Abstract
Background
Mutations in the ARV1 Homolog, Fatty Acid Homeostasis Modulator (ARV1), have recently been described in association with early infantile epileptic encephalopathy 38. Affected individuals presented with epilepsy, ataxia, profound intellectual disability, visual impairment, and central hypotonia. In S. Cerevisiae, Arv1 is thought to be involved in sphingolipid metabolism and glycophosphatidylinositol (GPI)-anchor synthesis. The function of ARV1 in human cells, however, has not been elucidated.
Methods
Mutations were discovered through whole exome sequencing and alternate splicing was validated on the cDNA level. Expression of the variants was determined by qPCR and Western blot. Expression of GPI-anchored proteins on neutrophils and fibroblasts was analyzed by FACS and immunofluorescence microscopy, respectively.
Results
Here we describe seven patients from two unrelated families with biallelic splice mutations in ARV1. The patients presented with early onset epilepsy, global developmental delays, profound hypotonia, delayed speech development, cortical visual impairment, and severe generalized cerebral and cerebellar atrophy. The splice variants resulted in decreased ARV1 expression and significant decreases in GPI-anchored protein on the membranes of neutrophils and fibroblasts, indicating that the loss of ARV1 results in impaired GPI-anchor synthesis.
Conclusion
Loss of GPI-anchored proteins on our patients’ cells confirms that the yeast Arv1 function of GPI-anchor synthesis is conserved in humans. Overlap between the phenotypes in our patients and those reported for other GPI-anchor disorders suggests that ARV1-deficiency is a GPI-anchor synthesis disorder.
Keywords: GPI-anchor synthesis, Early Infantile Epileptic Encephalopathy 38, Rare disease, endoplasmic reticulum
INTRODUCTION
Glycosylphosphatidylinositol (GPI) is a glycolipid that anchors proteins involved in signal transduction and immune response to the outer membrane of cells [1, 2]. The GPI-anchor consists of a phospholipid tail, a glycan core and a phosphoethanolamine linker, and is added to proteins as a post-translational modification [1, 2]. Biosynthesis of GPI-anchors, a process that involves at least 26 genes, occurs in the endoplasmic reticulum (ER) and Golgi [3, 4]. The process starts with the formation of a phospholipid tail on the cytoplasmic side of the ER, during which the first glycosylation steps occur; the GPI-precursor is then flipped into the ER lumen to access the mannosyltransferases [3–5]. After the GPI-anchor is formed and attached to the target protein, it travels through the Golgi, where additional modifications are made [3].
Thus far 19 genes involved in GPI-anchor biosynthesis have been associated with human disease [4, 6–27]; all present with a multitude of symptoms, including seizures, hematologic and metabolic abnormalities, developmental delays, intellectual disabilities and occasionally dysmorphic features and congenital anomalies [4, 28].
Studies in S. cerevisiae suggest that ACAT related enzyme 2 required for viability 1 (Arv1) may be involved in the process of flipping the GPI-precursors into the ER lumen [5], a role that is conserved in the human homolog ARV1 (Arv1 homolog, or Fatty Acid Homeostasis Modulator: MIM 611647; NM_022786.1) [29]. A homozygous missense mutation in ARV1 (c.565G>A, p.Gly189Arg) was first described in affected members of a consanguineous family who presented with phenotypes resembling GPI-anchor disorders, including severe intellectual disability, early onset seizures, poor head control and ataxia [6]. An additional homozygous splicing mutation (c.294+1G>A, p.59–98del) was described in a second report in an individual with a similar phenotype, including visual impairment, central hypotonia, and dystonia [27]; that condition was classified as early infantile epileptic encephalopathy 38 (EIEE38, MIM 617020). These cases did not include a description of GPI anchor abnormalities due to ARV1 mutations.
Here we describe seven additional patients from two unrelated families with homozygous splice mutations in ARV1 and EIEE38. In addition to what has been described in literature, we show that, similar to Arv1 in S. cerevisiae, GPI-anchor synthesis in humans is impaired by the loss of ARV1 protein, causing a significant decrease in membrane bound GPI-anchored proteins measured in neutrophils and fibroblasts.
METHODS
Subjects and samples
Patient 1 and 2 (Family I) were enrolled at the National Institutes of Health Clinical Center (NIH-CC) under the protocol 14-HG-0071: “Clinical and Basic Investigations Into Known and Suspected Congenital Disorders of Glycosylation” (NCT02089789) and protocol 15-HG-0130: “Clinical and Genetic Evaluation of Individuals With Undiagnosed Disorders Through the Undiagnosed Diseases Network” (NCT02450851) [30–33]. Both clinical protocols were approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board (IRB), and written informed consent was obtained from the patients’ parents. Genomic DNA was extracted from whole blood using the Gentra Puregene Blood kit (Qiagen, Valencia, CA) and primary fibroblasts were obtained from a forearm punch skin biopsy.
Patients 3 through 7 (Family II) were enrolled through a gene discovery project based at the Sydney Children’s Hospitals Network under the protocol approved by its IRB (HREC/10/CHW/114): “Use of High Throughput Screening Technologies for Gene Discovery in Mendelian Disorders”. Written informed consent was obtained from the parents of the relevant family members. DNA was prepared from blood and cultured skin fibroblasts were established as previously reported [34]. Pediatric control fibroblasts were obtained from Coriell (GM01852, GM01651 and GM01562; Camden, NJ) and adult control fibroblasts from ATCC (ATCC61683453 and ATCC59220266; Manassas, VA).
Mutation analysis
For patients 1 and 2, a clinical exome (Ambry’s ExomeNext™, San Antonio, TX), performed before their NIH admission, revealed a homozygous splice variant in ARV1 (NM_022786: c.674–2A>T). The variant was Sanger validated on genomic DNA derived from whole blood using Hotstart Mastermix with the primer pair 5’-GGAATCTGCCGCTTGATATG-3’ and 5’-TGTAAGGGCACCGACTTCTT-3’and standard recommended amplification conditions with annealing temperature at 55°C and extension at 72°C for 1 min. PCR products were treated with ExoSapIT and sent to Macrogen (Rockville, MD) for Sanger sequencing. Results were analyzed and interpreted using Sequencer Software version 5.0.1 (Gene Codes Corporation, Ann Arbor, MI).
Patients 4 and 5 underwent research-based exome analyses at the Centre for Applied Genomics (CAG) at the Children’s Hospital of Philadelphia (CHOP; Philadelphia, PA). The exome analyses revealed a previously described homozygous variant (NM_022786: c.294+1G>A). This variant was Sanger verified on genomic DNA of each affected child and their parents, and available unaffected family members.
The effects of the predicted splice mutations were evaluated on cDNA derived from cultured fibroblasts. Cells were grown in Dulbecco’s Minimum Essentials Medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with 10% v/v Fetal Bovine Serum (FBS; Life Technologies) and 1% v/v antibiotic-antimycotic (Life Technologies). RNA was extracted from ATCC adult control dermal fibroblast cells, and patient cells using RNeasy Mini kit following the manufacturer’s standard protocol with on-column DNAse digestion (Qiagen, Valencia, CA) and 2μg RNA was reverse transcribed using the Omniscript RT kit (Qiagen). The region around each predicted splice was PCR amplified using Multiplexing master mix (Qiagen) with addition of buffer Q and primer pair 5’-TGTTTGCGATTGCTGCTTTA-3’ and 5’-GCTTGTCTCTTGGGTGCATT-3’ for c.674–2A>T or primer pair 5’-GAGGCCAAAGAGTTGTACCG-3’ and 5’-CGTTCTACCCACAGGAAGGT-3’ for c.294+1G>A. Amplification conditions were as described above. For c.674–2A>T PCR products were separated on a 2% agarose gel and extracted using the QiaQuick Gel Extraction kit (Qiagen) and for c.294+1G>A, PCR products were treated with ExoSapIT before they were sent to Macrogen for Sanger sequencing. Results were interpreted using Sequencer Software version 5.0.1.
