Abstract
BACKGROUND
Disease-related single nucleotide polymorphisms (SNPs) based genetic risk score (GRS) has been proven to provide independent inherited risk other than family history in multiple cancer types.
AIM
To evaluate the potential of GRS in the prediction of pancreatic cancer risk.
METHODS
In this case-control study (254 cases and 1200 controls), we aimed to evaluate the association between GRS and pancreatic ductal adenocarcinoma (PDAC) risk in the Chinese population. The GRS was calculated based on the genotype information of 18 PDAC-related SNPs for each study subject (personal genotyping information of the SNPs) and was weighted by external odd ratios (ORs).
RESULTS
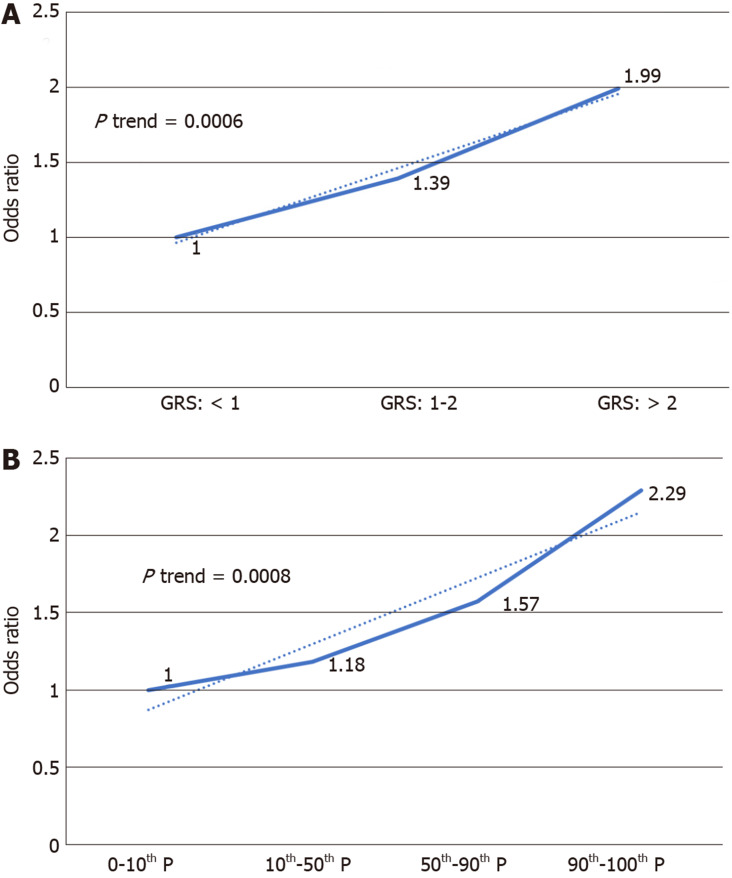
GRS was significantly different in cases and controls (1.96 ± 3.84 in PDACs vs 1.09 ± 0.94 in controls, P < 0.0001). Logistic regression revealed GRS to be associated with PDAC risk [OR = 1.23, 95% confidence interval (CI): 1.13-1.34, P < 0.0001]. GRS remained significantly associated with PDAC (OR = 1.36, 95%CI: 1.06-1.74, P = 0.015) after adjusting for age and sex. Further analysis revealed an association of increased risk for PDAC with higher GRS. Compared with low GRS (< 1.0), subjects with high GRS (2.0) were 99% more likely to have PDAC (OR: 1.99, 95%CI: 1.30-3.04, P = 0.002). Participants with intermediate GRS (1.0-1.9) were 39% more likely to have PDAC (OR: 1.39, 95%CI: 1.03-1.84, P = 0.031). A positive trend was observed (P trend = 0.0006).
CONCLUSION
GRS based on PDAC-associated SNPs could provide independent information on PDAC risk and may be used to predict a high risk PDAC population.
Keywords: Pancreatic cancer, Single nucleotide polymorphisms, Genetic risk score, Chinese population, Genome-wide association study
Core tip: Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant tumor with no effective method for early diagnosis and high-risk population screening. In this pioneer study, we evaluated single nucleotide polymorphisms based genetic risk score (GRS) in the prediction of PDAC risk. Our results revealed that GRS was significantly associated with PDAC. Compared with low GRS (< 1.0), subjects with high GRS (2.0) were 99% more likely to be PDAC. Although further verification is needed, our study suggested that GRS was an independent risk factor for PDAC.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies worldwide. With an estimated death toll of 330400 in 2012, it is the seventh leading cause of cancer death in both sexes[1]. In China, PDAC ranks among the top ten most common cancers. The estimated new cases and death toll in 2015 was 90100 and 79400, respectively[2,3]. In addition, data showed that PDAC mortalities have increased rapidly in China during the past decades. There were 27482 deaths in 1990, while the number increased to 59423 in 2013[4]. Therefore, PDAC has become a major public health issue both in China and worldwide.
As one of the most lethal cancers, early diagnosis of PDAC is essential to effective treatment and better prognosis. Unfortunately, due to the lack of symptoms at an early stage and no efficient way of screening, early diagnosis of PDAC remains challenging[5]. Carbohydrate antigen 19-9 and carcinoembryonic antigen are commonly used biomarkers. However, carbohydrate antigen 19-9 and carcino-embryonic antigen are not PDAC specific, and a small group of patients do not show an elevated level of these two biomarkers[6,7]. Computed tomography or other imaging technologies may increase diagnostic utilities for PDAC, but no evidence shows their benefits in screening. It will be less cost-effective to screen the entire population. A model to select a high-risk population group for screening would be of great clinical utilization and public health importance. Known risk factors for PDAC includes diabetes[8], smoking[9] and family history.
