Abstract
Pleiotropy and variable expressivity have been cited to explain the seemingly distinct neurodevelopmental disorders due to a common genetic etiology within the same family. Here we present a family with a de novo 1-Mb duplication involving 18 genes on Chromosome 19. Within the family there are multiple cases of neurodevelopmental disorders including autism spectrum disorder, attention deficit/hyperactivity disorder, intellectual disability, and psychiatric disease in individuals carrying this copy-number variant (CNV). Quantitative polymerase chain reaction (PCR) confirmed the CNV was de novo in the mother and inherited by both sons. Whole-exome sequencing did not uncover further genetic risk factors segregating within the family. Transcriptome analysis of peripheral blood demonstrated a ∼1.5-fold increase in RNA transcript abundance in 12 of the 15 detected genes within the CNV region for individuals carrying the CNV compared with their noncarrier relatives. Examination of transcript abundance across the rest of the transcriptome identified 407 differentially expressed genes (P-value < 0.05; adjusted P-value < 0.1) mapping to immune response, response to endoplasmic reticulum stress, and regulation of epithelial cell proliferation pathways. 16S microbiome profiling demonstrated compositional difference in the gut bacteria between the half-brothers. These results raise the possibility that the observed CNV may contribute to the varied phenotypic characteristics in family members through alterations in gene expression and/or dysbiosis of the gut microbiome. More broadly, there is growing evidence that different neurodevelopmental and psychiatric disorders can share the same genetic variant, which lays a framework for later neurodevelopmental and psychiatric manifestations.
Keywords: autism; borderline personality disorder; immune dysregulation; intellectual disability, severe; microretrognathia
INTRODUCTION
Genomic variation is an important contributor to observable phenotypic differences in human traits ranging from morphology, physiology, and behaviors to disease predisposition (Albert and Kruglyak 2015). Individuals differ by thousands to millions of DNA sequence variants, which include single-nucleotide polymorphisms (SNPs), small insertions and deletions (indels), and large structural variants such as block substitutions, inversions, and copy-number variants (CNVs) (Scherer et al. 2007; Frazer et al. 2009; Albert and Kruglyak 2015). Although SNPs are the most common genetic variant, comprising 78% of all variants, structural variants account for 74% of all variant nucleotides (Frazer et al. 2009; Gamazon and Stranger 2015). Recent efforts have made tremendous strides in identification, categorization, and mapping common and rare variants including the Single Nucleotide Polymorphism Database (dbSNP) (Sachidanandam et al. 2001), haplotype map (The International HapMap Consortium 2005; Frazer et al. 2007), 1000 Genomes Project (Abecasis et al. 2010), DECIPHER (Firth et al. 2009), Exome Aggregation Consortium (ExAC) (Lek et al. 2016), gnomAD (Karczewski et al. 2019), and the Database of Genomic Variants (DGV) (MacDonald et al. 2014). Although identification of variants has progressed rapidly over the past few years, the fundamental challenge to understand the effect of variants has remained (Zarrei et al. 2015).
Although some adaptive CNVs have been described (e.g., the higher copy number of salivary amylase gene (AMY1) in populations with a high-starch diet [Perry et al. 2007]), most CNVs are maladaptive and implicated in human disease (Coe et al. 2014; Sudmant et al. 2015; Gambin et al. 2017). CNVs have an established role in autism spectrum disorder (ASD), in which inherited CNVs contribute to an estimated 5%–15% of cases of ASD (Sudmant et al. 2015; Uddin et al. 2015). Likewise, novel deletions and duplications are observed in 15% of cases with schizophrenia (Walsh et al. 2008). CNVs have also been linked with hemiplegic cerebral palsy (Oskoui et al. 2015; Zarrei et al. 2018), developmental delay and intellectual disability (ID) (Cooper et al. 2011; Rees et al. 2016), epilepsy (Helbig et al. 2009), bipolar disorder (Green et al. 2016), and many other disorders (Fanciulli et al. 2010; Sudmant et al. 2015; Uddin et al. 2015; Zarrei et al. 2015; Gambin et al. 2017).