ARV1 expression analysis
mRNA expression of ARV1 was evaluated using quantitative RT-PCR on fibroblast-derived cDNA. The qPCR reactions were set up following the TaqMan gene expression protocol (Thermo Fisher Scientific, Waltham, MA) using the 20μL reaction in four-fold and the delta-delta CT (ΔΔCT) setting for TaqMan reagents on the 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA). Two TaqMan gene-expression assays were used to distinguish between wild-type ARV1 and the novel variant c.674–2A>T; one upstream of exon 5 (Hs00224467_m1: exon 3–4 junction) and one on including exon 5 (Hs00917617_m1: exon 5–6 junction). GAPDH (Hs03929097_g1) was used as an endogenous control and data were analyzed using the software (V.2.0.1) accompanying the qPCR machine (Applied Biosystems).
For analysis of the expression of ARV1 on protein level, fibroblasts were harvested with 0.25% trypsin (Life Technologies) and washed with 1x phosphate buffered saline (PBS). Cells were lysed in 300μL radio-immunoprecipitation assay buffer (RIPA; Sigma Aldrich, St. Louis, MO) supplemented with one protease inhibitor cocktail tablet (Roche, Manheim, Germany) and one phosphatase inhibitor tablet (Roche). After sonication, Laemmli buffer and β-mercaptoethanol were added, and 25μg lysate was loaded on a 4–15% polyacrylamide gel (Biorad, Hercules, CA) and run at a constant 100V. Proteins were transferred to a PVDF membrane using a Transblot Turbo transfer system (Biorad) with the 7-min standard protocol. The membrane was blocked for 1 h in Odyssey PBS-blocking buffer (Li-cor, Lincoln, NE) and was probed using mouse ARV1 (4G12) antibody (sc-517099; Santa Cruz Biotechnology, Dallas, TX) and rabbit anti-vinculin (ab73412; Abcam, Cambridge, UK) for 2 h at RT. After 3 washes PBS with Tween (PBST), the blot was probed with Li-cor secondary antibodies IRDye® 800CW donkey anti-mouse IgG (H + L) and IRDye® 680RD donkey anti-rabbit IgG (H + L), washed with PBST and scanned on the Li-Cor Odyssey Clx infrared imaging system (Li-cor). Bands were analyzed using Image Studio software (Li-cor).
GPI-anchored protein analysis by flow cytometry and immunofluorescence
Flow cytometry analysis of GPI-anchored proteins in neutrophils of patients 1 and 2 (Family I) was performed as described previously [4].
No neutrophils were available for Family II. Therefore, fibroblast expression of CD59 and uPAR (CD87) were analyzed by immunofluorescence confocal microscopy for patients 1, 2 and 7. Fibroblasts were seeded at a density of 5×104 cells per 12-mm round coverslip. After adhering overnight, cells were fixed with 4% paraformaldehyde in PBS, blocked in 4% bovine serum albumin (BSA) in PBS, incubated for 2 h in either anti-CD59 [MEM-43/5] antibody (ab9183, Abcam) or anti-CD87 antibody (anti-uPAR (R-4); Thermo Fisher Scientific), as described [35]. After washing with PBS, coverslips were incubated with donkey anti-mouse IgG (H+L) Alexa Fluor 488 secondary antibodies (Invitrogen), co-stained with Hoechst 33342 (Invitrogen) and mounted using Prolong Gold anti fade reagent (Thermo Fisher Scientific). Images were taken on a Zeiss LSM700 confocal laser-scanning microscope using a 20X objective (Carl Zeiss Microscopy GmbH, Jena, Germany). Mean fluorescence intensity was measured for 10 images per cell line using Zen black 2012 LSM software (Carl Zeiss Microscopy GmbH).
RESULTS
Clinical descriptions
Family I
Patient 1, is a 17 years old female, the first child to American-Mexican parents (Figure 1A, II.1) who was born after a 41-week gestation. At 6 months of age, she had her first seizure, followed by frequent episodes of both febrile and non-febrile seizures, some requiring hospitalization. Patient 1 never developed a social smile or met any motor milestones. She has been dependent on g-tube feeding since age 2 years and is wheelchair dependent. She has cortical blindness and bilateral retrocochlear hearing impairment.
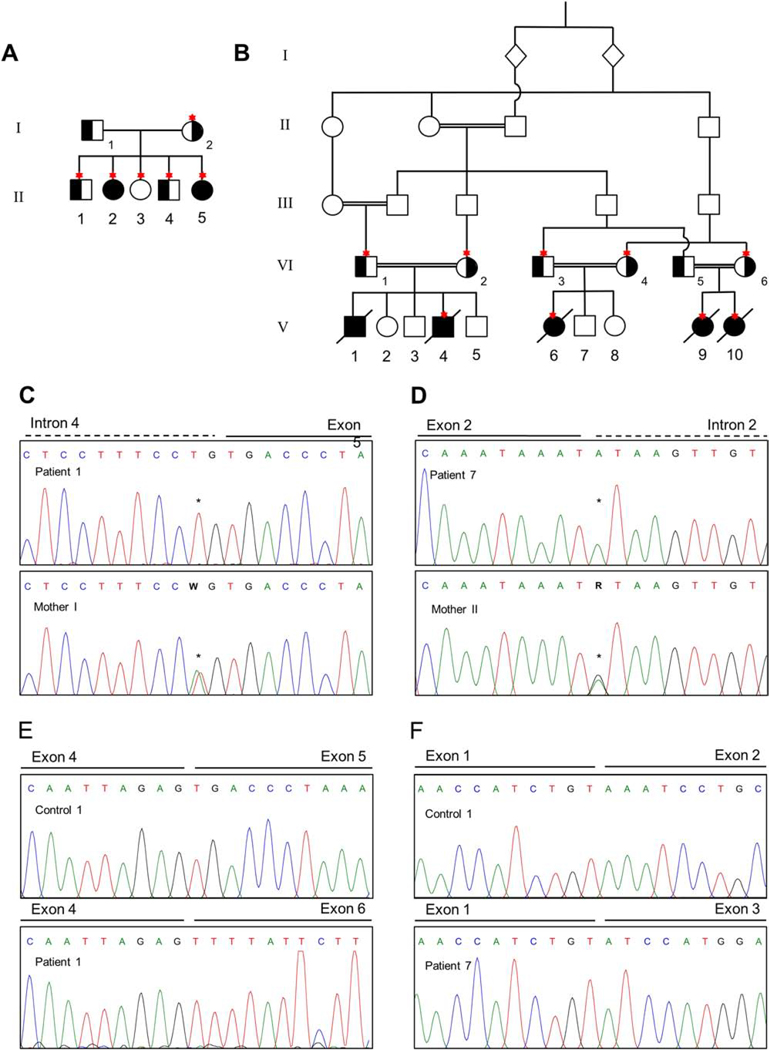
Figure 1. Pedigree and mutation analyses.
