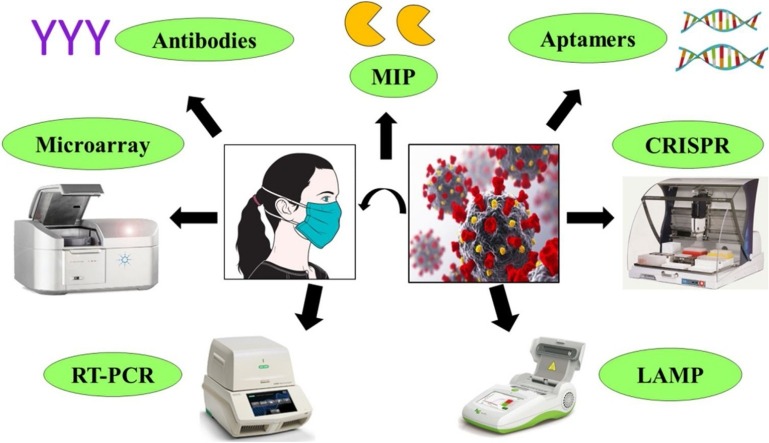
Graphical abstract
Keywords: Coronavirus, Diagnosis, PCR, COVID-19
Highlights
-
•
Multiple methods are reported to detect coronavirus.
-
•
Most of these methods are PCR and antibody-based.
-
•
Simple, low-cost and sensitive methods are required for large-scale testing.
-
•
WHO validated many in-house and commercial assays for the detection of COVID-19.
-
•
Vaccine for creating immunity against coronavirus is also an urgent requirement.
Abstract
To develop diagnostics and detection methods, current research is focussed on targeting the detection of coronavirus based on its RNA. Besides the RNA target, research reports are coming to develop diagnostics by targeting structure and other parts of coronavirus. PCR based detection system is widely used and various improvements in the PCR based detection system can be seen in the recent research reports. This review will discuss multiple detection methods for coronavirus for developing appropriate, reliable, and fast alternative techniques. Considering the current scenario of COVID-19 diagnostics around the world and an urgent need for the development of reliable and cheap diagnostic, various techniques based on CRISPR technology, antibody, MIP, LAMP, microarray, etc. should be discussed and tried.
1. Introduction
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected almost every country with 7,410,510 total cases and 418,294 deaths globally as of 12 June 2020 [1]. Since this virus is new and due to lack of any approved drug or vaccine, there is urgent need for a highly specific and sensitive diagnostic measures to identify infected people and isolate them to avoid the further spread of the virus [2,3]. Testing is paramount in the fight to slow down and minimize the spread of the virus. Effective diagnostic approaches need to be discussed and tried to detect the virus itself or the response of the host body to the virus. Currently, numerous diagnostic technologies are available for SARS-CoV-2 detection, and many are under development [4]. Recently, the World Health Organization (WHO) has enumerated the first two diagnostic tests for emergency use to detect COVID-19 to increase the accurate assessment of disease. The tests are genesis Real-Time polymerase chain reaction (RT-PCR) Corona virus and Cobas SARS-CoV-2, a qualitative assay for use on the Cobas® 6800/8800 Systems, and are for in-vitro diagnosis of COVID-19 [5]. Currently, the most popular diagnostic tool that identifies the viral RNA through amplification is RT-PCR. It is a sensitive technique that requires a small amount of input RNA but takes hours to get results [6]. Other tools that target virus includes microarray relying on the binding of the viral genome-specific probe and CRISPR technology that recruits Cas12/13 enzyme specific for viral genes for detection of SARS-CoV-2 [7,8]. As SARS−COV-2 infected individuals produce antibodies against the virus, these antibodies can be used for the identification of infected individuals. Antibody-based diagnostics give a positive result if they find binding antibodies that might have generated earlier during the infection and would serve to provide protection against reinfection thereby, limits their use in the detection of active diseases [9]. The sensors are another approach that presents low-cost promising diagnostic tools with high sensitivity to detect COVID-19 [10]. Most of the diagnostic techniques used now-a-days are time-consuming and require skilled expertise, but for large-scale screening, we need to develop more efficient methods that can be used for point-of-care (POC) detection to identify COVID-19 positive patients. Besides several diagnostic kits, US Food, and drug administration (USFDA) approved the first at home COVID-19 test kit with a home collection option to increase COVID-19 testing capacity [11]. In this crisis, where a collective pool of knowledge is a prerequisite, here, we recapitulate available updates on diagnostic methods such as PCR, microarray, molecularly imprinted polymer (MIP)-based sensor, CRISPR, etc. for COVID-19 detection.
2. Structure and genomics of SARS-CoV-2 are a vital premise for diagnostic development
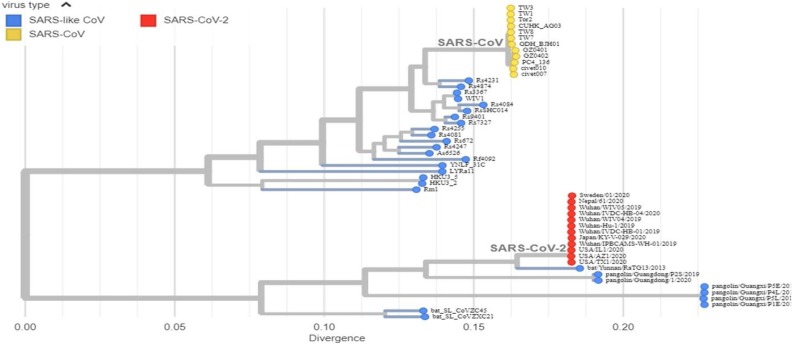
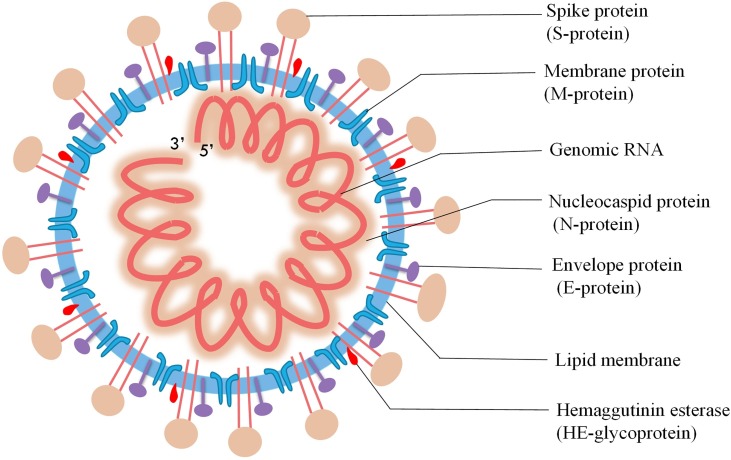
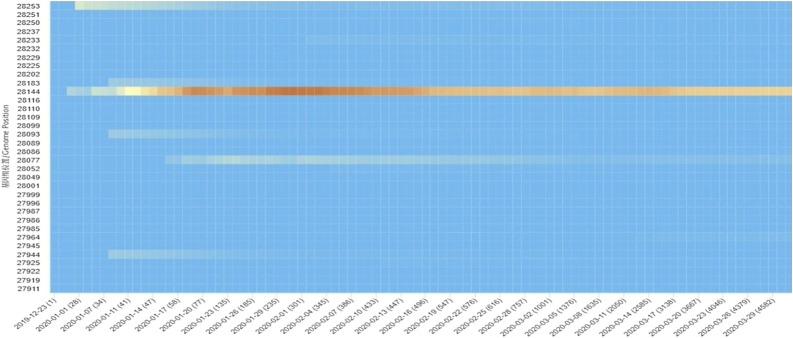
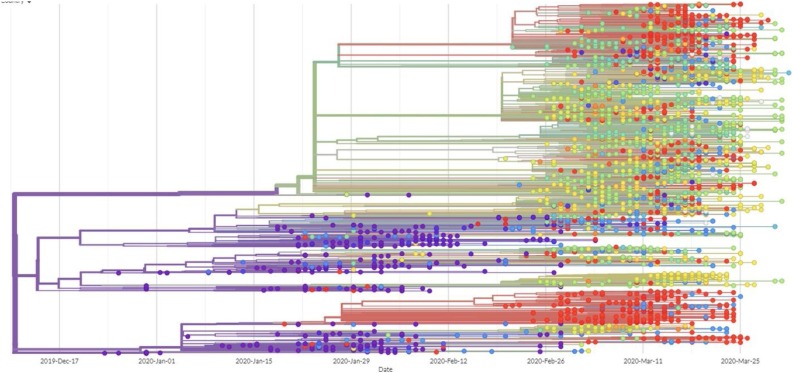
As current and future COVID-19 detection methods are based on genomics and structure of SARS-CoV-2, it is pertinent to review the recent progress on these aspects. SARS-CoV-2, a novel coronavirus species, placed under the betacoronavirus genus based on genomic similarity and phylogenetic relationship with SARS-CoV (Fig. 1 ). Genome sequence of SARS-CoV-2 has 88 % similarity with SARS-like bat derived coronaviruses, SL-CoVZC45, and SL-CoVZXC21. Among different coronaviruses, RNA-dependent RNA polymerase (RdRp) gene sequence is an extremely conserved sequence. According to the International Committee on Taxonomy of Viruses criteria, if a species shows less than 90 % similarity for conserved RdRp sequence, it would be considered as novel species. RdRp sequence of isolated strain in Wuhan, China exhibits 86 % similarity with existing SL-CoVZC45 