About 5%-10% of PDAC cases are familial pancreatic cancer, defined as the patient having two first degree relatives diagnosed with pancreatic cancer[10]. Those patients are usually linked with germline mutation (BRCA1, BRCA2)[11] or cancer syndrome (Lynch syndrome)[12]. However, with a trend of decreased family sizes[13], family history could be uninformative, especially in China where the “one-child policy” had been applied in the past decades. Disease-related single nucleotide polymorphisms (SNPs) based genetic risk score (GRS) could be a promising substitute. It has been proven to provide independent inherited risk information other than family history in multiple cancer types and can be associated with early onset of diseases[14-17]. PDAC risk associated SNPs were revealed by genome-wide association studies (GWAS)[18-22]. The application of these results needs further investigation. Therefore, we conducted the first study to evaluate the association between GRS and PDAC in the Chinese population.
MATERIALS AND METHODS
Study population and genotyping
This study included 254 pathologically confirmed PDAC patients of Chinese Han population from the Department of Pancreatic Surgery in Huashan Hospital diagnosed between March 2013 and August 2014. They had been recruited for a PDAC-associated SNP verification study[23]. The control population was a healthy community population from east China[24]. Written informed consent was obtained from each participant. Data was de-identified after collection. Genotype and phenotype information was retrospectively collected from the previous studies. This study was approved by the Institutional Review Board of Huashan Hospital affiliated to Fudan University.
Genotype data of 21 SNPs was obtained from our previous SNP evaluation study. These SNPs were reported to be associated with PDAC risk by GWAS or pathway study[18-22,25]. SNPs were genotyped by a Sequenom MassARRY iPLEX system (Sequenome Inc., San Diego, CA, United States) and Taqman PCR (rs4885093 and rs10919791)[23]. GWAS was performed by Illumina Human OmniExpress Bead Chips[24] on the control samples. Imputation was performed using IMPUTE 2.2.2 program based on 1000 Genomes Project CHB+JPT population data (Phase I version 3, release March 2012) if the SNPs were not included in the GWAS chip.
GRS calculation and statistical analysis
A GRS was calculated for each subject based on personal genotype of the SNPs and was weighted by external odd ratios (ORs from previous GWAS studies): Carrying two risk alleles = 1 * OR2; carrying one risk allele = 1 * OR; and not carrying risk allele = 1. The final GRS was also adjusted by minor allele frequency. A detailed method of GRS calculation was described in the previous study[17]. The OR used for calculations were from previous GWAS studies. In this study, 18 out of 21 SNPs were used in the final calculation of GRS. rs2736098 did not pass quality control. rs12413624 and rs792864 showed different minor allele between case and control group, possibly due to the difference between positive and negative chains of DNA during genotyping (the cases and controls were genotyped separately with different platforms). This was not a problem for OR calculations in the previous study. But for GRS calculation, we just ruled out these two SNPs for caution. Information of all 21 SNPs was displayed in Table 1.
Table 1.
Information of single nucleotide polymorphisms for calculating genetic risk score
| Chromosome | SNP | Region | Related-gene |
Previous GWAS study |
|||
| OP1 | RA | OR | RAF | ||||
| 1 | rs10919791 | 1q32.1 | NR5A2 | EU | A | 0.77 | 0.19 |
| 5 | rs2736098 | 5p15.33 | TERT, CLPTM1L | EU | T | 0.8 | NA1 |
| 5 | rs401681 | 5p15.33 | CLPTM1L | EU | T | 1.19 | 0.49 |
| 5 | rs2255280 | 5p13.1 | DAB2 | CH | G | 0.81 | 0.32 |
| 6 | rs2317900 | 6p25.3 | FOXQ1 | JA | C | 1.288 | 0.41 |
| 7 | rs6971499 | 7q32.3 | LINC-PINT | EU | C | 0.79 | NA1 |
| 7 | rs7779540 | 7q36.2 | DPP6 | JA | A | 3.69 | 0.17 |
| 7 | rs167020 | 7q36.3 | SHH | EU | A | 1.17 | 0.3 |
| 8 | rs1561927 | 8q24.21 | MIR1208, PVT1 | EU | C | 0.87 | NA1 |
| 9 | rs2073828 | 9q34.2 | ABO | EU | A | 0.85 | 0.37 |
| 9 | rs505922 | 9q34.2 | ABO | EU | C | 1.2 | 0.35 |
| 10 | rs12413624 | 10q26.11 | PRLHR | CH | T | 1.23 | 0.42 |
| 12 | rs792864 | 12p11.21 | BICD1 | JA | A | 0.71 | 0.24 |
| 13 | rs9581943 | 13q12.2 | PDX1 | EU | A | 1.46 | NA1 |
| 13 | rs4885093 | 13q22.1 | NA | CH | C | 1.25 | 0.5 |
| 13 | rs9543325 | 13q22.1 | NA | EU | C | 1.26 | 0.37 |
| 16 | rs7190458 | 16q23.1 | BCAR1, CTRB1, CTRB2 | EU | A | 1.46 | NA1 |
| 21 | rs372883 | 21q21.3 | BACH1 | CH | C | 0.79 | 0.39 |
| 21 | rs1547374 | 21q22.3 | TFF1 | CH | G | 0.79 | 0.4 |
| 22 | rs16986825 | 22q12.1 | ZNRF3 | EU | T | 1.18 | NA1 |
| 22 | rs5768709 | 22q13.32 | FAM19A5 | CH | G | 1.25 | 0.28 |
Not available. These loci were not included in the final calculation of genetic risk score. OP: Original genome-wide association study population; RA: Risk allele; OR: Odds ratio from genome-wide association studies; RAF: Risk allele frequency in our study; EU: European; JA: Japanese; CH: Chinese; GWAS: Genome-wide association studies.