A. Pedigree for Family I showing the two affected sisters II-2 (patient 1) and II-5 (patient 2) and their nuclear family. Carrier status of the variant c.674–2A>T is indicated in black and Sanger confirmation was done for the individuals depicted with an asterisk. B. Pedigree for the consanguineous Family II showing the five affected 3rd cousins V-1 (patient 3), V-4 (patient 4), V-6 (patient 5), V-9 (patient 6) and V-10 (patient 7). Carrier status of the variant c.294+1G>A is indicated in black and Sanger confirmation was performed in the individuals depicted with an asterisk. C. Sanger confirmation of c.674–2A>T in Family I showing the affected patient 1 with a homozygous variant and an unaffected family member with a heterozygous variant. D. Sanger confirmation for c.294+1G>A in Family II showing the patient 7 with a homozygous variant and an unaffected family member with heterozygous variant. E. Chromatograms showing the effect of the variant c.674–2A>T in Family I resulting to the loss of exon 5 on cDNA level. F. Chromatograms showing the effect of the variant c.294+1G>A in Family II resulting in the loss of exon 2 on cDNA level.
Craniofacial examination showed severe midfacial aplasia, coarse facial features, infraorbital flatness, bitemporal narrowing and an increased bizygomatic width as well as skin lesions, deep set eyes, long ears, low posterior hairline, thick lips, long lower third and short middle third of the face, and a right-deviating chin. Patient 1 demonstrated marked spasticity. On neuroimaging she had cortical and cerebellar atrophy, with undermyelination of the U-fibers suggestive of delayed myelination as well as foci of gliosis. Her musculoskeletal issues include contractures at the elbows leading to deforming and fixed pronation and internal rotation of both forearms. Her feet are rounded on the heels and have no arch. Skeletal radiographs showed significant dysplasia. She had an elongated mandible on lateral view, narrow elongated skull and thickened calvarium. Marked rotatory scoliosis of the thoracic or lumbar spine was observed, in both her and one of her unaffected brothers, with deformities and wedging of multiple lower lumbar spine vertebral bodies. The radial heads of both radii and ulnae were atrophic and misshapen with bowing bilaterally, and the right hand had a mild rotatory deformity at the wrist. The femoral heads were large bilaterally, and her pelvis was deformed.
She had generalized aminoaciduria, suggestive of a proximal kidney tubule dysfunction. Most of her liver tests were normal, but she had elevated gamma-glutamyl transferase activity with normal alkaline phosphatase activity. Analysis of esterification of cholesterol in response to LDL stimulation was depressed relative to normal controls, but filipin staining of free cholesterol is normal. Other remarkable labs included elevated iron levels and high levels of the allergy type antibodies IgE and ACA IgM (Table S1).
Patient 2, the youngest sibling of patient 1 (Figure 1A, II.5), was a 29-month-old girl when she presented to the NIH Clinical Center with global developmental delay, profound hypotonia, microcephaly, cortical visual impairment, epilepsy, borderline short stature and poor weight gain. Born vaginally at 39 weeks, she had abnormal eye movements first noted at 2 to 3 months of age and onset of seizures initially at 7 months of age. Her disease course was overall similar to that of her older sister, but lacked the bone malformations and her facial features were less coarse. On neuroimaging she had diffuse cortical and cerebellar atrophy and delayed myelination, with low N acetyl-asparatate on magnetic resonance spectroscopy. Like her sister, she has diffused osteopenia, elevated IgE, increased red cell turnover, increased serum iron levels (Table S1), and depressed levels of cholesterol esterification in response to LDL stimulation relative to normal controls.
Family II
Patient 3 was the first born male child to a consanguineous Lebanese parents (Figure 1B, V.1). After an uneventful pregnancy and normal 18-week morphology ultrasound scan, he was born through normal vaginal delivery. Birth weight was 4.3kg (90th %ile) and the newborn examination was unremarkable. At 11 weeks, he presented with visual inattentiveness and developmental delay; by age 4 months he had hypotonia and seizures. He had visual impairment but with normal appearing retina and was fed through a nasogastric tube. He deteriorated with increasing seizures and died at age 15 months as result of aspiration pneumonia.
Patient 4, the younger brother of patient 3 and the 4th child (Figure 1B, V.4), was born after a normal pregnancy with all birth parameters approximately 50th %ile (weight = 3.6kg; occipital-frontal circumference or OFC = 35cm; length = 50cm). Abnormal eye movements and poor head control were noted at 4 weeks. He was visually inattentive and had an intermittent convergent squint. Onset of seizures at 5 months prompted admission to hospital; a prolonged seizure at 7 months consisted of a choking episode leading to a 7 minute right sided tonic clonic seizure. An EEG showed short duration bursts of slow repetitive sharp and wave epileptic discharges over either hemisphere particularly in the central regions, but this was not accompanied by observable clinical events. Treatment with anti-epileptic drugs (AEDs) was commenced at that time.
Patient 4 had an abnormal neurological examination with increased extensor tone peripherally, fisting of hands and increased deep tendon reflexes. He was unable to feed orally; failure to thrive led to NG-tube feeding until age 2 years (weight =10.5kg; 3rd %ile). He had progressive microcephaly from 5 months (OFC 45.8cm; <2nd %ile at age 2 years). At age 2 years he was visually inattentive, consistent with cortical blindness; a formal ophthalmological examination showed normal fundoscopy and a normal electroretinogram (ERG), but visual evoked potential (VEP) responses to bright flashes under dark and light adapted conditions showed negative b-waves in both eyes.
He had severe global developmental delay. He rolled over at 18 months but was still unable to sit unsupported at age 3 years. He did make occasional vocalizations, but at age 3 years and 4 months he remained virtually unresponsive, not reaching for or holding objects in his hands. He required feeding with thickened feeds and continued to choke on thin fluids. He had intermittent myoclonic jerking movements despite AEDs and he died at 5 years of age.
Patient 5 is a first cousin of patients 3 and 4 (Figure 1B, V.6). She is the first of three children born to her consanguineous parents at full term after a pregnancy complicated by hyperemesis. She was delivered by lower segment caesarian section because of fetal macrosomia and failure to progress. Her birth parameters were weight 4.1kg (90th - 95th %ile); OFC = 39cm (>97th centile); and length = 56cm (>97th centile). From birth, she displayed poor feeding requiring NG-tube feeds and eventually a gastrostomy inserted at age 2 years for failure to thrive. She had severe central hypotonia and abnormal posturing of limbs in the newborn period, which persisted with poor head control. Reflexes were generally brisk, and her tone was increased peripherally. Head circumference at 6 months was 44 cm ( >98%), but progressively fell to 10–25% by 2 years and 3 months of age.
Roving eye movements were noted at birth. An ophthalmological examination confirmed visual inattention, nystagmus, roving eye movements and small optic discs on fundoscopy. At 4 months she was admitted to hospital with focal seizures, status epilepticus and severe developmental delay. Over the next two years she had a progressive course with ongoing seizures, poor development, and failure to thrive. Her newborn brain MRI showed a thin corpus callosum and an MRI at age 4 months showed small bilateral subdural collections. She died at 2 years and 4 months after presentation with fever, enterovirus infection, seizures, respiratory distress and liver dysfunction.
