Abstract
Background
Cambodia is the epicentre of the emergence of Plasmodium falciparum drug resistance. Much less is known regarding the drug susceptibility of the co-endemic Plasmodium vivax. Only in vitro drug assays can determine the parasite’s intrinsic susceptibility, but these are challenging to implement for P. vivax and rarely performed.
Objectives
To evaluate the evolution of Cambodian P. vivax susceptibility to antimalarial drugs and determine their association with putative markers of drug resistance.
Methods
In vitro response to three drugs used in the past decade in Cambodia was measured for 52 clinical isolates from Eastern Cambodia collected between 2015 and 2018 and the sequence and copy number variation of their pvmdr1 and pvcrt genes were analysed. pvmdr1 polymorphism was also determined for an additional 250 isolates collected in Eastern Cambodia between 2014 and 2019.
Results
Among the 52 cryopreserved isolates tested, all were susceptible to the three drugs, with overall median IC50s of 16.1 nM (IQR 11.4–22.3) chloroquine, 3.4 nM (IQR 2.1–5.0) mefloquine and 4.6 nM (IQR 2.7–7.0) piperaquine. A significant increase in chloroquine and piperaquine susceptibility was observed between 2015 and 2018, unrelated to polymorphisms in pvcrt and pvmdr1. Susceptibility to mefloquine was significantly lower in parasites with a single mutation in pvmdr1 compared with isolates with multiple mutations. The proportion of parasites with this single mutation genotype increased between 2014 and 2019.
Conclusions
P. vivax with decreased susceptibility to mefloquine is associated with the introduction of mefloquine-based treatment during 2017–18.
Introduction
Malaria remains one of the most common infectious diseases in the world and the rapid selection of Plasmodium falciparum parasites resistant to antimalarial drugs is threatening control efforts. Southeast Asia has been the hotspot of the emergence of P. falciparum resistance against virtually all antimalarials used. Regarding Plasmodium vivax, inherent constraints associated with parasite biology limit the capacity to investigate this phenomenon. Indeed, P. vivax cannot be continuously cultivated in vitro and only tedious in vitro short-term cultures are possible to determine the intrinsic susceptibility of the parasites. Besides, P. vivax parasites are typically asynchronous and multiple developmental stages can be present within an infected patient, which can affect the in vitro drug susceptibility of the parasites.1 Therefore, drug efficacy for P. vivax is mainly evaluated through clinical studies that are confounded by a number of biases of host origin (immunity, drug adsorption, metabolism and so on) and parasite factors (reinfection and relapses).2 Molecular markers of resistance are useful tools to rapidly assess changes in drug susceptibility within a given parasite population. While widely used for the surveillance of P. falciparum resistance to many antimalarials, there are still no validated molecular markers of resistance for P. vivax clinical isolates. Candidate markers have been proposed in some studies and, notably, polymorphism, amplification or expression of pvcrt and pvmdr1 are suspected to be involved in chloroquine and/or mefloquine resistance; however, formal evidence of their involvement in natural populations of P. vivax remains to be found.3–6
The treatment for P. vivax malaria in Cambodia until 2012 was chloroquine. In order to provide a unified drug policy for all species, the treatment changed to a combination of dihydroartemisinin/piperaquine in 2012. Following the emergence of P. falciparum resistance to piperaquine, the first-line treatment for all malaria species changed again in 2016 to a combination of artesunate/mefloquine, which was gradually implemented in the country mid-2017. While the susceptibility of P. falciparum to these different antimalarials is continuously monitored, very few data exist concerning the response of P. vivax.
This study aimed to determine the in vitro susceptibility of parasites collected in Eastern Cambodia over 4 years to three antimalarials used in the country in the past decade and discover any association of in vitro phenotypes to polymorphisms in the pvcrt and pvmdr1 genes.
