Abstract
Standardization of the use of next-generation sequencing for the diagnosis of rare neurological disorders has made it possible to detect potential disease-causing genetic variations, including de novo variants. However, the lack of a clear pathogenic relevance of gene variants poses a critical limitation for translating this genetic information into clinical practice, increasing the necessity to perform functional assays. Genetic screening is currently recommended in the guidelines for diagnosis of hypomyelinating leukodystrophies (HLDs). HLDs represent a group of rare heterogeneous disorders that interfere with the myelination of the neurons in the central nervous system. One of the HLD-related genes is HSPD1, encoding the mitochondrial chaperone heat shock protein 60 (HSP60), which functions as folding machinery for the mitochondrial proteins imported into the mitochondrial matrix space. Disease-causing HSPD1 variants have been associated with an autosomal recessive form of fatal hypomyelinating leukodystrophy (HLD4, MitCHAP60 disease; MIM #612233) and an autosomal dominant form of spastic paraplegia, type 13 (SPG13; MIM #605280). In 2018, a de novo HSPD1 variant was reported in a patient with HLD. Here, we present another case carrying the same heterozygous de novo variation in the HSPD1 gene (c.139T > G, p.Leu47Val) associated with an HLD phenotype. Our molecular studies show that the variant HSP60 protein is stably present in the patient's fibroblasts, and functional assays demonstrate that the variant protein lacks in vivo function, thus confirming its disease association. We conclude that de novo variations of the HSPD1 gene should be considered as potentially disease-causing in the diagnosis and pathogenesis of the HLDs.
Keywords: abnormal CNS myelination, cerebral hypomyelination
INTRODUCTION
Hypomyelinating leukodystrophies (HLDs) are neurodevelopmental disorders characterized by defective myelination in the central nervous system accompanied by developmental delay, spasticity, and intellectual disability (Charzewska et al. 2016). HLDs are genetically heterogeneous disorders, and 17 types of HLDs have been identified by variations in different genes thus far. These genes include those that are directly related to myelination (i.e., PLP1, GJC2, FAM126A, and TUBB4A) but also genes for which the involvement in myelination is not yet characterized (e.g., PYCR2, HIKESHI, and HSPD1) (Pouwels et al. 2014; Vanderver et al. 2015; Duncan and Radcliff 2016; Duncan et al. 2017; van der Knaap and Bugiani 2017).
Among different types of HLDs, HLD4, also known as MitCHAP60 disease (MIM #612233), is attributed to a biallelic pathogenic variant in the HSPD1 gene and is inherited in an autosomal recessive manner (Magen et al. 2008). HLD4 is a progressive neurodevelopmental disorder, presenting initially with central hypotonia, strabismus, and psychomotor delay. Patients develop acquired microcephaly, spasticity, and developmental regression. Seizures are also reported in some patients. Typically, the onset of the condition is within the first 3 months of life and is fatal within the first 2 decades. There is a wide range of reported intrafamilial and interfamilial phenotypic heterogeneity, particularly with respect to the degree of psychomotor impairment and the rate of neurologic and developmental deterioration. Brain magnetic resonance imaging (MRI) in these patients reveals diffuse hypomyelination, thin corpus callosum, thin brainstem, and varying degrees of ventricular enlargement. A subset of patients has markedly increased urinary secretion of ethylmalonic acid as well (Magen et al. 2008; Bross and Fernandez-Guerra 2016; Kusk et al. 2016).
The human HSPD1 and HSPE1 genes encode the essential mitochondrial HSP60 and HSP10 chaperones, respectively, which form a folding chamber to facilitate protein folding and assembly in the mitochondrial matrix space. Variations in HSPD1 and HSPE1 are implicated in the following neurological diseases (Table 1; Bross and Fernandez-Guerra 2016): (i) homozygosity for the HSP60:p.Asp29Gly variation causes the fatal hypomyelination disorder (HLD4; MitCHAP60 disease; MIM #612233) (Magen et al. 2008; Parnas et al. 2009; Kusk et al. 2016), (ii) heterozygosity for the HSP60:p.Val98Ile variation causes spastic paraplegia (SPG13; MIM #605280) (Hansen et al. 2002), (iii) heterozygosity for the HSP10:p.Leu73Phe variation causes a neurological and developmental disorder (Bie et al. 2016), and (iv) heterozygosity for the HSP60:p.Leu47Val variation causes hypomyelination disorder (Yamamoto et al. 2018). In addition to these genetic variations, another molecular mechanism can lead to HSP10 chaperone deficiency, without involving a genetic alteration in the HSPE1 gene. Specifically, Szego et al. (2019) demonstrated that newly synthesized HSP10 directly interacts with α-synuclein, forming α-synuclein aggregates that result in cytosolic trapping of HSP10. This prevents import of HSP10 into the mitochondrial matrix. The subsequent reduced levels of HSP10 in the mitochondrial matrix result in mitochondrial dysfunction. In agreement with this conclusion, Szego et al. showed that HSP10 overexpression delays α-synuclein pathology both in vitro and in vivo.