coronavirus; therefore, CoVs were declared as a new species (SARS-CoV-2). SARS-CoV-2 has a single-stranded positive helix RNA genome of 30 kb with a GC content of 38 % [12]. Whole-genome sequencing showed that the virus genome from different parts of the world exhibited sequence homology of more than 99.9 % with SARS-CoV-2 isolated from Wuhan, China [13]. Homology modeling showed that the receptor-binding domain of SARS-CoV-2 and SARS-CoV differs only in a few amino acid residues [14]. The genome of SARS-CoV-2 consists of an open reading frame (ORF) 1a/b-coding region and four protein-coding regions flanking with the non-coding region on both sides. Starting from 5′ end protein-coding region, an s-region coding for spike protein, e-region coding for envelope protein, m-region coding for a membrane protein, and n-region coding for nucleocapsid protein are present [15]. Structural and accessory proteins (S, M, E, N-proteins) are translated from sgRNAs (single guide RNAs). The most abundant structural protein in coronavirus is membrane glycoprotein (∼25−30 kDa), spans the lipid membrane thrice with the N-terminal domain on the outside and C-terminal domain inside the virion. S-protein (∼150 kDa) recognizes and binds to the receptor present on the host cell, thereby responsible for viral infectivity. Scanning electron micrograph of the virus revealed that it is oval or spherical with stalk-like projections ending in round structure (spike) like other viruses of coronaviridae family. Spikes are essential for viral infectivity and host specificity. While invading host cell, furin-like proteases cleave S-protein into two parts: a receptor binding unit (S1) and a membrane-anchored fusion unit (S2). Envelope protein (8−12 kDa) determines the formation and composition of the viral membrane. Nucleocapsid protein protects and enfolds the viral RNA [16] (Fig. 2 ). SARS-CoV-2 binds to receptors on the cell surface via receptor-binding domain (RBD) present in their S1 subunit. RBD of SARS-CoV-2 is an almost identical 3-D structure with that of SARS-CoV and 76.47 % amino acid sequence similarity, which uses spike proteins to bind with Angiotensin-Converting Enzyme 2 (ACE2) on host cell [17]. Thereby, it is believed that SARS-CoV-2 also enters cells by binding spike proteins to ACE2. SARS-CoV-2 contains ORF3 and whole ORF8 gene regions, which are characteristic features of bat-origin coronaviruses [12]. Scanning electron micrograph revealed that virus particle size ranges from 70−90 nm and invades various intracellular organelles, especially vesicles [13]. Immunofluorescent assays of the culture of Vero cells showing cytopathic effect with the convalescent serum from patients showed green signals in the cytoplasm; in contrast, no signal was detected in control serum. Though viruses recruit error-prone RNA polymerase for replication, so it is more likely to get mutated [18]. The emergence of new strains is a consequence of mutation. Mutation in SARS-CoV-2 enabled it to spread more efficiently from animal to humans and then from human to human. Mutation in ORF8 region at 28,144 and ORF1b region at 8872 occurred in the early stages of the outbreak of SARS-CoV-2 (SARS-CoV was also mutated in the ORF region during 2002−03 outbreak). Mutation in the ORF8 gene at 28,144 position is stable and increases population frequency from 0 to 29% as the epidemic progressed (Fig. 3 ). These mutations became fixed during the outbreak and increase population frequency gradually. The phylogenetic analysis shows SARS-CoV-2 is the mutant product of SARS-CoV. The phylogeny indicates that the initial emergence of novel strain in Wuhan, China gained transmission between humans and was followed by worldwide outbreak transmission (Fig. 4 ). All the research reports on genomics and structure of SARS-CoV-2 are essential, which can become the premise of the development of more accurate, sensitive, and effective detection methods.
Fig. 1.
Phylogeny of SARS-like betacoronaviruses including novel coronavirus SARS-CoV-2.
Phylogeny shows evolution of SARS-like betacoronaviruses. SARS-CoV-1 coronaviruses from the 2002−03 SARS outbreak are colored in yellow, SARS-CoV-2 coronaviruses from the COVID-19 epidemic are colored in red, while related SARS-like coronaviruses are colored in blue. Image has been taken from GISAID database (https://www.gisaid.org/).
Fig. 2.
Schematic of structure of SARS-CoV-2.
Diagram showing the single stranded RNA genome and proteins present in coronavirus.
Fig. 3.
Heatmap showing the mutation in ORF 8.
The trend of the mutation sites over time is displayed in the form of heatmap. Population frequency of site 28,144 located in gene ORF8 slowly raised from 0 to 35.98 %, and then dropped to 16.96 % as the epidemic progressed (see the area that gradually changes from blue to red). Image has been taken from http://bigd.big.ac.cn/ncov/variation/heatmap.
Fig. 4.
Tree showing genomes of SARS−COV-2 worldwide.
Phylogenetic tree shows the 4645 genomes sampled from December 2019 to April 2020 in different geographical regions. Each color represent specific country where genome has been sequenced. Image has been taken from GISAID database (https://www.gisaid.org/epiflu-applications/next-hcov-19-app/).
