Abstract
Objectives
To investigate the prevalence of the optrA, poxtA and cfr linezolid resistance genes in linezolid-resistant enterococci from Irish hospitals and to characterize associated plasmids.
Methods
One hundred and fifty-four linezolid-resistant isolates recovered in 14 hospitals between June 2016 and August 2019 were screened for resistance genes by PCR. All isolates harbouring resistance genes, and 20 without, underwent Illumina MiSeq WGS. Isolate relatedness was assessed using enterococcal whole-genome MLST. MinION sequencing (Oxford Nanopore) and hybrid assembly were used to resolve genetic environments/plasmids surrounding resistance genes.
Results
optrA and/or poxtA were identified in 35/154 (22.7%) isolates, the highest prevalence reported to date. Fifteen isolates with diverse STs harboured optrA only; one Enterococcus faecium isolate harboured optrA (chromosome) and poxtA (plasmid). Seven Enterococcus faecalis and one E. faecium harboured optrA on a 36 331 bp plasmid with 100% identity to the previously described optrA-encoding conjugative plasmid pE349. Variations around optrA were also observed, with optrA located on plasmids in five isolates and within the chromosome in three isolates. Nine E. faecium and 10 E. faecalis harboured poxtA, flanked by IS1216E, within an identical 4001 bp region on plasmids exhibiting 72.9%–100% sequence coverage to a 21 849 bp conjugative plasmid. E. faecalis isolates belonged to ST480, whereas E. faecium isolates belonged to diverse STs. Of the remaining 119 linezolid-resistant isolates without linezolid resistance genes, 20 investigated representatives all harboured the G2576T 23S RNA gene mutation associated with linezolid resistance.
Conclusions
This high prevalence of optrA and poxtA in diverse enterococcal lineages in Irish hospitals indicates significant selective pressure(s) for maintenance.
Introduction
Linezolid is an antibiotic used for infections caused by MDR Gram-positive bacteria, including VRE. Linezolid resistance was first reported in vancomycin-resistant Enterococcus faecium (VREfm) in Greece in 2004.1,2 Ireland had the highest rate of VREfm bloodstream infections in Europe between 2007 and 2017.3 Although no data are available on linezolid usage in Ireland, an almost 10% increase in the overall use of antimicrobials was noted between 2007 and 2017.4 A linezolid usage increase of 40% between 2012 and 2013 was reported in one Irish hospital.5 The emergence of linezolid-resistant enterococci (LRE) during or after linezolid exposure has been described.6–10
Linezolid binds in the V domain of the 23S rRNA component of the 50S ribosomal subunit and inhibits protein synthesis.11 Enterococcal linezolid resistance can be due to G2576T or G2505A mutations in the 23S rRNA binding site or mutations in the genes encoding ribosomal proteins L3 and/or L4.7 However, linezolid resistance can develop following acquisition of the resistance genes optrA, poxtA and variants of the cfr gene, which have been described in detail previously.12 Although the reported number of E. faecium and Enterococcus faecalis isolates harbouring these genes is low, optrA has been reported with increased frequency recently. In 2014, it was reported that 3/9 linezolid non-susceptible isolates (MIC ≥4 mg/L) were optrA-positive E. faecalis (two from Ireland).13 This increased to 8/17 in 2016.14 German researchers reported that 6% of 698 LRE recovered between 2007 and 2017 harboured optrA.15
The poxtA gene was originally identified in an Italian clinical MRSA in 2018 and subsequently in a porcine E. faecium.16,17 More recently, optrA and poxtA were co-located on a conjugative plasmid in a porcine E. faecalis.18 To date, poxtA has only been reported in a Greek clinical E. faecium in 2018.19 The cfr gene, and its variants cfr(B) and cfr(D) (GenBank: MG707078.1) have been reported in clinical E. faecium,5 whereas only cfr has been reported in E. faecalis.5,20,21 The first reported linezolid-resistant VREfm outbreak in Ireland occurred in 2014; it involved 15 patients and was identified as a clonal outbreak using PFGE.22 All isolates harboured the G2576T 23S mutation and were cfr negative. Other linezolid resistance genes were not investigated.22 In 2014, the first two optrA-positive VREfm were recovered in separate Irish hospitals.13 In 2016, the Irish Health Protection Surveillance Centre requested that all LRE identified in Irish hospitals should be sent to the National MRSA Reference Laboratory (NMRSARL) for linezolid resistance gene screening. In 2017, a VREfm clinical isolate harbouring a cfr- and optrA-encoding plasmid was reported from an Irish hospital.5
The purpose of this study was to investigate the molecular mechanisms and spread of linezolid resistance in LRE from Irish hospitals sent to the NMRSARL for linezolid resistance gene screening between June 2016 and August 2019. All isolates harbouring optrA, poxtA or cfr and a selection of isolates without these genes were investigated using WGS.