Table 1.
Genotype and clinical features of patients with HSPD1 and HSPE1 variations
| Disease association | Variations in HSPD1 or HSPE1 | Genotype | Age at onset | Gender | Symptoms | PolyPhen-2 predictiona | Growth in genetic complementation assay | ACADS polymorphism genotype | Urine ethylmalonic acid | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Hypomyelinating leukodystrophy 4 (HLD4;MitCHAP60; MIM 612233) | p.Asp29Gly c.[86A > G] | Homozygous | 3 mo | M | Progressive hypertonia and hyperreflexia, intermittent nystagmus, lack of head control, psychomotor developmental delay, no language, decreased social contact, progressive limb spasticity | Benign | Slow, temperature-sensitive | c.[625G > A]; [625G > A] | Elevated | Magen et al. 2008; Kusk et al. 2016; Parnas et al. 2009 |
| Hereditary spastic paraplegia (SPG13; MIM 605280) | p.Val98Ile c.[292G > A] | Heterozygous | 40 yr | F | Severe functional handicaps, weakness of the lower limbs, retinopathy, ataxia, and mental retardation | Possibly damaging | No growth | c.[625G > A]; [625=] | N/A | Hansen et al. 2002 |
| Neurological and developmental disorder | HSP10:p.Leu73Phe c.[217C > T] | Heterozygous (de novo) | 3 mo | M | Infantile spasms, hypotonia, developmental delay, a slightly enlarged liver, nystagmus, macrocephaly | Possibly damaging | N/A | c.[625=]; [625=] | Normal | Bie et al. 2016 |
| Hypomyelination | p.Leu47Val c.[139T > G] | Heterozygous (de novo) | 18 mo | M | Diffuse hypomyelination, gait instability, mild ataxia, dysmature motor coordination | Probably damaging | No growth | c.[625G > A]; [625=] | Elevated | Yamamoto et al. 2018 |
(M) Male, (F) female, (N/A) not available.
aPolyPhen-2 has three levels of damage prediction, from most damaging to the least: probably damaging, possibly damaging, and benign.
Here, we report a case of a 7-yr-old patient, surprisingly carrying the same heterozygous de novo variant reported by Yamamoto et al. in the HSPD1 gene, HSP60:p.Leu47Val, also presenting with an HLD phenotype. We share our functional protein assay results and discuss our findings in the context of the other phenotypes associated with HSPD1 and HSPE1 variations hitherto detected.
RESULTS
Clinical Presentation and Family History
The patient is a 7-yr-old boy who was initially referred for evaluation because of speech and motor delays. He was born by vacuum-assisted vaginal delivery at 38 wk gestation to a G1P0 woman following an uncomplicated pregnancy. His early gross motor milestones fell within the normal range, but delayed milestones became apparent by 18 mo of age. He did not walk independently until 22 mo of age. Neurological evaluation at 24 mo of age revealed mild bilateral postural tremor in the upper extremities, mild ataxia, brisk deep tendon reflexes, and abnormal motor coordination. He was also clumsy and showed gait instability. He had no history of developmental regression or seizures. His brain MRI recorded at 36 mo of age showed diffuse/delayed hypomyelination of the cerebral and cerebellar white matter suggestive of underlying HLD (Fig. 1A–C). However, the patient had normal corpus callosum and cerebellar vermis at this age (Fig. 1D). Patient had mild postural tremor at 4 yr and 2 mo; reflexes were 2+ bilaterally in the patella and the Achilles’ and 1+ bilaterally in the triceps. He was able to jump with two feet at 4 yr and 10 mo of age. Physical examination at 5 yr and 2 mo of age revealed normal growth parameters (height 85th percentile, weight 25–50th percentile, head circumference 75th percentile) and no dysmorphic features. No regression and no seizures were noted at this age. Examination at 7 yr and 9 mo revealed coordination and fatigue challenges, and motor development reaching to plateau. Urine metabolic screening detected increased excretion of ethylmalonic acid on three independent samples.