3. PCR-based detection
Due to the increasing incidence of SARS-CoV-2, PCR based detection is routinely used for the diagnosis of COVID-19. RT-PCR or qPCR is a molecular biology technique used to study gene expression at the transcript level. It involves the following steps: RNA isolation from samples and cDNA synthesis using reverse transcription kit. PCR was set by making a working mix of the buffer, dNTPs, primers of the target gene, Taq polymerase, cDNA template and, SYBR green dye. PCR mix incubated in a PCR machine and fluorescence generated by PCR amplification is measured by machine to give Cycle threshold (Ct) values. Ct values of control and experimental samples are compared, and relative expression is estimated (Fig. 5 ). On 14 January 2020, the protocol of RT-PCR for detection of 2019-nCoV was published on the WHO website for its use of COVID-19 detection [19]. Other than regularly used RT-PCR for diagnosis of COVID-19, some other studies used other diagnostic techniques like RT-insulated isothermal PCR (RT-iiPCR) [20], reverse transcription loop-mediated isothermal amplification (RT-LAMP) [21] and a one-step rRT-PCR assay [22] in which unique TaqMan probes is used. RT-LAMP has a high specificity for detection of MERS-CoV, and its sensitivity is comparable to RT-PCR. RT-iiPCR and a one-step rRT-PCR assay also show similar sensitivity and specificity, but RT-PCR is used mainly for the diagnosis of MERS-CoV. The selection of specimen type and time of taking that sample for RT-PCR is also a crucial point for the diagnosis of SARS-CoV-2.
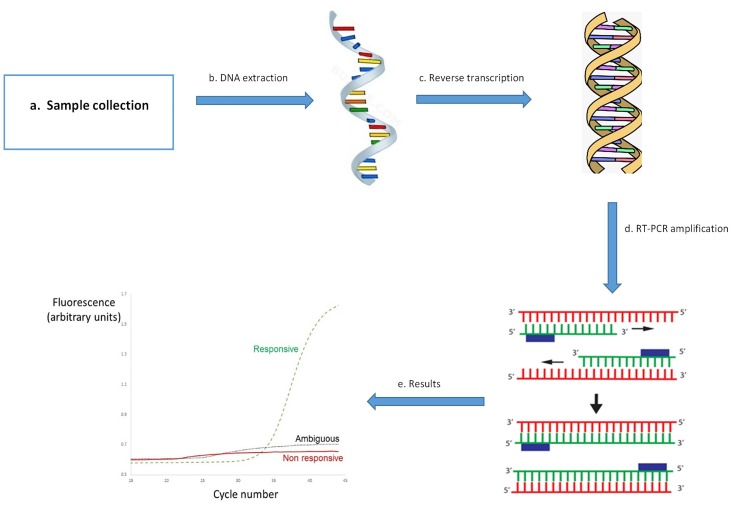
Fig. 5.
Diagnosis of COVID-19 using PCR.
a) Respiratory sample collection from patient b) Extraction of RNA from sample c) Reverse transcription for cDNA synthesis d) DNA amplification using RT-PCR e) COVID-19 positive DNA sample shows amplification, if sample is negative for COVID-19 then it doesn’t show any amplification
The PCR-based first test for diagnosis of SARS-CoV-2 was constructed in Germany by Corman et al., at first, before the release of sequence, RT-PCR assay was designed following SARS or SARS-like coronavirus because it was supposed that 2019-nCoV is SARS-related. After getting the sequence information, the RT-PCR assay was designed according to sequence similarity with SARS-CoV-2 after sequence alignment. Corman et al., developed a detection method that can differentiate between SARS-CoV and SARS-CoV-2. At first, three assays were done: E gene assay, N gene assay, and RdRp gene assay. But due to less sensitivity of N gene assay, only two assays were selected where E gene assay works as a first-line screening tool and RdRp gene assay was done for confirmation of this testing. They used two different probes: one binds with 2019-nCoV and SARS-CoV and 2nd one bind only with 2019-nCoV. Results show that 2019-nCoV related probe only binds with 2019-nCoV but not with SARS-CoV, so these assays are highly sensitive and specific [23]. Chu et al., designed two 1-step quantitative real-time reverse transcriptase PCR assays which detect two different regions: ORF1b and N as these are the highly conserved sequence in sarbecoviruses. Throat swab and sputum samples were tested from two patients along with positive and negative controls, and results are highly specific and sensitive. N gene RT-PCR works as a screening tool, and ORF1b used as confirmatory testing [24]. Konrad et al., done laboratory testing for comparing RT-PCR assays: QuantiTect Virus + Rox Vial kit (QIAGEN), SuperScript III One-Step RT-PCR Platinum TaqDNA Polymerase (Invitrogen, Darmstadt, Germany) and a commercial kit: RealStar SARS-CoV-2 RT-PCR kit 1.0 (Altona).
The protocol and thermal conditions of each assay were adjusted according to the manufacturer, but the concentration of probes was taken the same as published before by Corman et al., in 2020. These assays show unspecific E gene signals but using SuperScript III these unspecific signals reduce to 5%, so SuperScript III RT-PCR is better as given by Corman et al. This research also recommended RealStar SARS-CoV-2 RT-PCR kit 1.0 (Altona) as it does not give any unspecific E gene signals [25]. In a study, a combined approach was developed by using RT-PCR, CRISPER- based assay, and metagenomic next-generation sequencing (mNGS) for diagnosis of pneumonia patient, which later finds positive according to RT-PCR and CRISPER-based assay [26]. So, by combining these approaches, equitable diagnostic results can be found. An article published in a magazine named Semiconductor Digest stated that Cepheid, a California company, made a new point of care detection test: Xpert® XpressSARS-CoV-2. This test can give results only in 45 min, and it requires less than one min for sample preparation, so even healthcare workers can use it easily [27]. POC devices are easy to use and less time taking so these can be used primarily for diagnostic purposes in the next few months.
4. Antibody-based detection
Antibodies (Abs), also called immunoglobulins, are Y-shaped proteins produced by the immune system when a foreign substance (like a virus) enters the system, and then these molecules bind to the foreign substance and neutralize it. Simply put, they are the weapon used by the immune system to fight new infections [28]. The SARS-CoV-2 is being tested for an approach to treating new diseases that worked for the Ebola and Zika viruses, which involves using antibodies to the disease as a drug. Because of exquisitely affinity, target specificity, designed and engineered nature, they have played a significant role in the number of diagnostic approaches [29]. Currently, a variety of COVID-19 tests are already available that determine the viral genome from mucosal membrane swabs, till now, eleven POC diagnostics for COVID-19 are reported, which is mainly based on molecular and antibody-based tests. Various research groups and companies are trying hard to develop antibody tests which include lateral flow immunoassays (BioMedomics rapid test and Surescreen rapid test cassette), time-resolved fluorescence immunoassay (Goldsite diagnostics kit), colloidal gold immunoassay (Assay Genie POC kit and VivaDiag COVID-19 IgG-IgM test), ELISA, etc [30]. While a variety of antibody tests are also available in the market, there is a lot of confusion regarding the efficacy of such tests given the high percentage of the asymptomatic population and the fact that detectable antibodies typically develop later in the disease. Therefore, diagnostic approach proposals will have to describe their novelty in terms of process, sensitivity, specificity, the strength of risk stratification, and scalability.