Materials and methods
Isolates
Between June 2016 and August 2019, 154 LRE from patients in Irish hospitals were sent to the NMRSARL for linezolid resistance gene PCR screening (Table S1, available as Supplementary data at JAC Online). Thirty-five of these harboured at least one of the genes optrA and poxtA and were investigated in detail. Of the remaining 119 isolates without linezolid resistance genes, 20 representatives from a range of isolation dates and hospital locations were also investigated (Table S2). These 55 isolates investigated in detail were recovered in 14 Irish hospitals (H1–H14) (Figure S1). The remaining 99 LRE isolates lacking resistance genes were not investigated further.
Phenotypic and genotypic testing
All isolates were tested for susceptibility to linezolid, vancomycin, chloramphenicol and tetracycline using the VITEK®2 system (bioMérieux, France). MICs were interpreted using the EUCAST interpretative criteria.23 Etests (bioMérieux) were used to assess linezolid MICs between 8 and 256 mg/L. PCRs for enterococcal species and resistance genes (Table S1) were performed using GoTaq DNA polymerase and buffers (Promega Corporation, USA).
Conjugation
Conjugative transfer of plasmids encoding optrA and/or poxtA harboured by all 35 LRE was undertaken by filter mating using the plasmid-free rifampicin- and fusidic acid-resistant recipient strains E. faecium 64/3 and E. faecalis OG1RF.15 Putative transconjugants were screened for enterococcal species, optrA and poxtA by PCR. Transconjugants harbouring optrA or poxtA underwent WGS and genomes were assembled using SPAdes v3.7.1 and compared with the corresponding recipient strain genomes.
WGS
The 55 LRE and transconjugants underwent WGS using genomic DNA extracted using the S. aureus Genotyping Kit 2.0 [Abbott (Alere Technologies), Germany] and the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN, UK). Libraries prepared with the Nextera DNA Flex Library Preparation Kit (Illumina, The Netherlands) underwent paired-end sequencing using the 500-cycle MiSeq Reagent Kit v2 (Illumina). Libraries were scaled to yield ≥50× coverage.
For isolates selected for hybrid assembly, DNA was extracted using the GenFind v3 kit (Beckman Coulter, USA). Long-read sequencing was performed in multiplex using MinION sequencing (Oxford Nanopore Technologies, UK), the 1D Genomic DNA sequencing kit (SQK-LSK109) and 1D Native Barcoding Kit (EXP-NBD103). Libraries were sequenced on an Mk1B (MIN101B) MinION platform with a FLO-MIN106D (SpotON R9.4) flow cell and using MinKNOW v1.7.10 (Oxford Nanopore). Basecalls were performed on MinION FAST5 files using Guppy v3.1.5 (Oxford Nanopore) and demultiplexing was performed using qCat v1.0.1 (https://github.com/nanoporetech/qcat).
Analysis of WGS data
WGS data were analysed using the enterococcal whole-genome (wg) MLST schemes available in BioNumerics v7.7 (Applied Maths, Belgium). The E. faecium scheme consisted of 5489 wgMLST loci [including 1423 core-genome (cg) MLST loci], while the E. faecalis scheme consisted of 5285 wgMLST loci.24 Two BioNumerics algorithms were used to generate a consensus wgMLST profile for each isolate, one of which determined locus presence/absence and allelic identity using an assembly-free k-mer approach. The other, assembly-based, method used a BLAST approach to detect alleles on contigs assembled using SPAdes v3.7.1, all using default parameters. Minimum-spanning trees (MSTs) were created using BioNumerics based on allelic differences. Illumina WGS data for all isolates were examined for 23S rRNA mutations (G2576T and G2505A) using LRE-Finder (https://cge.cbs.dtu.dk/services/LRE-finder/).25
Assembly and analysis of plasmids encoding resistance genes
MinION-generated FASTQ files and MiSeq-generated FASTQ files were used to perform a hybrid assembly using Unicycler.26 The genetic organization of plasmids harbouring optrA or poxtA was determined following hybrid assembly and annotation using RAST v2.0 (http://rast.nmpdr.org/).27 These were used as reference sequences for further analysis. MiSeq reads were mapped against reference plasmid sequences and percentage depth and breadth of coverage was calculated using Burrows–Wheeler aligner, SAMtools and BEDTools coverage.28–30 Alignments were viewed for quality using Tablet.31
Sequences of plasmids resolved by hybrid assembly (pM16/0594, pM18/0011 and pM17/0314) and DNA regions encoding optrA variants have been deposited in GenBank under accession numbers MN831410, MN831411, MN831412, MN831413, MN831414, MN831415, MN831416, MN831417, MN831418 and MN831419.