Patient 6 is a first cousin of patients 3, 4 and 5, and first of two affected females born to her parents (Figure 1B, V.9) at full term after normal pregnancy with large birth weight (4.2kg; >97th %ile) and normal head circumference. Onset of seizures was at 4 months at which time she was admitted with generalized tonic clonic and focal motor seizures in status epilepticus. She received multiple AEDs (levetiracetam, clobazam, vitamin B6) with some success. At 4 months, she had reasonable head control except when unwell and febrile. Growth parameters were still within the normal range, with an OFC of 43cm (75%). She was visually inattentive with intermittent nystagmus and reduced response to bright light, but normal fundoscopy. At age 11 months she presented to hospital after a prolonged hypotonic, hyporesponsive cyanotic seizure at home. At that time, she was blind, not fixing and following, had no response to light, had horizontal nystagmus and no recordable VEP response, consistent with cortical blindness. She had severe global developmental delay and was not able to sit at age 2 years and continued to require NG-tube feeding. At age 2 years she was again admitted to hospital with high fever, increasing seizure frequency, decreased level of consciousness and was unresponsive. She died as result of rhinovirus respiratory infection and aspiration pneumonia at 2 years and 3 months.
Patient 7 is the younger sister of patient 6 (Figure 1B, V.10). She was born after a normal pregnancy at 38 weeks with normal birth parameters (weight = 2.98kg; 75th %ile). Her mother first became concerned at about 2.5 months due to central hypotonia and poor visual attention, similar to her sister. Eye exam at age 4 months showed no eye contact and no fixing and following, with intermittent exotropia and some response (blinks) to bright light in a dark room. Fine horizontal nystagmus was observed at times and fundoscopy showed normal macula and optic discs. VEP and ERG at age 6 months showed scotopic 24dB flash response present but delayed for her right eye and no recordable photopic response for her left eye.
At age 7 months she was admitted in status epilepticus with a febrile illness. She had repeated tonic clonic seizures and decerebrate posturing during interictal periods, and was treated with intravenous anti-epileptic drugs. Chorea and dystonic movements were noted during that admission. Thereafter she was noted to have epileptic encephalopathy, and poor head control and minimal purposeful movement of limbs with increased deep tendon reflexes. She smiled intermittently to her mother’s voice and touch but did not respond to visual stimulation. She was discharged on oral topiramate, clobazam, levetiracetam, vitamin B. Brain MRI in patient 7 showed normal myelination and absence of hyperintense regions.
At 9 months, she had severe global developmental delay with progressive postnatal microcephaly (OFC 43.1cm; 5%ile) and failure to thrive (weight 6.43 kg, 3%ile; and length 66.4cm, 25%ile). Examination showed spontaneous choreiform movements, dystonic posturing with variable tone in limbs, and central hypotonia with poor head control. There was little change in head circumference since age 17 months at which time the OFC was 44cm (5%ile). Poor feeding, choking episodes and probable aspiration pneumonia were managed with NG-feeding from age 17 months. Increased frequency of seizures occurred with recurrent respiratory illness due to aspiration and led to frequent hospital presentations in status epilepticus. At the time of manuscript preparation, she died at age 3 years and 2 months.
The phenotypes in these patients and in those previously reported are listed and compared in Table 1.
Table 1.
Patient phenotypes compared to those described in literature
| This report (Family I) | This report (Family I) | This report (Family II) | This report (Family II) | This report (Family II) | This report (Family II) | This report (Family II) | Palmer (2016) | Alazami (2015) | Alazami (2015) | Alazami (2015) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient number | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | IV:4 | DG08RC00075/X:2 | DG08RC00076/IX:8 | DG08RC00077I/IX:7 |
| Relation to the proband | Proband | Sibling | Cousin | Cousin | Cousin | Sister | Proband | Proband | Proband | 2nd-Cousin | 2nd-Cousin |
| Mutation (NM_022786.2)a | c.674−2A>T | c.674−2A>T | c.294+1G>Ab | c.294+1G>A | c.294+1G>A | c.294+1G>A | c.294+1G>A | c.294+1G>A | c.565G>A | c.565G>A | c.565G>A |
| Mutation (NP_073623.1) | p.Thr266_Phe271delc | p.Thr266_Phe271delc | p.Lys59_Asn98del | p.Lys59_Asn98del | p.Lys59_Asn98del | p.Lys59_Asn98del | p.Lys59_Asn98del | p.Lys59_Asn98del | p.Gly189Arg | p.Gly189Arg | p.Gly189Arg |
| Inheritance | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR |
| Gender | Female | Female | Male | Male | Female | Female | Female | Female | Male | Female | Male |
| Age | 17y | 2y 5mo | 15 mo (died) | 5y (died) | 2y 5mo (died) | 2y 3mo (died) | 3y 2mo (died) | 1y (died) | 25y | 8y | 13y |
| Ethnicity | Mexican American | Mexican American | Lebanese Australian | Lebanese Australian | Lebanese Australian | Lebanese Australian | Lebanese Australian | Lebanese Australian | Saudi | Saudi | Saudi |
| Consanguinity reported | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Epileptic encephalopathy | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Seizure onset | 6 months | 7 months | 11 weeks | 5 months | 4 months | 7 months | 7 months | 6 months | <1yr | <1yr | <1yr |
| Severe developmental delay | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Profound intellectual disability | Yes | ** | ** | ** | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Acquired microcephaly | Yes | Yes | - | Yes | Yes | No | Yes | No | No | - | - |
| Facial dysmorphisms | Yes | Yes | - | - | - | - | - | - | - | - | - |
| Visual impairment | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | - |
| Retinal dystrophy | No | No | - | Normal ERG, Abnormal VEP | - | Abnormal ERG, Abnormal VEP | Abnormal ERG, Abnormal VEP | Abnormal ERG | - | - | - |
| Hearing impairment | Yes | Yes | - | - | - | - | - | - | - | - | - |
| Severe speech delay | Yes | Yes | - | - | Yes | Yes | Yes | - | - | - | - |
| Central hypotonia | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | - | - | - |
| Peripheral hypertonia | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | - | - | - |
| Poor head control | Yes | Yes | Yes | Yes | Yes | Yes | Yes | - | Yes | Yes | Yes |
| Ataxia | Yes | Yes | - | - | - | - | - | - | Yes | - | Yes |
| Dystonia | Yes | Yes | - | - | Yes | Yes | Yes | Yes | - | - | - |
| Spasticity | Yes | - | - | - | - | Yes | Yes | - | - | - | - |
| Cerebral palsy | Yes | Yes | - | - | - | - | - | - | - | - | - |
| Severe scoliosis | Yesd | No | - | - | - | Yes | Yes | - | - | - | - |
| Diffuse osteopenia | Yes | Yes | - | - | - | - | - | - | - | - | - |
| Deformity of the hip | Yes | Yes | - | - | - | Yes | Yes | - | - | - | - |
| Feeding difficulties | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | - | - | - |
| Aspiration | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | - | - | - |
Mutations are homozygous/biallelic in all patients.
Mutations were confirmed by Sanger Sequencing in all patients except patient 3. Variant in this patient is presumed, not confirmed as patient died before DNA was obtained.
Variant on protein level is p.Thr266_Phe271delinsLeuPhePhePheThrIleCysGlyMetThrSerCysIle.
One of the unaffected siblings also has severe scoliosis.
“-“ indicates no information available
indicates likely, but not formally tested.