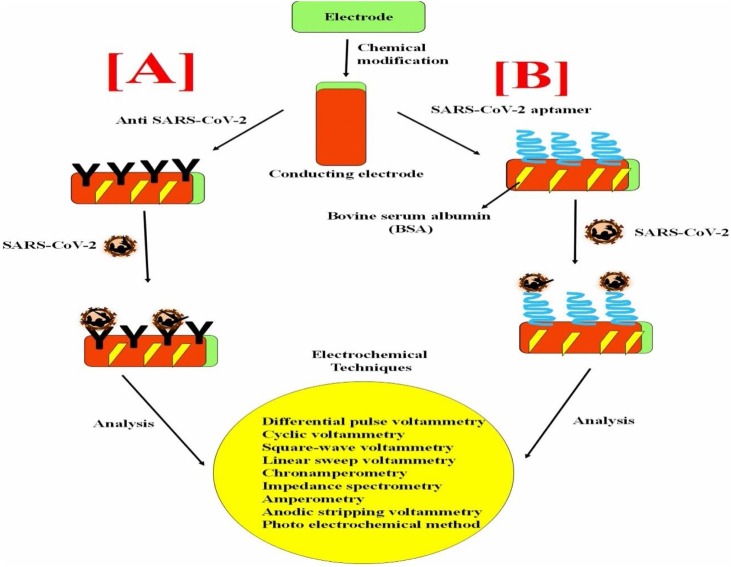
We believe that the electrochemical immunosensor technique could be a potential approach for the POC device application in the detection of SARS-CoV-2, as shown in (Fig. 6 A). There are several recent research studies about a variety of virus detection using immunosensors [[31], [32], [33], [34], [35], [36], [37], [38]]. For example, the electrochemical based influenza A virus H5N1 biosensors are used for the detection of influenza A virus H5N1 to prevent massive death and to control the transmission from one country to the other due to their sensitivity, selectivity and economically affordable than the conventional detection method [39]. In another study, reduced graphene oxide-based electrochemical immunosensor was developed using EDC-NHS coupling chemistry between the COOH group of graphene attached on the gold surface of the working electrode and NH2 of antibody specific for H1 of H1N1 influenza and the reported value of the limit of detection is 0.5 PFU ml−1 [40].
Fig. 6.
A hypothetical workflow of electrochemical SARS-CoV-2 sensor.
A conducting electrode can be altered with nanostructures for high loading of SARS-CoV-2 specific antibodies [A] and Aptamers [B] for detection of COVID-19 using specific transducer system.
Thus, several studies are still going on based on immunosensor, which aims to achieve good sensitivity and specificity via different electrodes surface modifications. In the overall view, the best advantage of these hypothesized assays is easy handling, no requirement of any sophisticated instruments, rapid detection efficacy, and cost-effectiveness than other molecular approaches. Identification of major antigenic and protective epitopes of target virus particles, is crucial in understanding the antibody response while developing these detection strategies, as antigenic variations make it difficult to generate similar antibodies, thus affecting the development of these diagnostic assays.
5. Aptamer-based detection
Deoxyribonucleic acid (DNA) is perhaps the most essential of all biomolecules and has been the foundation for genetic study over the past few years. However, the use of aptamers, which are small-sized, single-stranded artificial nucleotides (RNA or DNA), having 10–100 nucleotides, are now gaining enormous importance due to its specific binding just like in case of antibodies. The selection technique of aptamers is done via the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process that binds to a broad range of target analytes with high affinity and specificity. Aptasensors can measure analytes in tiny quantities where such small amounts cannot be identified with most of the other methods being used [41,42]. Furthermore, the low cost of the aptasensor design compared with other virus diagnostic methods should be regarded as one of the economic benefits [43,44]. Many vital benefits of aptasensors compared to other available diagnostic techniques include low detection time and a quick detection method [45,46]. Though aptamers have massive potential as a viable tool in therapeutics and virus detection, so aptamer-based biosensors also known as aptasensors, have reported the various methods for the immobilization of the DNA oligonucleotides sequences and have used different biomaterials for enhancing the stability and sensitivity of the electrode [47]. Several electrochemical DNA biosensors have been developed for the detection of various viruses [[48], [49], [50], [51], [52], [53]] and, based on that, a hypothetical schematic illustration of aptasensor based direct detection of SARS-CoV-2 using the different platform has been shown in (Fig. 6B). In a study, DNA influenza virus A H5N1 aptasensor has been developed, which is a reliable and suitable analytical device with newly emerged designed for the detection of AIV H5N1 [10]. The active biological element on the DNA biosensor is specific oligonucleotide sequences single-stranded DNA (ssDNA) for identifying the viral genome of complementary ssDNA during the hybridization process.
The standard diagnostic methods implemented for SARS-CoV-2 are often invasive and costly; these methods often cannot detect the presence of SARS-CoV-2 virus in tiny amounts. For the identification of the many viruses with high affinity, aptamers as molecular structures were used in the aptasensor system. The use of advanced nanostructures and the functionalized aptamer with certain organic materials will improve the aptasensor's diagnostic sensitivity and specificity towards SARS-CoV-2. As the aptasensor specificity and sensitivity are higher, they make them suitable commercial diagnostic devices for early diagnosis of SARS-CoV-2. The work is hoped to reduce the effects of interfering factors in SARS-CoV-2 aptasensors; the more reliable and compact SARS-CoV-2 diagnostic tools such as commercial aptasensors will also be developed by incorporating other technologies.
6. CRISPR-based approach
CRISPR is a well-known biotechnological technique for genome editing. Various research groups all over the world have developed a successful methods for the detection of SARS-CoV-2 using CRISPR. Zhang et al., developed a protocol for the detection of SARS-CoV-2 using CRISPR-based SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) technique. Protocol employed the use of Cas13 for targeting two genes in the virus genome, S gene, Cas 13 enzyme and ORF1ab gene. They used synthetic virus RNA fragments as input. The SHERLOCK detection protocol comprises of three steps: 1) isothermal amplification of RNA sample using recombinase polymerase amplification kit, 2) amplified viral RNA is incubated with Cas13 enzyme, guide RNA (gRNA) and reporter. With the help of gRNA, Cas13 enzyme recognizes viral RNA and starts cleaving nearby RNAs, including the reporter. As both ends of the reporter are differently labeled, cleavage creates a unique signature, and 3) cleaved reporter produces distinct band than intact reporter RNA, which can be visualized by eye using the paper dipstick. Using serial dilution of synthetic RNA fragment of the S-gene and ORF1ab gene, Zhang et al., have detected viral RNA in a range between 10–100 copies/μl [8]. This protocol is developed using synthetic RNA and has not validated using real patient samples. Unlike qPCR, which gives results in hours and requires skilled personnel. SHERLOCK technique is highly sensitive and robust and can give results in less than an hour using dipstick without any elaborate instrumentation.
MammothBiosciences published a white paper on the protocol for rapid detection of SARS-CoV-2 using CRISPR employing DNA endonuclease targeted CRISPR trans reporter (DETECTR). MammothBiosciences reconfigured DETECTR for quick and accurate detection of SARS-CoV-2 using the lateral flow strip. The technique utilizes Cas12 enzyme for targeting three genes, N gene (SARS-CoV-2 specific), E gene (SARS-CoV, bat-SARS-like-CoV, and SARS-CoV-2), and P gRNA (sample control). The protocol includes: 1) isothermal amplification of extracted RNA using RT-LAMP 2) Cas12 enzyme incubation to generate RNP complexes, one for each N-gene, E-gene, and P-gRNAs, and reporter probe and 3) visualize result using lateral flow strip. This method can discriminate between SARS-CoV-2 and other coronavirus strains. This assay takes 30 min from the sample to result in the detection limit of 70–300 copies/μl [54].