There are a variety of factors that complicate our understanding of the phenotypic consequence of carrying a CNV. First, CNVs in neurodevelopmental disorders are not consistently penetrant with individuals carrying an identical mutation displaying a large degree of phenotypic variability (Woodbury-Smith and Scherer 2018). Penetrance estimates for ASD vary from 5% for mutations in the dystrophin (DMD) and neurofibromin (NF1) genes to 80% for mutations involving the synaptic scaffold gene (SHANK3) and the calcium ion channel gene (CACNA1C) (Torre-Ubieta et al. 2016). Second, CNVs can have variation in expressivity resulting in different disease outcomes for the same CNV (Cooper et al. 2011). Stefansson et al. (2014) compared schizophrenia and control patients carrying the same CNV against the general population with a battery of cognitive tests. They showed that schizophrenia patients had the greatest impairment in attention, spatial working memory, executive function, cognitive flexibility, language, and processing speed, and that CNV control patients also showed impaired cognitive abilities in comparison to the general population. Third, the pleiotropic effect may result in multiple different clinical presentations. For example, the 1q21.1 duplication had been described in individuals with ASD, ID, epilepsy, schizophrenia, and other disorders (Bernier et al. 2016). Fourth, ASDs and other neurodevelopmental disorders have high rates of comorbidity: 40%–50% of individuals with an ASD also present symptoms of attention deficit/hyperactivity disorder (ADHD) and 70% present symptoms of ID (Matson and Nebel-Schwalm 2007). Finally, many neurodevelopmental disorders are clinically and genetically heterogeneous, with a complex etiology involving potential risk factors in hundreds of different genes (Damiano et al. 2014; De Rubeis and Buxbaum 2015).
As the number of studies describing novel de novo and inherited CNVs increase, genetic and phenotypic overlap between seemingly distinct neurodevelopmental disorders may begin to illustrate the phenotypic consequence of carrying a particular CNV. In this report we present a family with a 1.089-Mb duplication involving 18 genes on Chromosome 19. Within the family are multiple cases of neurodevelopmental disorders including ASD, ADHD, ID, and psychiatric disease. Quantitative polymerase chain reaction (PCR) confirmed that the CNV is de novo in the mother and inherited by both of her sons. Transcriptome data demonstrates a dosage effect of genes within the CNV and differential expression of 407 genes throughout the genome. Finally, we use gastrointestinal microbiome data to speculate on the phenotypic differences observed between the boys carrying the CNV.
RESULTS
Family 23538 (extended pedigree shown in Fig. 1A) has two maternal half-brothers diagnosed with neurodevelopmental disorders. The elder half-brother (47467) has diagnoses of autism, ID, and ADHD; his clinical history indicates he has been followed by pediatricians since the age of 2 yr old for “significant global developmental impairment” and “pervasive and incredible hyperactivity and overflow aggressive behaviors.” The younger half-brother (47468) has diagnoses of ADHD, ID, and suspected ASD; the Autism Diagnostic Observation Scale–2 (ADOS-2) showed that he met cutoff in the moderate severity range. The Wechsler Abbreviated Scale of Intelligence–Second Edition (WASI-II) showed both half-brothers’ IQ scores in the extremely low range (60 for 47467; 69 for 47468) and the mother's IQ score as 89. Both boys were taking stimulant medication to treat their ADHD (Adderall and quetiapine for 47467; Concerta and clonidine for 47468). Both boys had normal growth parameters, with the exception of truncal obesity in the older brother. Both boys had simple ears, tubular nose, high arched palate, and microretrognathia with prominent front teeth (Fig. 1B). On examination they both had bilateral fetal pads. Their mother (47464) has physical and psychiatric diagnoses that include borderline personality disorder, depression, anxiety, polycystic ovarian syndrome (PCOS), type I diabetes, speech delay, thyroid condition, and irritable bowel syndrome (IBS). The mother has self-reported ASD-like behaviors during childhood, which were confirmed by the maternal grandmother including rocking behaviors, difficulty interacting with her peers, and a lack of eye contact with others. The mother was recently assessed using the Diagnostic Interview for Social and Communication Disorders (DISCO) (Wing et al. 2002) and the International Personality Disorder Examination (Loranger et al. 1994). She fulfilled DSM-5 criteria for borderline personality disorder. She did not meet the criteria for ASD. In the extended family there are diagnoses of ADHD in the mother's half-brother and in maternal half-cousins. Some of the mother's conditions are shared by other family members, including PCOS, IBS, type I diabetes, and thyroid conditions. Furthermore, there is a history of mood disorders in the maternal family.
Figure 1.