Figure 1.

MRI scans of the patient at 3 yr and 11 mo of age indicates hypomyelination (A) and (B) axial FLAIR images at different levels and (C) coronal T2 image showing diffuse high signal intensity of the cerebral and cerebellar white matter including the subcortical U fibers. Abnormal hyperintensity was most notable in the peritrigonal regions bilaterally. (D) Sagittal T1 image showing corpus callosum and cerebellar vermis are normal at this stage of the disease.
The patient has not been on any medications. The family is of mixed Northern European, Italian, and French–Canadian descent. The family is nonconsanguineous, and its history is noncontributory.
Genomic Analyses
Trio whole-exome sequencing revealed a de novo heterozygous variant (c.139T > G, p.Leu47Val) in the HSPD1 gene. The variation was confirmed with Sanger sequencing. Subsequent multiplex ligation-dependent probe amplification (MLPA) analysis failed to detect any large deletions or duplications in the HSPD1 gene. Sequencing analysis of the short-chain acyl-CoA dehydrogenase gene (ACADS) showed heterozygosity for the c.625G > A common variant.
In Silico Analysis of the Variant Site
The HSP60 protein is highly conserved among species, and the sequence alignments of its first 100 amino acids, which contain the three disease-associated residues, confirms the complete sequence conservation of all known disease-associated variants from Escherichia coli to human (Fig. 2A). Moreover, the known disease-associated variants (Asp29, Leu47, and Val98) are closely localized and horizontally aligned in the HSP60 protein structure (Fig. 2B–D). The de novo HSPD1 c.139T > G, p.Leu47Val variant was not reported in the Genome Aggregation Database (gnomAD) v2.1.1 (Karczewski et al. 2019).
Figure 2.
The de novo HSP60:p.Leu47Val variation occurs at a conserved residue, and disease-associated HSPD1 variants are colocalized in the HSP60 protein structure. (A) Sequence alignment of the first 100 amino acids of the human HSP60 protein and its homologs illustrates the high sequence conservation among species. The patient variation occurs at the leucine residue at position 47 (red arrow), which is conserved from E. coli to human. Furthermore, other disease-causing variations occur at residues that are also conserved from E. coli to human (Asp29 and Val98). (B) The three disease-associated amino acids, Asp29, Leu47, and Val98, are shown on the seven-subunit HSP60 ribbon traces. Each color group highlights the three variant residues in their respective HSP60 subunit. ATP is shown as red sticks. (C) Disease-associated amino acids are localized not only in close proximity but also on the same horizontal plane. (D) The disease-associated variant residues are all in close proximity to the ATP-binding site; however, they are not in direct physical contact with the bound ATP molecule (marked as red sticks).
Functional Studies with the E. coli–Based Genetic Complementation System
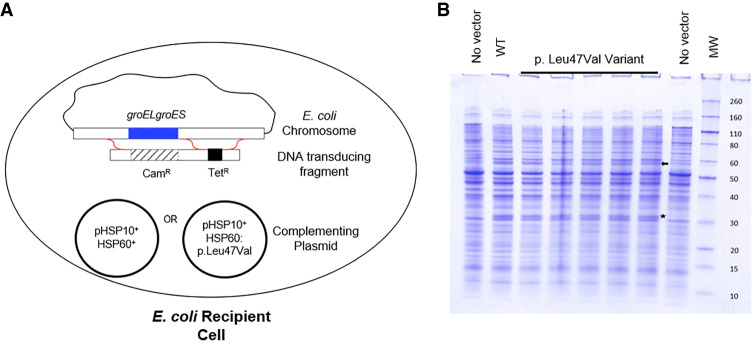
In this study, an E. coli–based in vivo complementation assay was performed to determine whether the HSP60:p.Leu47Val protein is functional. In a previous study we showed that the groEL and groES genes, the E. coli homologs of HSPD1 and HSPE1, are essential for bacterial growth but can be replaced by the human HSP60 and HSP10 homologs expressed from a complementing plasmid (Richardson et al. 2001; Magen et al. 2008). Thus, this system serves as a sensitive tool to check the in vivo function of various HSP60 variants and has been successfully used to determine the effects of patient variations on HSP60 function in vivo (Hansen et al. 2002). In these complementation studies, tetracycline resistance (TetR) is used to identify those bacterial transductants that have exchanged DNA in the groESgroEL region, whereas chloramphenicol resistance (CamR) is used to identify the subset that resulted in the simultaneous deletion of the entire groESgroEL operon. Thus, the frequency of TetRCamR transductants versus TetR-only transductants indicates the ability of the plasmid carrying the HSP10+ gene and either the HSP60 wild-type or HSP60:p.Leu47Val variant gene to substitute for the missing, essential E. coli GroES/GroEL function (Fig. 3A). Independent transformants carrying the HSP10+HSP60:p.Leu47Val plasmid were shown to express approximately equivalent HSP60 levels following isopropyl β-D-1-thiogalactopyranoside (IPTG) induction compared to transformants carrying the parental wild-type plasmid, as judged by total protein staining, suggesting that the variant protein is as stable as wild-type HSP60 in bacteria (Fig. 3B).