In the queue of detection of SARS-CoV-2 using CRISPR, Lucia et al., employed Cas12 enzyme for targeting RdRp, ORF1b, and ORF1ab genes, and used WH-human1 sequence as control. They employed synthetic RNA fragments and amplified isothermally using the RPA kit. Amplified fragments were mixed with Cas12, single guide RNA (sgRNA), and reporters. Detection of SARS-CoV-2 was performed using plate reader-based assay and paper strips. ssDNA reporters were labeled with fluorescein amidites (FAM) and biotin for fluorescence and paper assay, respectively. The limit of detection for both assays was 10 copies/μl. The study was carried out using synthetic fragments because of the non-availability of real samples in the nearby regions. The study represents the usefulness of CRISPR-Cas12 technology for the detection of the virus [55].
Besides detection, CRISPR/Cas13 approach can also be used for the treatment of COVID-9. Nguyen et al., proposed that CRISPR/Cas13d can be employed to specifically degrade the SARS-CoV-2 genome, thereby halting its ability to reproduce. For the specificity, they used gRNA containing spacer sequences complementary to ORF1ab and S-gene. With the help of gRNA, Cas13 recognizes the viral genome and chew it up without affecting the human transcriptome. To achieve higher specificity, for the delivery of Cas13d in COVID-19 patients, an adeno-associated virus (AAV) can be used as a vehicle. One AAV can carry up to three gRNAs targeting different genes, thereby increasing the efficiency of treatment. SARS-CoV-2 infects lungs primarily, so to ensure organ-specific delivery, lungs specific AAV serotype can be used. The proposed CRISPR/Cas13d approach is the potentially meteoric approach for the treatment of COVID-19. This approach is under review, and if proven to be effective, it will provide a worldwide weapon to fight against RNA virus infection [56].
Abbott et al., developed Prophylactic Antiviral CRISPR in human (PAC-MAN) strategy to define the conserved targetable region of SARS-CoV-2 using CRISPR/Cas13d. They screened and designed a panel of targetable coronavirus RNA. This study revealed that a group of 22 crRNA targets all coronaviruses, whereas a group of 6 crRNA can target 91 % of sequenced coronaviruses using CRISPR/Cas13d. Predicted crRNA pool was tested on lung epithelial cell lines using CRISPR because of a lack of access to live SARS-CoV-2 and observed manifold inhibition in viral replication. PAC-MAN strategy could be a powerful tool against COVID-19 [57].
7. Molecularly imprinted polymer (MIP)-based detection
While a variety of bioassays and biosensors have been developed, low-cost, disposable, or reusable biosensors are still needed that can quickly detect and accurately identify the SARS-CoV-2. The use of molecular imprinting technology as elements of biorecognition for the design of MIPs offers a real alternative to antibodies due to their robustness and reproducibility [58]. The molecular imprinting method allows the host components to selectively, sensitively, and rapidly identify and detect the numerous molecules. In the previous years, MIPs based sensors have formed an exciting horizon for surface modification methods by creating specific recognition cavities in the template molecules. They have to turned out to be fascinating, having sensitivity to small structural changes in the biomolecule structure. There are several research reports which are aimed at MIP based approach for medical diagnostics for the detection of various types of viruses such as Influenza virus, Dengue virus, Japanese encephalitis virus, human immunodeficiency virus, Hepatitis virus A and B, Adenovirus, Picornavirus, etc. [[59], [60], [61], [62], [63], [64], [65], [66]].
For example, a MIPs based nanosensor was developed to detect human papillomavirus derived E7 protein by Cai and co-workers [67]. Their EIS data analysis showed the detection of E7 protein could be as low as sub-pg L−1 levels. Ma et al., have fabricated an electrochemical biosensor based on multi-walled carbon nanotubes modified MIPs for detecting HIV-p24 in human serum samples whose linear range was in the range of 1.0 × 10-4 - 2.0 ng cm-3 and limit of detection (LOD) was found to be 0.083 pg cm-3 [68]. Recently, Tancharoen et al., have proposed an electrochemical sensor based on graphene oxide polymers imprinted for the detection of Zika virus. The detection limit of this sensor was then compared to the commercial method and found the observed value to be similar to the reverse transcription [69].
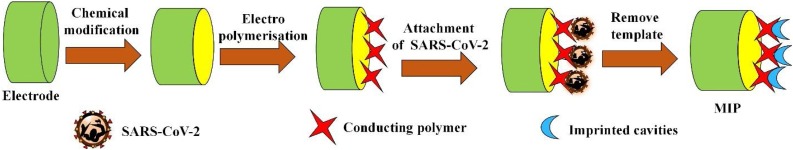
One of the key advantages of MIPs is their robustness, high selectivity, long-term stability, and cost-efficiency, which cannot be accomplished by using fragile biomolecules. MIPs were often used for selective and sensitive viral detection and the distinction of viral subtypes. Thus, it is anticipated that the many advantages of molecular imprinting technology should find its home in the frontlines of possible SARS-CoV-2 detection (Fig. 7 ).
Fig. 7.
Hypothetical illustration of MIP based electrochemical sensor for COVID-19 detection.
Electrode is fabricated and SARS-CoV-2 could be used as a template to create specific recognition cavities into polymers for their specific detection.
8. Microarray-based detection
Gene microarray technology can also be used for the diagnosis of SARS-CoV-2. In the microarray approach, there are two main steps: the formation of a specific probe and the production of targeted cDNA fragments. At first, specific probe sequences are immobilized on the glass slide called chip and then-unknown DNA molecules that are present in a sample are cut into fragments by restriction endonucleases. These fragments are attached with fluorescent marker and are allowed to bind with probesthat are present on the chip. Afterwards the target DNA fragments which have complementary sequences of probes will bind with DNA probe. Hybridization of the fluorescent DNA fragments with a DNA probe on the chip will release fluorescent signals, and analysis is done based on these signals for the identification of DNA fragments. Rong et al., developed a 60-mer oligonucleotide microarray for the detection of SARS coronavirus. Probes were designed, which screen the whole genome of the virus and immobilized on the microarray surface. Samples were taken from the throat swab and gargling fluid of SARS patients, and then RNA was extracted, and cDNA was synthesized by reverse transcription and fragmented through restriction display (RD) technique. Fragmented DNA was labeled with Cy5-universal primer through PCR, and then hybridization was done. There was no signal in negative and blank controls, and results showed that microarray could be used for the detection of SARS coronavirus [70].
As studies find out that there are many point mutations in SARS-CoV genome sequence, Long et al., designed a universal microarray system for detection and genotyping of SARS-CoV by targeting six single nucleotide polymorphisms (SNP) which are allocated through the whole genome. This microarray system was made by combining RT-PCR and ligase detection reaction (LDR). The Zip Codes were attached at the slide surface, and their complimentary Zip Codes were attached with targeted DNA fragments. These Zip Codes sequences were universal as these have no similarity with targeted sequence, human host, or SARS-CoV, so there is no chance of false-positive signal of mismatch hybridization. Twenty samples were tested using this assay, and results were confirmed by DNA sequencing [71]. Lu et al., done screening of specific antigens for the diagnosis of SARS-CoV by using protein microarray. In this study, structural proteins were expressed in E. coli as GST or TRX fusion protein and deposited on the microarray, and testing was done with serum samples from SARS patients. Results show that this GST-N2 fusion protein may prove an essential antigen for serological assay of SARS [72].
