Abstract
The clinical microbiology laboratory relies on traditional diagnostic methods such as culturing, Gram stains, and biochemical testing. Receipt of a high-quality specimen with an appropriate test order is integral to accurate testing. Recent technological advancements have led to decreased time to results and improved diagnostic accuracy. Examples of advancements discussed in this chapter include automation of bacterial culture processing and incubation, as well as introduction of mass spectrometry for the proteomic identification of microorganisms. In addition, molecular testing is increasingly common in the clinical laboratory. Commercially available multiplex molecular assays simultaneously test for a broad array of syndromic-related pathogens, providing rapid and sensitive diagnostic results. Molecular advancements have also transformed point-of-care (POC) microbiology testing, and molecular POC assays may largely supplant traditional rapid antigen testing in the future. Integration of new technologies with traditional testing methods has led to improved quality and value in the clinical microbiology laboratory.
Keywords: Microbiology, molecular, automation, point-of-care, syndromic testing
Learning objectives
After reviewing this chapter, the reader will be able to:
-
•
List key considerations for specimen collection for microbiology testing.
-
•
Discuss the advantages and limitations of automation in the clinical microbiology laboratory.
-
•
Describe the evolution of microorganism identification methods.
-
•
Discuss the benefits and limitations of molecular microbiology point-of-care testing.
-
•
Summarize currently available multiplex molecular microbiology testing options.
Specimen collection
Specimen collection in clinical microbiology is of utmost importance. The quality of the specimen determines the quality of the results. Proper specimen collection consists of (1) proper collection of the source; (2) proper container selection; and (3) proper transport conditions. Microbiology laboratory staff consists of technicians and technologists who are thoroughly trained on the appropriate processing and testing methodologies; however, even the most skilled microbiologists and the best laboratory practices cannot make up for a poor specimen. Although improperly collected specimens lead to unreliable results, they are received in the microbiology laboratory every day. Thus it is the job of the laboratory to convey information about proper collection techniques and containers to clinicians.
To communicate and highlight the importance of obtaining appropriate specimens for microbiological testing, the Infectious Diseases Society of America and the American Society for Microbiology (ASM) published a guidance document on utilization of the microbiology laboratory, which contains information on optimal test selection, optimal specimen collection approaches, and transport concerns [1]. In addition, the Guide to Specimen Management in Clinical Microbiology, now in its third edition, is available from ASM Press [2]. Both documents are critical resources for additional information regarding management of specimens in microbiology laboratories.
In general, aspirates, fluids, and tissue specimens are preferred over swabs for all microbiology culture testing due to their higher diagnostic yield. However, it is not always possible to obtain an aspirate, fluid, or tissue, so swabs are commonly accepted. Disinfection of the site (if applicable) must be carefully considered during specimen collection. For example, it is imperative to disinfect the skin prior to collection of blood cultures. Otherwise, skin flora may result in a false-positive culture, which can lead to an increase of $2923 to $5812 in hospital costs as well as exposure to unnecessary antimicrobials and increased length of stay [3]. Collection from the appropriate source is also an important consideration. If a nasopharyngeal (NP) swab is the preferred source for a test, it is important to collect a true NP swab, which is not an enjoyable experience for the patient, rather than a nasal swab. For example, Bordetella pertussis and many respiratory viruses are primarily found in the nasopharynx, so a properly collected NP swab is essential for detection and diagnosis. A swab of the nares or a mid-turbinate region may cause a false-negative result. Blood cultures and NP swabs are only such two examples to highlight the importance of appropriate source collection.
In comparison with the core laboratory, the microbiology laboratory receives a much wider variety of transport containers, which may include Tupperware containers, Ziploc bags, Mountain Dew bottles, etc. (all of which have been received in a clinical microbiology laboratory). Specimens received in such nonsterile containers are rejected, but the variety of acceptable sterile transport devices can still be overwhelming and collection containers are not standardized between the laboratories due to the extensive number of transport device manufacturers and variations in devices among manufacturers. One laboratory may use one set of collection devices, while other laboratories have their own sets. A standalone hospital microbiology lab may receive as few as 10–20 different types of containers, consisting of various sterile cups and tubes, preservative tubes, capped syringes, swabs, etc., while the number of collection devices received at the centralized and reference laboratories may be much larger. The variability in specimen collection containers was a major barrier to implementation of total laboratory automation (TLA) for clinical microbiology workflows.
Specimens for anaerobic culture should be submitted under conditions that allow recovery of anaerobes. For example, anaerobic transport containers that contain a semisolid reducing gel may be used for specimens submitted for anaerobic culture. In addition, capped syringes with excess air removed may be used. Dry swabs are not appropriate for anaerobic culture. Anaerobic culture testing should never be performed without an accompanying aerobic culture, unless selective culture for Clostridioides difficile is requested; however, this is uncommon. Specimens acceptable for anaerobic culture include aspirates, tissues, and deep wounds. Superficial wounds and other sites with normal anaerobic flora are not acceptable for anaerobic culture.
One of the major shifts in clinical microbiology was the development of the flocked swab in transport media. Historically, tightly wound cotton, rayon, and dacron swabs predominated, with some placed into a liquid or gel transport media while others were not. Traditional swabs performed poorly, because the vast majority of the organisms in the specimen remained trapped in the swab and were not released when the swabs were used to inoculate solid media (Petri dishes). Another drawback of traditional swabs is that if multiple plates need to be inoculated, the majority of the bacteria that are released were released onto the first plate, resulting in inconsistent inoculation across culture media. Compared with traditional swabs, flocked swabs contain fibers that radiate outward from the shaft, which allow for increased release of organisms from the swab. Once the specimen is collected, flocked swabs are placed into transport tubes, which contain liquid transport media. While dry swabs cannot be used for anaerobic culture due to anaerobes drying out and dying, the addition of transport media allows the swabs to be used for anaerobic culture in addition to aerobic culture. The transport media also permits a longer transit time to the lab due to improved specimen stability (24–48 hours). In addition, this setup allows for the release of organisms into the transport media, which can be used for the inoculation of plates rather than inoculating with a swab, allowing for consistent inoculation across culture media. Not only did the development of the flocked swab improve the quality of cultures, it also helped pave the way for automation in microbiology, because it is easier to automate the transfer of transport media to inoculate plates as compared with using a dry swab for plate inoculation. Commercially available flocked swab options include the Copan ESwab and Puritan PurFlock Ultra and HydraFlock swabs.
For urine testing in the microbiology laboratory, urine preservative tubes have become more common and can preserve organisms for up to 24–48 hours during transport. Preservative tubes are more standardized than sterile cups, which are different sizes and have different lid-threading properties. Transport media have been developed for stool, facilitating downstream automation and molecular testing of stool specimens. The shift toward transport media has allowed for improved culture results and support of automation. Although specimen containers have become more amenable to automation, there will always be containers that are not accommodated on automated instrumentation. For additional information, refer to the following section on laboratory automation.
After collection, the specimen should be transported to the laboratory in a timely manner. For off-site laboratories, specimens should be transported to maintain specimen integrity and quality. Excessive transport times may negatively affect results due to either the death of fastidious organisms or overgrowth of nonfastidious organisms. Generally, it is recommended that specimens are received in the laboratory within 1–2 hours after collection unless specimens are in a transport or preservative media, which permits longer transit times. Alternatively, some specimen sources, but not all (e.g., cerebrospinal fluid), can be refrigerated to preserve their quality. Transport time is also important, because the sooner the specimen arrives in the lab, the sooner the culture is inoculated and placed into an incubator. Delays in transport lead to delays in culture results.
Once specimens are received in the laboratory, they are processed in a biosafety cabinet to prevent staff exposure to infectious aerosols, which may be generated during specimen processing, and to prevent contamination from the environment. Only one specimen is processed at a time in order to prevent cross-contamination. This is important to ensure the quality of reported results, both for traditional microbiology and for molecular microbiology. Specimens processed in other areas of the laboratory outside of a biosafety cabinet should not be used for microbiology culture or molecular microbiology testing due to the possibility of false-positive results due to cross-contamination. Shared specimens should be processed initially in the microbiology laboratory prior to other testing (such as Core laboratory testing) or a separate specimen should be obtained.
A Gram stain is performed on the majority of specimens, with the exception of urines, stool, screening cultures [e.g., Group B Streptococcus and methicillin-resistant Staphylococcus (MRSA), etc.], and strep throat swabs. A Gram stain can be performed on direct specimens, such as respiratory specimens, fluids, and tissues and on swabs, such as wound swabs. However, Gram stains performed on specimens collected on swabs produce inferior results compared with those performed on direct specimens. The Gram stain provides the clinician with an early result while culture results are pending. In addition to the evaluation of bacteria, Gram stains are analyzed for the presence of white blood cells (WBCs) and squamous epithelial cells (SECs). The presence or absence of WBCs and SECs determines the quality of the specimen. The Gram stain plays a significant role in determining workup for respiratory and wound cultures. As an example, lower respiratory specimens with ≥25 SECs per low power field should be rejected and not cultured. The presence of SECs indicates that the specimen is from the oropharynx rather than the lower respiratory tract.
Rejection criteria are essential, and each microbiology laboratory should have a procedure that addresses specimen rejection in order to avoid providing inaccurate results. Examples of rejection criteria that are commonly used are:
-
•
Specimens submitted in nonapproved containers should be rejected.
-
•
Leaking specimens should be rejected.
-
•
Anaerobic culture, if requested, should not be performed if the specimen is not submitted under anaerobic conditions.
-
•
Specimens with excessive transport times (defined by the laboratory, will depend on source) should be rejected.
-
•
Respiratory specimens with ≥25 SECs per low power field on Gram stain should be rejected.
Laboratory automation in clinical microbiology
Several components in the clinical microbiology laboratory have been automated during the last few decades. Automation advances include automated blood culture instruments, automated antimicrobial susceptibility testing platforms, automated nucleic acid extraction, and others. Although components of the laboratory were automated, the primary role of the microbiology laboratory—specimen inoculation and culture automation—was not automated until recently due to a number of factors. The wide variety of specimens and specimen collection devices was a key factor as to why the development of TLA in microbiology took so long to become a reality. In addition to the variety of containers, specimens are processed in a variety of manners based on the specimen source and the test ordered. Specimens may be vortexed, centrifuged, minced, or ground. Moreover, once inoculated, plates are incubated in different temperatures under various atmospheric conditions. For decades, it seemed impossible to automate microbiology due to these aforementioned challenges. However, there was increasing need and demand for automation due to a nationwide shortage of medical technologists and due to the consolidation of microbiology laboratories, which has resulted in increased workloads. In addition, a greater emphasis has been placed on quality and standardization in healthcare. These factors, along with recent technological advances, led to the rapid evolution of laboratory automation in clinical microbiology. An overview of laboratory automation in clinical laboratories is further described in Chapter 14, Laboratory automation, of this book.
Currently, there are two commercially available laboratory automation systems for clinical microbiology: Becton Dickinson (BD)’s Kiestra TLA and Copan Diagnostic’s Walk Away Specimen Processor Laboratory automation system (WASPLab). Both systems automate all steps of the culture process, including specimen inoculation, incubation, and plate imaging (Fig. 55.1 ). The images are then presented on a computer monitor to technologists who read and interpret the cultures (Fig. 55.2 ). Both manufacturers are working to automate culture interpretation as well, but it is likely that there will always be a certain degree of human intervention required.
Figure 55.1.

Components of total laboratory automation in clinical microbiology.
Figure 55.2.

Total laboratory automation systems present high-resolution images on a computer monitor for technologists to read and interpret.
Components of automation
Inoculation unit
The processing components of the systems have been designed to accept multiple types of specimen containers and also allow for “offline” inoculation in case a nonstandard/nonprogrammed container is received. The systems vortex and centrifuge (available with the WASPLab system) specimens, uncap and recap specimen containers, and remove and replace lids on culture plates or broth-based media and inoculate specimens. After samples are added to plates, the systems streak plates according to programmed patterns that are preselected by the laboratory. In addition to processing and inoculation, both systems also prepare Gram stain slides for staining.