Figure 3.
E. coli functional study design and expression of wild-type HSP60 and the HSP60:p.Leu47Val variant. (A) Transduction experiment design to test complementation of E. coli GroEL with human HSP60. DNA transducing fragment carrying tetracycline resistance (TetR) and chloramphenicol resistance (CamR) is used to replace the endogenous groESgroEL operon in the presence of a complementing plasmid expressing HSP10+ and either HSP60+ or HSP60:p.Leu47Val. The red squiggles indicate potential recombination crossover pathways. The low frequency of deletion of the groESgroEL operon in the presence of the HSP10+HSP60: p.Leu47Val plasmid indicates the dysfunction of the disease-associated variant. (B) The wild-type HSP60 and the p.Leu47Val variant are expressed in E. coli cells. The arrow indicates HSP60 protein. The star indicates CamR protein expressed by the plasmid. “WT” is lysate from E. coli expressing wild-type HSP60. “p.Leu47Val Variant” are lysates from different clones expressing the HSP60:p.Leu47Val variant. HSP10 is not discernible on this gel.
The high linkage found between the TetR and CamR markers demonstrates that the plasmid pHSP10+HSP60+ encoding the wild-type HSP10 and HSP60 genes is able to substitute for E. coli GroES/GroEL function. In contrast, we found that the plasmid pHSP10+HSP60: p.Leu47Val does not complement the deletion of E. coli groESgroEL at 37°C (Table 2). The 11 unexpected CamR transductants isolated were shown to express the resident E. coli GroEL using functional tests and thus did not represent true groEL deletion variants. In conclusion, our results clearly demonstrate that wild-type HSP60, but not the HSP60: p.Leu47Val variant, can support the growth of E. coli cells.
Table 2.
Results of the complementation study show that the replacement of the E. coli groESgroEL operon by plasmid-encoded Hsp10+Hsp60:p.Leu47Val does not complement growth of E. coli
| Plasmid | TetR | CamR | Linkage (%) |
|---|---|---|---|
| pHsp10+Hsp60+ | 149 | 133 | 89b |
| pHsp10+Hsp60:p.Leu47Val | 312 | 11a | 3.5 |
A high linkage score indicates the groESgroEL operon can be readily deleted from the chromosome because the complementing plasmid encodes a functional protein.
aThe 11 CamR transductants were further tested and shown to be sensitive to bacteriophage T4 infection, indicating that they still possess GroEL function and most likely represent rare aberrant recombination/duplication events.
bBecause of the use of T4 as the transducing phage, the expected linkage between the TetR and CamR genes is higher than that reported by Hansen et al. (2002).
Quantification of Wild-Type and Variant HSP60 Peptide Levels
To assess whether the HSP60:p.Leu47Val variant protein was stably expressed in patient cells, we performed mass spectrometry analyses to determine the relative amounts of wild-type HSP60 and HSP60:p.Leu47Val variant protein present in the patient's fibroblasts. We detected a total of 29 different peptides (Fig. 4A), including the wild-type and variant peptide (Fig. 4B), of the HSP60 protein. Calculations based on our mass spectrometry data indicated similar amounts of variant and HSP60 wild-type proteins in the patient's fibroblasts (Fig. 4C). As the wild-type HSP60 protein constitutes 100% in the control cells, the percentage of the HSP60:p.Leu47Val protein in the patient cells is ∼50%, thus demonstrating that the mutant HSP60:p.Leu47Val protein is as stable as wild-type HSP60.
Figure 4.