The t-test was used to evaluate the differences of mean GRS mean between cases and controls. GRS data first underwent log transformation to achieve normality for the t-test. After the log transformation, GRS of both groups are normally distributed (Supplementary Figure 1). GRS were not log transformed for other analysis. A univariate and a multivariate mode controlled for age and sex were used to evaluate the association between GRS and PDAC risk. Chi-square trend tests and receiver operating characteristic curve was used to evaluate the performance of GRS in predicting PDAC risk. All statistical analyses were performed using SAS 9.3, and two-tailed P < 0.05 were considered as significant.
The statistical methods of this study were reviewed by Wang R from Yale School of Public Health, Department of Chronic Disease Epidemiology.
RESULTS
Table 2 shows the basic characteristics of the study population. The control group was significantly younger (case vs control: 63.31 ± 10.01 vs 48.80 ± 15.49, P < 0.01). All tested SNPs were polymorphic in the Chinese population. The prevalence of minor allele of SNPs in the Chinese 1000 genome project and the PDAC patients of our study were presented in our previous study[23]. The mean GRS for PDAC patients was 1.96 ± 3.84, and the mean for controls was 1.09 ± 0.94. The GRS for the two groups were significantly different (P < 0.0001). Further univariate logistic regression mode revealed that GRS was positively associated with PDAC risk [Table 3, OR = 1.23, 95% confidence interval (CI): 1.13-1.34, P < 0.0001]. Because the control group was significantly younger, a multivariate model was also conducted with age and sex as covariates. The GRS remained positively associated with PDAC (Table 3, OR = 1.36, 95%CI: 1.06-1.74, P = 0.015).
Table 2.
Characteristics of study population
| Variables | Case, n = 254 | Control, n = 1200 | P value | |||
| Sex | n | % | n | % | ||
| Male | 156 | 64.42 | 748 | 62.23 | 0.831 | |
| Female | 98 | 38.58 | 452 | 37.67 | ||
| Age at diagnosis, mean ± SD | 63.31 | 10.01 | 48.80 | 15.49 | 0.00012 | |
| GRS, mean ± SD | 1.96 | 3.84 | 1.09 | 0.94 | < 0.00013 | |
| GRS: < 1 | 127 | 50.00 | 723 | 60.25 | 0.0021 | |
| GRS: 1-2 | 91 | 35.83 | 374 | 31.17 | ||
| GRS: > 2 | 36 | 14.17 | 103 | 8.58 | ||
| < 10th P | 19 | 7.48 | 127 | 10.58 | 0.0071 | |
| 10th-50th P | 87 | 34.25 | 494 | 41.17 | ||
| 50th-90th P | 111 | 43.70 | 471 | 39.25 | ||
| 90th P | 37 | 14.57 | 108 | 9.00 | ||
Chi-square.
t-test.
Genetic risk score log transformed for t-test. GRS: Genetic risk score.
Table 3.
Univariate/multivariate logistic regression results
| Univariate logistic regression | OR (95%CI) | P value | |
| GRS | 1.23 (1.13-1.34) | < 0.0001 | |
| Multivariate logistic regression | OR (95%CI) | P value | |
| GRS | 1.36 (1.06-1.74) | 0.015 | |
| Age at diagnosis/enrollment | 1.14 (1.12-1.17) | < 0.0001 | |
| Sex | 0.84 (0.61-1.16) | 0.28 | |
| Subgroup analysis1 | OR (95%CI) | P value | |
| GRS: < 1 (Ref)2 | - | - | |
| GRS: 1-2 | 1.39 (1.03-1.84) | 0.031 | |
| GRS: > 2 | 1.99 (1.30-3.04) | 0.002 | |
| < 10th P (Ref)3 | - | - | |
| 10th-50th P | 1.18 (0.69-2.01) | 0.548 | |
| 50th-90th P | 1.57 (0.93-2.66) | 0.089 | |
| > 90th P | 2.29 (1.25-4.21) | 0.007 | |
Univariate mode.
P trend = 0.0006.
P trend = 0.0008. OR: Odds ratio; CI: Confidence interval; GRS: Genetic risk score.