Automated track
Once inoculated, plates are transported via automated track lines to automated or “smart” incubators, which can be programed at various temperatures and atmospheric conditions. Plates may then be summoned from the incubators to workbenches via the automated track for further workup by the technologist. Systems may have a unidirectional or bidirectional track. Systems with a unidirectional track allow plates to be summoned to workbenches, assuming workbenches are downstream of the incubators, but plates must be placed in a canister and manually returned to the track upstream of the incubators in order to be returned to the incubator. Systems with a bidirectional track allow plates to be transported back to incubators directly from the workbenches once workup is completed.
Automated “smart” incubators
Prior to automation, plates were stacked in racks and placed in traditional incubators. Plates toward the top of the rack were more exposed to the proper incubation conditions (temperature and atmospheric conditions) compared with plates in the middle and bottom of each rack. In addition, incubator doors were opened and closed each time a plate was added or removed, leading to inconsistency in thermal conditions and other incubator parameters.
“Smart” incubators can automate all forms of aerobic incubation. Plates enter and exit incubators via the automated track, so incubator doors remain closed throughout the process. This allows the incubators to maintain a constant temperature and environment. In addition, automated incubators contain individual shelves for each plate, which promotes faster, more consistent growth of colonies due to the consistent incubation conditions across all plates. Automated incubators can be programmed at a variety of temperatures and can maintain either an oxygen-rich or a carbon dioxide-enriched environment. Nonfastidious organisms, such as the Enterobacterales, grow well in an oxygen-rich environment, while a carbon dioxide environment promotes the growth of fastidious organisms. Moreover, carbon dioxide interferes with the performance of selective culture media. Selective media contain antibiotics and/or inhibitors to suppress the growth of certain organisms. An example is mannitol salt agar, which allows for the growth of Staphylococcus aureus while suppressing growth of other organisms. Such media should be incubated in an oxygen-rich environment. It is important to note that current systems do not offer an anaerobic incubator, so anaerobic culture plates must be incubated offline. Inside the incubators, a robotic arm facilitates the movement of the plates, by either placing them on a shelf or removing them for imaging or discarding. The imaging systems are a component either of the incubator or adjacent to the incubator, so plates are removed for a minimal amount of time when imaged.
Imaging
Automation systems are equipped with high-resolution cameras that capture images of plates at defined intervals, which are programmed by the laboratory. Imaging workflows vary based on culture type. Traditionally, technologists would hold plates up toward a light source and at various angles when viewing; this allowed easier detection of certain colony morphologies. Automated systems capture images using various lighting angles, including back lighting, front lighting, and side lighting to mimic manual viewing. Captured images are maintained on the system and can be viewed via remote access. In addition, the captured images allow technologists to go “back in time” when reading cultures. If growth on the second or third day differs from what was reported the first day, the technologist can review the previous days’ images for comparison. The storage of image files also creates an opportunity for training of new technologists or to emphasize concepts. Moreover, stored images create an opportunity for quality assurance review.
Workstations
After the images are captured, technologists read plates by viewing the images on a computer rather than viewing the actual plates. Individual plate images can be enlarged to the size of the computer screen and further magnified. Because technologists are never exposed to a plate before viewing an image, imaging could potentially prevent exposure to a select agent (i.e., Bacillus anthracis, Brucella species, and others) or other highly infectious organisms, such as Neisseria meningitidis, based on suspicious growth characteristics. When a plate is viewed, the technologist can virtually mark specific colony morphologies for additional workup processes, which may include identification, antimicrobial susceptibility testing, and subculturing. Additional workup can be completed at the same time by summoning the plate to the workbench or, alternatively, the culture can be added to a queue and batched for workup after viewing is completed.
With the onset of laboratory automation, many microbiology laboratories have adopted a reader/workup workflow where culture images are read by one technologist and annotated for workup and positive cultures are followed up by a second technologist. This differs from the traditional microbiology laboratory, where one technologist would perform all reading and workup for a given culture. The benefit of the reader/workup workflow is that two technologists work together on the same culture, which may lead to improved consistency. This workflow also allows for off-site reading, so technologists could read cultures while in another area of the lab or offsite.
Commercially available systems
The two commercially available laboratory automation systems for microbiology are the Copan WASPLab and BD Kiestra TLA, as mentioned above. Both systems are modular, scalable, and offer automated selection and labeling of media, inoculation, incubation, and plate imaging. The systems interface with the laboratory information system (LIS), and when a specimen barcode is scanned, the LIS is queried and communicates the type of culture that was ordered. The system then selects the corresponding media (previously programmed by the laboratory) based on the type of culture ordered, applies a specimen label to each piece of media and inoculates the specimen.
The WASPLab inoculation unit, the WASP DT, uses 1-, 10-, and 30-µL reusable inoculating loops. Loops are sterilized between the pieces of media or between the specimens, depending on which program the lab selects. The Kiestra inoculation unit, InoculA, uses a pipettor rather than loops for inoculation. The InoculA pipettor can pipette 10–250 µL, and once a specimen is plated, the InoculA uses a single-use sterile magnetic bead to streak each plate. In both cases, specimens must be relatively nonviscous and homogenous for automated inoculation. The Kiestra system has the added consumable cost due to the single-use nature of the beads, but produces more isolated colonies and more accurate colony counts than the WASPLab [4], [5]. Both systems allow for manual inoculation if tissues, viscous specimens, or unusual containers are received. The original WASPLab requires offline manual inoculation and streaking, and plates can subsequently be placed onto the automated track for transport to the incubators. Copan recently developed a robot, called the Collaborative Robot, which can be purchased as a separate unit to assist with manual cultures. Technologists manually inoculate plates, and the Collaborative Robot streaks and places them on the WASPLab automated track. The Kiestra inoculation unit has a biosafety cabinet, in which system-labeled plates are presented for the technologist to inoculate manually. This is referred to as a semiautomated mode and is equivalent to Copan’s Collaborative Robot. Once inoculated, the Kiestra system streaks the plates and processes them in the same manner as fully automated specimens. Both systems can create Gram stain slides, although staining and reading of smears are still performed manually.
While both systems have an automated track system, the Kiestra track is bidirectional, while the WASPLab track is unidirectional. The bidirectional track associated with the Kiestra TLA permits plates to be summoned to workbenches and subsequently returned to incubators directly from workbenches. Technologists at WASPLab workbenches can summon plates from incubators, assuming the benches are downstream of the incubators, but must manually place plates on the track upstream of the incubators for return. Both systems have the option of having workbenches that are separate from the automated track. This setup requires manual placement and retrieval of plates, but may be beneficial to labs with a smaller footprint.
Benefits of lab automation and future directions
Improved turnaround times
In traditional microbiology, there is significant variability in the inoculation efficiency between the microbiology staff members. The consistent inoculation of laboratory automation systems results in a higher number of isolated colonies, which are required for identification and susceptibility testing [5], [6]. In traditional microbiology, organisms often have to be subcultured to obtain isolated colonies for additional testing, which increases turnaround times. Additionally, staff inoculate several cultures before transporting plates to the incubator, which increases the time the plates are incubated in a suboptimal environment. With automated systems, inoculated plates are transported along the automated track directly to incubators, which provide ideal growth conditions. Moreover, with automation technologists read and interpret images, rather than actual plates, so plates remain in optimal incubation conditions, rather than sitting outside of the incubator for hours at a time.
Due to a combination of the factors listed above, several studies have shown a decrease in the turnaround times of 2–30 hours across multiple culture types on WASPLab and Kiestra [7], [8], [9], [10], [11]. However, automation alone is insufficient to improve turnaround times [12]. In addition to automation, workflow changes are required to achieve efficiencies. The majority of laboratories that have implemented automation have also implemented plate reading on the second and/or third shift rather than limiting plate reading to first shift as has traditionally been the case in clinical microbiology laboratories. This allows cultures to be read, as they are ready rather than batching cultures for a single shift.
Automated culture reading
Artificial intelligence (AI) software, PhenoMATRIX, is available for WASPLab. PhenoMATRIX performs automated reading, interpretation, and segregation of cultures into growth versus no growth categories [13], [14]. Results are grouped and presented to the technologist for final interpretation and resulting. The software is available for multiple culture types, but each version must be purchased separately. Multicenter studies using chromogenic agar for detection of MRSA and vancomycin-resistant Enterococcus (VRE) surveillance cultures demonstrated 100% sensitivity and 89.5%–96% specificity [13], [14]. AI software is not currently available for Kiestra but is in development. A proof-of-concept study demonstrated that the Kiestra OPTIS software is able to provide reliable quantitative urine colony counts [15]. In addition to current AI capabilities, there are plans to develop AI that can classify or preliminarily identify organisms based on their morphologies.
TLA for clinical microbiology is still in its early stages. As automation technologies continue to improve, the clinical microbiology laboratory will continue to optimize its operation and workflow. This will result in increased efficiencies that address the shortage of medical technologists, provide a consistent level of care in a healthcare environment that is increasingly focused on quality care, and will continue to improve turnaround times.
Methods for identification of microorganisms
Once an organism is recovered in culture, whether via traditional microbiology or via an automated system, it may be subjected to a downstream identification method. Such methods include biochemical testing, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), or DNA sequencing. DNA sequencing and MALDI-TOF MS can also directly identify a pathogen from a primary specimen.
DNA sequencing
DNA sequencing is the gold standard for microorganism identification. The 16S ribosomal RNA (rRNA) gene is the most common sequencing target for bacteria and is 1542 base pairs (bp) in length. Sequencing the first 500 bp often provides enough differentiation for identification purposes, but certain organism groups require sequencing of the full-length 16S rRNA gene for differentiation, while other genera are homologous across the entire gene and require sequencing of additional genes for differentiation. The additional gene(s) required depends on the genus, and examples include rpoB, recA, tuf, gyrA gyrB, and cpn60. Many clinical laboratories that perform 16S sequencing only sequence a portion of the 16S rRNA gene, while few sequence the entire 1542 bp. Most clinical laboratories do not perform sequencing of the supplemental genes. Once a sequence is obtained, it is compared with a public or private database for organism identification. Database selection is critical, as some databases are curated and routinely updated and others are static. For more information on selecting an appropriate database, refer to the Clinical and Laboratory Standards Institute (CLSI) MM18 document (Interpretive Criteria for Identification of Bacteria and Fungi by Targeted DNA Sequencing, second edition) [16].
Interpretive criteria are based on the aforementioned CLSI MM18 document [16]; ≥97% homology is required for genus-level identification of bacteria, while ≥99% homology is required for species-level identification. Less than 95% homology indicates an incomplete database or potentially a novel species. Although 16S rRNA sequencing is the most common, it does not provide sufficient differentiation for particular groups of bacteria, such as Escherichia coli and Shigella spp., which are essentially the same organism, and Bordetella spp. In such cases, sequencing of additional genes or biochemical reactions is required for complete identification.
For yeast and filamentous molds, the most common sequencing targets are the internal transcribed spacer regions (ITS), ITS1 and ITS2. ITS1 and ITS2 are variable regions located between the conserved rRNA genes. ITS2 alone is sufficient for discriminating multiple Candida spp. but not for other yeasts or molds. In addition to ITS1 and ITS2, the D1/D2 region of the 28S rRNA gene may be used. A consensus has not been reached in regard to cutoff values for genus- and species-level identification of fungi.
Although DNA sequencing is the gold standard, the methodology is technologically challenging, has a slow turnaround time, is not widely available, and is relatively expensive. Because of these drawbacks, it is not routinely used to identify microorganisms and is more commonly used as a backup or secondary method to other identification methods.