Detection and quantification of wild-type and variant HSP60 peptide levels by mass spectrometry demonstrate that the variant protein is present, suggesting that the mutation is causing a dominant negative effect on HSP60 function. The experiment was performed on fibroblast cultures from three nonrelated healthy control individuals and three independent cultivation samples from the patient. (A) The HSP60 peptide sequences detected and quantified by mass spectrometry are highlighted in yellow on the human HSP60 amino acid sequence. The peptide sequence detected containing the patient variation site (p.Leu47Val) is marked in red. (B) Variant peptide peak graphs extracted from the ion chromatogram of the mass spectrometry analysis show the detection of the variant peptide only in the patient's samples. (C) Calculation of the relative fraction of wild-type HSP60 and the p.Leu47Val variant based on mass spectrometry analyses (see Materials and Methods) reveals that the variant protein constitutes ∼50% of the total HSP60 protein in the patient fibroblasts.
DISCUSSION
The advancement of next-generation sequencing techniques has improved the detection of rare disease-causing genetic variations in the diagnosis of previously undiagnosed patients. These advancements considerably speed up the discovery of de novo mutations and mutational hotspots in the human genome. Detection of the same de novo variation in multiple patients with the same disease phenotypes contributes to the confirmation of the disease-causing nature of gene variations and in-depth functional studies of the variant proteins expand our knowledge on molecular functions (Acuna-Hidalgo et al. 2016).
As reported before, different disease-causing variations in the HSPD1 and HSPE1 genes, which respectively encode the mitochondrial HSP60 and HSP10 chaperones, have been found to result in a spectrum of neurodevelopmental diseases with different severities (summarized in Table 1) (Bross and Fernandez-Guerra 2016). Here, we propose a possible novel autosomal dominant mechanism for HLD4 and suggest that a subset of autosomal dominant pathogenic variants in the HSPD1 gene produces an HLD phenotype. We based this mechanism on our investigations of a case of the 7-yr-old patient that presented with a brain MRI finding consistent with HLD and a heterozygous p.Leu47Val de novo variation in the HSPD1 gene, together with the case previously reported (Yamamoto et al. 2018).
To evaluate the disease-causing nature of the p.Leu47Val variation, we performed in silico analyses and in vivo functional assays. The sequence alignment of the human HSP60 protein and its homologs shows the strict conservation of Leu47, together with the other disease-associated amino acid residues—Asp29 and Val98—from E. coli to humans (Fig. 2A). Our structural analysis suggests that these amino acids could be involved in stabilization of the seven-subunit HSP60 structure. Indeed, a recent publication has shown that the p.Asp29Gly and p.Val98Ile variations interfere with functional conformational changes in the HSP60 chaperone complex (Wang et al. 2019). These disease-associated amino acids are located in close proximity to each other and the ATP-binding domain, yet are not directly in contact with the bound ATP (Fig. 2B–D). This further supports the possibility that replacement of the linear carbon side chain of leucine with the β-branched valine in this densely packed part of the HSP60 structure may also disturb transmission of the conformational changes triggered by ATP hydrolysis, and thus impair protein function.
Urine metabolic screening detected increased excretion of ethylmalonic acid, which is an indicator of short chain acyl-CoA dehydrogenase (SCAD, ACADS) deficiency, an inborn error of mitochondrial fatty acid metabolism. Elevated urine ethylmalonic acid has also been observed in patients in whom homozygosity for the polymorphic c.624G > A variation was found as the only variation in the ACADS gene (Gregersen et al. 1998; Pedersen et al. 2008). Presence of intermittent increase of urinary ethylmalonic acid excretion has been described in patients with HLD4 (Magen et al. 2008; Kusk et al. 2016). Earlier studies have shown functional interaction of SCAD with the HSP60 chaperone (Pedersen et al. 2003), and therefore elevated levels of ethylmalonic acid, as seen in the current patient, may be the result of the heterozygosity for the c.625G > A common variant in the ACADS gene, in connection with the impaired HSP60 function.
Discrepancies between in silico analysis and in vivo functional assays for HSP60 variants have shown the importance of evaluating the activity of the HSP60/HSP10 complex in each case (Bross and Fernandez-Guerra 2016). Here we used both an E. coli–based genetic complementation system and patient's fibroblasts for in vivo functional analyses. The genetic complementation system showed abolished bacterial cell growth and a decreased linkage score for the HSP60:p.Leu47Val variant, indicating its failure to complement the deletion of the E. coli groESgroEL operon (Table 2), demonstrating a loss of protein function. Moreover, in the patient's fibroblasts we detected a 1:1 ratio of wild-type to variant peptide, indicating that it is stable (Fig. 4C). In the previous report of this variation, Yamamato et al. discusses the possibility of compound heterozygosity for the HSPD1 variant. Detection of both the variant and the wild-type peptides in equal amounts in patient's fibroblasts excludes the possibility of compound heterozygosity through variations in the noncoding regions of the wild-type allele that might affect its expression. Taken together, our functional results are consistent with the conclusion that the HSP60:p.Leu47Val variant exerts an autosomal dominant disease mechanism.