Results and discussion
Linezolid-resistant isolates
A total of 35/154 (22.7%) LRE (23 E. faecalis and 12 E. faecium) submitted to the NMRSARL between June 2016 and August 2019 harboured optrA (2 E. faecium and 13 E. faecalis), poxtA (9 E. faecium and 10 E. faecalis) or both optrA and poxtA (1 E. faecium). All 35 isolates were from hospitalized patients in 11 Irish hospitals (H1–H11) (Figure S1) and were phenotypically resistant to linezolid, chloramphenicol and tetracycline (Table S2). This is the highest prevalence of optrA and/or poxtA in human enterococci reported to date. Six E. faecium isolates were vancomycin resistant and harboured vanA. The remaining 29 LRE isolates were vancomycin susceptible and lacked vanA (Table S2). The majority (19/35) of LRE harboured poxtA only; the largest collection of poxtA-positive human isolates reported to date. The remaining LRE harboured optrA only (14/35), optrA and poxtA (1/35) or optrA and cfr(D) (1/35). One VREfm (M19/0595) harboured poxtA and the G2576T 23S mutation (4/6 copies mutated) and is the first report of mutational and gene-encoded linezolid resistance in a single enterococcal isolate. The remaining 34 LRE lacked 23S mutations.
All 20/119 LRE investigated that lacked cfr, optrA and/or poxtA (2 E. faecalis, 18 E. faecium) were phenotypically resistant to linezolid and chloramphenicol (Table S2). All 20 LRE exhibited a varying copy number (1–5) of the G2576T 23S mutation. The majority of the LRE (15/18 E. faecium) harboured vanA and exhibited vancomycin MICs ≥32 mg/L. The remaining five vanA-negative isolates (two E. faecalis and three E. faecium) were vancomycin susceptible (Table S2).
Relatedness of LRE based on WGS
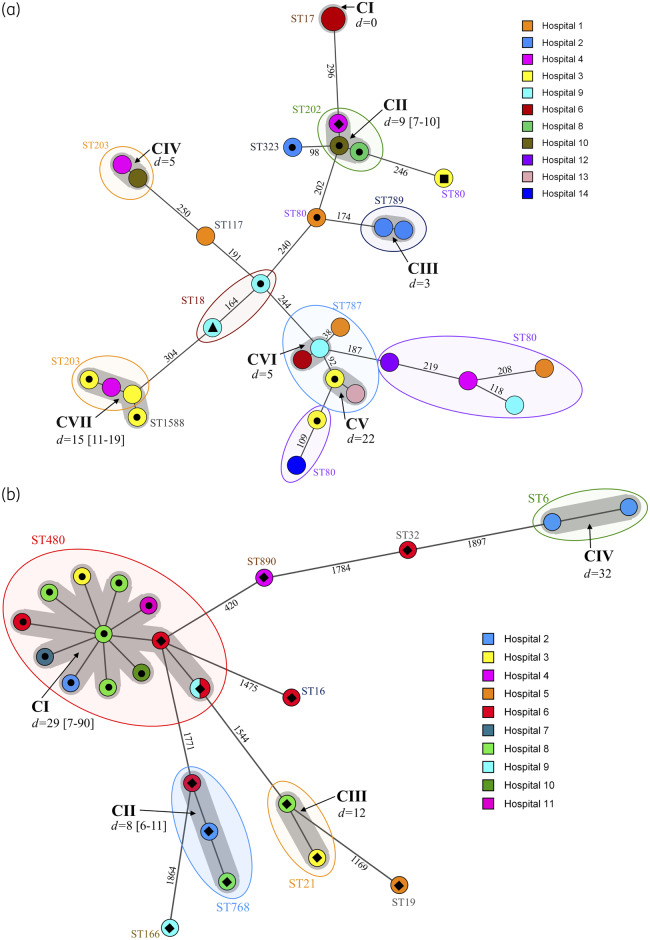
Of the 55 LRE investigated, the 30 E. faecium included from 11 hospitals were assigned to 10 STs using traditional MLST, with ST80 predominating (8/30, 26.6%). Seventeen E. faecium isolates were differentiated into seven clusters (CI–CVII) using cgMLST (Figure 1a). Clusters CI–CVI contained isolates of the same STs (ST17, ST787, ST789, ST202 and ST203), while cluster CVII consisted of ST203 and ST1588 (a single locus variant of ST203) isolates (Figure 1a). Clusters were differentiated by intracluster and intercluster allelic differences of 0–22 and 38–394, respectively. Isolates exhibiting ≤22 allelic differences were deemed closely related, based on previous work.24 Clusters CI and CIII contained isolates from the same hospitals (H2 and H6), all with the G2576T 23S mutation. The remaining five clusters consisted of isolates from two or more hospitals and a mixture of isolates exhibiting linezolid resistance associated with G2576T mutations or a resistance gene (Figure 1a).