Individuals in Family 23538 carry a 1.089-Mb duplication on Chromosome 19. (A) Pedigree showing two half-brothers and their mother that carry a copy-number variant (CNV) and extended family members that do not carry the CNV. All family members shown with a sample number were evaluated for the CNV. (B) Face, profile, and hand pictures of both boys. (C) View of the CNV, which spans 18 genes, in genome browser, as well as previously reported CNVs (Database of Genomic Variants [DGV]). (D) Quantitative polymerase chain reaction (qPCR) identifies copy number of genes within the CNV region (ARRCD5, DPP9, LONP1, SAFB2) and copy number of the X Chromosome (HPRT1).
A clinical microarray report provided by the family identified a 1.089-Mb duplication in both half-brothers (47467 and 47468). The CNV was located on Chromosome 19 spanning locations 4,660,013–5,749,143 (hg19 coordinates) and impacts 18 genes ARRDC5, MYDGF, C19orf70, DPP9-AS1, DPP9, FEM1A, HSD11B1L, KDM4B, LONP1, PLIN3, PTPRS, RPL36, SAFB, SAFB2, TICAM1, CATSPERD, UHRF1, and ZNRF4 (Fig. 1C). Because both half-brothers carried the CNV, it seemed to be inherited from the mother, although she was not formally tested. We used quantitative PCR (qPCR) to measure copy number of four genes within the CNV (ARRDC5, DPP9, LONP1, SAFB2), two genes outside the CNV as a control for copy number (SDHA, DYRK1A), and a gene on the sex chromosome (HPRT1) to validate that the method was effective at detecting copy number. We validated that the CNV was present in the mother and both half-brothers but absent in all other family members and unrelated controls. Figure 1D shows that the maternal grandfather (18326) and father (47466) carry two copies (gray) of the genes in the CNV region and one copy (red) of the gene on the X Chromosome and the maternal grandmother (18334) and maternal aunt (18402) carry two copies of the CNV genes and HPRT1, whereas the mother (47464) and both half-brothers carries three copies (blue) of the CNV genes and the expected copy number of the HPRT1 gene for their sex. Further genetic testing by whole-exome sequencing (WES) for both boys and their mother identified 10 rare, heterozygous variants that were predicted by in silico models to be damaging to the function of that gene (Table 1). The four variants shared by at least both boys were also observed in the neurotypical, maternal aunt (Table 1).
Table 1.
Variant discovery by whole-exome sequencing (WES) in Family 23538
| Gene | Chr | HGVS DNA reference | HGVS protein reference | Variant type | dbSNP | Genotype | pLI score | Carriersa |
|---|---|---|---|---|---|---|---|---|
| RAB3GAP1 | 2 | c.C2011T | p.R671X | Stop-gain | rs137853052 | Het | 0.00 | O |
| ITPR1 | 3 | c.C4218G | p.H1406Q | Nonsynonymous | rs61757110 | Het | 1.00 | MOYA |
| FLNB | 3 | c.G2935A | p.V979M | Nonsynonymous | rs376511120 | Het | 0.00 | Y |
| HTR1B | 6 | c.T212C | p.I71T | Nonsynonymous | rs199727324 | Het | 0.65 | MOYA |
| EPHB6 | 7 | c.G364A | p.G122S | Nonsynonymous | rs8177173 | Het | 0.25 | MOAC |
| DLGAP2 | 8 | c.C2039T | p.T680M | Nonsynonymous | rs759725513 | Het | 0.95 | MOYA |
| LAMC3 | 9 | c.C2282T | p.S761L | Nonsynonymous | rs376240028 | Het | 0.00 | MO |
| AAAS | 12 | c.T1145C | p.M382T | Nonsynonymous | rs200871966 | Het | 0.00 | Y |
| SCN4A | 17 | c.G3313A | p.V1105M | Nonsynonymous | N/A | Het | 0.01 | MYAC |
| PTPRT | 20 | c.G541A | p.E181K | Nonsynonymous | rs868295248 | Het | 1.00 | MOYAC |
aIndicates family members that carry the variant: Mother-47464; Older sibling-47467; Younger sibling-47468; maternal Aunt-18402, maternal Cousin-18424.