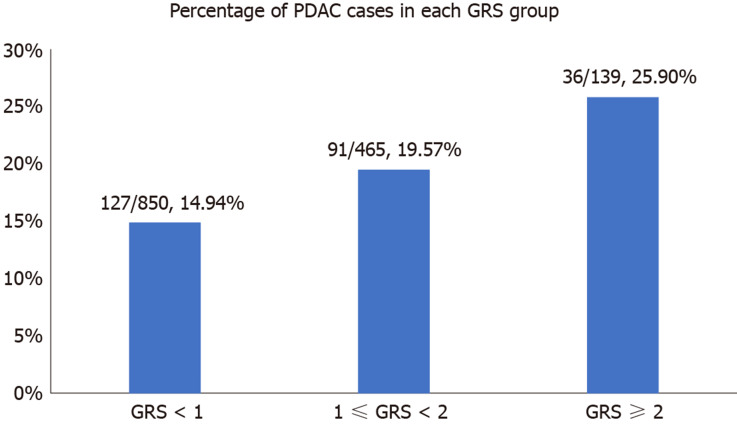
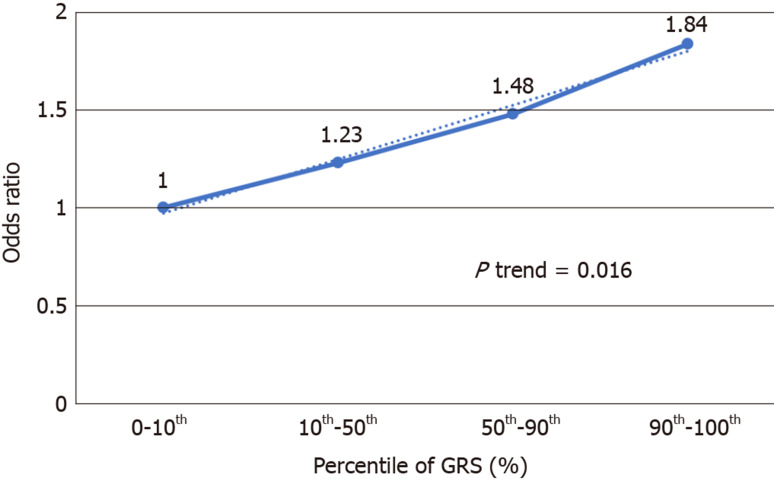
We further analyzed the risk of PDAC in different GRS groups among the study population. Compared to those with low GRS (< 1), participants with intermediate GRS (1-2) were 39% more likely to have PDAC (OR: 1.39, 95%CI: 1.03-1.84, P = 0.031). Those with high GRS were nearly two times more likely to have PDAC (OR: 1.99, 95%CI: 1.30-3.04, P = 0.002). A positive trend was also observed (P trend = 0.0006, Figure 1A). Figure 1B showed the increased risks of PDAC in patients with increased GRS by different percentiles (P trend = 0.0008). Patients with GRS ≥ 90th percentile would have a 2.29-fold increased risk for PDAC (95%CI: 1.25-4.21, P = 0.007). The receiver operating characteristic curve analysis showed that the area under the receiver operating characteristic curve (AUC) was 0.5675 for predicting PDAC risk (Supplementary Figure 2). A positive trend was also observed in the multivariate model, which adjusted for age and sex. The percentage of PDAC cases with low (< 1.0), intermediate (1.0-1.9) and high (≥ 2.0) GRS were 14.49%, 19.57% and 25.90%, respectively (Figure 2). Patients with GRS ≥ 90th percentile showed a 1.84-fold increased risk for pancreatic cancer in the multivariate mode (Figure 3).
Figure 1.
Increased risk of pancreatic ductal adenocarcinoma with increased genetic risk score in a univariate model. A: 1, 2 being genetic risk score cut-off value; B: Percentile as cut-off value. GRS: Genetic risk score.
Figure 2.
Percentage of pancreatic ductal adenocarcinoma cases in different genetic risk score groups. PDAC: Pancreatic ductal adenocarcinoma; GRS: Genetic risk score.
Figure 3.
Increased risk of pancreatic ductal adenocarcinoma with increased genetic risk score in multivariate model adjusted for age and sex. GRS: Genetic risk score.
DISCUSSION
To our best knowledge, this was the first study to evaluate the performance of SNPs based GRS for predicting PDAC risk in the Chinese population. In this case-control study, we found that: (1) GRS was an independent predictor of PDAC; and (2) As reflecting inherited risks, patients with higher GRS would have higher risks of PDAC in the study population.
Chronic disease such as heart disease and cancer have a complex etiology. Genetic factors and lifestyle factors both contribute to the development of these chronic diseases[26,27]. With the surge of GWAS, common germ line variations have been studied for their association with heart disease and many cancers. GRS is a disease risk prediction system based on disease risk related SNPs. It has been applied in the prediction of heart disease[28,29], obesity[30], prostate cancer[15,16,31] and colorectal cancer (CRC)[32,33].
Ripatti et al[28] and Thanassoulis et al[29] both reported GRS to be an independent risk factor for cardiovascular disease. However, GRS did not improve the risk stratification system with traditional risk factors and family history in their studies. Belsky et al[30] reported GRS to be a statistically significant predictor of body mass index and obesity in the white population of the Atherosclerosis Risk in Communities cohort (AUC: 0.57, 95%CI: 0.55-0.58)[30].
In CRC, Weigl et at[33] reported GRS, independent of family history, was associated with CRC risk (OR = 3.00, 95%CI: 2.24-4.02, highest vs lowest decile). More importantly, their study revealed that patients with both GRS in the highest decile and a family history had a 6-fold increased risk for CRC compared those who with no family history and a GRS in the lowest decile. Similar results were also reported by Jo et al[32]. Individuals with a family history of CRC in the highest quartile of GRS when compared to subjects without a family history of CRC in the lowest quartile of GRS had a significantly increased risk for CRC [OR: 47.9, 95%CI: 4.9-471.8 (men); OR: 22.3, 95%CI: 1.4-344.2 (women)].
In prostate cancer, researchers focused on the implementation of GRS to reduce unnecessary biopsies. Aly et al[15] reported the use of GRS could avoid 480 biopsies (22.7%) at a cost of missing a prostate cancer diagnosis in 3% of patients characterized as having an aggressive disease. Kader et al[31] reported adding the genetic score to the best clinical model improved the AUC from 0.62 to 0.66 (P < 0.001). Sun et al[16]compared the family history and GRS in five different populations and found that the AUC of GRS for predicting positive prostate cancer biopsy was significantly higher (0.58-0.62) than family history (0.51-0.55) in each study population (P < 0.05).
For PDAC, previous studies suggested genetic inheritance contributes to the risk of developing PDAC. Lochan et al[34] reported that individuals with first degree relatives of any malignancy would have 1.98-fold increased risk of PDAC. In another study, individuals with positive family history of pancreatic cancer were reported to have a 2.2-fold increased risk compared to those with negative family history[35]. Mucci et al[36] reported that when one twin had PDAC, the risks of PDAC increased by 4.3-fold for monozygotic twin and 3.7-fold for dizygotic twins. However, familial pancreatic carcinoma only counts for 5%-10% of total cases. Furthermore, family history may be influenced by family size, age and survival status of male relatives, recall ability and prevalence of the disease in populations[16]. The overall family history reported rate was only 64.3% for cases and 62.5% for controls in the PanScan Consortium[37].