Figure 1.
Minimum spanning trees based on (a) cgMLST data from 30 linezolid-resistant clinical E. faecium isolates and (b) wgMLST data from 25 linezolid-resistant E. faecalis clinical isolates. All isolates were recovered between June 2016 and August 2019 from 14 Irish hospitals, as denoted in the legends. The numbers on the branches represent the number of cgMLST/wgMLST allelic differences. STs are shown in coloured ovals. Grey shadowing around nodes indicates clusters of related isolates, which are labelled in bold and denoted CI–VII; ‘d=’ values indicate average allelic differences and the range in square brackets. Isolate designations are as follows: filled black circle, poxtA positive; filled black diamond, optrA positive; filled black square, optrA positive and cfr(D) positive; and filled black triangle, optrA positive and poxtA positive. Isolates not marked with a symbol were negative for linezolid resistance genes and harboured various copy numbers (1–5) of the G2576T 23S mutation associated with linezolid resistance.
Of the 55 LRE investigated, the 25 E. faecalis included originated from 10 hospitals and belonged to nine STs using traditional MLST, with ST480 predominating (13/25). Twenty of the E. faecalis isolates differentiated into four clusters (CI–CIV) using wgMLST (Figure 1b). The remaining five isolates were distantly related to any other isolate, exhibiting between 339 and 1897 allelic differences. Clusters were defined by the contrasting tight value of intracluster differences (0–43). Each cluster contained isolates from the same STs (ST6, ST21, ST480 and ST768). Only one cluster, CIV, contained two isolates from the same hospital (H2), both with the G2576T 23S mutation. The remaining three clusters contained isolates from 2–8 hospitals. Clusters CII (n = 3) and CIII (n = 2) contained only optrA-positive isolates, whereas cluster CI (n = 13) contained only ST480 isolates, 10 and 3 of which harboured poxtA or optrA, respectively. Isolates within cluster CI exhibited an average allelic difference of 29 (range 7–90) (Figure 1b).
Overall the population structure of LRE was polyclonal, but the presence of highly related strains in different hospitals was evident.
