, from wright, Fisher and lush to genomic prediction. Genetics. 2014;196:1–16. doi: 10.1534/genetics.112.147850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G. Is continued genetic improvement of livestock sustainable? Genetics. 2016;202:877–881. doi: 10.1534/genetics.115.186650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G., Kirkpatrick M. What animal breeding has taught us about evolution. Annu. Rev. Ecol. Evol. Syst. 2010;41:1–20. [Google Scholar]
- Jeong J.H., Lee D.K., Jo Y.H. Cholinergic neurons in the dorsomedial hypothalamus regulate food intake. Mol. Metab. 2017;6:306–312. doi: 10.1016/j.molmet.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B., Middleton J.L., Ernest B., Saxton A.M., Lamont S.J., Campagna S.R., Voy B.H. Molecular and metabolic profiles suggest that increased lipid catabolism in adipose tissue contributes to leanness in domestic chickens. Physiol. Genomics. 2014;46:315–327. doi: 10.1152/physiolgenomics.00163.2013. [DOI] [PubMed] [Google Scholar]
- Kershaw E.E., Flier J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- Laland K., Uller T., Feldman M., Sterelny K., Müller G.B., Moczek A., Jablonka E., Odling-Smee J., Wray G.A., Hoekstra H.E. Does evolutionary theory need a rethink? Nature. 2014;514:161–164. doi: 10.1038/514161a. [DOI] [PubMed] [Google Scholar]
- Lamichhaney S., Berglund J., Almén M.S., Maqbool K., Grabherr M., Martinez-Barrio A., Promerová M., Rubin C.J., Wang C., Zamani N. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- Lamichhaney S., Han F., Berglund J., Wang C., Almén M.S., Webster M.T., Grant B.R., Grant P.R., Andersson L. A beak size locus in Darwin's finches facilitated character displacement during a drought. Science. 2016;352:470–474. doi: 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- Lamichhaney S., Han F., Webster M.T., Andersson L., Grant B.R., Grant P.R. Rapid hybrid speciation in Darwin's finches. Science. 2018;359:224–228. doi: 10.1126/science.aao4593. [DOI] [PubMed] [Google Scholar]
- Lillie M., Sheng Z.Y., Honaker C.F., Andersson L., Siegel P.B., Carlborg Ö. Genomic signatures of 60 years of bidirectional selection for 8-week body weight in chickens. Poult. Sci. 2018;97:781–790. doi: 10.3382/ps/pex383. [DOI] [PubMed] [Google Scholar]
- Long A., Liti G., Luptak A., Tenaillon O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 2015;16:567–582. doi: 10.1038/nrg3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F. Mutations and quantitative genetic variation: lessons from Drosophila. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:1229–1239. doi: 10.1098/rstb.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E., Graff M., Medina-Gomez C., Lo K.S., Wood A.R., Kjaer T.R., Fine R.S., Lu Y., Schurmann C., Highland H.M. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton N.M., Beltram J., Carter R.N., Michailidou Z., Gorjanc G., McFadden C., Barrios-Llerena M.E., Rodriguez-Cuenca S., Gibbins M.T., Aird R.E. Genetic identification of thiosulfate sulfurtransferase as an adipocyte-expressed antidiabetic target in mice selected for leanness. Nat. Med. 2016;22:771–779. doi: 10.1038/nm.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naval-Sanchez M., Nguyen Q., McWilliam S., Porto-Neto L.R., Tellam R., Vuocolo T., Reverter A., Perez-Enciso M., Brauning R., Clarke S. Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds. Nat. Commun. 2018;9:859. doi: 10.1038/s41467-017-02809-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCD Risk Factor Collaboration (NCD-RisC) Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet. 2016;387:1377–1396. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L., Petersen K.F., Kibbey R.G., Goodman A.L., Shulman G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M.E., Johansson A.M., Siegel P.B., Carlborg O. Dynamics of adaptive alleles in divergently selected body weight lines of chickens. G3 (Bethesda) 2013;3:2305–2312. doi: 10.1534/g3.113.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.J., Megens H.J., Martinez Barrio A., Maqbool K., Sayyab S., Schwochow D., Wang C., Carlborg Ö., Jern P., Jørgensen C.B. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U S A. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Kofler R., Versace E., Tobler R., Franssen S.U. Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity (Edinb) 2015;114:431–440. doi: 10.1038/hdy.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M.E., Honaker C.F., Siegel P.B., Carlborg Ö. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome Biol. 2015;16:219. doi: 10.1186/s13059-015-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp S.L., Cline M.A., Gilbert E.R. Recent advances in the understanding of how neuropeptide Y and α-melanocyte stimulating hormone function in adipose physiology. Adipocyte. 2016;5:333–350. doi: 10.1080/21623945.2016.1208867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel P.B. Evolution of the modern broiler and feed efficiency. Annu. Rev. Anim. Biosci. 2014;2:375–385. doi: 10.1146/annurev-animal-022513-114132. [DOI] [PubMed] [Google Scholar]
- Stenkamp-Strahm C.M., Nyavor Y.E., Kappmeyer A.J., Horton S., Gericke M., Balemba O.B. Prolonged high fat diet ingestion, obesity, and type 2 diabetes symptoms correlate with phenotypic plasticity in myenteric neurons and nerve damage in the mouse duodenum. Cell Tissue Res. 2015;361:411–426. doi: 10.1007/s00441-015-2132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R., Dijk W., van Beek L., Jansen H., Heemskerk M., Houtkooper R.H., Denis S., van Harmelen V., Willems van Dijk K., Tack C.J. Mannose-binding lectin is required for the effective clearance of apoptotic cells by adipose tissue macrophages during obesity. Diabetes. 2014;63:4143–4153. doi: 10.2337/db14-0256. [DOI] [PubMed] [Google Scholar]
- Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016;87:159–167. doi: 10.1111/asj.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Barrick J.E., Ribeck N., Deatherage D.E., Blanchard J.L., Dasgupta A., Wu G.C., Wielgoss S., Cruveiller S., Médigue C. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hu X., Wang Z., Zhang Y., Wang S., Wang N., Ma L., Leng L., Wang S., Wang Q. Selection signature analysis implicates the PC1/PCSK1 region for chicken abdominal fat content. PLoS One. 2012;7:e40736. doi: 10.1371/journal.pone.0040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Cheng B.H., Yang L.L., Wang Z.P., Zhang H.L., Xu S.S., Wang S.Z., Wang Y.X., Zhang H., Li H. Identification of a potential functional single nucleotide polymorphism for fatness and growth traits in the 3'-untranslated region of the PCSK1 gene in chickens. J. Anim. Sci. 2017;95:4776–4786. doi: 10.2527/jas2017.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li J., Guo Y., Zhang L., Xu L., Gao X., Zhu B., Gao H., Ni H., Chen Y. Multi-strategy genome-wide association studies identify the DCAF16-NCAPG region as a susceptibility locus for average daily gain in cattle. Sci. Rep. 2016;6:38073. doi: 10.1038/srep38073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FL, fat line; LL, lean line
Data Availability Statement
The accession numbers for the genome re-sequencing data reported in this paper are NCBI SRA: PRJNA353057 and PRJNA354990. Other data that support the findings of this study are available from the corresponding authors on request.