Molecular analysis of splice mutation c.674–2A>T
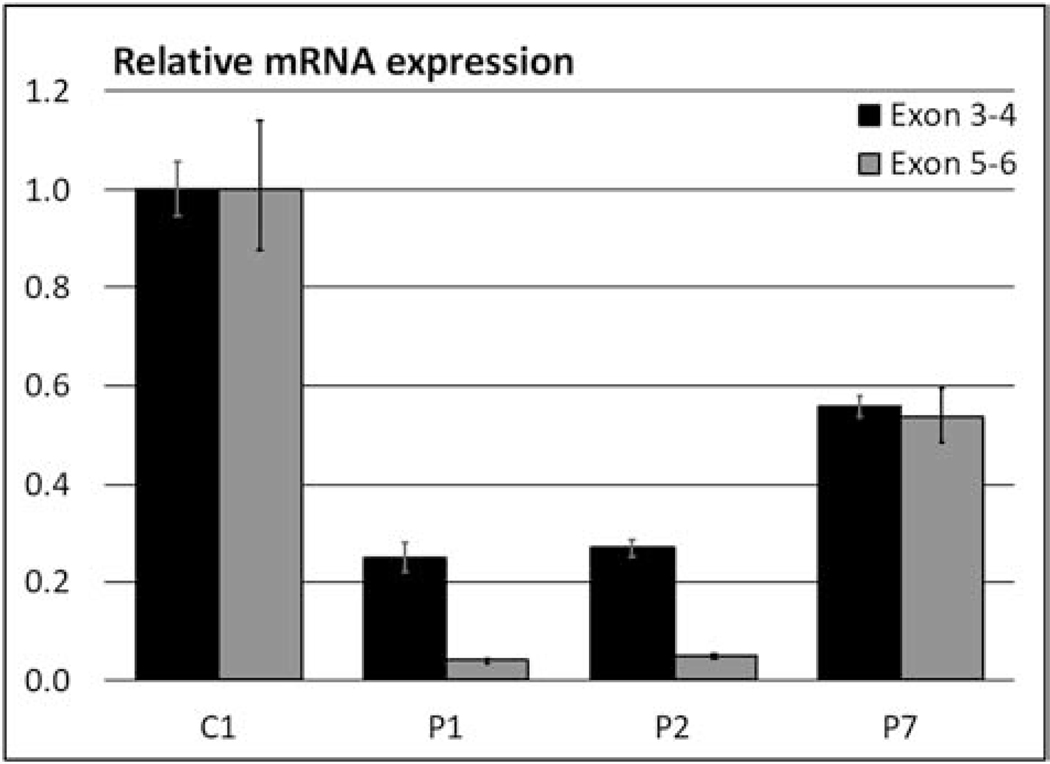
Sanger sequencing of genomic DNA confirmed the homozygous variant in the affected siblings of Family I that was identified by whole exome sequencing (Figure 1C). PCR amplification of cDNA around the predicted splice showed a ∼600nt band in the control and a ∼450nt band in both patients which would be consistent with the skipping of exon 5 (Figure S1A); this too was confirmed by Sanger sequencing of the gel-extracted bands (Figure 1E). Because the skipped exon 5 contains the predicted stop-codon, an alternate stop is utilized from exon 6, which could lead to the formation of either a shorter protein or to nonsense mediated decay. qPCR analysis targeting the exon 3–4 exon junction revealed that there is only 25% residual expression of this part of the gene, and an assay targeting the skipped exon showed that there was no residual expression of the exon 5–6 junction (Figure 2). A western blot also showed a 40–50% reduction in ARV1 protein (Figure S2).
Figure 2. mRNA expression analyses.
Relative mRNA expression at the exon 3–4 junction (black bars) and exon 5–6 junction (grey bars) showed reduced levels of ARV1 mRNA in all patients compared to a pediatric control (C1). For patients 1 and 2 the residually expressed mRNA did not contain exon 5.
Molecular analysis of the previously reported splice mutation c.294+1G>A
Sanger sequencing on genomic DNA confirmed the homozygous variant in the affected cousins of Family II that was identified by whole exome sequencing (Fig. 1D). PCR amplification of fibroblast cDNA around the predicted splice showed a ∼400nt band in the control and a ∼250nt band in patient 7, which is consistent with the skipping of exon 2 (Figure S1B). Sanger sequences of the PCR products confirmed that exon 1 and exon 3 are joined in this patient (Figure 1F), confirming the in-frame deletion of 40 amino acids (p.59–98) that was reported previously [27]. qPCR analysis showed a 45% reduction in mRNA expression by both assays targeting the exon 3–4 and exon 5–6 junctions (Figure 2); western blot analysis showed a 20% decrease in protein levels (Figure S2).
GPI-anchor analysis
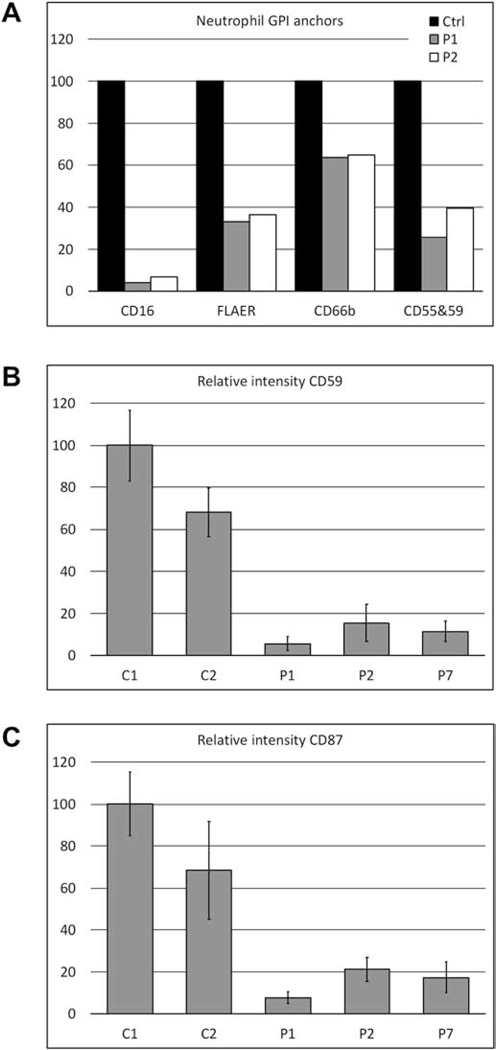
GPI-anchored proteins CD16, CD66b, a combination of CD55 & 59 and FLAER were measured by flow cytometry on neutrophils of patients 1 and 2 (Family I) and revealed significant reductions compared to an unaffected control (Figure 3A and S3). The largest reduction was observed for CD16 (∼5% residual expression); CD66b was the least affected (∼65% residual expression). Since no neutrophils were available for patient 7, fibroblasts were analyzed for the presence of CD59 and CD87, two GPI-anchored proteins expressed on fibroblasts [36]. Both proteins were significantly reduced (<20% residual expression) compared to the control in all three tested patients (Figure 3B, 3C and S4).
Figure 3. GPI-anchor expression on neutrophils and fibroblasts.
A. FACS analysis of the GPI-anchored proteins CD16, FLAER, CD66b and the combined measurement of CD55 and CD59 on neutrophils reveals a significant reduction in mean fluorescence intensity (MFI) for patients 1 and 2 compared to control. B. MFI of CD59 on patient fibroblasts, measured by immunofluorescence confocal microscopy, was less than 20% of the MFI of the adult control (C1). C2 is a pediatric control that had lower CD59 expression C. MFI of CD87 (uPAR) on patient fibroblasts, measured by immunofluorescence confocal microscopy, was less than 25% of the MFI of an adult control (C1). C2 is a pediatric control that had lower CD87 expression.
DISCUSSION
Here we describe seven patients from two unrelated families with early infantile epileptic encephalopathy 38 due to homozygous splice mutations in ARV1. Similar to two previous reports on patients with variants in ARV1, our patients present with early onset seizures, central hypotonia, severe developmental delay, visual impairment and feeding difficulties, supporting the clinical diagnosis of IEE38. The probands in this study had severe speech delay, spasticity, scoliosis and deformity of the hip. The neurodegenerative course (increasing seizures, central hypotonia, peripheral spasticity, ataxia and dystonia) appeared to be progressive in Family II and there was a lack of developmental progress in both families.