Various loci related to PDAC risk from different populations were reported[18-22]. However, the practical value of those loci from GWAS has not been fully studied. In this study, we explored the potential association between GRS and PDAC risk to find a novel method to define a pancreatic cancer high risk group. As mentioned above, there is no effective screening method for pancreatic cancer, and the downsize of households often leaves family history uninformative. Similar to previous studies in prostate cancer and CRC[15,16,31-33], we found an association between GRS and PDAC (OR = 1.36). More importantly, an increasing trend was observed. Higher GRS was associated with a higher risk of PDAC in our study population. The only other study that constructed a PDAC risk prediction model using SNPs was reported by Klein et al[37]. Three SNPs from PanScan population based GWAS (rs9543325, rs401681, rs3790844) were included in the model[22]. Other nongenetic factors included in the model were smoking, diabetes, alcohol consumption, ABO blood type, body mass index and family history. In Klein et al[37], the AUC of the risk model with only SNPs was 0.57, while the number was 0.58 for risk model with only nongenetic factors. A model with both genetic and nongenetic factors had a significantly larger AUC (0.61) than any other model (P < 0.0001). Due to the retrospective design of our study, we were not able to collect nongenetic risk factor from de-identified data. We included all loci from precious GWAS to construct a model with 18 SNPs. The AUC of our SNPs model (0.57) was close to the PanScan study. This may indicate that some common variation is shared by populations with different ancestors, and GRS could provide independent information on PDAC risk in the Chinese population. Although the result may need further confirmation, our study showed the potential of GRS in a PDAC high risk population selection. GRS could be applied in selecting high risk individuals for further tests, such as computed tomography or endoscopic ultrasound.
Several limitations should be noticed. First, this is a retrospective case-control study with a relatively small sample size of PDAC patients. The results of the study should be validated in a larger cohort before being applied in clinical use. Second, we were unable to evaluate the relationship of family history and GRS due to the uninformative family history in this Chinese population and the nature of our study design. However, based on the reported studies of other cancers, we believed that GRS could provide independent inherited risks supplementary to family history. Third, the cutoff values of GRS in the trend analysis (e.g., GRS = 1 or 2; 10th percentile, 50th percentile, 90th percentile) were chosen based on the frequency distribution of GRS. The GRS was normally distributed after log transformation. However, from the aspect of application, it is better to choose the cut-off value on the original GRS. Due to the relatively small sample size, our GRS frequency distribution may not represent the true distribution, and one may argue our cutoff values to be relatively subjective. This should not discourage the use of GRS because population average risks were considered when calculating GRS (carrying two risk alleles = 1 * OR2, carrying one risk allele = 1 * OR, not carrying risk allele = 1). GRS over one would indicate an increased inherited risk while GRS less than one would indicate a decreased inherited risk. In fact, population average level of GRS would always be slightly over one. The definition of controls is population with the potential to become cases. The GRS of our control group was 1.09 ± 0.94. The result fits the epidemiology principle and may indicate that a small number of individuals had increased inherited risks, but the disease had not occurred at the time of enrollment.
In conclusion, PDAC-associated SNPs based GRS could provide information on PDAC risk. This finding might be applied in clinical use for personal screening of PDAC after a validation in a larger cohort.
ARTICLE HIGHLIGHTS
Research background
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies worldwide. Lacking effective methods for screening, the early diagnosis of PDAC remains challenging leading to an extremely poor prognosis of PDAC.
Research motivation
Single nucleotide polymorphisms based genetic risk score (GRS) has been proven to provide independent inherited risk information in other cancers. GRS may be a promising way to select a high risk PDAC population for further screening.
Research objectives
We constructed a GRS based on 18 PDAC related single nucleotide polymorphisms, and we evaluated the effectiveness of GRS in the prediction of PDAC risk.
Research methods
We used personal genotyping data to calculate individual GRS. GRS was also weighted by population odds ratio. Final GRS was evaluated for the prediction of PDAC risk in the general Chinese population.
Research results
GRS was significantly associated with PDAC risk after being adjusted for age and sex (odds ratio = 1.36, 95% confidence interval: 1.06-1.74, P = 0.015). Higher GRS indicated a higher risk for PDAC (odds ratio = 2.29, 95% confidence interval: 1.25-4.21, P = 0.007, highest decile vs lowest decile). The area under the curve for GRS for PDAC risk was 0.5675.
Research conclusions
GRS was an independent predictor of PDAC. As reflecting inherited risks, patients with higher GRS would have higher risks of PDAC in the study population.
Research perspectives
GRS could provide independent risk information for PDAC. Further cohort study with a larger sample size may focus on the optimal PDAC risk prediction model built with both GRS and nongenetic factors.
ACKNOWLEDGEMENTS
We thank all participants who contributed to this research.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Shanghai Huashan Hospital Institutional Review Board.
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
STROBE statement: The authors have read the STROBE Statement Checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Manuscript source: Invited manuscript
Peer-review started: December 30, 2019
First decision: January 11, 2020
Article in press: May 13, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dambrauskas Z, Mastoraki A, Sameer S S-Editor: Yan JP L-Editor: Filipodia E-Editor: Liu JH
Contributor Information
Xiao-Yi Wang, Department of Pancreatic Surgery, Pancreatic Disease Institute, Huashan Hospital, Fudan University, Shanghai 200040, China.