Biochemical/phenotypical methods
Biochemical reactions and phenotypic characteristics have historically been used to identify microorganisms. Initially, biochemical reactions were carried out and analyzed in separate reaction tubes. However, this was burdensome, and some of the reactions required a significant amount of time. Commercial vendors miniaturized biochemical testing, which allowed multiple biochemical reactions to be combined into a panel. Commercial biochemical panels are interpreted manually by the technologist or automatically by an instrument. Examples of manual panels include ThermoFisher’s Remel RapID series and bioMerieux’s API series (Fig. 55.3 ). Automated instruments include bioMerieux’s VITEK 2 (Fig. 55.4A and B ), BD’s Phoenix, and Beckman Coulter’s MicroScan. For panels that require manual interpretation, the technologist visually determines the reactions and enters the results into a web-based database. Both manual and automated systems compare the reactions to an organism database and provide an identification with a confidence score, which is reported as a percentage.
Figure 55.3.

A bioMerieux API panel, an example of a miniaturized biochemical identification panel.
Figure 55.4.
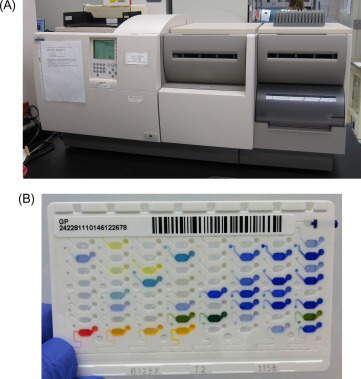
(A) VITEK 2 instrument and (B) VITEK 2 Gram-positive identification panel are shown. Each well on the identification panel represents a unique biochemical reaction.
Biochemical methods are inexpensive and easy to perform, but they are generally slow and have variable performance. Correct genus- and species-level identification rates for aerobic Gram-negative rods are 83% and 75%, respectively [17]. While biochemical methods perform relatively well for commonly isolated aerobic bacteria, they are not reliable for inert, slowly growing or infrequently isolated bacteria, including anaerobes. Correct genus-level identification rates range from 71% to 87% for anaerobes, while correct species-level identification rates are a mere 50% to 60% [18], [19], [20], [21]. Identification rates for infrequently isolated aerobic bacteria are even more dismal and have been reported to be 52% for genus level and 34% for species level [17]. During the past decade, biochemical methods are being replaced by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), which is discussed in the following section.
Mycobacteria should be cultured in a biosafety level 3 or BSL2+ (BSL2 plus BSL 3 practices) laboratory due to the potential aerosolization of Mycobacterium tuberculosis (MTB). Mycobacteria were traditionally identified by biochemical methods. However, this process required weeks for the slowly growing mycobacterial species, including MTB. Improvements in time to identification were made with the implementation of high-performance liquid chromatography (HPLC), DNA probes, MALDI-TOF MS, and polymerase chain reaction (PCR) for MTB complex. Biochemical methods and HPLC are no longer recommended due to their poor performance and slow turnaround times compared with newer methods [22]. Although DNA probes provide a same day turnaround time once growth is detected, they are labor-intensive and only available for four species: MTB complex, Mycobacterium avium complex, Mycobacterium kansasii, and Mycobacterium gordonae. In addition to routine bacteria, MALDI-TOF MS is also replacing traditional methods for identification of mycobacteria.
Yeasts have historically been identified using biochemical methods, which require up to 72 hours. The phenotypic methods commonly employed by clinical laboratories for yeast identification have variable accuracy, with most methods performing well for common species; however, accuracy may decrease with infrequently encountered isolates [23], [24], [25], [26], [27]. One of the notable drawbacks of biochemical methods for yeast testing is the misidentification of Candida auris [28]. C. auris has gained international attention due to isolates frequently demonstrating multidrug resistance. Molds, on the other hand, have historically been identified using macroscopic and microscopic morphologies. The surface color and texture of the mold colony, the color of the reverse of the colony, and the microscopic morphology using lactophenol cotton blue (LPCB) tape preparation are combined to identify the mold. LPCB tape preparation is performed by touching a piece of clear tape to the top of the mold colony and applying it to a microscope slide with a drop of LPCB dye, which stains the fungal elements and allows visualization. MALDI-TOF MS is replacing biochemical methods for yeast. MALDI-TOF MS has also been applied to mold identification. However, there are some drawbacks specific to molds, which have resulted in limited usage compared with other organism groups and are discussed in the following section.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
MALDI-TOF MS is a rapid and inexpensive method for identification of bacteria, acid-fast bacilli (Mycobacterium spp.), yeasts, and molds in the clinical microbiology laboratory. MALDI-TOF MS uses proteomic profiling to assign and identification, which primarily includes ribosomal proteins due to their relative abundance. Thus MALDI-TOF MS is the closest identification to the gold standard of 16S ribosomal DNA sequencing. MALDI-TOF MS measures proteins in the range of 2–20 kilodaltons. Bruker’s Biotyper system (Fig. 55.5A ) and bioMerieux’s VITEK MS (Fig. 55.5B) are the two commercially available MALDI-TOF MS instruments available in the United States for microorganism identification. The Bruker is a bench top model, while the VITEK MS is a larger floor model. Both systems have Food and Drug Administration (FDA)-cleared in vitro diagnostic (IVD) organism libraries as well as more comprehensive research use only (RUO) libraries. Although the majority of clinically encountered bacteria and yeast can be identified using the IVD libraries, many laboratories have validated RUO libraries for clinical use to provide complete coverage of organism identification. Mycobacteria and molds are included in the most recent VITEK MS IVD library v.3.0, while the Bruker has RUO mycobacterial and mold libraries that must be purchased separately from the bacterial library.
Figure 55.5.

(A) Bruker’s Biotyper system and (B) bioMerieux’s VITEK MS are the two commercially available matrix-assisted laser desorption/ionization time-of-flight mass spectrometry instruments available in the United States for microorganism identification.
For bacteria and yeasts, a thin layer of organism is applied to the target plate using a toothpick or inoculation loop and 1 µL of formic acid overlay may be added, depending on the organism group. The organism±formic acid is subsequently overlaid with matrix (α-cyano-4-hydroxycinnamic acid), allowed to dry, and subjected to MALDI-TOF analysis. The process is more complicated for mycobacteria and molds, which require extraction and inactivation prior to MALDI-TOF MS analysis. Of note, both commercially available laboratory automation systems are developing modules that will automate inoculation of MALDI-TOF targets. Once the organism’s mass spectrum is obtained, it is compared with the manufacturer’s library/database, and a confidence score is provided. The Bruker uses a logarithmic scale, where, per the manufacturer, a score ≥1.7 is sufficient for genus-level identification and a score of ≥2.0 is sufficient for species-level identification. Some laboratories have validated lower thresholds off-label for species-level identification. The VITEK MS system reports, on the other hand, identifications with a percent confidence score up to 99.9%.
MALDI-TOF MS has a turnaround time of approximately 40 minutes for identification of bacteria and yeasts [8], [29]. This is a significant improvement compared with 1.5–72 hours for traditional biochemical methods [8], [29]. In addition to a faster turnaround time, MALDI requires less organism mass. A single isolated colony is sufficient for MALDI-TOF MS analysis, while biochemical methods require sufficient colonies to create a 0.5–3 McFarland turbidity standard, depending on the organism group.
MALDI-TOF MS has demonstrated excellent performance for aerobic bacteria with genus- and species-level identification rates of 98.6% and 96.5% for Gram-positives and 98.5% and 96.8% for Gram-negatives [30]. One of the biggest improvements over biochemical methods is the identification of anaerobes. Genus- and species-level identification rates for anaerobes by MALDI-TOF MS have been reported to be 98.5% and 85.8%–97.4%, as compared with 71%–87% and 50%–60%, respectively, by biochemical methods [18], [19], [20], [21], [30]. Approximately 60% of College of American Pathologists (CAP)-accredited laboratories are using MALDI-TOF MS for the identification of bacteria (CAP DEX-B 2019 survey).
MALDI-TOF MS has a few limitations with regard to bacterial identification. It is unable to separate E. coli and Shigella spp., which are essentially the same organism. The technology also has challenges with discrimination and accurate detection of Streptococcus pneumoniae and Streptococcus mitis/oralis group due to the high degree of relatedness between the two organisms; however, this has improved with the release of updated libraries. MALDI-TOF MS typically does not identify or misidentifies select agents, such as Brucella spp., B. anthracis, Burkholderia pseudomallei, etc. even when they are present in the library being used [31]. Therefore, it is imperative that microbiology technologists are familiar with recognizing select agents and recognizing common misidentifications by MALDI-TOF MS. It is essential that technologists are aware of the limitations and maintain competency for recognizing patterns and questioning things that do not fit. For microorganisms not identified by MALDI-TOF, DNA sequencing should be used if identification is necessary.
A recent CAP proficiency testing survey (E-B 2019) revealed that 20% of laboratories are using MALDI-TOF MS for identification of mycobacteria. MALDI-TOF MS performs well for mycobacteria, although more hand-on time is required than bacteria and yeast. The Bruker extraction/inactivation method requires approximately 1 hours, while the VITEK MS extraction method requires 30–60 minutes. However, a recent study published a method that requires only 10 minutes, decreasing the hands-on time and turnaround time [32]. Species-level identification rates are 84.7%–100% [32], [33], [34].
Notable drawbacks of MALDI-TOF are that it cannot differentiate members of the MTB complex nor can it differentiate Mycobacterium abscessus complex to subspecies-level (M. abscessus subsp. abscessus, M. abscessus subsp. bolletii and M. abscessus subsp. massiliense). The former is significant because Mycobacterium bovis, a member of the MTB complex, is intrinsically resistant to pyrazinamide, a first-line drug for tuberculosis treatment. The latter is significant, because M. abscessus subsp. abscessus and M. abscessus subsp. bolletii have a functional erm gene, encoding macrolide resistance, whereas M. abscessus subsp. massilience has a nonfunctional erm gene. MALDI-TOF MS also cannot differentiate Mycobacterium chimaera from Mycobacterium intracellulare. While this is normally not a concern, sequencing should be performed if the infection is associated with cardiothoracic surgery, as there is currently a global outbreak of M. chimaera associated with heater-cool units [35].
Currently, 48% of CAP-accredited laboratories are using MALDI-TOF MS for identification of yeasts (CAP F-B 2019 survey). Several studies have demonstrated species-level identification rates of 83%–99% (reviewed in [36]), and the majority of yeasts require formic acid overlay or extraction with formic acid and acetonitrile for successful identification. MALDI-TOF MS identification of molds, on the other hand, has been less widespread. While 48% of laboratories are using MALDI-TOF MS for yeasts, only 5% are using it for molds (CAP F-B 2019 survey). There are no molds in the most recent FDA-cleared library (IVD Claim 4) for Bruker, and there are 79 species in the recent VITEK MS library (v3.0). The RUO libraries for both instruments, however, contain numerous species. Although libraries are available, there are two issues with identification of molds. Mold colonies are not homogeneous the way bacterial, mycobacterial, or yeast colonies are. Protein expression varies across mold colonies, with a subset of proteins being more highly expressed at the inner portion of the colony, whereas another subset of proteins may be more highly expressed at the outer edge of the colony, etc. Thus the sampling location has a significant impact on which proteins are present in the spectra. Moreover, in the case of the Bruker, the original mold library was created using molds that had been grown in broth rather than on plates. Although the broth method standardizes protein expression, molds are not grown in broth in clinical laboratories.
The difficulty in mold identification is demonstrated by publications reporting 78%–83.5% identification to genus level and 54%–79% identification to species level (reviewed in [36]). Some laboratories have created their own user-developed libraries, either for all molds or for a particular subset of molds. User-developed libraries have demonstrated species-level identification rates of 89%–98% [36]; however, many labs do not have the capacity to create their own library. A recent study evaluating the VITEK MS v3.0 library demonstrated 86.6% correct identification to the species level [37]. If only the species present in the library were included, 91% were correctly identified to species level. These are the most promising data using a commercially developed library thus far. MALDI-TOF MS identification of molds is an ongoing area of research.