Genetic environment surrounding optrA
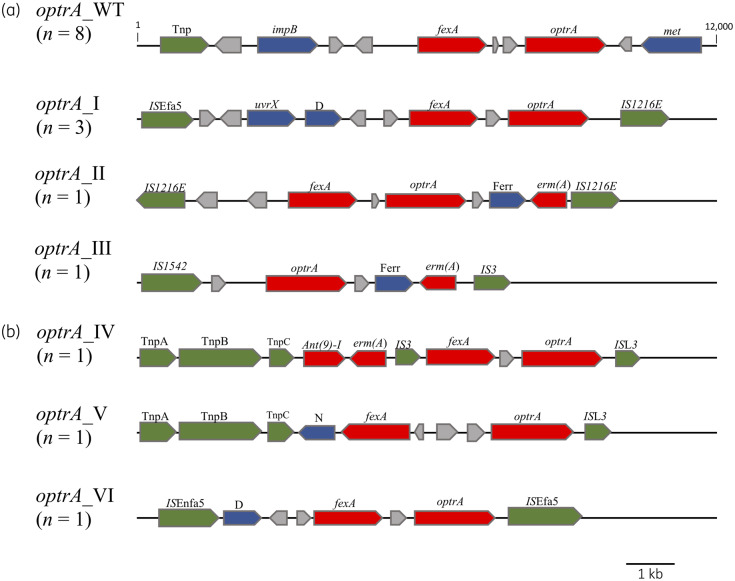
The WGS data of a selection of isolates encoding optrA underwent hybrid assembly, namely two E. faecium (M16/0594 and M17/0314) and seven E. faecalis (M17/0149, M17/0240, M18/0173, M18/0497, M18/0582, M18/0906 and M19/0596). The first plasmid resolved was a 36 331 bp optrA-encoding plasmid (pM17/0149) from E. faecalis M17/0149, which was the same size and exhibited 100% DNA sequence identity to plasmid pE349, first described from a clinical E. faecalis in China and subsequently identified in E. faecium and E. faecalis from humans and food-producing animals throughout European, American and Asian countries.7,9,13,14,32 Of the 16 optrA-positive isolates, half (one E. faecium and seven E. faecalis) harboured plasmids exhibiting 100% sequence coverage to pM17/0149 (pE349-like). Multiple E. faecalis STs (ST19, ST21, ST166 and ST768) harboured this pE349-like plasmid, suggesting independent acquisition by different genetic backgrounds (Table S2). For these eight isolates, the genetic environment surrounding optrA was designated optrA_WT (Figure 2). The remaining eight optrA-positive isolates exhibited <35% sequence identity to pM17/0149 and were selected for WGS hybrid assembly. The genetic environment surrounding optrA was examined and optrA was identified on plasmids in five isolates (M17/0240, M18/0582, M19/0596, M18/0173 and M17/0314) and within the chromosome in three isolates (M18/0497, M16/0594 and M18/0906) (Figure 2). Three variations surrounding optrA were evident on the plasmid-encoded regions: optrA_I (M17/0240, M18/0582 and M19/0596); optrA_II (M18/0173); and optrA_III (M17/0314). In optrA_I and optrA_II, fexA was encoded around 750 bp upstream of optrA, similarly to optrA_WT, whereas optrA_III lacked fexA. Variations were distinguished by various flanking IS elements and other genes encompassed between these elements (Figure 2a). Interestingly, isolate M17/0314, harbouring optrA_III, also encoded the linezolid resistance gene cfr(D) and the macrolide, lincosamide and streptogramin B resistance genes erm(A) and erm(B), on a 103 600 bp plasmid (pM17/0314) (Figure S2). In the case of 1/8 optrA_WT isolates (M16/0427), 2/3 isolates harbouring optrA_I (M17/0240 and M19/0596) and 1 optrA_III isolate (M17/0314), optrA was successfully conjugated into a recipient strain of the same species (Table S3).
Figure 2.
Schematic representation of the optrA gene loci in E. faecalis and E. faecium clinical isolates from Ireland. Different variants of the genetic environment surrounding optrA were detected in LRE encoded on plasmids (a) and within the chromosome (b) and were aligned against the prototype optrA_WT, first described by Wang et al. in 2015.9 Genes of interest and their orientation are shown with arrows and labelled; red indicates antibiotic resistance genes, green indicates ISs/transposases, blue indicates known proteins and grey indicates hypothetical proteins. Tnp, transposase; met, truncated DNA adenine methylase; uvrX, putative UV-damage repair protein; D, DNA-directed RNA polymerase β subunit; Ferr, ferredoxin; N, NAD(P)H oxidoreductase.
Three variations of the genetic environment surrounding optrA in the chromosome, designated optrA_IV, optrA_V and optrA_VI, were identified in isolates M18/0906, M16/0594 and M18/0497, respectively (Table S2). Both the optrA_IV and optrA_V variants harboured optrA, flanked by tnpA and tnpB from Tn554 on one end and by ISL3 on the other end. The optrA_IV variant encoded fexA and optrA in the same orientation as optrA_WT, but in optrA_V, fexA was encoded around 2800 bp upstream from optrA in the opposite orientation to the arrangement in optrA_WT (Figure 2b). In optrA_VI, optrA was flanked by ISEnfa5 and ISEfa5 and exhibited the same optrA and fexA arrangement present in optrA_WT (Figure 2b). Attempts to transfer optrA in the three isolates encoding chromosomal optrA by conjugation were unsuccessful.
The variation surrounding optrA in the investigated LRE, combined with the identification of multiple linezolid resistance genes [optrA/poxtA and optrA/cfr(D)] in individual isolates, is indicative of an environment with a high selective pressure for linezolid resistance.