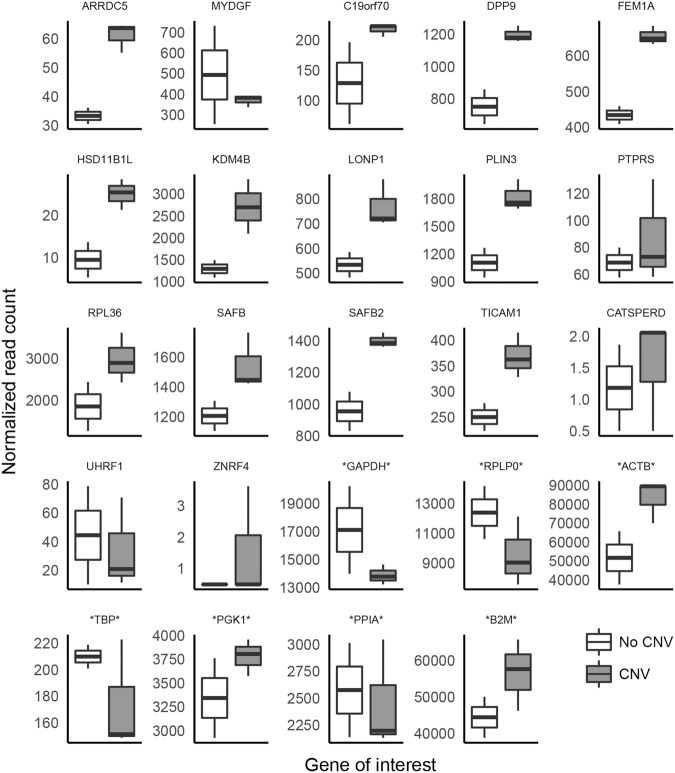
Whole-transcriptome data was analyzed for the three individuals with the CNV and two individuals without the CNV; the maternal aunt (18402), who is an approximate age and gender match for the mother, and a maternal first cousin (18424), who is an approximate age and gender match for the two half-brothers. We detected reads for 15 of the 18 genes located within the CNV region (Fig. 2); reads of the pseudogene DPP9-AS1 were not detected, and the number of reads aligned to CATSPERD and ZNRF4 were almost negligible. Of the remaining 15 genes in the CNV, 12 were expressed ∼1.5-fold higher in the individuals with the CNV when compared to the individuals without the CNV. When considering the whole transcriptome, principal components analysis clustered the individuals carrying the CNV together (Fig. 3A), and differential gene expression identified 407 genes that were differentially expressed in the individuals carrying the CNV versus those without the CNV (Fig. 3B: −1.5 ≤ FC ≥ 1.5; P-value <0.05; P-adj ≤ 0.1). Mapping the 407 genes to gene ontology pathways using WebGestalt identified three overrepresented biological processes including immune response, response to endoplasmic reticulum stress, and regulation of epithelial cell proliferation (Fig. 3C).
Figure 2.
Normalized read count of 17 genes within the CNV and seven housekeeping genes (denoted by *) in family members carrying the CNV (47464, 47467, 47468) and their relatives without the CNV (18402, 18424). Individuals carrying the CNV show an ∼1.5-fold increase in transcript abundance in 12 of the genes contained within the CNV region when compared to their relatives not carrying the CNV. DPP9-AS1 is not shown on this figure because no reads mapping to this gene were observed in the transcriptome data set.
Figure 3.
Whole-transcriptome and pathway analysis of genes differentially expressed in the peripheral blood of family members carrying the CNV. (A) Principal components analysis (PCA) for the first two axes explain 82% of the variability in the data set. (B) Volcano plot shows 245 up-regulated and 162 down-regulated genes in the family members carrying the CNV compared with noncarrier family members. (C) Gene Ontology pathway analysis using WebGestalt identified three overrepresented biological processes including immune response, response to endoplasmic reticulum stress, and regulation of epithelial cell proliferation.