In summary, this case study reveals a de novo variant in the HSPD1 gene with an autosomal dominant disease mechanism. As such, it is important to consider the possibility that other heterozygous de novo variants in the HSPD1 gene may exist, resulting in neurodevelopmental disorder manifestations such as HLD. The presence of the same de novo variation in two different patients with similar clinical phenotypes might suggest that the p.Leu47Val variant is the consequence of a mutational hotspot in the HSPD1 gene. However, the mutated base is not in a CpG site that would increase susceptibility to mutation. It is likely that variation of this amino acid comes to clinical attention because this position in the protein is especially crucial. The basis for the mechanistic role of HSPD1 in myelination is yet to be understood. These developments reveal the therapeutic potential of HSP60-targeting compounds and contribute to the continuing interest of developing small molecules to modulate HSP60 function in a variety of diseases (Meng et al. 2018). Finally, this study further underlines the potential of whole-exome sequencing in the diagnosis of rare diseases, and the necessity of performing functional assays in order to confirm disease-causing potential of de novo variants.
METHODS
Genomic Analyses
Whole-exome sequencing was performed by Centogene AG, and 100% coverage for coding regions of the HSPD1 gene was reported by the company (Supplemental Table S1). Sanger sequencing confirmation was performed on genomic DNA isolated from the patient's fibroblasts and blood samples from the patient and his parents. Exon 2 of the HSPD1 gene was amplified by polymerase chain reaction (PCR), and bidirectional sequencing of the PCR product was performed with BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) and using an Applied Biosystems 3500 Dx Genetic Analyzer.
E. coli Functional Studies
E. coli cells were grown to exponential phase in LB medium at 37°C; plasmid-encoded gene expression was induced by the addition of 4 mM IPTG and grown for an additional 2 h. Equal volumes of cell lysate were resolved on a Novex NuPAGE 4%–12% Bis-Tris/MES Protein Gel (Invitrogen) and stained with Coomassie blue for total protein detection. Independent transformants carrying Hsp60:p.Leu47Val-coding plasmids induced approximately equivalent HSP60 levels following IPTG induction as the parental wild-type plasmid, as judged by protein staining (Fig. 3B). The transduction procedure was essentially as described by Hansen et al. (2002) except that bacteriophage T4 was used to prepare the lysate on the donor strain AR1893, in which the essential groESgroEL operon is deleted and replaced by the CamR gene on the bacterial chromosome. A TetR gene is inserted downstream as an outside marker. These cells are kept alive by the presence of a plasmid expressing the wild-type E. coli groESgroEL operon. Wild-type E. coli cells transformed with a kanamycin-resistant (KanR) plasmid encoding HSP10+ and either HSP60+ or HSP60:p.Leu47Val were infected with the T4 donor lysate. The resulting transductants were selected first for TetR, followed by screening for the simultaneous acquisition of the nearby CamR marker. Our previous work showed that pHSP10+HSP60+ supports deletion of groESgroEL, resulting in transductants that are both TetR and CamR. The red squiggles in Figure 3A indicate potential recombination crossover pathways. DNA sequencing of the plasmids confirmed that no other mutations were present in the insert.
Quantification of Wild-Type and Variant HSP60 Peptide Levels
Human dermal fibroblasts from the patient, as well as three nonrelated healthy individuals as controls, were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% (v/v) fetal bovine serum, 29 mg/mL of L-glutamine (Sigma-Aldrich), and 0.1% penicillin/streptomycin (Sigma-Aldrich) at 37°C and 5% CO2 in Mycoplasma sp.-free conditions. Fibroblast pellets were dissolved in 0.35 M Tris-HCl pH 6.8, 0.60 M dithiothreitol, and 10% (w/v) sodium dodecyl sulfate and ultrasonicated (Branson Sonifier 250, Branson Ultrasonics Corp) at 30% duty cycle for three cycles of five pulses, with 1 min of ice incubation between each cycle. Lysates were centrifuged at 14,000g for 30 min at 4°C, and 30 µg of extracted proteins were separated by polyacrylamide gel electrophoresis on a BioRad AnykD Criterion TGX protein gel and stained with Coomassie blue. Each sample was excised from the stained gel and samples were further prepared for nano Liquid-Chromatography coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) as described previously (Hansen et al. 2011). Relative amounts of wild-type and variant HSP60 proteins in control and patient fibroblasts were quantified with mass spectrometry. HSP60 peptides were identified and quantitated using Proteome Discoverer 2.2 and MaxQuant 1.5.3.30 software. The fraction of wild-type and variant HSP60 protein in the control and patient samples was calculated by quantifying the ratio of the area under the curve of the wild-type peptide containing the mutation site to the average areas under the curve of 29 HSP60 peptides (Fig. 4).