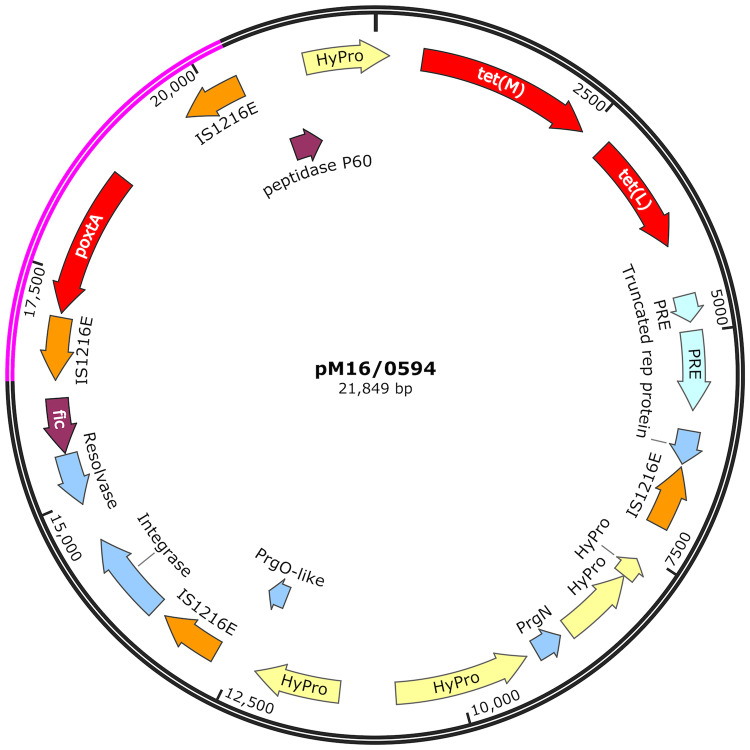
Characterization of isolates encoding poxtA
The E. faecium isolate M16/0594 harboured optrA_V in its chromosome and poxtA, tet(M) and tet(L) on a 21 849 bp plasmid (pM16/0594) (Figure 3). Plasmid pM16/0594 was conjugative and transconjugant derivatives were obtained with the E. faecium 64/3 recipient. Of the remaining 19 poxtA-positive LRE (9 E. faecium and 10 E. faecalis), 5/19 (all E. faecium) exhibited 100% sequence coverage to pM16/0594, while 14/19 (4 E. faecium and 10 E. faecalis) exhibited between 72.9% and 99.3% coverage. All isolates exhibited 100% sequence coverage to a 4001 bp poxtA-encoding region, flanked by two IS1216E elements in parallel orientation; this conserved poxtA element has previously been shown to be responsible for horizontal gene transfer of poxtA.16,33,34 These findings indicate the spread of poxtA in human E. faecium and E. faecalis in Ireland via this 4001 bp element or by a very similar poxtA plasmid. One limitation of this method of read mapping against pM16/0594 is the risk of missing larger plasmids, as 100% coverage will indicate the query sequence is identical to the reference used but cannot indicate if the plasmid is larger. Therefore, isolates with 100% sequence coverage to pM16/0594 have at least a 21 849 bp plasmid region identical to that of pM16/0594, which may be contained within larger plasmids.
Figure 3.
Schematic diagram of the structural organization of plasmid pM16/0594 from E. faecium isolate M16/0594, encoding the poxtA linezolid resistance gene resolved by hybrid assembly of paired-end Illumina MiSeq short reads with Oxford Nanopore Technologies long reads. The 4001 bp highly conserved region around poxtA, flanked by two IS1216E in parallel orientation, is highlighted in pink. Genes of interest and their orientation are represented by arrows as follows: red indicates antibiotic resistance genes, orange indicates ISs, blue indicates rep proteins, purple indicates other known proteins and yellow indicates hypothetical proteins. The plasmid size is labelled (bp). PRE, plasmid recombination enzyme; fic, cell filamentation protein; HyPro, hypothetical protein.
The poxtA-positive E. faecalis isolate M18/0011 exhibiting 73.7% sequence coverage to pM16/0594 was also resolved using hybrid assembly. The plasmid encoding poxtA in M18/0011 (pM18/0011) was 3570 bp smaller than pM16/0594 (18 279 bp) and lacked tet(M) and tet(L). An identical 4001 bp region encoding poxtA, flanked by two IS1216E elements in parallel orientation, was observed in pM18/0011 (Figure S3), albeit in the reverse orientation to the identical region in pM16/0594. Plasmid pM18/0011 was also conjugative and transconjugant derivatives were obtained with the E. faecalis OG1RF and E. faecium 64/3 recipients (Table S3).