Finally, we collected environmental and dietary information relevant to bacterial composition of the gastrointestinal tract and performed microbiota profiling for the two half-brothers using 16S rDNA sequencing. The boys were raised together and are cohabiting, so they share many environmental factors and dietary habits. Several differences reported in their current dietary habits include higher consumption of fish and lower consumption of dairy, artificial sweeteners, and fruit/vegetables by the elder half-brother (47467) compared with the younger half-brother (47268). 16S rDNA sequencing generated 75658 and 83038 mapped reads for individuals 47467 and 47468, respectively. Alpha diversity was similar between the half-brothers with 26 families of bacteria in the stool of individual 47467 and 29 families of bacteria in 47468. Comparing the relative abundance of bacteria at the phylum taxonomic level (Fig. 4A) demonstrates the composition within participant 47467 includes predominantly Firmicutes (85.2%) but also Bacteroidetes (9.9%), Proteobacteria (4.14%), and Actinobacteria (0.75%); whereas 47468 has approximately equal abundance of Firmicutes (41.5%) and Bacteroidetes (54.8%) but also Proteobacteria (2.83%), Tenericutes (0.67%), and Actinobacteria (0.28%). Notable observations at the family taxonomic level include the presence of Christensenellaceae in 47468 (Fig. 4B), increased abundance of Bifidobacteriaceae in 47467 (Fig. 4C), and presence of Hyphomicrobiaceae in 47467 but Desulfovibrionaceae and Enterobacteriaceae in 47468.
Figure 4.
Bacterial composition of the gastrointestinal tract of the two half-brothers carrying the CNV as determined by 16S rRNA sequencing. (A) Relative abundance of bacteria at the phylum taxonomic level. (B–D) Relative abundance of the bacteria at the family taxonomic level within the (B) Firmicutes, (C) Actinobacteria, (D) Proteobacteria, and (E) Bacteroidetes phylum.
DISCUSSION
Family 23538 enrolled in our research registry because both children had a diagnosis of a neurodevelopment disorder. The family provided clinical reports demonstrating that both children had a 1.089-Mb duplication on Chromosome 19 detected by microarray. Using qPCR, we validated that the CNV was carried by both boys and their mother. This CNV was absent in the father, who provided a sample, and the rest of the extended maternal family indicating that the CNV occurred de novo in the mother and was inherited by both sons. To identify other potential contributing genetic factors, we used WES to identify 10 rare, damaging variants in the family. A nonsynonymous variant in the DLGAP2 gene was confirmed by Sanger sequencing in all three family members. DLGAP2 encodes a membrane-associated protein that has a role in synaptic organization and neuronal signaling and has evidence as an ASD candidate gene (Marshall et al. 2008; Pinto et al. 2010; Chien et al. 2013). However, the variant in DLGAP2, and five other variants in the ITPR1, HTR1B, EPHB6, PRPRT, and SCN4A genes, are also carried by the neurotypical aunt, suggesting that they are not be responsible for the phenotypes observed in Family 23538. A stop-gain variant in RAB3GAP1 was observed only in the older brother, indicating that this variant may be de novo or inherited from his father. RAB3GAP1 encodes a subunit of RAB GTPase activating protein and is thought to be involved in the neurotransmitter exocytosis which is important during eye and brain development (Aligianis et al. 2005). Inactivation of RAB3GAP via homozygous mutations in RAB3GAP has been implicated in Warburg Micro syndrome (Aligianis et al. 2005), a developmental disorder that often includes severe intellectual disability (Bokhoven 2011; Rump et al. 2016). The RAB3GAP1 variant reported in participant 47467 is a heterozygous variant, and the pLI score of RAB3GAP1 is 0.00, indicating that this heterozygous variant is likely not responsible for the deficiencies observed in this family. Therefore, the 1.089-Mb duplication on Chromosome 19 was the only detected genetic risk factor that segregates with disease in the family, raising the possibility that the duplication contributes to the observed phenotypic deficiencies and neurodevelopmental disability diagnoses observed in this nuclear family.
The Female Protective Model
Levy et al. (2011) reported that females with ASDs have a higher frequency of de novo events than males, but males are more likely to be referred for genetic testing (Jacquemont et al. 2014). The female protective model (or male susceptibility model) suggests that females have a protective mechanism that shields them from the effects of genetic variation (Jacquemont et al. 2014), and a mother can pass dominant mutations that have reduced penetrance in females to multiple affected male children (Torre-Ubieta et al. 2016). This model is supported by the 4:1 male to female gender disparity in ASD prevalence (Newschaffer et al. 2007). The 1.089-Mb CNV reported in this family supports the female protective mechanism. The mother displays less severe phenotypic characteristics, such as an IQ that is in the low average range and gene expression more similar to family members without the CNV, as seen in both the PCA plot (Fig. 3A) and heatmap (Fig. 3C).