Materials and methods
P. vivax clinical isolate collection
Samples were collected from symptomatic patients seeking treatment in Eastern Cambodia between 2014 and 2019. Venous blood was collected in EDTA tubes for molecular analysis and immediately stored at −20°C. Additionally, for in vitro assays a heparin tube was collected from which leucocytes were depleted using non-woven fabric (NWF) filters7 (Zhi Xing Bio S&T Co. Ltd, China) and RBCs were immediately cryopreserved in Glycerolyte 57 (Baxter Healthcare Corporation, USA) following published procedures.8
DNA extraction and PCR detection of P. vivax
DNA was extracted from 200 μL of whole blood using the QIAamp DNA Blood Mini Kit (QIAGEN, Courtaboeuf, France), according to the manufacturer’s instructions. Molecular detection and identification of Plasmodium parasites were performed by real-time PCR as previously described.9
Drug plate preparation and in vitro susceptibility assay
P. vivax susceptibility to drugs was measured using a protocol modified from Suwanarusk et al.3 in which 96-well plates of drugs were prepared in advance and stored at −20°C. Chloroquine, mefloquine and piperaquine [obtained from the WorldWide Antimalarial Resistance Network (WWARN)] were tested, each at eight different concentrations. Cryopreserved isolates associated with PCR-confirmed P. vivax monoinfection, containing more than 75% of ring stages determined by microscopy, were thawed and cultured into schizont stage (42–48 h) in Iscove’s Modified Dulbecco’s Medium (IMDM)-based medium following established protocols.8,10 Incubation was stopped when >40% of mature schizonts in the drug-free control wells was reached and thick blood films were stained with 5% Giemsa (Merck, Germany). The number of schizonts (>4 nuclei visible) per 300 asexual parasites (free merozoites and gametocytes excluded) was determined and normalized to the no-drug controls. IC50 values were determined using ICEstimator online software (http://www.antimalarial-icestimator.net).
Gene copy number determination of pvmdr1 and pvcrt
The number of copies of pvmdr1 and pvcrt was measured by quantitative PCR relative to the single-copy β-tubulin gene using a CFX96 real-time PCR instrument (Bio-Rad) (primers listed in Table 1). PCRs were conducted in 20 μL volumes in a 96-well plate containing 1× HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Tartu, Estonia), 0.5 μM each forward and reverse primers and 2 μL of template DNA. Amplifications were performed under the following conditions: 95°C for 15 min, followed by 45 cycles of 95°C for 15 s, 60°C for 20 s and 72°C for 20 s. The number of pvmdr1 copies for each sample was measured in triplicate relative to a standard curve made of synthetic β-tubulin and pvmdr1 genes, each cloned in a pEX-A2 vector (Eurofins Genomics, Greece) and mixed at different ratios from 1:1 up to 1:5 (one copy of β-tubulin and up to five copies of pvmdr1) (Table 1). The number of pvcrt copies for each sample was measured in triplicate relative to an internal calibrator control. The ΔΔCT method (where CT is the cycle threshold) was used to determine the number of copies in each sample.
Table 1.
List of primers and synthetic genes used for pvcrt and pvmdr1 gene copy number determination and sequencing
| Name | Sequence (5′→3′) | Reference |
|---|---|---|
| Gene copy number determination | ||