In total, 5/20 poxtA-positive LRE yielded transconjugant derivatives including E. faecium M16/0594 (harbouring pM16/0594) and E. faecalis M18/0011 (harbouring pM18/0011), two E. faecalis donors (M16/0419, 75.2% like pM16/0594; and M16/0633, 74.5% like pM16/0594) conjugated to E. faecalis OG1RF and one E. faecium donor (M19/0357, 77.9% like pM16/0594) conjugated to E. faecium 64/3 (Table S3). For the remaining 15 poxtA-positive LRE, conjugation was unsuccessful. The spread of a single clone was indicated in all 10 identified poxtA-positive E. faecalis LRE, as all belonged to ST480 and were closely related, with an average of 22 (range 7–54) wgMLST allelic differences (Figure 1b). The ST480 clone was recovered in seven hospitals (Table S2). ST480 is one of the predominant optrA-positive clinical E. faecalis clones in France and Germany.15,35 In contrast, poxtA was found to have spread in E. faecium via the 4001 bp mobile element or a promiscuous plasmid to multiple STs (ST18, ST80, ST202, ST203, ST323, ST787 and ST1588).
Conclusions
The results of this study revealed the high prevalence and spread of optrA and poxtA among enterococci in Irish hospitals. A major cause for concern is that 26.3% (5/19) of isolates harbouring a poxtA-encoding plasmid also harboured vanA, which poses a significant risk for hospitalized patients, as treatment options for such strains are limited. Linezolid consumption in Irish hospitals is currently not specifically recorded as part of annual national hospital antimicrobial consumption surveillance.4 This needs to change and linezolid consumption in hospitals needs to be more rigidly controlled.
Supplementary Material
Acknowledgements
We thank all the staff of the National MRSA Reference Laboratory for their support in facilitating and managing the collection of linezolid-resistant clinical isolates from Irish hospitals. We thank the clinical microbiology staff from all the Irish hospitals that sent clinical enterococcal isolates for investigation of linezolid resistance genes. We wish to acknowledge Dr Mette Pinholt, Professor Henrik Westh and the bioinformatic staff of the Hvidovre University Hospital, Copenhagen, Denmark, for their assistance and guidance in the bioinformatic analysis of clinical enterococci. We thank Dr Peter Flanagan for support with Oxford Nanopore MinION sequencing and analysis. We would also like to thank Dr Jennifer Bender of the Robert Koch Institute for providing us with the recipient strains for filter mating experiments.
Funding
This work was supported by the Dublin Dental University Hospital Microbiology Research Unit and by the Irish Health Research Board grant number ILP-POR-2019-010.
Transparency declarations
None to declare.
References
- 1. Zahedi Bialvaei A, Rahbar M, Yousefi M. et al. Linezolid: a promising option in the treatment of Gram-positives. J Antimicrob Chemother 2017; 72: 354–64. [DOI] [PubMed] [Google Scholar]
- 2. Bersos Z, Maniati M, Kontos F. et al. First report of a linezolid-resistant vancomycin-resistant Enterococcus faecium strain in Greece. J Antimicrob Chemother 2004; 53: 685–6. [DOI] [PubMed] [Google Scholar]
- 3.ECDC. The European Antimicrobial Resistance Surveillance System EARS-Net Results. 2017. https://ecdc.europa.eu/en/antimicrobial-resistance/surveillance-and-disease-data/data-ecdc.
- 4.HSE/HPSC. Hospital Antimicrobial Consumption Surveillance. 2018. http://www.hpsc.ie/a-z/microbiologyantimicrobialresistance/europeansurveillanceofantimicrobialconsumptionesac/PublicMicroB/SACHC/SACHC_Current.pdf.