A recent study demonstrated no significant difference in the Adult Autism-Spectrum Quotient score between individuals diagnosed with borderline personality disorder and ASD (Dudas et al. 2017), suggesting that there may be overlap between the two disorders or that individuals (especially females) with borderline personality disorder may be misdiagnosed (their ASD was overlooked), and a proportion may have both conditions. The mother presented in this report was diagnosed with borderline personality disorder and has self-reported ASD-like behaviors during childhood. Further testing demonstrated that the mother does not have ASD supporting the presence of distinct but possibly etiologically related disorders.
Gene Dosage Effect
The 1.089-Mb duplication on Chromosome 19 is much larger than previously reported CNVs in this region. The DGV contains 331 CNVs in this region of Chromosome 19 (Fig. 1C) including 280 deletions and 44 insertions ranging in size from single-nucleotide indels to a 362,050-bp insertion (Coe et al. 2014), which overlaps the genes at the 3′ end of this CNV. The gnomAD-SV browser (Collins et al. 2019) contains 145 CNVs in this region, including 77 deletions and 62 duplications/insertions ranging from 51 bp to 80,417 bp. CNVs in this region of Chromosome 19 have been reported in CNV maps for individuals with ASD, although they did not maintain statistical significance following Benjamini–Hochberg correction (Cooper et al. 2011). Although dysfunction of each gene may not result in psychiatric disability, the cumulative involvement of multiple genes within this large CNV may reflect an accumulated risk for presentation of several psychiatric disorders. We measured the expression levels of the 18 genes in the peripheral blood for the three individuals carrying the CNV as well as controls from the extended maternal family that were approximate age and gender matched and found that 15 genes were expressed in the peripheral blood. Of the 15 genes whose expression was detectable in the blood, 12 genes were expressed ∼1.5-fold higher in individuals carrying the CNV than their relatives, indicating the CNV may have a dosage effect on the expression of the genes contained within the CNV. The Clinical Genome Resource (ClinGen) consortium has developed a community resource designed to improve our understanding of genomic variation and its use in clinical care (Rehm et al. 2015) and are currently curating genes and regions of the genome to assess whether there is evidence to support that these genes/regions are dosage-sensitive. However, the genes in this reported CNV have not yet been curated by ClinGen. Although there is not an obvious connection between the genes differentially expressed in the peripheral blood and neurodevelopmental disability, the CNV dosage effect may also affect other tissues and other periods of development that may impede typical neurological development. For example, PTPRS is a member of the protein tyrosine phosphatase (PTP) family, which is involved in cell–cell interaction, primary axonogenesis, axon guidance during embryogenesis (Shen et al. 2009), and molecular control of adult nerve repair (Lang et al. 2015). Several rodent studies have shown that 60% of Ptprs double-knockouts are lethal, whereas the surviving mice show severe developmental delays, craniofacial defects, and neurologic defects (Wallace et al. 1999; Uetani et al. 2009). Although overexpression of PTPRS in the human brain has not been reported to date, regulated expression of PTPRS appears to be very important for typical embryonic and brain development.
When considering the whole transcriptome, the family members carrying the CNV demonstrated differential expression of 407 genes, 245 overexpressed and 162 underexpressed, compared with their typical relatives. Pathway analysis maps the genes to three overrepresented pathways including immune response, response to endoplasmic reticulum stress, and regulation of epithelial cell proliferation. There is extensive evidence of immune system implications in neurological processes, and research supporting immune system dysfunction in ASDs (Onore et al. 2012). Dysfunctional immune activity has been observed in ASD spanning both the innate and adaptive immune systems, which can impact neurodevelopment, cognitive function, and behavior (Onore et al. 2012). CD38 has an important role in social recognition through its role as a transmembrane protein engaged in oxytocin secretion (Munesue et al. 2010; Sauer et al. 2012). The three individuals carrying the CNV demonstrated a reduction in peripheral blood expression levels of CD38 (Fig. 3C). Reduced CD38 expression has been reported previously in immortalized lymphocytes derived from an individual with ASD (Lerer et al. 2010). Furthermore, two CD38 SNPs have been associated with high functioning cases of ASD (Munesue et al. 2010), in which the genotype of one of these CD38 SNPs (rs3796863) modulates oxytocin activation of the fusiform gyrus (Sauer et al. 2012). Recently, Martucci et al. (2019) reported that CD38 knockout male mice show an abnormal cortex development, a shifted excitation–inhibition balance, and impaired synaptic plasticity in the prefrontal cortex such as those observed in various mouse models of ASD.