| pvmdr1 synthetic gene | GCAACTCCATAAAGAACAACATCAAGTATAGTTTGTACAGCCTGAAAGATTTAGAAGCCTTATCGGAGGAGTCGAACGAAGATGGTTTTTCTTCTCAAA | this work |
| β-TubulinPv synthetic gene | CAGGAGTTACATGTTCGTTAAGATTTCCTGGTCAGTTAAATTCTGATTTGAGAAAATTAGCTGTCAATTTAATTCCCTTCCCAAGACTCCACTTTTTTATGATTGGTTTTGCACCACTAACAAGCAGAGG | 20 |
| PvMDR1_F | GCAACTCCATAAAGAACAACATC | this work |
| PvMDR1_R | TTTGAGAAGAAAAACCATCTTCG | this work |
| PvCRT_F | GGGAGTCCCCAAATAACCCC | this work |
| PvCRT_R | GTTGTCTCGCCACTCTCCTG | this work |
| CN_β-tubulin_F | CATGTTCGTTAAGATTTCCTGGT | 20 |
| CN_β-tubulin_R | GTTAGTGGTGCAAAACCAATCA | |
| Sequence polymorphism | ||
| Pvmdr976 F | GGATAGTCATGCCCCAGGATTG | 3 |
| Pvmdr976 R | CATCAACTTCCCGGCGTAGC | |
| Pvcrt-o F1 (sense) | AAGAGCCGTCTAGCCATCC | 11 |
| Pvcrt-o R3 | AGTTTCCCTCTACACCCG | |
Sequence polymorphism of pvmdr1 and pvcrt genes
PCR and Sanger sequencing (Macrogen, Seoul, South Korea) were used to determine pvmdr1 and pvcrt sequences using the following conditions. For pvmdr1, the PCR was conducted in 30 μL reactions consisting of 3 μL of DNA, 0.15 μM primers and 1× HOT FirePol Blend MasterMix (Solis BioDyne) under the following conditions: 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 64°C for 90 s, 72°C for 45 s and a final extension at 72°C for 10 min. For pvcrt, the PCR was conducted in 30 μL reactions consisting of 3 μL of DNA, 0.25 μM primers and 1× HOT FirePol Blend MasterMix under the following conditions: 94°C for 10 min, followed by 35 cycles of 94°C for 50 s, 65°C for 60 s, 72°C for 90 s and a final extension at 72°C for 10 min. Primers used are described in Table 1. Expected amplicon sizes are 604 bp and 1186 bp, respectively, for pvmdr1 and pvcrt.3,11 Nucleotides and corresponding amino acids were analysed using MEGA 7 software (Beckman). The sequences generated were compared with AY571984.1 for pvmdr1 and with AF314649.1 for pvcrt.
Statistical analysis
Comparison of mean of non-Gaussian data was done by the Mann–Whitney test. Multiple comparisons of means of non-Gaussian data were performed by Kruskal–Wallis and Dunn’s post hoc tests. Proportions were compared by Fisher’s exact test. All analyses were performed using GraphPad Prism (v7.00). A P value of <0.05 was considered significant.
Ethics
The research was conducted in accordance with the Declaration of Helsinki and national and institutional standards. Ethics clearance for the samples used in this study was obtained from the Ministry of Health National Ethics Committee in Cambodia (364NECHR, 478NECHR, 270NECHR and 317NECHR). All patients or their parents/guardians provided informed written consent.
Results
Evolution of Cambodian P. vivax in vitro susceptibility to three antimalarials
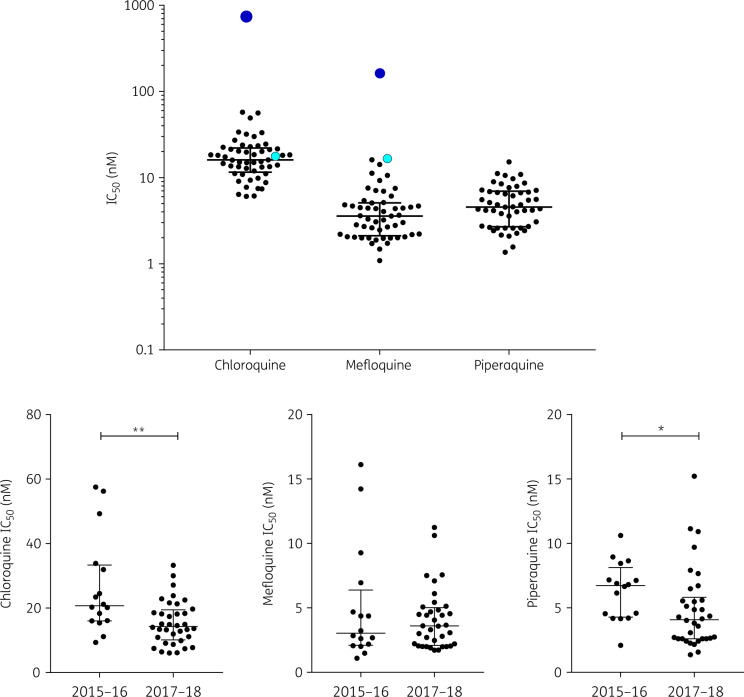
The susceptibility of 52 P. vivax cryopreserved clinical isolates from Eastern Cambodia collected between 2015 and 2018 was assessed by a schizont maturation assay validated using susceptible and resistant strains of P. falciparum (Table S1, available as Supplementary data at JAC Online). The IC50 values for all 52 isolates were low and homogeneous for the three drugs, indicating the absence of high-grade resistant parasites. Median IC50 values of 16.1 nM (IQR 11.4–22.3), 3.4 nM (IQR 2.1–5.0) and 4.6 nM (IQR 2.7–7.0) were observed for chloroquine, mefloquine and piperaquine, respectively (Figure 1).