Beyond colony identification
To date, the majority of studies evaluating MALDI-TOF MS have focused on identification of organisms growing on solid media; a smaller number of studies have evaluated MALDI-TOF MS analysis on positive blood culture broth. An initial extraction is required when testing positive blood cultures to eliminate interfering substances, including blood and broth. Time to identification ranges from 30 minutes to 2 hours, depending on the method used. Of note, this method applies to bacteria and yeasts, which grow well in routine blood culture bottles. Mycobacteria and molds are rarely recovered in routine blood culture bottles and require alternative methods for detection. Correct identification rates vary based on which extraction method is utilized but appear most optimized with the use of a commercially available Sepsityper kit (Bruker-Daltonics) or saponin [38], [39], [40], [41], [42]. The majority of studies have evaluated the Bruker Biotyper for this analysis. One drawback is that MALDI-TOF cannot accurately identify multiple species in polymicrobial blood cultures, though it frequently identifies the predominant organism from mixed cultures [43], [44], [45], [46]. The issue with polymicrobial specimens has also been reported with molecular methods [47], [48].
MALDI-TOF MS has been shown to correctly identify 84%–99% of Gram-negative bacteria from positive blood culture broth, with most studies reporting rates from 90% to 99% (reviewed in [49]). The rate varies from 65% to 96% for Gram-positive bacteria, with most studies reporting identification rates of 80% [49]. In regard to the identification of yeasts, one study showed concordance rates of 95.9% for Candida albicans and 86.5% for non-albicans Candida species [50].
Rather than using lysis or extraction methods, some labs have opted to test microcolonies or “scum” growth. Positive blood culture broth is inoculated to solid media and incubated, and MALDI-TOF MS analysis is performed after 2–6 hours of incubation. Although the turnaround time is longer than the extraction methods, this method is less labor-intensive and fits better into a laboratory workflow. Species-level identification rates have been reported to be in the range of 81.8%–95.5% [49]. Anaerobes and slow growing bacteria have lower identification rates, and polymicrobial cultures cannot be differentiated after 2–6 hours of growth.
Direct identification from urine via MALDI has also been investigated. One study used diafiltration and concentration to obtain bacterial identification in 2–3 hours. Detection limits were 5×104 to 106 colony forming units/mL [51]. A second study inoculated urine into broth media, incubated the broth for 3 hours, and checked the density. If the density was ≥0.3 on the McFarland turbidity scale, the broth was centrifuged, and the bacterial pellet was used to spot the MALDI target. The study demonstrated 96.5% sensitivity and 71.4% specificity [52]. Neither workflow is ideal for a clinical microbiology laboratory, so MALDI-TOF MS of urine cultures is not widely performed. In addition, urine cultures have a high rate of contamination and are often polymicrobial in nature, further limiting the utility of this method.
Point-of-care microbiology
Point-of-care (POC) testing generally refers to testing that is performed near the site of patient care (such as a physician office, clinic, or hospital unit) by nonlaboratory trained individuals. POC tests often have a rapid turnaround time, providing actionable results that facilitate immediate patient management decisions. This advantage is particularly pronounced in the emergency department and in cases where the traditional laboratory is off-site. A more expanded definition of POC testing may include testing performed in small laboratories that serve as a satellite laboratory for a main central laboratory. Centralization of laboratory testing, where a large centralized laboratory performs the majority of testing for hospitals and clinics within a region, has led to increased turnaround times for results when transport times are taken into account. Smaller satellite laboratories may be staffed by general laboratory technologists without specialized microbiology training. Testing performed in these types of laboratories may confer the turnaround time benefits of POC testing by eliminating or reducing transit times while offering an expanded menu of assays. In this context, many rapid sample-to-answer microbiology tests may be considered POC tests, such as syndromic multiplex molecular testing platforms. For remote clinics or hospitals, simple and rapid POC assays are essential for optimal patient management.
Most POC tests receive waived status by the United States FDA. Laboratories or clinics exclusively performing waived tests are required to obtain a Certificate of Waiver, which is issued by the Centers for Medicare and Medicaid Services under Clinical Laboratory and Improvement Amendments of 1988 (CLIA). Waived tests are considered low risk for errors, are performed using simple and straightforward protocols, require minimal instrument maintenance, and require minimal training for test interpretation. Although these assays are simple, appropriate quality control and user training is essential to preventing diagnostic errors.
POC microbiology testing has historically been limited to lateral flow immunoassays (LFAs). LFA tests are performed on a lateral flow strip and are often double-antibody sandwich assays. A capture antibody is immobilized on the membrane and the clinical specimen is flowed over the strip. The target antigen, if present, is bound by conjugate-labeled antibodies, such as colloidal gold-labeled antibodies, and the target antigen–antibody complex binds the immobilized capture antibody, producing a visible line. The sample flows further along the membrane where excess conjugate binds the control antibodies, producing a visible control line. For some assays, digital analyzers have been introduced to eliminate the subjective nature of visual interpretation of LFA assays. Examples of LFA POC tests in use include assays for the diagnosis of S. pyogenes [Group A Streptococcus (GAS)], infectious mononucleosis, influenza A/B viruses, respiratory syncytial virus (RSV), HIV-1 and HIV-2, and adenovirus.
Although LFA assays are rapid, inexpensive, and simple, these assays have variable clinical performance and often suffer from suboptimal sensitivity. Metaanalysis of the diagnostic accuracy of rapid antigen tests for GAS found a sensitivity of 86% and specificity of 95%, although the reported sensitivities in the included studies were highly variable, ranging from 44% to 98% [53]. Owing to the low sensitivity of these tests, negative rapid GAS tests are reflexed to culture for confirmation testing.
Rapid influenza diagnostic tests (RIDTs) are immunoassays that detect influenza viral antigens. Several metaanalyses have evaluated the sensitivity of RIDTs, reporting a pooled sensitivity of 51%–68% and lower sensitivity for adult patients compared with pediatric patients due to the relatively lower viral load [54], [55], [56]. When seasonal influenza activity is high, the negative predictive value of RIDTs is low, due to the substantial number of falsely negative results. Because of these poor performance characteristics, the Centers for Disease Control and Prevention recommends that all negative samples be tested by a more sensitive assay, although adherence to this recommendation is likely low. Further, antiviral therapy is commonly not withheld based on a negative RIDT alone, particularly if there is a high clinical suspicion for influenza illness. Conversely, when influenza activity is low to moderate, the positive predictive value of RIDTs is poor. Due to low sensitivity for the detection of influenza viruses, especially novel or emerging influenza viruses, in 2017 the FDA reclassified RIDTs from class I to class II devices, which requires these devices meet minimum criteria for sensitivity and specificity [57].
Development of POC molecular techniques has the potential to meet the clinical need for highly sensitive, rapid POC microbiology diagnostics. The first POC molecular test to be granted a CLIA waiver was the Alere i influenza A & B test (Alere Inc.), which received waived status in 2015. Subsequently, Abbott purchased Alere and renamed the Alere i to the infectious disease (ID) NOW platform. It is CLIA-waived for influenza A/B virus, RSV, and GAS testing, with results reported in 2–15 minutes. The ID NOW system uses strand displacement amplification technology, where target nucleic acid is amplified in an isothermal reaction using a nicking endonuclease and a strand-displacing DNA polymerase in addition to target primers. Fluorescently labeled molecular beacon probes are used to monitor amplification in real time. Three additional molecular platforms have subsequently received CLIA-waived status. The Cobas Liat (Roche Diagnostics) is CLIA-waived for influenza A/B virus, RSV, and GAS testing, with results in 15–20 minutes. Amplification occurs using real-time PCR technology and results are interpreted based on real-time detection of fluorescently labeled probes. The GeneXpert Xpress platform (Cepheid) is CLIA-waived for influenza/RSV and GAS testing, with results in 18–30 minutes. The Xpress system uses real-time PCR and fluorescently labeled probes for the amplification and detection of target nucleic acid. The Accula system (Mesa Biotech) is CLIA-waived for influenza and RSV detection, with results in 30 minutes. The Accula system uses reverse-transcription PCR technology to amplify target RNA, which is then complexed to oligonucleotide probes conjugated to dye-labeled microspheres. Using technology similar LFAs, the amplicon-microsphere complex is flowed across a detection membrane where complementary oligonucleotide capture probes are bound. Hybridization of capture probes to the amplicon-microsphere complex is visualized on the membrane as a colored line. Additional molecular POC assays are currently under development and in clinical trials.
Molecular POC tests have demonstrated clinical performance characteristics similar to molecular testing performed in the clinical microbiology laboratory. GAS molecular POC assays have a reported sensitivity of 99%–100% and specificity of 91%–97% when compared with culture [58], [59]. POC molecular assays for RSV and influenza A/B viruses also demonstrate high sensitivity. Reported sensitivity of the Liat is >97% for influenza A virus, influenza B virus, and RSV targets [60], [61], [62], [63]. Similarly, evaluation of the GeneXpert influenza A/B assay demonstrated high sensitivity (>96%) and specificity (>97%) for both influenza A and influenza B viruses [63], [64]. Studies of the Alere i have reported lower sensitivity for all targets when compared with the Liat, with overall influenza A/B sensitivity as low as 64% in one study, although the majority of studies report sensitivities >90% [60], [63], [64], [65], [66]. Metaanalysis of the reported diagnostic accuracy of influenza POC nucleic acid amplification tests found a pooled sensitivity 92% for influenza A virus and 95% for influenza B virus [67]. This was a marked increase compared with the pooled sensitivity of newer generation influenza antigen immunoassays, which had a pooled sensitivity of 80% for influenza A virus and 77% for influenza B virus.
Risk of contamination is a significant concern for molecular POC assays, as amplicon contamination can lead to false-positives and pseudooutbreaks. Because these tests are performed by nonlaboratory staff that are inexperienced in performing highly sensitivity laboratory assays, proper training and quality control, such as frequent testing of the setup area for target contamination, are essential. In addition, improved test utilization education and stewardship strategies are a focus across all areas of laboratory medicine. As increasingly expensive and sensitive tests are introduced, education on which patients are appropriate to test will be essential. For example, overdiagnosis of pediatric patients colonized with GAS is a concern, as up to 26% of healthy children may be colonized with GAS [68], [69]. Testing only patients with clinically significant signs and symptoms of GAS is imperative to prevention of overdiagnosis.
A drawback of molecular POC testing is increased cost per test compared with traditional antigen tests. Benefits that may offset cost increases include the potential for improved antibiotic and antiviral use. In the cases of GAS, patients that tested antigen-negative but culture-positive would potentially receive results and antibiotic therapy 1–2 days sooner if molecular testing was used. In addition, molecular GAS POS testing does not require reflex of negative results to culture, reducing the workflow burden in the clinical microbiology laboratory and simplifying the overall operational process. In the cases of influenza A/B testing, the increased negative predictive value of molecular testing over antigen testing during influenza virus season may facilitate appropriate use of antiviral medications and judicious use of antibiotics.
Molecular POC platforms will allow for expansion of the POC microbiology test menu. Numerous assays are currently in development and many are currently CE-marked for use in European laboratories. Promising new assays include HIV-1 viral load testing and diagnostic assays for sexually transmitted infections. In 2019 the first assay for molecular POC STI detection received FDA 510(k) clearance. The binx io platform is a rapid molecular assay for the diagnosis of Chlamydia trachomatis and/or Neisseria gonorrhoeae from clinician or self-collected vaginal swab specimens, with results in less than 30 minutes.
Introduction of molecular microbiology into the POC testing environment represents a paradigm shift and has led to increased versatility, scope, and availability of POC rapid testing. POC molecular testing is also accompanied by an increase in cost per test, which may limit uptaking these assays, especially if viewed independent of changes to cost utilization of other resources, such as administration of antibiotics.