The transcriptome data presented has two key limitations: the use of peripheral blood to understand the biological consequence of carrying a mutation and the small sample size. Though gene expression in the peripheral blood may indicate some effects of the CNV, the observed gene expression profile reported here may not represent gene expression in the brain or during development. Comparing the transcriptomes of three individuals carrying a CNV with two individuals without the CNV does not allow for a proper statistical power analysis, so these results only have exploratory value (Conesa et al. 2016).
The Role of the Gut Microbiome
A female protective model may account for the phenotypic differences observed between the mother and her two sons (discussed above), but it does not account for the differences between the two boys. Although some of these differences may be accounted for by their different fathers, there may be other factors to consider. Emerging studies have demonstrated the gut microbiome is an integral component of human health by contributing to the host's digestion, nutrition, and immune system where dysbiosis of the gut microbiome has been implicated in a wide range of diseases including ASD (DeGruttola et al. 2016). Several of these dysbiosis trends are observed in the elder half-brother (47467) who presents with a more severe phenotype when compared with the younger half-brother (47468), including lower Bacteroidetes/Firmicutes ratio (Strati et al. 2017) and increased Clostridium (Eshraghi et al. 2018) and Sutterella (Williams et al. 2012; Wang et al. 2013b). Although both half-brothers display neurodevelopmental disorders, potentially as a result of the CNV, we propose that the observed differences in phenotype may be, in part, a result of differences in the composition of the gut microbiome. The microbiome data has the same sample size limitation as the transcriptome data set (described above) and should be viewed as exploratory. Future studies correlating gut microbiota and blood metabolites may provide a better view of which bacterial compounds make it into circulation and may impact host metabolism and health (Wilmanski et al. 2019).
In conclusion, we present a family in which the mother carries a de novo 1.089-Mb duplication on Chromosome 19 and passed the CNV to both of her sons. All three individuals display neurodevelopmental disorders including ASD, ADHD, ID, and psychiatric disorders. WES sequencing did not identify any additional variants that segregate within the family, which may account for the observed phenotype. As extended, neurotypical family members carry the same damaging variants but not the CNV, we propose that the CNV may contribute to the etiology underlying these disorders. Exploratory analysis comparing the transcriptome of individuals carrying the CNV with relatives without the CNV demonstrated a dosage effect on the expression of the 18 genes within the CNV region and altered transcriptome in the peripheral blood including dysregulation of many genes involved in immune response. Finally, microbiome differences between the half-brothers may contribute to the observed phenotypic differences. These results raise the possibility that the observed CNV may contribute to varied phenotypic characteristics in different family members through altered gene expression or through altered gut microbiota composition, and more broadly that different psychiatric disorders can share genetic variants that lay a framework for later psychiatric dysfunction.
METHODS
Sample Collection
Family members carrying the CNV (47464, 47467, 47468) and two members not carrying the CNV, a maternal aunt (18402) and a maternal cousin (18424), supplied whole blood in EDTA tubes for DNA extraction, and whole blood in Biomatrica's RNAgard tubes for RNA extraction. One of the fathers (47466), the maternal grandparents (18326, 18334), and other members of the extended family supplied saliva for DNA extraction. The other father was unavailable to provide a sample. Additionally, both half-brothers provided stool samples to represent the gastrointestinal microbiome community. DNA was isolated from saliva following QIAGEN's FlexiGene DNA Handbook and from whole blood in EDTA tubes using Affymetrix's PrepEase Genomic DNA Isolation Kit. Total RNA was isolated from whole blood collected in RNAgard tubes following Biometrica's RNAgard Blood RNA Purification Kit User Manual. Bacterial DNA was isolated from 200 mg of stool using QIAGEN's Allprep PowerFecal Kit. DNA quantity was determined using the Qubit Fluorometer 2.0 dsDNA Broad Range Assay Kit (ThermoFisher Scientific) and quality via gel electrophoresis. RNA quantity was determined using the Qubit 2.0 RNA Assay Kit and quality on an RNA 6000 Pico Kit on the Agilent Technologies Bioanalyzer 2100.