Nonetheless, there are clinical features that are different among the probands even within families who all share the same genetic change; hearing impairment, facial dysmorphisms and delayed myelination were only observed in Family I, which may either be a part of the disease spectrum or can be due to other unknown genetic factors. Brain MRI in patient 7 showed normal myelination and absence of hyperintense regions as shown in the patient reported with a similar mutation [27]. In addition, the affected individuals from Family I had osteopenia, although this might be a nonspecific finding, especially in severely delayed, non-weight bearing patients. Identification of more patients with biallelic mutations in ARV1 will likely help clarify the spectrum of this disorder.
A few papers have been published describing mouse models for Arv1 deficiency. Mice that lacked Arv1 constitutively had reduced body weight, fat mass and plasma lipids, even though they consumed more food [37]. In contrast, most of our patients with ARV1 deficiency were above the 90th %-ile for weight at birth but had poor weight gain associated with feeding difficulties and were treated with GI-tube feeding. Tissue-specific (neuronal) deletion of Arv1 in mice, however, resulted to a circling behavior and seizures [27], which can be comparable to what is seen in patients with ARV1 deficiency, and suggest the importance of ARV1 in neuronal cells.
In S. cerevisiae, the function of Arv1 has been studied extensively and has been shown to play roles in lipid and cholesterol homeostasis [38], sphingolipid distribution [39] and glycophosphatidylinositol (GPI)-anchor synthesis [5, 29]. The latter was of specific interest because the phenotypes overlap between our patients and those with other GPI-anchor synthesis disorders [4]. The elevated gamma-glutamyl transferase activity with normal alkaline phosphatase activity seen in Family I has been reported in other GPI-anchor synthesis disorders [4] and the generalized aminoaciduria, suggestive for a proximal kidney tubule dysfunction, has been reported in a variety of congenital disorders of glycosylation [40, 41]. Interestingly, iron-overload, which was documented in the siblings from Family I, was also seen in PIGA- related GPI-anchor disease [42].
Studies in S. cerevisiae pinpointed a major role for Arv1 in the delivery of early GPI-intermediates to the lumen of the ER [5] acting either as the “flippase” or contributing to its function [43]. As a result, loss of Arv1 function may lead to deficiencies in GPI-anchor synthesis and the accumulation of immature GPI-anchors and GPI-anchor proteins in the ER, potentially inducing ER-stress [44]. Lack of GPI-anchor synthesis may also lead to failure of localization and anchoring of GPI-anchored proteins to the membranes at their proper destination. Complementation analysis with human ARV1 suggested conservation of this function [29], which we now confirm with the significant decreases in levels of GPI-anchored proteins on fibroblasts and/or neutrophils in both families described herein. Nonetheless, complementation of the patient fibroblasts with full-length ARV and replication experiments using fibroblasts or blood cells from patients with ARV1 mutations will further solidify the results of this current report.
Although the variants in both families affected the levels of GPI-anchored proteins to a similar degree, all affected children of Family II died before the age of 5, whereas both affected sisters of Family I are still alive at age 4.5 and 17, suggesting that the disease course of Family II is more severe than that of Family I. This may be related to the location of the variants. The loss of exon 5 in Family I removes a predicted helical transmembrane domain, which may lead to failure of proper ARV1 localization to the ER. However, the loss of exon 2 in Family II overlaps with the ARV1 homology domain (AHD), which is a cysteine-rich domain that contains a putative zinc-binding motif [38, 45]. This domain is critical for the function of ARV1 in maintenance of lipid homeostasis and prevention of free sterol toxicity [38] and the loss of this domain may lead to a more severe disease course by affecting multiple functions of ARV1. In conclusion, the phenotypes of patients with ARV1 mutations are consistent with those of other GPI-anchor synthesis disorders and the loss of GPI-anchored proteins in our patients confirms that this function of Arv1 in yeast is conserved in humans. Our results suggest that ARV1-deficiency may be a GPI-anchor synthesis disorder. Further studies that will include a larger number of samples and additional experiments interrogating accumulation of GPI biosynthetic intermediates will be needed to solidify this hypothesis.
Supplementary Material
Table S1: Abnormal laboratory results
Figure S1: Agarose gels for splicing analysis
Figure S2: Western blot analysis comparing ARV1 expression between fibroblasts from patients and controls.
Figure S3: Flow cytometry data of GPI-anchor proteins on neutrophils for patient 1
Figure S4: Representative images for GPI-anchor proteins on fibroblasts by confocal microscopy
ACKNOWLEDGEMNTS
The authors would like to thank all patients and their families for their participation in this research. We also thank Yan Huang for establishing primary fibroblast cultures and for sample preparations. This research was in part supported by the Intramural Research Program of the National Human Genome Research Institute and the NIH Office of the Director’s Common Fund. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. Whole Exome Sequencing of two case samples (Family II) and Sanger validation of eight samples, including relevant bioinformatics data analysis was supported by an Institutional Development Award to the Center of Applied Genomics from The Children’s Hospital of Philadelphia.
Footnotes
DISCLOSURES
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Nosjean O, Briolay A, Roux B, Mammalian GPI proteins: sorting, membrane residence and functions Biochimica et biophysica acta 1331 (1997) 153–186. [DOI] [PubMed] [Google Scholar]
- [2].Paulick MG, Bertozzi CR, The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins Biochemistry 47 (2008) 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kinoshita T, Biosynthesis and deficiencies of glycosylphosphatidylinositol Proceedings of the Japan Academy. Series B, Physical and biological sciences 90 (2014) 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lam C, Golas GA, Davids M, Huizing M, Kane MS, Krasnewich DM, Malicdan MC, Adams DR, Markello TC, Zein WM, Gropman AL, Lodish MB, Stratakis CA, Maric I, Rosenzweig SD, Baker EH, Ferreira CR, Danylchuk NR, Kahler S, Garnica AD, Bradley Schaefer G, Boerkoel CF, Gahl WA, Wolfe LA, Expanding the clinical and molecular characteristics of PIGT-CDG, a disorder of glycosylphosphatidylinositol anchors Molecular genetics and metabolism 115 (2015) 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kajiwara K, Watanabe R, Pichler H, Ihara K, Murakami S, Riezman H, Funato K, Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum Molecular biology of the cell 19 (2008) 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS, Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families Cell reports 10 (2015) 148–161. [DOI] [PubMed] [Google Scholar]