Hai-Tao Chen, State Key Laboratory of Organ Failure Research, Guangdong Key Laboratory of Viral Hepatitis Research, Department of Infectious Diseases and Hepatology Unit, Nanfang Hospital, Southern Medical University, Guangzhou 510515, Guangdong Province, China.
Rong Na, Shanghai Medical College, Fudan University, Shanghai 200043, China; Department of Urology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200025, China.
De-Ke Jiang, State Key Laboratory of Organ Failure Research, Guangdong Key Laboratory of Viral Hepatitis Research, Department of Infectious Diseases and Hepatology Unit, Nanfang Hospital, Southern Medical University, Guangzhou 510515, Guangdong Province, China.
Xiao-Ling Lin, State Key Laboratory of Genetic Engineering, School of Life Science, Fudan University, Shanghai 200433, China.
Feng Yang, Department of Pancreatic Surgery, Pancreatic Disease Institute, Huashan Hospital, Fudan University, Shanghai 200040, China.
Chen Jin, Department of Pancreatic Surgery, Pancreatic Disease Institute, Huashan Hospital, Fudan University, Shanghai 200040, China.
De-Liang Fu, Department of Pancreatic Surgery, Pancreatic Disease Institute, Huashan Hospital, Fudan University, Shanghai 200040, China. fudeliang@huashan.org.cn.
Jian-Feng Xu, Program for Personalized Cancer Care and Department of Surgery, North Shore University Health System, Evanston, IL 60201, United States.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zheng R, Zeng H, Zhang S, Chen T, Chen W. National estimates of cancer prevalence in China, 2011. Cancer Lett. 2016;370:33–38. doi: 10.1016/j.canlet.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, Wang H, Zhu J, Chen W, Wang L, Liu S, Li Y, Wang L, Liu Y, Yin P, Liu J, Yu S, Tan F, Barber RM, Coates MM, Dicker D, Fraser M, González-Medina D, Hamavid H, Hao Y, Hu G, Jiang G, Kan H, Lopez AD, Phillips MR, She J, Vos T, Wan X, Xu G, Yan LL, Yu C, Zhao Y, Zheng Y, Zou X, Naghavi M, Wang Y, Murray CJ, Yang G, Liang X. Cause-specific mortality for 240 causes in China during 1990-2013: a systematic subnational analysis for the Global Burden of Disease Study 2013. Lancet. 2016;387:251–272. doi: 10.1016/S0140-6736(15)00551-6. [DOI] [PubMed] [Google Scholar]
- 5.Society AC. Cancer Facts and Figures 2016. 2016. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2016/cancer-facts-and-figures-2016.pdf. [Google Scholar]
- 6.Ballehaninna UK, Chamberlain RS. Biomarkers for pancreatic cancer: promising new markers and options beyond CA 19-9. Tumour Biol. 2013;34:3279–3292. doi: 10.1007/s13277-013-1033-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee KJ, Yi SW, Chung MJ, Park SW, Song SY, Chung JB, Park JY. Serum CA 19-9 and CEA levels as a prognostic factor in pancreatic adenocarcinoma. Yonsei Med J. 2013;54:643–649. doi: 10.3349/ymj.2013.54.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: risk factors, screening, and early detection. World J Gastroenterol. 2014;20:11182–11198. doi: 10.3748/wjg.v20.i32.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulte A, Pandeya N, Tran B, Fawcett J, Fritschi L, Risch HA, Webb PM, Whiteman DC, Neale RE Queensland Pancreatic Cancer Study Group. Cigarette smoking and pancreatic cancer risk: more to the story than just pack-years. Eur J Cancer. 2014;50:997–1003. doi: 10.1016/j.ejca.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Yeo TP. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin Oncol. 2015;42:8–18. doi: 10.1053/j.seminoncol.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Verna EC, Hwang C, Stevens PD, Rotterdam H, Stavropoulos SN, Sy CD, Prince MA, Chung WK, Fine RL, Chabot JA, Frucht H. Pancreatic cancer screening in a prospective cohort of high-risk patients: a comprehensive strategy of imaging and genetics. Clin Cancer Res. 2010;16:5028–5037. doi: 10.1158/1078-0432.CCR-09-3209. [DOI] [PubMed] [Google Scholar]
- 12.Salo-Mullen EE, O'Reilly EM, Kelsen DP, Ashraf AM, Lowery MA, Yu KH, Reidy DL, Epstein AS, Lincoln A, Saldia A, Jacobs LM, Rau-Murthy R, Zhang L, Kurtz RC, Saltz L, Offit K, Robson ME, Stadler ZK. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer. 2015;121:4382–4388. doi: 10.1002/cncr.29664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liefbroer AC. Changes in Family Size Intentions Across Young Adulthood: A Life-Course Perspective. Eur J Popul. 2009;25:363–386. doi: 10.1007/s10680-008-9173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu L, Jeon J, Brenner H, Gruber SB, Schoen RE, Berndt SI, Chan AT, Chang-Claude J, Du M, Gong J, Harrison TA, Hayes RB, Hoffmeister M, Hutter CM, Lin Y, Nishihara R, Ogino S, Prentice RL, Schumacher FR, Seminara D, Slattery ML, Thomas DC, Thornquist M, Newcomb PA, Potter JD, Zheng Y, White E, Peters U Colorectal Transdisciplinary (CORECT) Study; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology. 