Figure 1.
Overall susceptibility of 52 cryopreserved Cambodian P. vivax isolates to chloroquine, mefloquine and piperaquine measured by in vitro schizont maturation assays (top panel). Black circles represent individual IC50 values of P. vivax isolates (the horizontal line shows the median and the whiskers show the IQR). For comparison, plain light blue circles represent IC50 values of the susceptible P. falciparum 3D7 reference strain and plain dark blue circles the chloroquine or mefloquine IC50 values of resistant P. falciparum clinical isolates. Susceptibility to chloroquine, mefloquine and piperaquine of isolates collected when the first-line treatment was dihydroartemisinin/piperaquine (2015–16) or artesunate/mefloquine (2017–18) (bottom panels). The susceptibility of isolates to chloroquine and piperaquine significantly decreased over time (Mann–Whitney test, **P = 0.0023 and *P = 0.0213, respectively). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The susceptibility data for the three drugs were analysed according to the time of collection of the isolates. While for mefloquine there was no significant difference in IC50 depending on the collection period, the response of parasites to chloroquine and piperaquine was significantly different. The IC50 of chloroquine decreased between 2015–16 [median 20.7 nM (IQR 16.0–33.44)] and 2017–18 [median 14.3 nM (IQR 10.1–19.4), P = 0.0023] in a fashion similar to the IC50 of piperaquine [median 6.7 nM (IQR 4.3–8.1) versus median 4.1 nM (IQR 2.6–5.3), P = 0.0213].
Polymorphisms of pvcrt and pvmdr1 and association with drug susceptibility of isolates
Sequences of pvmdr1 were interpretable for the 52 cryopreserved isolates. All isolates had the T958M mutation. Two other mutations were detected: Y976F (in 28/52, 54% of isolates) and F1076L (in 45/52, 86.5% of isolates). Three different pvmdr1 genotypes were observed, the most frequent being the triple T958M-Y976F-F1076L mutant (28/52, 54%), followed by the double T958M-F1076L mutant (17/52, 33%) and the single T958M mutant (7/52, 13%) (Table S2). All isolates with the Y976F mutation had also the F1076L mutation. No isolate had the WT pvmdr1 Sal-1 sequence. Sequences of pvcrt were interpretable for 45 isolates, of which 69% (31/45) had a lysine insertion at codon 10 (K10insert). In addition, two newly described mutations were observed, albeit at low frequency: one isolate (1/45, 2%) had an A107V mutation and four (4/45, 9%) an S163N mutation. Overall, four different pvcrt genotypes were detected, the most frequent being the K10insert alone (noted as genotype 1, 27/45, 60%), followed by the Sal-1 WT (13/45, 29%), the K10insert/S163N (genotype 2, 3/45, 7%) and one isolate (1/45, 2%) for each of the S163N (genotype 3) and K10insert/A107V (genotype 4) genotypes. All isolates had a single copy of both pvmdr1 and pvcrt genes.
The in vitro susceptibility of isolates to all drugs was analysed according to their polymorphisms in pvmdr1 and pvcrt. IC50 values were compared between the different genotypes. Among all the pvmdr1 and pvcrt polymorphisms, the only significant association was between pvmdr1 and the response of isolates to mefloquine (Figure S1).
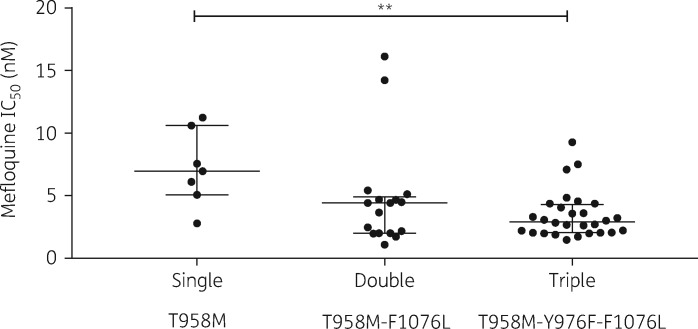
Multiple comparison analysis of the susceptibility to mefloquine between the three pvmdr1 genotypes showed a significantly lower IC50 for the triple T958M-Y976F-F1076L mutants [median 2.9 nM (IQR 2.1–4.3)] compared with the single T958M mutants [median 7.0 nM (IQR 5.0–10.6), P = 0.0072] (Figure 2). The double T958M-F1076L mutant parasites had a median IC50 of 4.4 nM (IQR 2.0–4.9), intermediate between single and triple mutants, although the differences did not reach statistical significance.