- [7].Almeida AM, Murakami Y, Layton DM, Hillmen P, Sellick GS, Maeda Y, Richards S, Patterson S, Kotsianidis I, Mollica L, Crawford DH, Baker A, Ferguson M, Roberts I, Houlston R, Kinoshita T, Karadimitris A, Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency Nature medicine 12 (2006) 846–851. [DOI] [PubMed] [Google Scholar]
- [8].Barone R, Aiello C, Race V, Morava E, Foulquier F, Riemersma M, Passarelli C, Concolino D, Carella M, Santorelli F, Vleugels W, Mercuri E, Garozzo D, Sturiale L, Messina S, Jaeken J, Fiumara A, Wevers RA, Bertini E, Matthijs G, Lefeber DJ, DPM2-CDG: a muscular dystrophy-dystroglycanopathy syndrome with severe epilepsy Annals of neurology 72 (2012) 550–558. [DOI] [PubMed] [Google Scholar]
- [9].Chiyonobu T, Inoue N, Morimoto M, Kinoshita T, Murakami Y, Glycosylphosphatidylinositol (GPI) anchor deficiency caused by mutations in PIGW is associated with West syndrome and hyperphosphatasia with mental retardation syndrome Journal of medical genetics 51 (2014) 203–207. [DOI] [PubMed] [Google Scholar]
- [10].Edvardson S, Murakami Y, Nguyen TT, Shahrour M, St-Denis A, Shaag A, Damseh N, Le Deist F, Bryceson Y, Abu-Libdeh B, Campeau PM, Kinoshita T, Elpeleg O, Mutations in the phosphatidylinositol glycan C (PIGC) gene are associated with epilepsy and intellectual disability Journal of medical genetics 54 (2017) 196–201. [DOI] [PubMed] [Google Scholar]
- [11].Howard MF, Murakami Y, Pagnamenta AT, Daumer-Haas C, Fischer B, Hecht J, Keays DA, Knight SJ, Kolsch U, Kruger U, Leiz S, Maeda Y, Mitchell D, Mundlos S, Phillips JA 3rd, Robinson PN, Kini U, Taylor JC, Horn D, Kinoshita T, Krawitz PM, Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation American journal of human genetics 94 (2014) 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ilkovski B, Pagnamenta AT, O’Grady GL, Kinoshita T, Howard MF, Lek M, Thomas B, Turner A, Christodoulou J, Sillence D, Knight SJ, Popitsch N, Keays DA, Anzilotti C, Goriely A, Waddell LB, Brilot F, North KN, Kanzawa N, Macarthur DG, Taylor JC, Kini U, Murakami Y, Clarke NF, Mutations in PIGY: expanding the phenotype of inherited glycosylphosphatidylinositol deficiencies Human molecular genetics 24 (2015) 6146–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johnston JJ, Gropman AL, Sapp JC, Teer JK, Martin JM, Liu CF, Yuan X, Ye Z, Cheng L, Brodsky RA, Biesecker LG, The phenotype of a germline mutation in PIGA: the gene somatically mutated in paroxysmal nocturnal hemoglobinuria American journal of human genetics 90 (2012) 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnstone DL, Nguyen TT, Murakami Y, Kernohan KD, Tetreault M, Goldsmith C, Doja A, Wagner JD, Huang L, Hartley T, St-Denis A, le Deist F, Majewski J, Bulman DE, Kinoshita T, Dyment DA, Boycott KM, Campeau PM, Compound heterozygous mutations in the gene PIGP are associated with early infantile epileptic encephalopathy Human molecular genetics 26 (2017) 1706–1715. [DOI] [PubMed] [Google Scholar]
- [15].Krawitz PM, Murakami Y, Hecht J, Kruger U, Holder SE, Mortier GR, Delle Chiaie B, De Baere E, Thompson MD, Roscioli T, Kielbasa S, Kinoshita T, Mundlos S, Robinson PN, Horn D, Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation American journal of human genetics 91 (2012) 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Krawitz PM, Murakami Y, Riess A, Hietala M, Kruger U, Zhu N, Kinoshita T, Mundlos S, Hecht J, Robinson PN, Horn D, PGAP2 mutations, affecting the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation syndrome American journal of human genetics 92 (2013) 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, Isau M, Fischer A, Dahl A, Kerick M, Hecht J, Kohler S, Jager M, Grunhagen J, de Condor BJ, Doelken S, Brunner HG, Meinecke P, Passarge E, Thompson MD, Cole DE, Horn D, Roscioli T, Mundlos S, Robinson PN, Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome Nature genetics 42 (2010) 827–829. [DOI] [PubMed] [Google Scholar]
- [18].Kvarnung M, Nilsson D, Lindstrand A, Korenke GC, Chiang SC, Blennow E, Bergmann M, Stodberg T, Makitie O, Anderlid BM, Bryceson YT, Nordenskjold M, Nordgren A, A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT Journal of medical genetics 50 (2013) 521–528. [DOI] [PubMed] [Google Scholar]
- [19].Makrythanasis P, Kato M, Zaki MS, Saitsu H, Nakamura K, Santoni FA, Miyatake S, Nakashima M, Issa MY, Guipponi M, Letourneau A, Logan CV, Roberts N, Parry DA, Johnson CA, Matsumoto N, Hamamy H, Sheridan E, Kinoshita T, Antonarakis SE, Murakami Y, Pathogenic Variants in PIGG Cause Intellectual Disability with Seizures and Hypotonia American journal of human genetics 98 (2016) 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Marcelis CL, Rieu P, Beemer F, Brunner HG, Severe mental retardation, epilepsy, anal anomalies, and distal phalangeal hypoplasia in siblings Clinical dysmorphology 16 (2007) 73–76. [DOI] [PubMed] [Google Scholar]
- [21].Martin HC, Kim GE, Pagnamenta AT, Murakami Y, Carvill GL, Meyer E, Copley RR, Rimmer A, Barcia G, Fleming MR, Kronengold J, Brown MR, Hudspith KA, Broxholme J, Kanapin A, Cazier JB, Kinoshita T, Nabbout R, Bentley D, McVean G, Heavin S, Zaiwalla Z, McShane T, Mefford HC, Shears D, Stewart H, Kurian MA, Scheffer IE, Blair E, Donnelly P, Kaczmarek LK, Taylor JC, Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis Human molecular genetics 23 (2014) 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Messina S, Tortorella G, Concolino D, Spano M, D’Amico A, Bruno C, Santorelli FM, Mercuri E, Bertini E, Congenital muscular dystrophy with defective alpha-dystroglycan, cerebellar hypoplasia, and epilepsy Neurology 73 (2009) 1599–1601. [DOI] [PubMed] [Google Scholar]
- [23].Murakami Y, Tawamie H, Maeda Y, Buttner C, Buchert R, Radwan F, Schaffer S, Sticht H, Aigner M, Reis A, Kinoshita T, Jamra RA, Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy PLoS genetics 10 (2014) e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ng BG, Hackmann K, Jones MA, Eroshkin AM, He P, Wiliams R, Bhide S, Cantagrel V, Gleeson JG, Paller AS, Schnur RE, Tinschert S, Zunich J, Hegde MR, Freeze HH, Mutations in the glycosylphosphatidylinositol gene PIGL cause CHIME syndrome American journal of human genetics 90 (2012) 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nguyen TTM, Murakami Y, Sheridan E, Ehresmann S, Rousseau J, St-Denis A, Chai G, Ajeawung NF, Fairbrother L, Reimschisel T, Bateman A, Berry-Kravis E, Xia F, Tardif J, Parry DA, Logan CV, Diggle C, Bennett CP, Hattingh L, Rosenfeld JA, Perry MS, Parker MJ, Le Deist F, Zaki MS, Ignatius E, Isohanni P, Lonnqvist T, Carroll CJ, Johnson CA, Gleeson JG, Kinoshita T, Campeau PM, Mutations in GPAA1, Encoding a GPI Transamidase Complex Protein, Cause Developmental Delay, Epilepsy, Cerebellar Atrophy, and Osteopenia American journal of human genetics 101 (2017) 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ohba C, Okamoto N, Murakami Y, Suzuki Y, Tsurusaki Y, Nakashima M, Miyake N, Tanaka F, Kinoshita T, Matsumoto N, Saitsu H, PIGN mutations cause congenital anomalies, developmental delay, hypotonia, epilepsy, and progressive cerebellar atrophy Neurogenetics 15 (2014) 85–92. [DOI] [PubMed] [Google Scholar]