Zhang et al., designed a novel method for detection SARS-CoV specific genes by using novel asymmetric multiplex PCR for amplification of DNA with universal primers and labeled PCR products are hybridized with oligonucleotide microarray. Results were analyzed by a fluorescent scanner [73]. Guo et al., developed a novel method: SNP DNA microarray for detection and genotyping of SARS-CoV by targeting 24 SNPs. Sample for testing was taken from 19 SARS-CoV patients,and PCR amplified product was hybridized with the microarray. Results of hybridization show that all samples were detected and genotyped accurately with 100 % accuracy [74]. So, the microarray is a better technique for the detection of the virus with a high mutation rate. Microarray-based diagnostics can be a better approach for detection of SARS-CoV-2 because PCR based methods can detect only a few numbers of genes per experiment, but microarray can detect a higher number of DNA fragments synchronously.
9. Loop-mediated isothermal amplification (LAMP)-based detection
In 2000, Notomi et al., had developed LAMP technique that amplifies DNA quickly with high specificity and simplicity at a constant temperature [75]. Now, after two decades of its development, LAMP seems a most promising approach to tackle the burden of COVID-19 testing as it is easy to use and does not require specialized machines such as PCR and costly reagents kits. The LAMP test can be done in a single tube at a constant temperature, hence very economical. DNA amplification can be detected by colorimetric, turbidity, or fluorescent-based methods [76]. There are increasing research reports which strongly hint at the possibility of employing the LAMP test for COVID-19 detection. Naama Geva-Zatorsky group from Israel developed a test for COVID-19 detection using Reverse Transcribed Loop-Mediated Isothermal Amplification (RT-LAMP) technique. From the nose and throat swabs, SARS-CoV-2 can be detected by the developed test without RNA purification step [77]. The test was applied to 180 suspected patients, and results were found comparable to standard detection methods for COVID-19.
Another fascinating research report can also be seen on the preprint server (bioRxiv) in which a group from ‘Weill Cornell Medicine’ designed a 30-minute colorimetric LAMP test for COVID-19 detection and also developed a shotgun metatranscriptomic profiling platform technology for nose swabs [78]. Simplicity and low cost of the LAMP technique will increase its chance to be chosen as a favored diagnostic tool for COVID-19 detection, and this can be used as a POC device at workplaces, clinics, and entry points.
10. Conclusion
Testing is a valuable tool to control the spread of COVID-19. In our fight with the COVID-19 pandemic, testing is playing a crucial role in public health monitoring. Research reports are evolving rapidly about new diagnostics for COVID-19 detection. Detection technologies based on PCR and antibody are helping us to identify infected people and isolate them for stopping the spread of infection. Currently, detection of COVID-19 is dominated by PCR and antibody-based diagnostics, but alternative technologies such as LAMP, RT-LAMP, CRISPR, etc. are under development and may hit the diagnostic market of COVID-19 soon. Most of the detection technologies use the nasopharyngeal samples for SARS-CoV-2 detection, but oral and blood samples seem more appropriate for upcoming technologies. For sensitivity, reproducibility, and reliability, all COVID-19 testing technologies need to be studied and compared. The next few months will witness the comparison, advantage, and disadvantage of these technologies and how suitable they are for mass screening of healthy and suspected patients. Techniques such as LAMP, which requires minimum resources and reagents may be favoured and increase the testing capabilities of hospitals and clinics. In the coming months, we can witness more comprehensive technologies that will provide more insights for viral and host biology besides just detection. MIP, rolling circle amplification [79] real-time NASBA [80] AI, and smart phone are some examples of future technologies that can be pursued and optimized for the detection of COVID-19. Besides, development of therapeutics and vaccines for the control of COVID-19, the development of new testing technologies is equally important to fight against COVID- and lessen the socio-economic harm, we all are facing in current time due to this pandemic.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgment
The NII Core grant supported this work. This work was also financially supported by the Indo-Russia project (DBT/IC-2/Indo-Russia/2017-19/02), Government of India.
Contributor Information
Anil Kumar, Email: anilk@nii.ac.in.
Pratima R. Solanki, Email: partima@mail.jnu.ac.in.
References
- 1.World Health Organization (WHO). Novel-coronavirus-2019. Situation report.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed 13 June 2020.
- 2.Patel R., Babady E., Theel E.S., Storch G.A., Pinsky B.A., George K.St., Smith T.C., Bertuzzi S. Report from the american society for microbiology COVID-19 international summit, 23 March 2020: value of diagnostic testing for SARS–CoV-2/COVID-19. MBio. 2020;11 doi: 10.1128/mBio.00722-20. mBio.00722-20, e00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellewell J., Abbott S., Gimma A., Bosse N.I., Jarvis C.I., Russell T.W., Munday J.D., Kucharski A.J., Edmunds W.J., Funk S., Eggo R.M., Sun F., Flasche S., Quilty B.J., Davies N., Liu Y., Clifford S., Klepac P., Jit M., Diamond C., Gibbs H., van Zandvoort K. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health. 2020;8:e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization; 2020. WHO Lists Two COVID-19 Tests for Emergency Use.https://www.who.int/news-room/detail/07-04-2020-who-lists-two-covid-19-tests-for-emergency-use Retrived 10 April. [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Hou X., Wu X., Liang T., Zhang X., Wang D., Teng F., Dai J., Duan H., Guo S., Li Y., Yu X. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. Biochemistry. 2020 doi: 10.1101/2020.03.26.994756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.F. Zhang, O.O. Abudayyeh, J.S. Gootenberg, A protocol for detection of COVID-19 using CRISPR diagnostics, https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf.
- 9.Will antibody tests for the coronavirus really change everything? Nature. 2020;20(April) doi: 10.1038/d41586-020-01115-z. https://media.nature.com/original/magazine-assets/d41586-020-01115-z/d41586-020-01115-z.pdfRetrieved [DOI] [PubMed] [Google Scholar]
- 10.Jarocka U., Sawicka R., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecki J., Radecka H. An immunosensor based on antibody binding fragments attached to gold nanoparticles for the detection of peptides derived from avian influenza hemagglutinin H5. Sensors. 2014;14:15714–15728. doi: 10.3390/s140915714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-test-patient-home-sample-collection.