2015;148:1330–9.e14. doi: 10.1053/j.gastro.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D'Amato M, Adolfsson J, Grönberg H. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–28. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Na R, Hsu FC, Zheng SL, Wiklund F, Condreay LD, Trent JM, Xu J. Genetic score is an objective and better measurement of inherited risk of prostate cancer than family history. Eur Urol. 2013;63:585–587. doi: 10.1016/j.eururo.2012.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Zhang N, Li K, Chen H, Lin X, Yu Y, Gou Y, Hou J, Jiang D, Na R, Wang X, Ding Q, Xu J. Genetic scores based on risk-associated single nucleotide polymorphisms (SNPs) can reveal inherited risk of renal cell carcinoma. Oncotarget. 2016;7:18631–18637. doi: 10.18632/oncotarget.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, Jacobs EJ, LaCroix A, Zheng W, Albanes D, Bamlet W, Berg CD, Berrino F, Bingham S, Buring JE, Bracci PM, Canzian F, Clavel-Chapelon F, Clipp S, Cotterchio M, de Andrade M, Duell EJ, Fox JW Jr, Gallinger S, Gaziano JM, Giovannucci EL, Goggins M, González CA, Hallmans G, Hankinson SE, Hassan M, Holly EA, Hunter DJ, Hutchinson A, Jackson R, Jacobs KB, Jenab M, Kaaks R, Klein AP, Kooperberg C, Kurtz RC, Li D, Lynch SM, Mandelson M, McWilliams RR, Mendelsohn JB, Michaud DS, Olson SH, Overvad K, Patel AV, Peeters PH, Rajkovic A, Riboli E, Risch HA, Shu XO, Thomas G, Tobias GS, Trichopoulos D, Van Den Eeden SK, Virtamo J, Wactawski-Wende J, Wolpin BM, Yu H, Yu K, Zeleniuch-Jacquotte A, Chanock SJ, Hartge P, Hoover RN. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, Canzian F, Duell EJ, Gallinger S, Giles GG, Goodman GE, Goodman PJ, Jacobs EJ, Kamineni A, Klein AP, Kolonel LN, Kulke MH, Li D, Malats N, Olson SH, Risch HA, Sesso HD, Visvanathan K, White E, Zheng W, Abnet CC, Albanes D, Andreotti G, Austin MA, Barfield R, Basso D, Berndt SI, Boutron-Ruault MC, Brotzman M, Büchler MW, Bueno-de-Mesquita HB, Bugert P, Burdette L, Campa D, Caporaso NE, Capurso G, Chung C, Cotterchio M, Costello E, Elena J, Funel N, Gaziano JM, Giese NA, Giovannucci EL, Goggins M, Gorman MJ, Gross M, Haiman CA, Hassan M, Helzlsouer KJ, Henderson BE, Holly EA, Hu N, Hunter DJ, Innocenti F, Jenab M, Kaaks R, Key TJ, Khaw KT, Klein EA, Kogevinas M, Krogh V, Kupcinskas J, Kurtz RC, LaCroix A, Landi MT, Landi S, Le Marchand L, Mambrini A, Mannisto S, Milne RL, Nakamura Y, Oberg AL, Owzar K, Patel AV, Peeters PH, Peters U, Pezzilli R, Piepoli A, Porta M, Real FX, Riboli E, Rothman N, Scarpa A, Shu XO, Silverman DT, Soucek P, Sund M, Talar-Wojnarowska R, Taylor PR, Theodoropoulos GE, Thornquist M, Tjønneland A, Tobias GS, Trichopoulos D, Vodicka P, Wactawski-Wende J, Wentzensen N, Wu C, Yu H, Yu K, Zeleniuch-Jacquotte A, Hoover R, Hartge P, Fuchs C, Chanock SJ, Stolzenberg-Solomon RS, Amundadottir LT. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46:994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low SK, Kuchiba A, Zembutsu H, Saito A, Takahashi A, Kubo M, Daigo Y, Kamatani N, Chiku S, Totsuka H, Ohnami S, Hirose H, Shimada K, Okusaka T, Yoshida T, Nakamura Y, Sakamoto H. Genome-wide association study of pancreatic cancer in Japanese population. PLoS One. 2010;5:e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, Yang X, Cao G, Hu Z, Zhou Y, Zuo C, Wang C, Zhang X, Zhou Y, Yu X, Dai W, Li Z, Shen H, Liu L, Chen Y, Zhang S, Wang X, Zhai K, Chang J, Liu Y, Sun M, Cao W, Gao J, Ma Y, Zheng X, Cheung ST, Jia Y, Xu J, Tan W, Zhao P, Wu T, Wang C, Lin D. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011;44:62–66. doi: 10.1038/ng.1020. [DOI] [PubMed] [Google Scholar]
- 22.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M, Helzlsouer K, Holly EA, Jacobs EJ, Klein AP, LaCroix A, Li D, Mandelson MT, Olson SH, Risch HA, Zheng W, Albanes D, Bamlet WR, Berg CD, Boutron-Ruault MC, Buring JE, Bracci PM, Canzian F, Clipp S, Cotterchio M, de Andrade M, Duell EJ, Gaziano JM, Giovannucci EL, Goggins M, Hallmans G, Hankinson SE, Hassan M, Howard B, Hunter DJ, Hutchinson A, Jenab M, Kaaks R, Kooperberg C, Krogh V, Kurtz RC, Lynch SM, McWilliams RR, Mendelsohn JB, Michaud DS, Parikh H, Patel AV, Peeters PH, Rajkovic A, Riboli E, Rodriguez L, Seminara D, Shu XO, Thomas G, Tjønneland A, Tobias GS, Trichopoulos D, Van Den Eeden SK, Virtamo J, Wactawski-Wende J, Wang Z, Wolpin BM, Yu H, Yu K, Zeleniuch-Jacquotte A, Fraumeni JF Jr, Hoover RN, Hartge P, Chanock SJ. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Lin X, Na R, Jiang D, Zhang P, Li J, Jin C, Fu D, Xu J. An evaluation study of reported pancreatic adenocarcinoma risk-associated SNPs from genome-wide association studies in Chinese population. Pancreatology. 