- 5. Lazaris A, Coleman DC, Kearns AM. et al. Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J Antimicrob Chemother 2017; 72: 3252–7. [DOI] [PubMed] [Google Scholar]
- 6. Gonzales RD, Schreckenberger PC, Graham MB. et al. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 2001; 357: 1179. [DOI] [PubMed] [Google Scholar]
- 7. Bi R, Qin T, Fan W. et al. The emerging problem of linezolid-resistant enterococci. J Glob Antimicrob Resist 2018; 13: 11–9. [DOI] [PubMed] [Google Scholar]
- 8. Bai B, Hu K, Zeng J. et al. Linezolid consumption facilitates the development of linezolid resistance in Enterococcus faecalis in a tertiary-care hospital: a 5-year surveillance study. Microb Drug Resist 2019; 25: 791–8. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Lv Y, Cai J. et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 2015; 70: 2182–90. [DOI] [PubMed] [Google Scholar]
- 10. Cai J, Wang Y, Schwarz S. et al. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010-2014. Clin Microbiol Infect 2015; 21: 1095.e1–4. [DOI] [PubMed] [Google Scholar]
- 11. Swaney SM, Aoki H, Ganoza MC. et al. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother 1998; 42: 3251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bender JK, Cattoir V, Hegstad K. et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist Updat 2018; 40: 25–39. [DOI] [PubMed] [Google Scholar]
- 13. Mendes RE, Hogan PA, Jones RN. et al. Surveillance for linezolid resistance via the Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS) programme (2014): evolving resistance mechanisms with stable susceptibility rates. J Antimicrob Chemother 2016; 71: 1860–5. [DOI] [PubMed] [Google Scholar]
- 14. Mendes RE, Deshpande L, Streit JM. et al. ZAAPS programme results for 2016: an activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J Antimicrob Chemother 2018; 24: 328–37. [DOI] [PubMed] [Google Scholar]
- 15. Bender JK, Fleige C, Lange D. et al. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int J Antimicrob Agents 2018; 52: 819–27. [DOI] [PubMed] [Google Scholar]
- 16. Antonelli A, D’Andrea MM, Brenciani A. et al. Characterization of poxtA, a novel phenicol–oxazolidinone–tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother 2018; 73: 1763–9. [DOI] [PubMed] [Google Scholar]
- 17. Brenciani A, Fioriti S, Morroni G. et al. Detection in Italy of a porcine Enterococcus faecium isolate carrying the novel phenicol-oxazolidinone-tetracycline resistance gene poxtA. J Antimicrob Chemother 2019; 74: 817–8. [DOI] [PubMed] [Google Scholar]
- 18. Hao W, Shan X, Li D. et al. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother 2019; 74: 1771–5. [DOI] [PubMed] [Google Scholar]
- 19. Papagiannitsis CC, Tsilipounidaki K, Malli E. et al. Detection in Greece of a clinical Enterococcus faecium isolate carrying the novel oxazolidinone resistance gene poxtA. J Antimicrob Chemother 2019; 74: 2461–2. [DOI] [PubMed] [Google Scholar]
- 20. Diaz L, Kiratisin P, Mendes RE. et al. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother 2012; 56: 3917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bender JK, Fleige C, Klare I. et al. Detection of a cfr(B) variant in German Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS One 2016; 11: e0167042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O’Driscoll C, Murphy V, Doyle O. et al. First outbreak of linezolid-resistant vancomycin-resistant Enterococcus faecium in an Irish hospital, February to September 2014. J Hosp Infect 2015; 91: 367–70. [DOI] [PubMed] [Google Scholar]
- 23.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0. 2019. http://www.eucast.org.
- 24. de Been M, Pinholt M, Top J. et al. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 2015; 53: 3788–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hasman H, Clausen P, Kaya H. et al. LRE-Finder, a Web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J Antimicrob Chemother 2019; 74: 1473–6. [DOI] [PubMed] [Google Scholar]
- 26. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Overbeek R, Olson R, Pusch GD. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014; 42: D206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Handsaker B, Wysoker A. et al. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan AR, Hall IM.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26: 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Milne I, Stephen G, Bayer M. et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 2013; 14: 193–202. [DOI] [PubMed] [Google Scholar]
- 32. Cavaco LM, Bernal JF, Zankari E. et al. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother 2016; 72: 678–83. [DOI] [PubMed] [Google Scholar]
- 33. Lei C-W, Kang Z-Z, Wu S-K. et al. Detection of the phenicol-oxazolidinone-tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J Antimicrob Chemother 2019; 74: 2459–61. [DOI] [PubMed] [Google Scholar]
- 34. Huang J, Wang M, Gao Y. et al. Emergence of plasmid-mediated oxazolidinone resistance gene poxtA from CC17 Enterococcus faecium of pig origin. J Antimicrob Chemother 2019; 74: 2524–30. [DOI] [PubMed] [Google Scholar]
- 35. Sassi M, Guérin F, Zouari A. et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006–16. J Antimicrob Chemother 2019; 74: 1469–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.