- 12.Ren L.L., Wang Y.M., Wu Z.Q., Xiang Z.C., Guo L., Xu T., Jiang Y.Z., Xiong Y., Li Y.J., Li X.W., Li H., Fan G.H., Gu X.Y., Xiao Y., Gao H., Xu J.Y., Yang F., Wang X.M., Wu C., Chen L., Liu Y.W., Liu B., Yang J., Wang X.R., Dong J., Li L., Huang C.L., Zhao J.P., Hu Y., Cheng Z.S., Liu L.L., Qian Z.H., Qin C., Jin Q., Cao B., Wang J.W. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;1 doi: 10.1097/CM9. 0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J.M., Chung Y.S., Jo H.J., Lee N.J., Kim M.S., Woo S.H., Park S., Kim J.W., Kim H.M., Han M.G. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis, journal of microbiology. Immunol. Infection. 2020 doi: 10.1016/j.jmii.2020.03.022. S1684118220300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y., Ahn J.W., Hwang S., Sung K.S., Lim J., Kwack K. Structural similarity analysis of spike proteins of SARS-CoV-2 and other SARS-related coronaviruses. Life Sci. 2020 doi: 10.20944/preprints202003.0409.v2. [DOI] [Google Scholar]
- 18.Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization (WHO) Geneva: WHO; 2020. Novel Coronavirus. (2019-nCoV) Technical Guidance: Laboratory Guidance.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance Retrieved 09 April. [Google Scholar]
- 20.Go Y.Y., Kim Y.S., Cheon S., Nam S., Ku K.B., Kim M., Cho N.H., Park H., Alison Lee P.Y., Lin Y.C., Tsai Y.L., Thomas Wang H.T., Balasuriya U.B.R. Evaluation and Clinical Validation of Two Field–Deployable Reverse Transcription-Insulated Isothermal PCR Assays for the Detection of the Middle East Respiratory Syndrome–Coronavirus. J. Mol. Diagn. 2017;19:817–827. doi: 10.1016/j.jmoldx.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashemzadeh M.S., Rasouli R., Zahraei B., Izadi M., Tat M., Saadat S.H., Najarasl M., KhansariNejad B., Dorostkar R. Development of dual TaqMan based one-step rRT-PCR assay panel for rapid and accurate diagnostic test of MERS-CoV: a novel human coronavirus, ahead of hajj pilgrimage. Iran. Red Crescent Med. J. 2016;18 doi: 10.5812/ircmj.23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/15607917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konrad R., Eberle U., Dangel A., Treis B., Berger A., Bengs K., Fingerle V., Liebl B., Ackermann N., Sing A. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai J.W., Zhang Y., Zhang H.C., Xu T., Zhang W.H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microbes Infect. 2020;9:597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MEMS at the Forefront of SARS-CoV-2 Testing, https://www.semiconductor-digest.com/2020/04/08/mems-at-the-forefront-of-sars-cov-2-testing/.
- 28.Nicholson L.B. The immune system. Essays Biochem. 2016;60:275–301. doi: 10.1042/EBC20160017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S., Byrne H., O’Kennedy R.J. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016;60:9–18. doi: 10.1042/EBC20150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.K. Green, S. Graziadio, P. Turner, T. Fanshawe, J. Allen, Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19, (n.d.) 12.
- 31.Ahmed S.R., Kim J., Tran V.T., Suzuki T., Neethirajan S., Lee J., Park E.Y. In situ self-assembly of gold nanoparticles on hydrophilic and hydrophobic substrates for influenza virus-sensing platform. Sci. Rep. 2017;7:44495. doi: 10.1038/srep44495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nidzworski D., Pranszke P., Grudniewska M., Król E., Gromadzka B. Universal biosensor for detection of influenza virus. Biosens. Bioelectron. 2014;59:239–242. doi: 10.1016/j.bios.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 33.Lee D., Chander Y., Goyal S.M., Cui T. Carbon nanotube electric immunoassay for the detection of swine influenza virus H1N1. Biosens. Bioelectron. 2011;26:3482–3487. doi: 10.1016/j.bios.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafiee H., Lidstone E.A., Jahangir M., Inci F., Hanhauser E., Henrich T.J., Kuritzkes D.R., Cunningham B.T., Demirci U. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci. Rep. 2015;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzun L., Say R., Ünal S., Denizli A. Production of surface plasmon resonance based assay kit for hepatitis diagnosis. Biosens. Bioelectron. 2009;24:2878–2884. doi: 10.1016/j.bios.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Yanik A.A., Huang M., Kamohara O., Artar A., Geisbert T.W., Connor J.H., Altug H. An optofluidic nanoplasmonic biosensor for direct detection of live viruses from biological media. Nano Lett. 2010;10:4962–4969. doi: 10.1021/nl103025u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.-Z., Nair M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018;8:9700. doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashiba H., Sugiyama Y., Wang X., Shirato H., Higo-Moriguchi K., Taniguchi K., Ohki Y., Fujimaki M. Detection of norovirus virus-like particles using a surface plasmon resonance-assisted fluoroimmunosensor optimized for quantum dot fluorescent labels. Biosens. Bioelectron. 2017;93:260–266. doi: 10.1016/j.bios.2016.08.099. [DOI] [PubMed] [Google Scholar]
- 39.Yang J.M., Kim K.R., Kim C.S. Biosensor for rapid and sensitive detection of influenza virus. BiotechnolBioproc E. 2018;23:371–382. doi: 10.1007/s12257-018-0220-x. [DOI] [Google Scholar]
- 40.Singh R., Hong S., Jang J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017;7:42771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng L., Zhang J., Lin Y., Wang Q., Zhang X., Ding Y., Cui H., Fan H. An electrochemical molecular recognition-based aptasensor for multiple protein detection. Anal. Biochem. 2015;491:31–36. doi: 10.1016/j.ab.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Sattarahmady N., Rahi A., Heli H. A signal-on built in-marker electrochemical aptasensor for human prostate-specific antigen based on a hairbrush-like gold nanostructure. Sci. Rep. 2017;7:11238. doi: 10.1038/s41598-017-11680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negahdary M., Behjati-Ardakani M., Sattarahmady N., Yadegari H., Heli H. Electrochemical aptasensing of human cardiac troponin I based on an array of gold nanodumbbells-Applied to early detection of myocardial infarction. Sens. Actuators B Chem. 2017;252:62–71. doi: 10.1016/j.snb.2017.05.149. [DOI] [Google Scholar]
- 44.Negahdary M., Behjati-Ardakani M., Sattarahmady N., Heli H. An aptamer-based biosensor for troponin I detection in diagnosis of myocardial infarction. J. Biomed. Phys. Eng. 2018;8 doi: 10.31661/jbpe.v8i2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Zhang Q., Qian C., Hao N., Xu L., Yao C. Electrochemical aptasensor for mucin 1 based on dual signal amplification of poly(o-phenylenediamine) carrier and functionalized carbon nanotubes tracing tag. Biosens. Bioelectron. 2015;64:485–492. doi: 10.1016/j.bios.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 46.Negahdary M., Behjati-Ardakani M., Heli H. An electrochemical troponin T aptasensor based on the use of a macroporous gold nanostructure. Microchim Acta. 2019;186:377. doi: 10.1007/s00604-019-3472-z. [DOI] [PubMed] [Google Scholar]
- 47.Grabowska I., Singleton D.G., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecka H., Stulz E., Radecki J. A highly sensitive electrochemical genosensor based on Co-porphyrin-labelled DNA. Chem. Commun. 2014;50:4196–4199. doi: 10.1039/C4CC00172A. [DOI] [PubMed] [Google Scholar]
- 48.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Shariati M., Ghorbani M., Sasanpour P., Karimizefreh A. An ultrasensitive label free human papilloma virus DNA biosensor using gold nanotubes based on nanoporous polycarbonate in electrical alignment. Anal. Chim. Acta. 2019;1048:31–41. doi: 10.1016/j.aca.2018.09.062. [DOI] [PubMed] [Google Scholar]
- 50.Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- 51.Singhal C., Dubey A., Mathur A., Pundir C.S., Narang J. Paper based DNA biosensor for detection of chikungunya virus using gold shells coated magnetic nanocubes. Process. Biochem. 2018;74:35–42. doi: 10.1016/j.procbio.2018.08.020. [DOI] [Google Scholar]
- 52.Ilkhani H., Farhad S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018;557:151–155. doi: 10.1016/j.ab.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 54.J.P. Broughton, W. Deng, C.L. Fasching, J. Singh, J.S. Chen, A protocol for rapid detection of the 2019 novel coronavirus SARS-CoV-2 using CRISPR diagnostics: SARS-CoV-2 DETECTR, (n.d.) 9.