Syndromic-based multiplex molecular testing
Introduction
Multiplex testing was developed for the detection of pathogens associated with clinical syndromes of the bloodstream, respiratory, gastrointestinal, and central nervous systems (CNS). Since their introduction, these panels have become increasingly common with improved workflows and decreased turnaround times. Syndromic-based testing has streamlined ordering workflows, as providers need to select only one test order to analyze the most common organisms causing a specific syndrome. Moreover, some platforms offer flexibility in ordering, allowing the provider to order more specific targets based on clinical presentation, which can help limit unnecessary testing and costs. Molecular syndromic testing is more rapid than conventional microbiology methods, reducing time to diagnosis and, if the result is acted upon, potentially reducing time to appropriate therapy.
Early multiplex testing platforms, such as the Luminex xTAG, had cumbersome workflows and testing was often batched and performed once per day or once per shift. Subsequently, sample-to-answer platforms were developed and have replaced many of the older platforms. Sample-to-answer platforms require minimal set time, leading to simplified workflow as compared with traditional microbiology methods. A major limitation to adoption of multiplex testing is cost, as these panels incur an increased cost to both the laboratory as well as the patient compared with conventional methods. Institutional savings outside the clinical laboratory may be observed if utilization of these panels results in reduced antibiotic use, decreased length of stay, and promotion of appropriate infection prevention and control isolation precautions. In order to maximize institutional and patient benefits, multiplex testing should optimally be performed 24 hours per day and 7 days per week. In addition, test results are most optimally acted upon when accompanied by an antimicrobial stewardship intervention, either via direct contact with a member of an institutional antimicrobial stewardship team or via laboratory comments appended to results. Finally, test utilization strategies specific to each panel and patient population should be adopted to minimize inappropriate costly testing and target these assays to patients who will see the most benefit. Antimicrobial stewardship and test utilization opportunities specific to each syndromic panel are discussed below.
Currently, sample-to-answer multiplex testing platforms are commercially available via several vendors, including BioFire Diagnostics, the Luminex Corporation, and GenMark Diagnostics. GenMark panels are offered on the ePlex system, with a throughput of up to 96 samples within 8 hours. Following an amplification step, target DNA is hybridized to a signal probe, which reacts with a capture probe attached to a gold electrode. Application of voltage leads to electrochemical detection of the target DNA/signal probe complex. Current panels available on the ePlex instrument include respiratory pathogen and blood culture identification (BCID) panels. BioFire panels are offered on the FilmArray system. The FilmArray 2.0 is a single-bay instrument that is scalable by connecting up to eight instruments to a single processing computer. The FilmArray Torch is a fully integrated system with up to 12 modules in a tower configuration. In the FilmArray cartridge, nucleic acid is extracted and purified, followed by a nested PCR reaction and then a single-plex reaction for each target. Target detection is determined by endpoint melting curve analysis. BioFire currently has the largest menus of available panels, with testing available for infections of the upper respiratory, lower respiratory, gastrointestinal, and CNSs, as well as BCID. Luminex panels are performed on the Verigene system, consisting of a Verigene Processor and a Verigene reader. Specimen processing and target detection is performed on the Verigene Processor using a microarray format and gold nanoparticle probe-based technology. The test cartridge is then read by the Verigene reader, which offers scalable reading. Syndromic panels available on the Verigene system include respiratory, gastrointestinal, and BCID panels.
Respiratory panels
The differential diagnosis for upper respiratory infections is often broad, with nonspecific clinical signs and symptoms. Most illness is cause by viral infections, which do not have targeted individual FDA-approved/cleared assays. Multiplex molecular testing for respiratory pathogens was first approved in 2008. Most panels target common respiratory viruses, with some including atypical bacterial targets (e.g., Chlamydophila pneumoniae, Mycoplasma pneumoniae, and Bordetella species). NP swabs are the only approved specimen type for these panels, although many laboratories have performed validation studies to allow for testing of lower respiratory specimens such as bronchoalveolar fluid. Several multiplex panels designed to detect respiratory pathogens have been FDA-cleared, including more traditional assays such as Luminex xTAG Respiratory Viral Panel (RVP; Luminex Corporation), the Luminex xTAG RVP Fast (Luminex Corporation), and the Luminex xTAG Respiratory Pathogen Panel (RPP; Luminex Corporation); newer sample-to-answer assays such as FilmArray Respiratory Panel (RP; BioFire Diagnostics) and RP2; Verigene Respiratory Pathogens Flex test (Luminex Corporation); and ePlex RPP (GenMark Diagnostics). Panels include 8–20 respiratory pathogen targets and turnaround time ranges from 1 to 8 hours.
Clinical evaluations of syndromic respiratory panels have demonstrated overall sensitivities >85% and specificities >99% [70], [71], [72], [73]. However, suboptimal performance characteristics have been described for specific targets on individual panels. In one study, the FilmArray RP exhibited a sensitivity of 57% for adenovirus and 77% for influenza B virus when compared with a reference standard [70]. A redesign of the FilmArray RP, the RP2, has improved sensitivity for both adenovirus and influenza B virus [73]. The xTAG RVP Fast demonstrated <50% sensitivity for the detection of influenza B virus [70], [74]. The xTAG-RPP demonstrated lower sensitivity for human coronaviruses OC43 (67%) and HKU1 (67%) than other assays [72]. Low sensitivity for viral pathogens with treatment options, such as influenza B virus, may negatively affect patient outcome; for this reason, a higher sensitivity assay should be used for the primary diagnosis of these viruses. For viruses not historically detected by traditional testing and for which no targeted therapy is available (such as coronaviruses), the impact of missed diagnosis is unclear. Nonetheless, many of these older assays have been supplanted by newer versions of sample-to-answer assays with improved overall sensitivity.
Implementation of multiplex respiratory testing has demonstrated clinical and financial benefits. Studies have documented a decreased mean time to diagnosis compared with conventional testing [75], [76], [77]. In addition, because multiplex panels include pathogens for which targeted testing is not routinely available (e.g., coronaviruses), use of multiplex panels increased the overall number of patients with a microbiological diagnosis [76]. Additional benefits include decreased use of antibiotics in viral illness, decreased admission rates, and decreased length of stay [75], [76]. Although clinical benefits are promising, cost of implementation of multiplex panels is a significant concern. Mahony et al. [78] performed a cost evaluation of the xTAG RVP assay compared with conventional methods and determined that multiplex testing was cost-effective if the prevalence of respiratory viral illness was >11%. Availability and turnaround time of targeted assays in addition to the specific patient population (e.g., immunocompromised, pediatric, etc.) must be considered when evaluating the utility of multiplex respiratory pathogen tests.
The FilmArray Pneumonia (PN) panel received FDA approval/clearance in 2018. The PN panel includes targets for eight respiratory viruses, 18 bacteria, and seven antimicrobial resistance markers. This panel is approved for bronchoalveolar lavage (BAL), mini-BAL, sputum, and endotracheal aspirates. For 15 of the 18 bacterial targets, the target is reported as a semiquantitative result, which allows for interpretation of the results in a similar manner as traditional quantitative culture. Positive targets are reported within a 1 log bin, ranging from <104 to ≥107 copies/mL. At the time of this writing, no peer-reviewed studies have been published on the performance of the PN panel.
Gastroenteritis panels
Traditional diagnostic testing for infectious diarrhea has included bacterial culture, antigen detection, microscopy, and targeted molecular tests. These assays require that clinicians select the appropriate test and misordering may lead to missed diagnoses. In addition, traditional testing for gastroenteritis (GI) pathogens in the microbiology laboratory is time-consuming, often requiring multiple days, and involves complicated workflows. Detection of some pathogens, such as Giardia lamblia, may require collection of multiple samples for maximum sensitivity. Advantages of syndromic panel testing for the diagnosis of diarrheal illness include decreased turnaround time, increased sensitivity, and, in some cases, decreased cost. Three multiplex assays are currently FDA-approved/cleared for the detection of enteric pathogens: Luminex xTAG Gastrointestinal Pathogen Panel (GPP; Luminex Corporation), FilmArray Gastrointestinal Panel (GIP: BioFire Diagnostics), and Verigene Enteric Pathogens Panel (Luminex Corporation). These multiplex tests detect 9–22 targets and on-instrument time is 1–5 hours. All panels detect the most common bacterial pathogens associated with GI (Campylobacter spp., Salmonella spp., Shigella spp., Vibrio spp., Yersinia enterocolitica, and Shiga toxin). The Luminex GPP and FilmArray GIP additionally detect enterotoxigenic E. coli, E. coli O:157, and C. difficile. The FilmArray GIP is the only assay that detects enteropathogenic and enteroaggregative E. coli as well as Plesiomonas shigelloides. No multiplex GI panel detects Aeromonas species, an additional possible bacterial cause of infectious diarrhea. Of the viral causes of infectious diarrhea, all panels detect norovirus GI/II and rotavirus A. The Luminex GPP also detects adenovirus 40/41, while the FilmArray GIP also detects adenovirus 40/41, astrovirus, and sapovirus. Finally, the Luminex GPP and FilmArray GIP also include parasitic species, with the GPP detecting Cryptosporidium species, Entamoeba histolytica, and G. lamblia and the GIP detecting Cryptosporidium species, E, histolytica, G. lamblia, and Cyclospora cayetanensis. Stool submitted in Cary-Blair transport media is the acceptable specimen for all panels.
Multiplex panels offer significant sensitivity increases over conventional testing methods. Across multiple studies, all panels detect an enteric pathogen in 20%–50% of specimens, compared with detection of a pathogen in 8%–18% of samples tested by traditional methods [79], [80], [81], [82]. Increased overall detection is likely due to increased sensitivity for specific targets as well as increased number of targets tested. The most commonly detected organisms are C. difficile, enteropathogenic E. coli, Salmonella species, norovirus, rotavirus, sapovirus, and Cryptosporidium species [79], [80], [81], [82]. Sensitivity of all targets is generally >90%, although reduced sensitivities have been reported for Salmonella and Y. enterocolitica for the Luminex GPP and Campylobacter, Salmonella, and rotavirus for the Verigene EP [81], [83], [84]. Evaluation of multiplex molecular GI panels has repeatedly demonstrated that detection of >1 target is more frequent than previously appreciated. Codetections were observed in 16% of specimens tested by the BioFire GI Panel [79]. Among specimens with >1 targets detected, enteroaggregative and enteropathogenic E. coli were commonly detected [79], [80], [81].
Implementation of multiplex testing for the detection of GI pathogens has been associated with financial, clinical, and infection control benefits. In addition to increases in sensitivity and detection of coinfections, results are available in <6 hours when GI panel testing is performed, in contract with culture-based methods which require several days before a result is available. Although multiplex testing is associated with increased costs to the clinical laboratory when compared with traditional testing methods, in hospitalized patients, these costs may be offset by reduced overall institutional costs [85]. Improvement in utilization of patient isolation precautions associated with implementation of GI panel testing, including both increasing numbers of patients in appropriate isolation as well as discontinuing isolation precautions for patients with negative GI panel results, has important cost, patient satisfaction, and infection control implications [85], [86]. Consideration should be given to test utilization and interpretation strategies when implementing syndromic GI testing for optimal cost and clinical utility.
Blood culture identification panels
Early antimicrobial administration is an important predictor of morbidity and mortality associated with sepsis. Diagnosis of bacteremia and fungemia is predicated on appropriate collection of blood cultures, with at least two sets of aerobic and anaerobic blood culture bottles filled with 10 mL of blood per bottle or 5 mL of blood per bottle for pediatric bottles. Blood cultures are incubated for five days on a continuously monitored blood culture instrument. When a bottle signals positive, a Gram stain is performed and the results are immediately called to the clinical care team. The blood culture broth is then inoculated to solid media and organism identification and antimicrobial susceptibility testing is performed on culture growth, requiring an additional 18–48 hours. From the time of initial positivity, workup of positive blood culture bottles requires 48–72 hours before the report is finalized. Multiplex molecular testing of positive blood culture bottles may decrease both the time to organism identification and time to preliminary antimicrobial susceptibility results.