- [27].Palmer EE, Jarrett KE, Sachdev RK, Al Zahrani F, Hashem MO, Ibrahim N, Sampaio H, Kandula T, Macintosh R, Gupta R, Conlon DM, Billheimer JT, Rader DJ, Funato K, Walkey CJ, Lee CS, Loo C, Brammah S, Elakis G, Zhu Y, Buckley M, Kirk EP, Bye A, Alkuraya FS, Roscioli T, Lagor WR, Neuronal deficiency of ARV1 causes an autosomal recessive epileptic encephalopathy Human molecular genetics 25 (2016) 3042–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pagnamenta AT, Murakami Y, Taylor JM, Anzilotti C, Howard MF, Miller V, Johnson DS, Tadros S, Mansour S, Temple IK, Firth R, Rosser E, Harrison RE, Kerr B, Popitsch N, Kinoshita T, Taylor JC, Kini U, Analysis of exome data for 4293 trios suggests GPI-anchor biogenesis defects are a rare cause of developmental disorders European journal of human genetics : EJHG 25 (2017) 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ikeda A, Kajiwara K, Iwamoto K, Makino A, Kobayashi T, Mizuta K, Funato K, Complementation analysis reveals a potential role of human ARV1 in GPI anchor biosynthesis Yeast (Chichester, England: ) 33 (2016) 37–42. [DOI] [PubMed] [Google Scholar]
- [30].Gahl WA, Tifft CJ, The NIH Undiagnosed Diseases Program: lessons learned JAMA 305 (2011) 1904–1905. [DOI] [PubMed] [Google Scholar]
- [31].Gahl WA, Mulvihill JJ, Toro C, Markello TC, Wise AL, Ramoni RB, Adams DR, Tifft CJ, Udn, The NIH Undiagnosed Diseases Program and Network: Applications to modern medicine Molecular genetics and metabolism 117 (2016) 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gahl WA, Markello TC, Toro C, Fajardo KF, Sincan M, Gill F, Carlson-Donohoe H, Gropman A, Pierson TM, Golas G, Wolfe L, Groden C, Godfrey R, Nehrebecky M, Wahl C, Landis DMD, Yang S, Madeo A, Mullikin JC, Boerkoel CF, Tifft CJ, Adams D, Progr NCS, The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases Genetics in Medicine 14 (2012) 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ramoni RB, Mulvihill JJ, Adams DR, Allard P, Ashley EA, Bernstein JA, Gahl WA, Hamid R, Loscalzo J, McCray AT, Shashi V, Tifft CJ, Wise AL, Network UD, The Undiagnosed Diseases Network: Accelerating Discovery about Health and Disease American journal of human genetics 100 (2017) 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nafisinia M, Guo Y, Dang X, Li J, Chen Y, Zhang J, Lake NJ, Gold WA, Riley LG, Thorburn DR, Keating B, Xu X, Hakonarson H, Christodoulou J, Whole Exome Sequencing Identifies the Genetic Basis of Late-Onset Leigh Syndrome in a Patient with MRI but Little Biochemical Evidence of a Mitochondrial Disorder JIMD Rep 32 (2017) 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pode-Shakked B, Heimer G, Vilboux T, Marek-Yagel D, Ben-Zeev B, Davids M, Ferreira CR, Philosoph AM, Veber A, Pode-Shakked N, Kenet G, Soudack M, Hoffmann C, Vernitsky H, Safaniev M, Lodzki M, Lahad A, Shouval DS, Levinkopf D, Weiss B, Barg AA, Daka A, Amariglio N, Malicdan MCV, Gahl WA, Anikster Y, Cerebral and portal vein thrombosis, macrocephaly and atypical absence seizures in Glycosylphosphatidyl inositol deficiency due to a PIGM promoter mutation Molecular genetics and metabolism 128 (2019) 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Knaus A, Awaya T, Helbig I, Afawi Z, Pendziwiat M, Abu-Rachma J, Thompson MD, Cole DE, Skinner S, Annese F, Canham N, Schweiger MR, Robinson PN, Mundlos S, Kinoshita T, Munnich A, Murakami Y, Horn D, Krawitz PM, Rare Noncoding Mutations Extend the Mutational Spectrum in the PGAP3 Subtype of Hyperphosphatasia with Mental Retardation Syndrome Human mutation 37 (2016) 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lagor WR, Tong F, Jarrett KE, Lin W, Conlon DM, Smith M, Wang MY, Yenilmez BO, McCoy MG, Fields DW, O’Neill SM, Gupta R, Kumaravel A, Redon V, Ahima RS, Sturley SL, Billheimer JT, Rader DJ, Deletion of murine Arv1 results in a lean phenotype with increased energy expenditure Nutr Diabetes 5 (2015) e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tinkelenberg AH, Liu Y, Alcantara F, Khan S, Guo Z, Bard M, Sturley SL, Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1 The Journal of biological chemistry 275 (2000) 40667–40670. [DOI] [PubMed] [Google Scholar]
- [39].Swain E, Stukey J, McDonough V, Germann M, Liu Y, Sturley SL, Nickels JT Jr., Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism. Complementation by human ARV1 The Journal of biological chemistry 277 (2002) 36152–36160. [DOI] [PubMed] [Google Scholar]
- [40].de Lonlay P, Seta N, Barrot S, Chabrol B, Drouin V, Gabriel BM, Journel H, Kretz M, Laurent J, Le Merrer M, Leroy A, Pedespan D, Sarda P, Villeneuve N, Schmitz J, van Schaftingen E, Matthijs G, Jaeken J, Korner C, Munnich A, Saudubray JM, Cormier-Daire V, A broad spectrum of clinical presentations in congenital disorders of glycosylation I: a series of 26 cases Journal of medical genetics 38 (2001) 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Scott K, Gadomski T, Kozicz T, Morava E, Congenital disorders of glycosylation: new defects and still counting Journal of inherited metabolic disease 37 (2014) 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Swoboda KJ, Margraf RL, Carey JC, Zhou H, Newcomb TM, Coonrod E, Durtschi J, Mallempati K, Kumanovics A, Katz BE, Voelkerding KV, Opitz JM, A novel germline PIGA mutation in Ferro-Cerebro-Cutaneous syndrome: a neurodegenerative X-linked epileptic encephalopathy with systemic iron-overload Am J Med Genet A 164A (2014) 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fujita M, Kinoshita T, Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins FEBS letters 584 (2010) 1670–1677. [DOI] [PubMed] [Google Scholar]
- [44].Shechtman CF, Henneberry AL, Seimon TA, Tinkelenberg AH, Wilcox LJ, Lee E, Fazlollahi M, Munkacsi AB, Bussemaker HJ, Tabas I, Sturley SL, Loss of subcellular lipid transport due to ARV1 deficiency disrupts organelle homeostasis and activates the unfolded protein response The Journal of biological chemistry 286 (2011) 11951–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fores O, Arro M, Pahissa A, Ferrero S, Germann M, Stukey J, McDonough V, Nickels JT Jr., Campos N, Ferrer A, Arabidopsis thaliana expresses two functional isoforms of Arvp, a protein involved in the regulation of cellular lipid homeostasis Biochimica et biophysica acta 1761 (2006) 725–735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Abnormal laboratory results
Figure S1: Agarose gels for splicing analysis
Figure S2: Western blot analysis comparing ARV1 expression between fibroblasts from patients and controls.
Figure S3: Flow cytometry data of GPI-anchor proteins on neutrophils for patient 1
Figure S4: Representative images for GPI-anchor proteins on fibroblasts by confocal microscopy