- 55.Lucia C., Federico P.-B., Alejandra G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. Mol. Biol. (N.Y.) 2020 doi: 10.1101/2020.02.29.971127. [DOI] [Google Scholar]
- 56.Nguyen T.M., Zhang Y., Pandolfi P.P. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., Endy D., La Russa M., Lewis D.B., Qi L.S. Development of CRISPR as a prophylactic strategy to combat novel coronavirus and influenza. Bioengineering. 2020 doi: 10.1101/2020.03.13.991307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye L. Molecularly imprinted polymers with multi-functionality. Anal. Bioanal. Chem. 2016;408:1727–1733. doi: 10.1007/s00216-015-8929-2. [DOI] [PubMed] [Google Scholar]
- 59.Wangchareansak T., Thitithanyanont A., Chuakheaw D., Gleeson M.P., Lieberzeit P.A., Sangma C. A novel approach to identify molecular binding to the influenza virus H5N1: screening using molecularly imprinted polymers (MIPs) Med. Chem. Res. 2014;5:617–621. doi: 10.1039/C3MD00272A. [DOI] [Google Scholar]
- 60.Tai D.F., Lin C.Y., Wu T.Z., Chen L.K. Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 2005;77:5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- 61.He K., Chen C., Liang C., Liu C., Yang B., Chen X., Cai C. Highly selective recognition and fluorescent detection of JEV via virus-imprinted magnetic silicon microspheres. Sens. Actuators B Chem. 2016;233:607–614. doi: 10.1016/j.snb.2016.04.127. [DOI] [Google Scholar]
- 62.Lu C.H., Zhang Y., Tang S.F., Fang Z.B., Yang H.H., Chen X., Chen G.N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012;31:439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 63.Yang B., Gong H., Chen C., Chen X., Cai C. A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus. Biosens. Bioelectron. 2017;87:679–685. doi: 10.1016/j.bios.2016.08.087. [DOI] [PubMed] [Google Scholar]
- 64.Uzun L., Say R., Ünal S., Denizli A. Hepatitis B surface antibody purification with hepatitis B surface antibody imprinted poly(hydroxyethyl methacrylate-N-methacryloyl-l-tyrosine methyl ester) particles. J. Chromatogr. B. 2009;877:181–188. doi: 10.1016/j.jchromb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Altintas Z., Pocock J., Thompson K.A., Tothill I.E. Comparative investigations for adenovirus recognition and quantification: Plastic or natural antibodies? Biosens. Bioelectron. 2015;74:996–1004. doi: 10.1016/j.bios.2015.07.076. [DOI] [PubMed] [Google Scholar]
- 66.Jenik M., Schirhagl R., Schirk C., Hayden O., Lieberzeit P., Blaas D., Paul G., Dickert F.L. Sensing picornaviruses using molecular imprinting techniques on a Quartz Crystal Microbalance. Anal. Chem. 2009;81:5320–5326. doi: 10.1021/ac8019569. [DOI] [PubMed] [Google Scholar]
- 67.Cai D., Ren L., Zhao H., Xu C., Zhang L., Yu Y., Wang H., Lan Y., Roberts M.F., Chuang J.H., Naughton M.J., Ren Z., Chiles T.C. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nature Nanotech. 2010;5:597–601. doi: 10.1038/nnano.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma Y., Shen X.L., Zeng Q., Wang H.S., Wang L.S. A multi-walled carbon nanotubes based molecularly imprinted polymers electrochemical sensor for the sensitive determination of HIV-p24. Talanta. 2017;164:121–127. doi: 10.1016/j.talanta.2016.11.043. [DOI] [PubMed] [Google Scholar]
- 69.Tancharoen C., Sukjee W., Thepparit C., Jaimipuk T., Auewarakul P., Thitithanyanont A., Sangma C. Electrochemical biosensor based on surface imprinting for zika virus detection in serum. ACS Sens. 2019;4:69–75. doi: 10.1021/acssensors.8b00885. [DOI] [PubMed] [Google Scholar]
- 70.Shi R. Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chin. Sci. Bull. 2003;48:1165. doi: 10.1360/03wc0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Long W.H., Xiao H.S., Gu X.M., Zhang Q.H., Yang H.J., Zhao G.P., Liu J.H. A universal microarray for detection of SARS coronavirus. J. Virol. Methods. 2004;121:57–63. doi: 10.1016/j.jviromet.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu D.D., Chen S.H., Zhang S.M., Zhang M.L., Zhang W., Bo X.C., Wang S.Q. Screening of specific antigens for SARS clinical diagnosis using a protein microarray. Analyst. 2005;130:474. doi: 10.1039/b415888a. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Z., Zhou Y., Zhang Y., Guo Y., Tao S., Li Z., Zhang Q., Cheng J. Sensitive detection of SARS coronavirus RNA by a novel asymmetric multiplex nested RT-PCR amplification coupled with oligonucleotide microarray hybridization. In: Joos T.O., Fortina P., editors. Microarrays in Clinical Diagnostics. Humana Press; Totowa, NJ: 2005. pp. 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo X., Geng P., Wang Q., Cao B., Liu B. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J. Microbiol. Biotechnol. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 75.Notomi T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63e–663e. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 77.Ben-Assa N., Naddaf R., Gefen T., Capucha T., Hajjo H., Mandelbaum N., Elbaum L., Rogov P., King D.A., Kaplan S., Rotem A., Chowers M., Szwarcwort-Cohen M., Paul M., Geva-Zatorsky N. Public and Global Health; 2020. SARS-CoV-2 On-the-Spot Virus Detection Directly From Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butler D.J., Mozsary C., Meydan C., Danko D.C., Foox J., Rosiene J., Shaiber A., Afshinnekoo E., MacKay M., Sedlazeck F.J., Ivanov N.A., Sierra M.A., Pohle D., Zietz M., Gisladottir U., Ramlall V., Westover C.D., Ryon K., Young B., Bhattacharya C., Ruggiero P., Langhorst B.W., Tanner N.A., Gawrys J., Meleshko D., Xu D., Steel P.A.D., Shemesh A.J., Xiang J., Thierry-Mieg J., Thierry-Mieg D., Schwartz R.E., Iftner A., Bezdan D., Sipley J., Cong L., Craney A., Velu P., Melnick A., Hajirasouliha I., Horner S.M., Iftner T., Salvatore M., Loda M., Westblade L.F., Levy S., Cushing M., Wu S., Tatonetti N.P., Imielinski M., Rennert H., Mason C. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Mol. Biol. (N.Y.) 2020 doi: 10.1101/2020.04.20.048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martel N., Gomes S.A., Chemin I., Trépo C., Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J. Virol. Methods. 2013;193:653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 80.Wat D., Gelder C., Hibbitts S., Cafferty F., Bowler I., Pierrepoint M., Evans R., Doull I. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros. 2008;7:320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]