ADDITIONAL INFORMATION
Data Deposition and Access
The HSPD1 variant (c.139T > G, p.Leu47Val) was submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV001164600. Authors declare no further data to be deposited, as consent for publicly reporting the full exome sequencing data was not obtained.
Ethics Statement
Informed consent for using the photos, clinical data, biological material, and scientific publication was obtained from the patient's parents. This case report was determined not to be a Human Health Research Study by the Central Denmark Region Committees on Health Research Ethics, and IRB approval was not required. This study conformed to the Declaration of Helsinki.
Acknowledgments
We thank our patient and his family for their generous participation in this study. We acknowledge the technical assistance of Helle Highland Nygaard at the Research Unit for Molecular Medicine, Aarhus University and Aarhus University Hospital.
Author Contributions
C.C. wrote the first draft of the manuscript and performed the genetic and protein analyses. L.B., M.K., and B.F.M. provided the clinical evaluation and whole-exome sequencing. D.A. and C.G. performed the in vivo E. coli functional studies. J.P. performed and supervised the mass spectrometry analysis. P.F.-G. provided critical feedback on the results. P.B. provided conceptual design and supervised the study. All authors discussed the results and contributed to the final manuscript.
Funding
C.C. is supported by a PhD fellowship from the Graduate School of Health, Aarhus University, Denmark. This work was supported by the Eva & Henry Frænkels Mindefond and John & Birthe Meyer Foundation.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Acuna-Hidalgo R, Veltman JA, Hoischen A. 2016. New insights into the generation and role of de novo mutations in health and disease. Genome Biol 17: 241 10.1186/s13059-016-1110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bie AS, Fernandez-Guerra P, Birkler RI, Nisemblat S, Pelnena D, Lu X, Deignan JL, Lee H, Dorrani N, Corydon TJ, et al. 2016. Effects of a mutation in the HSPE1 gene encoding the mitochondrial co-chaperonin HSP10 and its potential association with a neurological and developmental disorder. Front Mol Biosci 3: 59–73. 10.3389/fmolb.2016.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross P, Fernandez-Guerra P. 2016. Disease-associated mutations in the HSPD1 gene encoding the large subunit of the mitochondrial HSP60/HSP10 chaperonin complex. Front Mol Biosci 3: 92–98. 10.3389/fmolb.2016.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charzewska A, Wierzba J, Izycka-Swieszewska E, Bekiesinska-Figatowska M, Jurek M, Gintowt A, Klosowska A, Bal J, Hoffman-Zacharska D. 2016. Hypomyelinating leukodystrophies—a molecular insight into the white matter pathology. Clin Genet 90: 293–304. 10.1111/cge.12811 [DOI] [PubMed] [Google Scholar]
- Duncan ID, Radcliff AB. 2016. Inherited and acquired disorders of myelin: the underlying myelin pathology. Exp Neurol 283: 452–475. 10.1016/j.expneurol.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan ID, Bugiani M, Radcliff AB, Moran JJ, Lopez-Anido C, Duong P, August BK, Wolf NI, van der Knaap MS, Svaren J. 2017. A mutation in the Tubb4a gene leads to microtubule accumulation with hypomyelination and demyelination. Ann Neurol 81: 690–702. 10.1002/ana.24930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen N, Winter VS, Corydon MJ, Corydon TJ, Rinaldo P, Ribes A, Martinez G, Bennett MJ, Vianey-Saban C, Bhala A, et al. 1998. Identification of four new mutations in the short-chain acyl-CoA dehydrogenase (SCAD) gene in two patients: one of the variant alleles, 511C→T, is present at an unexpectedly high frequency in the general population, as was the case for 625G→A, together conferring susceptibility to ethylmalonic aciduria. Hum Mol Genet 7: 619–627. 10.1093/hmg/7.4.619 [DOI] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, et al. 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70: 1328–1332. 10.1086/339935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Palmfeldt J, Vang S, Corydon TJ, Gregersen N, Bross P. 2011. Quantitative proteomics reveals cellular targets of celastrol. PLoS ONE 6: e26634 10.1371/journal.pone.0026634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 [DOI] [Google Scholar]