Currently, several assays have received FDA approval/clearance for testing positive blood culture bottles. The FilmArray BCID panel tests for 19 bacterial targets (Gram-positive and Gram-negative), five yeast targets, and four resistance genes. The Luminex Verigene assay is divided into two panels based on Gram stain results. The Gram-positive blood culture (BC-GP) panel tests for 12 Gram-positive bacterial targets and three resistance genes. The Gram-negative blood culture (BC-GN) panel contains eight Gram-negative bacterial targets and six resistance markers. The GenMark ePlex system offers three panels. The Gram-positive panel contains 20 bacterial targets and four resistances gene; the Gram-negative panel tests for 21 bacterial targets and six resistance genes; and the fungal panel tests for 15 fungal targets. Finally, the Accelerate Pheno system (Accelerate Diagnostics) uses gel electrofiltration and fluorescence in situ hybridization for the identification of six Gram-positive bacteria, eight Gram-negative bacteria, or two yeast targets within 90 minutes and subsequent phenotypic susceptibility testing within 7 hours. The Accelerate Pheno system is the only multiplex system that offers phenotypic susceptibility testing.
Performance characteristics of multiplex molecular testing have been evaluated across multiple studies. Implementation of molecular testing of positive blood culture bottles reduced time to organism identification by 20–30 hours [87], [88]. In monomicrobial positive blood culture bottles, an organism was correctly identified in 90%–100% of bottles [48], [87], [88], [89]. In addition, select resistance genes have been incorporated into these panels for a preliminary indication of antimicrobial resistance, including mecA detection for the preliminary identification of MRSA species, vanA/B detection for the preliminary identification of VRE species, bla CTX-M, the most commonly detected extended spectrum beta-lactamase in Enterobacterales, and the five most common carbapenemase genes detected in the United States. Assays from each manufacturer have different combinations of genetic resistance markers. Assessment of performance characteristics of the susceptibility markers on each panel have shown high rates of agreement between assay detection and conventional testing and/or sequencing [47], [48], [89].
False-positive results have been reported using the FilmArray BCID, possibly due to microbial DNA contamination of blood culture broth. P. aeruginosa DNA has been detected in bioMerieux BacT/Alert standard anaerobic bottles when tested by the FilmArray BCID [87]. Moreover, false identification of Candida parapsilosis and Proteus species has been reported using BD Bactec blood culture bottles when testing by the FilmArray BCID. The Verigene BC-GN and BC-GP assays, on the other hand, have not been affected by contaminating DNA in blood culture bottles, because the assays do not include an amplification step. Due to the possibility of false-positive target detection, results from molecular testing should be correlated with the bottle Gram stain and discordant results warrant further testing and investigation.
Numerous studies have evaluated the clinical impact of implementation of rapid BCID. Differences in study population, individualized institutional practices, and study design lead to differences in outcome metrics across studies. Decreased time to appropriate antibiotic therapy was observed in most studies following implementation of rapid blood culture panels [90], [91], [92], [93], [94], [95], [96]. In addition, some but not all studies have demonstrated decreased length of hospital stay and decreased overall institution costs [90], [94], [96]. No mortality benefit was noted following implementation in most clinical studies; however, a few studies reported a statistically significant reduction in 30-day mortality rate [95], [96].
Maximum impact of these panels is appreciated when results are coupled with antibiotic stewardship interventions, providing real-time guidance to clinicians to promote optimization of antibiotic choice and dose [97], [98]. Antibiotic stewardship interventions may include real-time communication of BCID results from the microbiology laboratory to a member to the antimicrobial stewardship team (such as an ID pharmacist or ID physician). In such interventions, real-time antibiotic decision support is then provided to the primary clinical team by an ID expert. While real-time in-person interventions are likely to result in optimal antibiotic therapy, institutions may not have sufficient resources to implement fully these labor-intensive interventions. Alternatively, laboratories may release BCID testing results with evidence-based antibiotic stewardship recommendations in the form of comments appended to test results. Such comments should be carefully crafted in consultation with ID specialists. Although BCID reporting comments provide decision support and are more easily implemented compared with real-time expert advice, comments do not provide individualized guidance and are easily overlooked by physicians. Ultimately, improved communication between primary clinical providers, antibiotic experts, and the clinical microbiology laboratory is essential for optimal utilization of BCID panel results and improved patient care.
Meningitis and encephalitis panel
Infections of the CNS are associated with significant morbidity and mortality. Establishing a specific diagnosis is integral to administering appropriate therapy, as the etiologic agents of meningitis and encephalitis vary. Clinical presentation may be nonspecific, with many patients experiencing headache, alternated mental status, and potentially nuchal rigidity. WBC count and differential in cerebrospinal fluid, in addition to protein and glucose concentrations, may be indicative of the infecting agent. In the cases of bacterial meningitis, WBC counts are typically elevated with a neutrophilic predominance. Moreover, CSF protein concentrations are usually elevated and glucose concentrations are usually below normal limits. In the cases of viral meningitis, WBC counts are also elevated, with a lymphocytic predominance, although neutrophils may be present early in infection. Protein concentration is normal to elevated and glucose concentrations are generally normal. In the cases of fungal or tubercular CNS infections, WBC counts are elevated with lymphocytosis, protein concentrations are normal to elevated, and glucose concentration is normal to low. Traditional microbiologic testing of CSF is necessary to determine the infecting agent. Bacterial meningitis is generally diagnosed by Gram stain and bacterial culture. Viral meningitis requires molecular testing of CSF for the diagnosis of enterovirus and herpes simplex virus 1 and 2 (HSV-1 and HSV-2). In some cases of viral CNS infections, such as in cases of vector-borne encephalitis, serological testing may be warranted. Tubercular and fungal CNS infections are diagnosed by molecular and culture-based methods and, in the case of cryptococcal meningitis, by antigen detection in the CSF. Molecular detection of CNS pathogens may allow for a more rapid diagnosis, especially in the cases of bacterial meningitis. In addition, molecular testing is less likely to be affected by prior antimicrobial therapy that may affect culture sensitivity.
At this time, only one panel has received FDA approval/clearance for the diagnosis of CNS infections, the FilmArray Meningitis/Encephalitis (ME) panel (BioFire Diagnostics). This panel tests for 14 pathogens, including enterovirus, human parechovirus, HSV-1 and -2, cytomegalovirus, human herpesvirus 6, varicella zoster virus, E. coli K1, Haemophilus influenzae, Listeria monocytogenes, N. meningitidis, Streptococcus agalactiae, S. pneumoniae, and Cryptococcus neoformans/gattii. Several studies have evaluated the performance of the ME Panel and found high overall positive and negative agreement rates. In a large prospective evaluation of CSF specimens, the authors found 84.4% and >99.9% positive and negative agreement between the FilmArray ME Panel and conventional microbiology testing [99]. In a smaller study of pediatric patients, authors found 92.5% positive agreement and 96.2% negative agreement [100].
False-positive results from these panels are a matter of concern and prevention of contamination during specimen collection and handling is integral to appropriate utilization of the FilmArray ME Panel. Several studies have documented concerning rates of S. pneumoniae false-positive detection [99], [101]. For these reasons, many laboratories have placed testing restrictions on these panels. In some cases, a laboratory director may opt to evaluate CSF parameters prior to performing the FilmArray ME Panel. Alternatively, some laboratory directors may evaluate all positive results prior to release to clinicians. False-negative results are also concerning. Cases of false-negative results for cryptococcal meningitis have been documented when compared with cryptococcal culture results [101], [102]. Finally, some studies have reported false-negative HSV-1 and HSV-2 results, possibly due to viral burden below the panel limit of detection in Refs. [100] and [101].
Additional studies are necessary to determine the clinical and economic benefits of syndromic panel testing for the diagnosis of meningitis and encephalitis. Pre-/postintervention studies evaluating the impact of FilmArray ME Panel testing on length of stay and duration of antibiotic treatment have shown variable results, with reduced length of stay documented in one study but not observed in another [103], [104]. Similar to outcome studies associated with other syndromic panels, it is likely that antibiotic stewardship intervention at the time of result reporting will be helpful in realizing the maximum impact of these results.
Conclusion
The clinical microbiology laboratory has undergone a period of rapid growth and evolution during the past 10–15 years. Significant strides in advancement continue to be made as more rapid and automated technologies obtain FDA-clearance.
References
- 1.Miller J.M., Binnicker M.J., Campbell S., Carroll K.C., Chapin K.C., Gilligan P.H. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin. Infect. Dis. 2018;67:e1–e94. doi: 10.1093/cid/ciy381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller J.M., Miller S.A. A Guide to Specimen Management in Clinical Microbiology. third ed. ASM Press; 2017. [Google Scholar]
- 3.Dempsey C., Skoglund E., Muldrew K.L., Garey K.W. Economic health care costs of blood culture contamination: a systematic review. Am. J. Infect. Control. 2019;47:963–967. doi: 10.1016/j.ajic.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Iversen J., Stendal G., Gerdes C.M., Meyer C.H., Andersen C.O., Frimodt-Moller N. Comparative evaluation of inoculation of urine samples with the Copan WASP and BD Kiestra InoqulA instruments. J. Clin. Microbiol. 2016;54:328–332. doi: 10.1128/JCM.01718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croxatto A., Dijkstra K., Prod’hom G., Greub G. Comparison of inoculation with the InoqulA and WASP automated systems with manual inoculation. J. Clin. Microbiol. 2015;53:2298–2307. doi: 10.1128/JCM.03076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quiblier C., Jetter M., Rominski M., Mouttet F., Bottger E.C., Keller P.M. Performance of Copan WASP for routine urine microbiology. J. Clin. Microbiol. 2016;54:585–592. doi: 10.1128/JCM.02577-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherkaoui A., Renzi G., Vuilleumier N., Schrenzel J. Copan WASPLab automation significantly reduces incubation times and allows earlier culture readings. Clin. Microbiol. Infect. 2019;25:1430.e5–1430.e12. doi: 10.1016/j.cmi.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Theparee T., Das S., Thomson R.B., Jr. Total laboratory automation and matrix-assisted laser desorption ionization-time of flight mass spectrometry improve turnaround times in the clinical microbiology laboratory: a retrospective analysis. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01242-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froment P., Marchandin H., Vande Perre P., Lamy B. Automated versus manual sample inoculations in routine clinical microbiology: a performance evaluation of the fully automated InoqulA instrument. J. Clin. Microbiol. 2014;52:796–802. doi: 10.1128/JCM.02341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham M., Tilson L., Streitberg R., Hamblin J., Korman T.M. Improved standardization and potential for shortened time to results with BD Kiestra total laboratory automation of early urine cultures: a prospective comparison with manual processing. Diagn. Microbiol. Infect. Dis. 2016;86:1–4. doi: 10.1016/j.diagmicrobio.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Mutters N.T., Hodiamont C.J., de Jong M.D., Overmeijer H.P., van den Boogaard M., Visser C.E. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann. Lab. Med. 2014;34:111–117. doi: 10.3343/alm.2014.34.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarbrough M.L., Lainhart W., McMullen A.R., Anderson N.W., Burnham C.D. Impact of total laboratory automation on workflow and specimen processing time for culture of urine specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:2405–2411. doi: 10.1007/s10096-018-3391-7. [DOI] [PubMed] [Google Scholar]
- 13.Faron M.L., Buchan B.W., Coon C., Liebregts T., van Bree A., Jansz A.R. Automatic digital analysis of chromogenic media for vancomycin-resistant-enterococcus screens using Copan WASPLab. J. Clin. Microbiol. 2016;54:2464–2469. doi: 10.1128/JCM.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faron M.L., Buchan B.W., Vismara C., Lacchini C., Bielli A., Gesu G. Automated scoring of chromogenic media for detection of methicillin-resistant Staphylococcus aureus by use of WASPLab image analysis software. J. Clin. Microbiol. 2016;54:620–624. doi: 10.1128/JCM.02778-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croxatto A., Marcelpoil R., Orny C., Morel D., Prod’hom G., Greub G. Towards automated detection, semi-quantification and identification of microbial growth in clinical bacteriology: a proof of concept. Biomed. J. 2017;40:317–328. doi: 10.1016/j.bj.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. Clinical Laboratory and Standards Institute (CLSI) Guideline MM18: Interpretive Criteria for Identification of Bacteria and Fungi byTargeted DNA Sequencing, second ed., 2018, Clinical and Laboratory Standards Institute, PA, USA.