Figure 2.
Associations between pvmdr1 polymorphism and susceptibility to mefloquine. Median mefloquine IC50s of the different genotypes are shown. Genotypes are described under each panel. Multiple comparison of mean IC50 of the three genotypes was assessed by the Kruskal–Wallis test and Dunn’s correction. **P < 0.01.
Evolution of pvmdr1 polymorphism in Eastern Cambodia following successive changes in national treatment guidelines
The polymorphism of pvmdr1 was then analysed according to the therapeutic strategy implemented in Eastern Cambodia. To increase the sample size and identify significant changes in frequency, we included a larger number of isolates (total N = 301 isolates) collected in the same area between May 2014 and September 2019 for which DNA only was available. No other genotypes were observed among those additional isolates. During the period of dihydroartemisinin/piperaquine deployment (2014–16), the frequency of the single mutant (T958M) was 8% (N = 9/113). A significant increase (P = 0.005) to 20% (N = 38/188) was observed after adoption of artesunate/mefloquine (Figure 3). No significant difference in frequency over time was observed for the double and triple mutants. The copy number of the pvmdr1 gene could be determined for 241 clinical isolates and all had a single copy of the gene.
Figure 3.
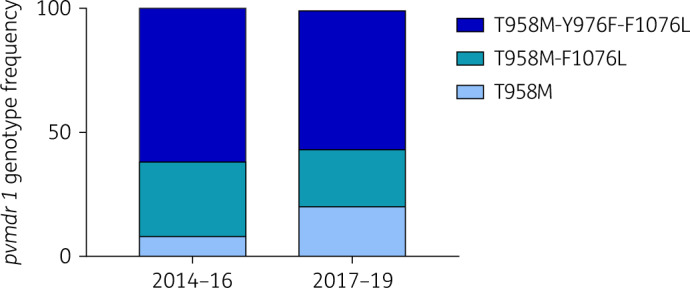
Evolution over time of the frequency of the different pvmdr1 genotypes among clinical isolates collected in Eastern Cambodia from 2014 to 2019. Dihydroartemisinin/piperaquine was the first-line treatment from 2012 to 2016 and artesunate/mefloquine was introduced in 2017. The frequency of the less-susceptible single mutant (T958M, light blue) significantly increased from 8% to 20% after the introduction of artesunate/mefloquine treatment (Fisher’s exact test, P = 0.0050) while no significant changes were observed for the double (T958M-F1076L, medium blue) and triple (T958M-Y976F-F1076L, dark blue) mutants. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
The aim of this study was to assess the evolution of P. vivax in response to antimalarials used in Cambodia following the successive changes in first-line treatment. We therefore determined the in vitro susceptibility to drugs of cryopreserved parasites collected in Eastern Cambodia between 2015 and 2018 and tested for associations between susceptibility and polymorphisms in the pvmdr1 and pvcrt genes. Overall, no isolate showed susceptibility above the assumed resistance threshold of 220 nM chloroquine.3 Currently there is no established cut-off for piperaquine or mefloquine resistance in P. vivax. Future studies that associate in vitro drug response of parasites with clinical outcomes (adequate clinical and parasitological response versus treatment failure) are needed to determine clinically relevant thresholds of resistance. However, the IC50 values observed in this study are very low (median <5 nM for both mefloquine and piperaquine). It is thus highly unlikely that those susceptibilities are of any therapeutic concern. Additionally, the values obtained here are among the lowest reported in the literature for Southeast Asian P. vivax, whether for chloroquine, mefloquine or piperaquine.3,4,12 It should be noted that in this work we used cryopreserved isolates instead of fresh parasites, which could, in theory, have introduced some biases in the IC50 measurements. Future studies examining differences in IC50 between paired isolates determined before and after cryopreservation should be conducted to assess any effect of freeze–thaw procedures on the drug response of parasites.