2017;17:931–935. doi: 10.1016/j.pan.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Jiang DK, Ma XP, Yu H, Cao G, Ding DL, Chen H, Huang HX, Gao YZ, Wu XP, Long XD, Zhang H, Zhang Y, Gao Y, Chen TY, Ren WH, Zhang P, Shi Z, Jiang W, Wan B, Saiyin H, Yin J, Zhou YF, Zhai Y, Lu PX, Zhang H, Gu X, Tan A, Wang JB, Zuo XB, Sun LD, Liu JO, Yi Q, Mo Z, Zhou G, Liu Y, Sun J, Shugart YY, Zheng SL, Zhang XJ, Xu J, Yu L. Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology. 2015;62:118–128. doi: 10.1002/hep.27794. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Duell EJ, Yu K, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Dong X, Wheeler B, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross M, Hartge P, Hoover RN, Holly EA, Jacobs EJ, Klein AP, LaCroix A, Mandelson MT, Petersen G, Zheng W, Agalliu I, Albanes D, Boutron-Ruault MC, Bracci PM, Buring JE, Canzian F, Chang K, Chanock SJ, Cotterchio M, Gaziano JM, Giovannucci EL, Goggins M, Hallmans G, Hankinson SE, Hoffman Bolton JA, Hunter DJ, Hutchinson A, Jacobs KB, Jenab M, Khaw KT, Kraft P, Krogh V, Kurtz RC, McWilliams RR, Mendelsohn JB, Patel AV, Rabe KG, Riboli E, Shu XO, Tjønneland A, Tobias GS, Trichopoulos D, Virtamo J, Visvanathan K, Watters J, Yu H, Zeleniuch-Jacquotte A, Amundadottir L, Stolzenberg-Solomon RZ. Pathway analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinogenesis. 2012;33:1384–1390. doi: 10.1093/carcin/bgs151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.KANNEL WB, DAWBER TR, KAGAN A, REVOTSKIE N, STOKES J., 3rd Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 27.Hammond EC, Garfinkel L. Coronary heart disease, stroke, and aortic aneurysm. Arch Environ Health. 1969;19:167–182. [PubMed] [Google Scholar]
- 28.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D'Agostino RB, Hwang SJ, O'Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, Caspi A. Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol. 2013;59:85–100. doi: 10.1080/19485565.2013.774628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kader AK, Sun J, Reck BH, Newcombe PJ, Kim ST, Hsu FC, D'Agostino RB Jr, Tao S, Zhang Z, Turner AR, Platek GT, Spraggs CF, Whittaker JC, Lane BR, Isaacs WB, Meyers DA, Bleecker ER, Torti FM, Trent JM, McConnell JD, Zheng SL, Condreay LD, Rittmaster RS, Xu J. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. Eur Urol. 2012;62:953–961. doi: 10.1016/j.eururo.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jo J, Nam CM, Sull JW, Yun JE, Kim SY, Lee SJ, Kim YN, Park EJ, Kimm H, Jee SH. Prediction of Colorectal Cancer Risk Using a Genetic Risk Score: The Korean Cancer Prevention Study-II (KCPS-II) Genomics Inform. 2012;10:175–183. doi: 10.5808/GI.2012.10.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigl K, Chang-Claude J, Knebel P, Hsu L, Hoffmeister M, Brenner H. Strongly enhanced colorectal cancer risk stratification by combining family history and genetic risk score. Clin Epidemiol. 2018;10:143–152. doi: 10.2147/CLEP.S145636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lochan R, Daly AK, Reeves HL, Charnley RM. Family history of cancer and tobacco exposure in index cases of pancreatic ductal adenocarcinoma. J Oncol. 2011;2011:215985. doi: 10.1155/2011/215985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulte A, Pandeya N, Fawcett J, Fritschi L, Klein K, Risch HA, Webb PM, Whiteman DC, Neale RE. Association between family cancer history and risk of pancreatic cancer. Cancer Epidemiol. 2016;45:145–150. doi: 10.1016/j.canep.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, Graff RE, Holst K, Möller S, Unger RH, McIntosh C, Nuttall E, Brandt I, Penney KL, Hartman M, Kraft P, Parmigiani G, Christensen K, Koskenvuo M, Holm NV, Heikkilä K, Pukkala E, Skytthe A, Adami HO, Kaprio J Nordic Twin Study of Cancer (NorTwinCan) Collaboration. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein AP, Lindström S, Mendelsohn JB, Steplowski E, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross M, Helzlsouer K, Holly EA, Jacobs EJ, Lacroix A, Li D, Mandelson MT, Olson SH, Petersen GM, Risch HA, Stolzenberg-Solomon RZ, Zheng W, Amundadottir L, Albanes D, Allen NE, Bamlet WR, Boutron-Ruault MC, Buring JE, Bracci PM, Canzian F, Clipp S, Cotterchio M, Duell EJ, Elena J, Gaziano JM, Giovannucci EL, Goggins M, Hallmans G, Hassan M, Hutchinson A, Hunter DJ, Kooperberg C, Kurtz RC, Liu S, Overvad K, Palli D, Patel AV, Rabe KG, Shu XO, Slimani N, Tobias GS, Trichopoulos D, Van Den Eeden SK, Vineis P, Virtamo J, Wactawski-Wende J, Wolpin BM, Yu H, Yu K, Zeleniuch-Jacquotte A, Chanock SJ, Hoover RN, Hartge P, Kraft P. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PLoS One. 2013;8:e72311. doi: 10.1371/journal.pone.0072311. [DOI] [PMC free article] [PubMed] [Google Scholar]