- 17.Saffert R.T., Cunningham S.A., Ihde S.M., Jobe K.E., Mandrekar J., Patel R. Comparison of Bruker biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 2011;49:887–892. doi: 10.1128/JCM.01890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barba M.J., Fernandez A., Oviano M., Fernandez B., Velasco D., Bou G. Evaluation of MALDI-TOF mass spectrometry for identification of anaerobic bacteria. Anaerobe. 2014;30:126–128. doi: 10.1016/j.anaerobe.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Blairon L., Maza M.L., Wybo I., Pierard D., Dediste A., Vandenberg O. Vitek 2 ANC card versus BBL crystal anaerobe and RapID ANA II for identification of clinical anaerobic bacteria. Anaerobe. 2010;16:355–361. doi: 10.1016/j.anaerobe.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Lee E.H., Degener J.E., Welling G.W., Veloo A.C. Evaluation of the Vitek 2 ANC card for identification of clinical isolates of anaerobic bacteria. J. Clin. Microbiol. 2011;49:1745–1749. doi: 10.1128/JCM.02166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Sanchez B., Alcala L., Marin M., Ruiz A., Alonso E., Bouza E. Evaluation of MALDI-TOF MS (Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry) for routine identification of anaerobic bacteria. Anaerobe. 2016;42:101–107. doi: 10.1016/j.anaerobe.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 22.CLSI. Clinical and Laboratory Standards Institute (CLSI) Guideline M48: Laboratory Detection and Identification of Mycobacteria, second ed., 2018, Clinical and Laboratory Standards Institute, PA, USA.
- 23.Hata D.J., Hall L., Fothergill A.W., Larone D.H., Wengenack N.L. Multicenter evaluation of the new VITEK 2 advanced colorimetric yeast identification card. J. Clin. Microbiol. 2007;45:1087–1092. doi: 10.1128/JCM.01754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meletiadis J., Arabatzis M., Bompola M., Tsiveriotis K., Hini S., Petinaki E. Comparative evaluation of three commercial identification systems using common and rare bloodstream yeast isolates. J. Clin. Microbiol. 2011;49:2722–2727. doi: 10.1128/JCM.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanheira M., Woosley L.N., Diekema D.J., Jones R.N., Pfaller M.A. Candida guilliermondii and other species of candida misidentified as Candida famata: assessment by vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 2013;51:117–124. doi: 10.1128/JCM.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijgen S., Nys S., Naesens R., Magerman K., Boel A., Cartuyvels R. Comparison of Vitek identification and antifungal susceptibility testing methods to DNA sequencing and Sensititre YeastOne antifungal testing. Med. Mycol. 2011;49:107–110. doi: 10.3109/13693786.2010.494255. [DOI] [PubMed] [Google Scholar]
- 27.Posteraro B., Ruggeri A., De Carolis E., Torelli R., Vella A., De Maio F. Comparative evaluation of BD Phoenix and vitek 2 systems for species identification of common and uncommon pathogenic yeasts. J. Clin. Microbiol. 2013;51:3841–3845. doi: 10.1128/JCM.01581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spivak E.S., Hanson K.E. Candida auris: an emerging fungal pathogen. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhiman N., Hall L., Wohlfiel S.L., Buckwalter S.P., Wengenack N.L. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 2011;49:1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson D.A., Young S., Timm K., Novak-Weekley S., Marlowe E.M., Madisen N. Multicenter evaluation of the Bruker MALDI biotyper CA system for the identification of clinically important bacteria and yeasts. Am. J. Clin. Pathol. 2017;147:623–631. doi: 10.1093/ajcp/aqw225. [DOI] [PubMed] [Google Scholar]
- 31.Rudrik J.T., Soehnlen M.K., Perry M.J., Sullivan M.M., Reiter-Kintz W., Lee P.A. Safety and accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of highly pathogenic organisms. J. Clin. Microbiol. 2017;55:3513–3529. doi: 10.1128/JCM.01023-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotcheewaphan S., Lemon J.K., Desai U.U., Henderson C.M., Zelazny A.M. Rapid one-step protein extraction method for the identification of mycobacteria using MALDI-TOF MS. Diagn. Microbiol. Infect. Dis. 2019;94:355–360. doi: 10.1016/j.diagmicrobio.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Wilen C.B., McMullen A.R., Burnham C.A. Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant mycobacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2015;53:2308–2315. doi: 10.1128/JCM.00567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo L., Cao W., Chen W., Zhang R., Jing L., Chen H. Evaluation of the VITEK MS knowledge base version 3.0 for the identification of clinically relevant Mycobacterium species. Emerg. Microbes Infect. 2018;7:114. doi: 10.1038/s41426-018-0120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasperbauer S.H., Daley C.L. Mycobacterium chimaera infections related to the heater-cooler unit outbreak: a guide to diagnosis and management. Clin. Infect. Dis. 2019;68:1244–1250. doi: 10.1093/cid/ciy789. [DOI] [PubMed] [Google Scholar]
- 36.Patel R. A moldy application of MALDI: MALDI-ToF mass spectrometry for fungal identification. J. Fungi (Basel). 2019;5 doi: 10.3390/jof5010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rychert J., Slechta E.S., Barker A.P., Miranda E., Babady N.E., Tang Y.W. Multicenter evaluation of the Vitek MS v3.0 system for the identification of filamentous fungi. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01353-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loonen A.J., Jansz A.R., Stalpers J., Wolffs P.F., van den Brule A.J. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1575–1583. doi: 10.1007/s10096-011-1480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juiz P.M., Almela M., Melcion C., Campo I., Esteban C., Pitart C. A comparative study of two different methods of sample preparation for positive blood cultures for the rapid identification of bacteria using MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1353–1358. doi: 10.1007/s10096-011-1449-x. [DOI] [PubMed] [Google Scholar]
- 40.Meex C., Neuville F., Descy J., Huynen P., Hayette M.P., De Mol P. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J. Med. Microbiol. 2012;61:1511–1516. doi: 10.1099/jmm.0.044750-0. [DOI] [PubMed] [Google Scholar]
- 41.Klein S., Zimmermann S., Kohler C., Mischnik A., Alle W., Bode K.A. Integration of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in blood culture diagnostics: a fast and effective approach. J. Med. Microbiol. 2012;61:323–331. doi: 10.1099/jmm.0.035550-0. [DOI] [PubMed] [Google Scholar]
- 42.Buchan B.W., Riebe K.M., Ledeboer N.A. Comparison of the MALDI Biotyper system using Sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J. Clin. Microbiol. 2012;50:346–352. doi: 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson L.G., Drake S.K., Murray P.R. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2010;48:444–447. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Scola B., Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4:e8041. doi: 10.1371/journal.pone.0008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moussaoui W., Jaulhac B., Hoffmann A.M., Ludes B., Kostrzewa M., Riegel P. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry identifies 90% of bacteria directly from blood culture vials. Clin. Microbiol. Infect. 2010;16:1631–1638. doi: 10.1111/j.1469-0691.2010.03356.x. [DOI] [PubMed] [Google Scholar]
- 46.Prod’hom G., Bizzini A., Durussel C., Bille J., Greub G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J. Clin. Microbiol. 2010;48:1481–1483. doi: 10.1128/JCM.01780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledeboer N.A., Lopansri B.K., Dhiman N., Cavagnolo R., Carroll K.C., Granato P. Identification of gram-negative bacteria and genetic resistance determinants from positive blood culture Broths by use of the Verigene gram-negative blood culture multiplex microarray-based molecular assay. J. Clin. Microbiol. 2015;53:2460–2472. doi: 10.1128/JCM.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altun O., Almuhayawi M., Ullberg M., Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol. 2013;51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faron M.L., Buchan B.W., Ledeboer N.A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for use with positive blood cultures: methodology, performance, and optimization. J. Clin. Microbiol. 2017;55:3328–3338. doi: 10.1128/JCM.00868-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spanu T., Posteraro B., Fiori B., D’Inzeo T., Campoli S., Ruggeri A. Direct maldi-tof mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 2012;50:176–179. doi: 10.1128/JCM.05742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demarco M.L., Burnham C.A. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am. J. Clin. Pathol. 2014;141:204–212. doi: 10.1309/AJCPQYW3B6JLKILC. [DOI] [PubMed] [Google Scholar]
- 52.Montgomery S., Roman K., Ngyuen L., Cardenas A.M., Knox J., Tomaras A.P. Prospective evaluation of light scatter technology paired with matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid diagnosis of urinary tract infections. J. Clin. Microbiol. 2017;55:1802–1811. doi: 10.1128/JCM.00027-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen J.F., Bertille N., Cohen R., Chalumeau M. Rapid antigen detection test for group A streptococcus in children with pharyngitis. Cochrane Database Syst. Rev. 2016;7:CD010502. doi: 10.1002/14651858.CD010502.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babin S.M., Hsieh Y.H., Rothman R.E., Gaydos C.A. A meta-analysis of point-of-care laboratory tests in the diagnosis of novel 2009 swine-lineage pandemic influenza A (H1N1) Diagn. Microbiol. Infect. Dis. 2011;69:410–418. doi: 10.1016/j.diagmicrobio.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu H., Lofgren E.T., Halloran M.E., Kuan P.F., Hudgens M., Cole S.R. Performance of rapid influenza H1N1 diagnostic tests: a meta-analysis. Influenza Other Respir. Viruses. 2012;6:80–86. doi: 10.1111/j.1750-2659.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chartrand C., Leeflang M.M., Minion J., Brewer T., Pai M. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann. Intern. Med. 2012;156:500–511. doi: 10.7326/0003-4819-156-7-201204030-00403. [DOI] [PubMed] [Google Scholar]
- 57.Green D.A., StGeorge K. Rapid antigen tests for influenza: rationale and significance of the FDA reclassification. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.00711-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berry G.J., Miller C.R., Prats M.M., Marquez C., Oladipo O.O., Loeffelholz M.J. Comparison of the Alere i Strep A Test and the BD Veritor System in the detection of group A streptococcus and the hypothetical impact of results on antibiotic utilization. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01310-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker K.G., Gandra S., Matushek S., Beavis K.G., Tesic V., Charnot-Katsikas A. Comparison of 3 nucleic acid amplification tests and a rapid antigen test with culture for the detection of group A streptococci from throat swabs. J. Appl. Lab. Med. 2019;4:164–169. doi: 10.1373/jalm.2018.028696. [DOI] [PubMed] [Google Scholar]
- 60.Young S., Illescas P., Nicasio J., Sickler J.J. Diagnostic accuracy of the real-time PCR cobas Liat Influenza A/B assay and the Alere i Influenza A&B NEAR isothermal nucleic acid amplification assay for the detection of influenza using adult nasopharyngeal specimens. J. Clin. Virol. 2017;94:86–90. doi: 10.1016/j.jcv.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 61.Binnicker M.J., Espy M.J., Irish C.L., Vetter E.A. Direct detection of influenza A and B viruses in less than 20 minutes using a commercially available rapid PCR assay. J. Clin. Microbiol. 2015;53:2353–2354. doi: 10.1128/JCM.00791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson J., Schechter-Perkins E.M., Mitchell P., Mace S., Tian Y., Williams K. Multi-center evaluation of the cobas Liat Influenza A/B & RSV assay for rapid point of care diagnosis. J. Clin. Virol. 2017;95:5–9. doi: 10.1016/j.jcv.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Valentin T., Kieslinger P., Stelzl E., Santner B.I., Groselj-Strele A., Kessler H.H. Prospective evaluation of three rapid molecular tests for seasonal influenza in patients presenting at an emergency unit. J. Clin. Virol. 2019;111:29–32. doi: 10.1016/j.jcv.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Chen J.H., Lam H.Y., Yip C.C., Cheng V.C., Chan J.F., Leung T.H. Evaluation of the molecular Xpert Xpress Flu/RSV assay vs. Alere i Influenza A & B assay for rapid detection of influenza viruses. Diagn. Microbiol. Infect. Dis. 2018;90:177–180. doi: 10.1016/j.diagmicrobio.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 65.Bell J., Bonner A., Cohen D.M., Birkhahn R., Yogev R., Triner W. Multicenter clinical evaluation of the novel Alere i Influenza A&B isothermal nucleic acid amplification test. J. Clin. Virol. 2014;61:81–86. doi: 10.1016/j.jcv.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Peters R.M., Schnee S.V., Tabatabai J., Schnitzler P., Pfeil J. Evaluation of Alere i RSV for rapid detection of respiratory syncytial virus in children hospitalized with acute respiratory tract infection. J. Clin. Microbiol. 2017;55:1032–1036. doi: 10.1128/JCM.02433-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merckx J., Wali R., Schiller I., Caya C., Gore G.C., Chartrand C. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann. Intern. Med. 2017;167:394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 68.Martin J.M., Green M., Barbadora K.A., Wald E.R. Group A streptococci among school-aged children: clinical characteristics and the carrier state. Pediatrics. 2004;114:1212–1219. doi: 10.1542/peds.2004-0133. [DOI] [PubMed] [Google Scholar]
- 69.AAP, Red Book, Report of the Committee on Infectious Diseases, thirty-first ed., American Association of Pediatrics, 2018, pp 750, Itasca, IL, USA.