Even though these parasites cannot be categorized as resistant, this study demonstrates an interesting evolution in their drug susceptibility profiles. First, P. vivax susceptibility to chloroquine increased significantly between 2015 and 2018, with the average IC50 decreasing by almost half within 4 years. This observation is consistent with the withdrawal of chloroquine from guidelines since 2012. Indeed, in a study conducted between 2008 and 2010, up to 18% of treatment failure following chloroquine treatment was reported in Eastern Cambodia, where the cryopreserved isolates used in our work originated.13 However, another therapeutic efficacy study conducted in the same area, but 2 years after the replacement of chloroquine with dihydroartemisinin/piperaquine, did not find any sign of treatment failure.14 Given those previous clinical results and the decrease in IC50 reported here, we hypothesize that there was indeed some chloroquine resistance before the change in the treatment guidelines, following which parasites gradually regained susceptibility. Importantly, none of the polymorphisms in pvcrt or pvmdr1 detected in this work could be linked to the changes in susceptibility to chloroquine observed over time. This suggests that determinants other than pvcrt and pvmdr1 are involved in the modulation of chloroquine susceptibility in P. vivax.
Because we did not identify any isolate highly resistant to chloroquine, pvcrt and pvmdr1 polymorphisms detected in our work can definitely be excluded as markers of resistance. Other mutations in these genes (or their amplification) not observed in our study could, however, still be related to chloroquine resistance. Besides, our work cannot exclude a link between increased pvcrt expression and chloroquine resistance.6 Similar genotype–phenotype association studies in areas where high-grade chloroquine-resistant P. vivax are reported are needed to provide conclusive data on the basis of chloroquine resistance for this species.
Interestingly, the same trend for piperaquine susceptibility evolution was observed, as IC50s were significantly lower after artesunate/mefloquine implementation. This increase in piperaquine susceptibility is probably the consequence of the withdrawal of dihydroartemisinin/piperaquine as first-line treatment in the country but could also result from mefloquine pressure on parasite populations. Indeed, in P. falciparum, piperaquine and mefloquine exert opposite pressure on parasites and increased susceptibility to piperaquine is linked to decreased susceptibility to mefloquine.15 Of note, only normal dose–response curves for piperaquine (and other drugs) and no paradoxical parasite growth at high drug concentrations were observed (as seen with P. falciparum piperaquine-resistant strains), further confirming the absence of resistance of P. vivax in Cambodia.16
No significant evolution of mefloquine susceptibility over time was observed but polymorphism in pvmdr1 was associated with differences in susceptibility to mefloquine. Indeed, the single T958M mutant was significantly less susceptible than the triple T958M-Y976F-F1076L mutant, though none of the isolates had an IC50 high enough to indicate high-grade mefloquine resistance. Additionally, the frequency of this less-susceptible T958M genotype doubled between 2015 and 2019 in Eastern Cambodia following the introduction of artesunate/mefloquine. Altogether, these results suggest ongoing selection of decreased susceptibility to mefloquine. Notably, high IC50 values of mefloquine were previously associated with pvmdr1 gene amplification4 and parasites with multiple copies of pvmdr1 did not carry the Y976F mutation, suggesting a fitness cost associated with having both the mutation and the gene amplification.17 In our current work, none of the isolates tested had pvmdr1 amplification and none had high mefloquine resistance. We believe we are currently observing changes in genotypes and phenotypes predating the emergence of clinically relevant resistance to mefloquine with parasites currently losing the mutations within pvmdr1 prior to acquiring multiple copies of the gene.