- Kusk MS, Damgaard B, Risom L, Hansen B, Ostergaard E. 2016. Hypomyelinating leukodystrophy due to HSPD1 mutations: a new patient. Neuropediatrics 47: 332–335. 10.1055/s-0036-1584564 [DOI] [PubMed] [Google Scholar]
- Magen D, Georgopoulos C, Bross P, Ang D, Segev Y, Goldsher D, Nemirovski A, Shahar E, Ravid S, Luder A, et al. 2008. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet 83: 30–42. 10.1016/j.ajhg.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Li BX, Xiao X. 2018. Toward developing chemical modulators of Hsp60 as potential therapeutics. Front Mol Biosci 5: 74–84. 10.3389/fmolb.2018.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas A, Nadler M, Nisemblat S, Horovitz A, Mandel H, Azem A. 2009. The MitCHAP-60 disease is due to entropic destabilization of the human mitochondrial Hsp60 oligomer. J Biol Chem 284: 28198–28203. 10.1074/jbc.M109.031997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CB, Bross P, Winter VS, Corydon TJ, Bolund L, Bartlett K, Vockley J, Gregersen N. 2003. Misfolding, degradation, and aggregation of variant proteins. The molecular pathogenesis of short chain acyl-CoA dehydrogenase (SCAD) deficiency. J Biol Chem 278: 47449–47458. 10.1074/jbc.M309514200 [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Kølvraa S, Kølvraa A, Stenbroen V, Kjeldsen M, Ensenauer R, Tein I, Matern D, Rinaldo P, Vianey-Saban C, et al. 2008. The ACADS gene variation spectrum in 114 patients with short-chain acyl-CoA dehydrogenase (SCAD) deficiency is dominated by missense variations leading to protein misfolding at the cellular level. Hum Genet 124: 43–56. 10.1007/s00439-008-0521-9 [DOI] [PubMed] [Google Scholar]
- Pouwels PJ, Vanderver A, Bernard G, Wolf NI, Dreha-Kulczewksi SF, Deoni SC, Bertini E, Kohlschutter A, Richardson W, Ffrench-Constant C, et al. 2014. Hypomyelinating leukodystrophies: translational research progress and prospects. Ann Neurol 76: 5–19. 10.1002/ana.24194 [DOI] [PubMed] [Google Scholar]
- Richardson A, Schwager F, Landry SJ, Georgopoulos C. 2001. The importance of a mobile loop in regulating chaperonin/co-chaperonin interaction: humans versus Escherichia coli. J Biol Chem 276: 4981–4987. 10.1074/jbc.M008628200 [DOI] [PubMed] [Google Scholar]
- Szego EM, Dominguez-Meijide A, Gerhardt E, Konig A, Koss DJ, Li W, Pinho R, Fahlbusch C, Johnson M, Santos P, et al. 2019. Cytosolic trapping of a mitochondrial heat shock protein is an early pathological event in synucleinopathies. Cell Rep 28: 65–77 e66. 10.1016/j.celrep.2019.06.009 [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Bugiani M. 2017. Leukodystrophies: a proposed classification system based on pathological changes and pathogenetic mechanisms. Acta Neuropathol 134: 351–382. 10.1007/s00401-017-1739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderver A, Prust M, Tonduti D, Mochel F, Hussey HM, Helman G, Garbern J, Eichler F, Labauge P, Aubourg P, et al. 2015. Case definition and classification of leukodystrophies and leukoencephalopathies. Mol Genet Metab 114: 494–500. 10.1016/j.ymgme.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Enriquez AS, Li J, Rodriguez A, Holguin B, Von Salzen D, Bhatt JM, Bernal RA. 2019. MitCHAP-60 and hereditary spastic paraplegia SPG-13 arise from an inactive hsp60 chaperonin that fails to fold the ATP synthase β-subunit. Sci Rep 9: 12300 10.1038/s41598-019-48762-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Yamamoto-Shimojima K, Ueda Y, Imai K, Takahashi Y, Imagawa E, Miyake N, Matsumoto N. 2018. Independent occurrence of de novo HSPD1 and HIP1 variants in brothers with different neurological disorders: leukodystrophy and autism. Hum Genome Var 5: 18 10.1038/s41439-018-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HSPD1 variant (c.139T > G, p.Leu47Val) was submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV001164600. Authors declare no further data to be deposited, as consent for publicly reporting the full exome sequencing data was not obtained.