- 70.Popowitch E.B., O’Neill S.S., Miller M.B. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J. Clin. Microbiol. 2013;51:1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Babady N.E., England M.R., Jurcic Smith K.L., He T., Wijetunge D.S., Tang Y.W. Multicenter evaluation of the ePlex respiratory pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J.H.K., Lam H.Y., Yip C.C.Y., Wong S.C.Y., Chan J.F.W., Ma E.S.K. Clinical evaluation of the new high-throughput luminex NxTAG respiratory pathogen panel assay for multiplex respiratory pathogen detection. J. Clin. Microbiol. 2016;54:1820–1825. doi: 10.1128/JCM.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leber A.L., Everhart K., Daly J.A., Hopper A., Harrington A., Schreckenberger P. Multicenter evaluation of BioFire FilmArray respiratory panel 2 for detection of viruses and bacteria in nasopharyngeal swab samples. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01945-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pabbaraju K., Wong S., Tokaryk K.L., Fonseca K., Drews S.J. Comparison of the Luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J. Clin. Microbiol. 2011;49:1738–1744. doi: 10.1128/JCM.02090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rappo U., Schuetz A.N., Jenkins S.G., Calfee D.P., Walsh T.J., Wells M.T. Impact of early detection of respiratory viruses by multiplex PCR assay on clinical outcomes in adult patients. J. Clin. Microbiol. 2016;54:2096–2103. doi: 10.1128/JCM.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Subramony A., Zachariah P., Krones A., Whittier S., Saiman L. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J. Pediatr. 2016;173:196–201. doi: 10.1016/j.jpeds.2016.02.050. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogers B.B., Shankar P., Jerris R.C., Kotzbauer D., Anderson E.J., Watson J.R. Impact of a rapid respiratory panel test on patient outcomes. Arch. Pathol. Lab. Med. 2015;139:636–641. doi: 10.5858/arpa.2014-0257-OA. [DOI] [PubMed] [Google Scholar]
- 78.Mahony J.B., Blackhouse G., Babwah J., Smieja M., Buracond S., Chong S. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. 2009;47:2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spina A., Kerr K.G., Cormican M., Barbut F., Eigentler A., Zerva L. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin. Microbiol. Infect. 2015;21:719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 80.Buss S.N., Leber A., Chapin K., Fey P.D., Bankowski M.J., Jones M.K. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J. Clin. Microbiol. 2015;53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khare R., Espy M.J., Cebelinski E., Boxrud D., Sloan L.M., Cunningham S.A. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J. Clin. Microbiol. 2014;52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Halligan E., Edgeworth J., Bisnauthsing K., Bible J., Cliff P., Aarons E. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin. Microbiol. Infect. 2014;20:O460–O467. doi: 10.1111/1469-0691.12476. [DOI] [PubMed] [Google Scholar]
- 83.Huang R.S., Johnson C.L., Pritchard L., Hepler R., Ton T.T., Dunn J.J. Performance of the Verigene(R) enteric pathogens test, Biofire FilmArray gastrointestinal panel and Luminex xTAG(R) gastrointestinal pathogen panel for detection of common enteric pathogens. Diagn. Microbiol. Infect. Dis. 2016;86:336–339. doi: 10.1016/j.diagmicrobio.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 84.Wessels E., Rusman L.G., van Bussel M.J., Claas E.C. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG(R) GPP) testing in the diagnosis of infectious gastroenteritis. Clin. Microbiol. Infect. 2014;20:O182–O187. doi: 10.1111/1469-0691.12364. [DOI] [PubMed] [Google Scholar]
- 85.Goldenberg S.D., Bacelar M., Brazier P., Bisnauthsing K., Edgeworth J.D. A cost benefit analysis of the Luminex xTAG Gastrointestinal Pathogen Panel for detection of infectious gastroenteritis in hospitalised patients. J. Infect. 2015;70:504–511. doi: 10.1016/j.jinf.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 86.Rand K.H., Tremblay E.E., Hoidal M., Fisher L.B., Grau K.R., Karst S.M. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn. Microbiol. Infect. Dis. 2015;82:154–157. doi: 10.1016/j.diagmicrobio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 87.Ward C., Stocker K., Begum J., Wade P., Ebrahimsa U., Goldenberg S.D. Performance evaluation of the Verigene(R) (Nanosphere) and FilmArray(R) (BioFire(R)) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- 88.Bhatti M.M., Boonlayangoor S., Beavis K.G., Tesic V. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J. Clin. Microbiol. 2014;52:3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang T.D., Melnik E., Bogaerts P., Evrard S., Glupczynski Y. Evaluation of the ePlex blood culture identification panels for detection of pathogens in bloodstream infections. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.01597-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Box M.J., Sullivan E.L., Ortwine K.N., Parmenter M.A., Quigley M.M., Aguilar-Higgins L.M. Outcomes of rapid identification for gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy. 2015;35:269–276. doi: 10.1002/phar.1557. [DOI] [PubMed] [Google Scholar]
- 91.MacVane S.H., Nolte F.S. Benefits of adding a rapid PCR-based blood culture identification panel to an established antimicrobial stewardship program. J. Clin. Microbiol. 2016;54:2455–2463. doi: 10.1128/JCM.00996-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Messacar K., Hurst A.L., Child J., Campbell K., Palmer C., Hamilton S. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J. Pediatric Infect. Dis. Soc. 2017;6:267–274. doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neuner E.A., Pallotta A.M., Lam S.W., Stowe D., Gordon S.M., Procop G.W. Experience with rapid microarray-based diagnostic technology and antimicrobial stewardship for patients with gram-positive bacteremia. Infect. Control. Hosp. Epidemiol. 2016;37:1361–1366. doi: 10.1017/ice.2016.175. [DOI] [PubMed] [Google Scholar]
- 94.Pardo J., Klinker K.P., Borgert S.J., Butler B.M., Giglio P.G., Rand K.H. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn. Microbiol. Infect. Dis. 2016;84:159–164. doi: 10.1016/j.diagmicrobio.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 95.Suzuki H., Hitomi S., Yaguchi Y., Tamai K., Ueda A., Kamata K. Prospective intervention study with a microarray-based, multiplexed, automated molecular diagnosis instrument (Verigene system) for the rapid diagnosis of bloodstream infections, and its impact on the clinical outcomes. J. Infect. Chemother. 2015;21:849–856. doi: 10.1016/j.jiac.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 96.Walker T., Dumadag S., Lee C.J., Lee S.H., Bender J.M., Cupo Abbott J. Clinical impact of laboratory implementation of verigene BC-GN microarray-based assay for detection of gram-negative bacteria in positive blood cultures. J. Clin. Microbiol. 2016;54:1789–1796. doi: 10.1128/JCM.00376-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Timbrook T.T., Morton J.B., McConeghy K.W., Caffrey A.R., Mylonakis E., LaPlante K.L. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin. Infect. Dis. 2017;64:15–23. doi: 10.1093/cid/ciw649. [DOI] [PubMed] [Google Scholar]
- 98.Banerjee R., Teng C.B., Cunningham S.A., Ihde S.M., Steckelberg J.M., Moriarty J.P. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin. Infect. Dis. 2015;61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leber A.L., Everhart K., Balada-Llasat J.M., Cullison J., Daly J., Holt S. Multicenter evaluation of BioFire FilmArray Meningitis/Encephalitis Panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J. Clin. Microbiol. 2016;54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Graf E.H., Farquharson M.V., Cardenas A.M. Comparative evaluation of the FilmArray meningitis/encephalitis molecular panel in a pediatric population. Diagn. Microbiol. Infect. Dis. 2017;87:92–94. doi: 10.1016/j.diagmicrobio.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 101.Liesman R.M., Strasburg A.P., Heitman A.K., Theel E.S., Patel R., Binnicker M.J. Evaluation of a commercial multiplex molecular panel for diagnosis of infectious meningitis and encephalitis. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01927-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lewis P.O., Lanier C.G., Patel P.D., Krolikowski W.D., Krolikowski M.A. False negative diagnostic errors with polymerase chain reaction for the detection of cryptococcal meningoencephalitis. Med. Mycol. 2019 doi: 10.1093/mmy/myz064. [DOI] [PubMed] [Google Scholar]
- 103.DiDiodato G., Bradbury N. Cerebrospinal fluid analysis with the BioFire FilmArray Meningitis/Encephalitis Molecular Panel reduces length of hospital stay in patients with suspected central nervous system infections. Open. Forum Infect. Dis. 2019;6:ofz119. doi: 10.1093/ofid/ofz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dack K., Pankow S., Ablah E., Zackula R., Assi M. Contribution of the BioFire FilmArray Meningitis/Encephalitis Panel: assessing antimicrobial duration and length of stay. Kans. J. Med. 2019;12:1–3. [PMC free article] [PubMed] [Google Scholar]
Self-assessment questions
-
1.What is the primary benefit of flocked vs. traditional swabs?
-
a.Flocked swabs are larger and allow for collection of a greater number of organisms
-
b.Flocked swabs allow a greater number of organisms to be released from the swab
-
c.Specimens collected using flocked swabs can be transported at any temperature
-
d.Flocked swabs are less expensive
-
a.
-
2.Which of the following is not a component of current microbiology laboratory automation systems?
-
a.Specimen inoculation
-
b.Automated track
-
c.Anaerobic incubation
-
d.Digital imaging
-
a.
-
3.Which of the following is the gold standard method for identification of microorganisms?
-
a.PCR
-
b.Biochemical methods
-
c.MALDI
-
d.DNA sequencing
-
a.
-
4.Clinics in your hospital system perform rapid influenza diagnostic tests for the detection of influenza A and B viral antigens, which have a low negative predictive value (NPV) during influenza virus season. Which test performance characteristic is associated with the low NPV of influenza antigen testing?
-
a.High cross-reactivity with other viruses
-
b.Poor reproducibility
-
c.Poor sensitivity
-
d.Poor specificity
-
a.
-
5.Several multiplex molecular panels are commercially available for syndromic-based microbiology diagnosis. Which of the following infections cannot be diagnosed by a syndromic-based test?
-
a.Central nervous system infection
-
b.Gastrointestinal infection
-
c.Prosthetic joint infection
-
d.Upper respiratory infection
-
a.
Answers
|