The results of this work indicate that the susceptibility of P. vivax parasites to antimalarials evolves in response to the drugs used in the country. The difference with P. falciparum parasites from the same area is, however, striking. Indeed, while for P. vivax only modest changes in susceptibility were observed, for P. falciparum in the same time frame and the same locations, emergence of fully resistant parasites was thoroughly documented. This difference most probably results from several different factors and among these, the production of gametocytes by P. vivax (and its transmission to the vectors) before patients become symptomatic (and thus exposed to the drugs) certainly limits the selective pressure exerted on the parasite populations.18,19 In addition, a significant proportion of individuals infected by P. vivax are asymptomatic and therefore undetected and untreated. Despite these factors, changes in susceptibility over time and selection of specific genotypes are observed, indicating that some selective pressure does occur. This selection is probably driven by the pressure exerted by lingering antimalarials after treatment on reactivating hypnozoites during relapsing episodes. Decreased susceptibility to mefloquine would allow parasites to survive lingering subtherapeutic drug concentrations upon relapses.
In conclusion, this work warrants further monitoring of the evolution of P. vivax susceptibility to mefloquine and pvmdr1 polymorphism in Cambodia in order to allow early detection of the emergence of fully resistant mefloquine isolates.
Supplementary Material
Acknowledgements
We thank all patients for participating in this study.
Funding
This study was supported by internal funding. J.P. is partly supported by the International Centers of Excellence for Malaria Research program from the National Institutes of Health, grant 1U19AI129392-0.
Transparency declarations
None to declare.
References
- 1. Thomson-Luque R, Adams JH, Kocken CHM. et al. From marginal to essential: the golden thread between nutrient sensing, medium composition and Plasmodium vivax maturation in in vitro culture. Malar J 2019; 18: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Popovici J, Ménard D.. Challenges in antimalarial drug treatment for vivax malaria control. Trends Mol Med 2015; 21: 776–88. [DOI] [PubMed] [Google Scholar]
- 3. Suwanarusk R, Russell B, Chavchich M. et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One 2007; 2: e1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suwanarusk R, Chavchich M, Russell B. et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis 2008; 198: 1558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silva SR, Almeida ACG, da Silva GAV. et al. Chloroquine resistance is associated to multi-copy pvcrt-o gene in Plasmodium vivax malaria in the Brazilian Amazon. Malar J 2018; 17: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sá JM, Kaslow SR, Moraes Barros RR. et al. Plasmodium vivax chloroquine resistance links to pvcrt transcription in a genetic cross. Nat Commun 2019; 10: 4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J, Tao Z, Li Q. et al. Further evaluation of the NWF filter for the purification of Plasmodium vivax-infected erythrocytes. Malar J 2017; 16: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russell B, Suwanarusk R, Borlon C. et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 2011; 118: e74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canier L, Khim N, Kim S. et al. An innovative tool for moving malaria PCR detection of parasite reservoir into the field. Malar J 2013; 12: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urusova D, Carias L, Huang Y. et al. Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP. Nat Microbiol 2019; 4: 1486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu F, Lim CS, Nam DH. et al. Genetic polymorphism in pvmdr1 and pvcrt-o genes in relation to in vitro drug susceptibility of Plasmodium vivax isolates from malaria-endemic countries. Acta Trop 2011; 117: 69–75. [DOI] [PubMed] [Google Scholar]
- 12. Chaorattanakawee S, Lon C, Chann S. et al. Measuring ex vivo drug susceptibility in Plasmodium vivax isolates from Cambodia. Malar J 2017; 16: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leang R, Barrette A, Bouth DM. et al. Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob Agents Chemother 2013; 57: 818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Popovici J, Pierce-Friedrich L, Kim S. et al. Recrudescence, reinfection, or relapse? A more rigorous framework to assess chloroquine efficacy for Plasmodium vivax malaria. J Infect Dis 2019; 219: 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Witkowski B, Duru V, Khim N. et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis 2017; 17: 174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duru V, Khim N, Leang R. et al. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 2015; 13: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Auburn S, Serre D, Pearson RD. et al. Genomic analysis reveals a common breakpoint in amplifications of the Plasmodium vivax multidrug resistance 1 locus in Thailand. J Infect Dis 2016; 214: 1235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bousema T, Drakeley C.. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev 2011; 24: 377–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kitchen SF, Boyd MF.. On the infectiousness of patients infected with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med Hyg 1937; s1-17: 253–62. [Google Scholar]
- 20. Roesch C, Popovici J, Bin S. et al. Genetic diversity in two Plasmodium vivax protein ligands for reticulocyte invasion. PLoS Negl Trop Dis 2018; 12: e0006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.