Confirmation of CNV
Forward and reverse primers were designed using NCBI PrimerBlast for four genes in the duplicated region (ARRDC5, DPP9, LONP1, and SAFB2), two autosomal genes for copy-number controls (SDHA and DYRK1A), and a sex chromosome gene (HPRT1) (Table 2). qPCR was performed on Applied Biosystems Viia7 Real-Time PCR machine using ThermoFisher Scientific's 1X Power SYBR Green Master Mix, 100 nM of each primer, and 3 ng of gDNA with the following cycling conditions: initial 10 min denaturation at 95°C, 40 cycles of 15 sec at 95°C, 1 min at 60°C with data collection, and melt curve at 0.5°C/sec increments from 60°C to 95°C. The qPCR data was analyzed using qbase+ v3.1 (www.qbaseplus.com).
Table 2.
Sequence and location of primers used for quantitative polymerase chain reaction (qPCR) validation of the copy-number variant (CNV) on Chromosome 19 in members of family 23538
| Gene name | Primer sequence (5′–3′) | Chromosome:location |
|---|---|---|
| DPP9 | Forward: ATGTGGCCCTTAGCCTCTC | Chr 19:4,689,228–4,689,312 |
| Reverse: CTGACTCGTGCCCTGGACTA | ||
| LONP1 | Forward: TTCCAGCACGGGATTTGTTG | Chr 19:5,707,215–5,707,325 |
| Reverse: GAGGTTGGGGGAAAATGCG | ||
| SAFB2 | Forward: TTACCGGAATCAACCACCTAATG | Chr 19:5,610,429–5,610,541 |
| Reverse: TGTGCTTTTCACCATCCCATAG | ||
| ARRDC5 | Forward: AGTGCTGCCCGAGGATAGAA | Chr 19:4,902,710–4,902,811 |
| Reverse: TCCACCTTCACTATGGGGTC | ||
| SDHA | Forward: CAATATACACGTCTTTCCCCTTC | Chr 5:223,240–223,326 |
| Reverse: CTCAGATACGAAATGAAAAAGGCAC | ||
| DYRK1A | Forward: CTGGGCCTTGTTTTTACCCTC | Chr 21:38,794,783–38,794,888 |
| Reverse: TGAACCTGTGCCTCATCAAG | ||
| HPRT1 | Forward: TACGGGCCAACCTGACAA | Chr X:133,604,318–133,604,403 |
| Reverse: GGTTTGTGCTGTCTTTCAGTC |
Whole-Exome Sequencing
Exome sequencing and variant discovery were performed for both half-brothers (47467 and 47468) and their mother (47464) as we have described previously (Sjaarda et al. 2019), with the exception that we used an Ion 550 Chip on the Ion GeneStudio S5 Plus Semiconductor Sequencer. WES generated an average of 36,858,736 reads and base coverage depth of 94.36 (Table 3). The 10 reported variants were validated in the three family members and the maternal aunt and cousin by Sanger sequencing as previously described (Sjaarda et al. 2019).
Table 3.
Exome sequencing coverage
| Sample | Mapped reads | Average base coverage depth | Uniformity of base coverage |
|---|---|---|---|
| 47464 | 34,050,284 | 86.2 | 92.71% |
| 47467 | 40,538,962 | 106.9 | 94.38% |
| 47468 | 35,986,961 | 90.0 | 94.43% |
Whole-Transcriptome Sequencing
mRNA was isolated from total RNA using ThermoFisher Scientific's Dynabeads mRNA DIRECT Micro Kit recommended protocol for low total RNA input. Libraries were constructed using ThermoFisher Scientific's Ion Total RNA-seq Kit v2 for Whole-Transcriptome Libraries using IonXpress RNA 3′ Barcode Primers to allow for multiplex sequencing. Library yield and size distribution were assessed with the Bioanalyzer 2100 system with the High Sensitivity DNA Kit. ThermoFisher Scientific's Ion OneTouch 2 System was used for templating following Ion PI Hi-Q OT2 200 Kit protocol. Whole-transcriptome sequencing runs were performed with the Ion Proton System following Ion PI Hi-Q Sequencing 200 Kit protocol on an Ion PI Chip v3 (ThermoFisher Scientific).
Transcriptome Data Analysis
Raw sequencing reads were exported from the Ion Torrent Suite and analyzed using Partek Flow software, version 7.0.18.0401 (Partek Inc.). Reads were aligned to the hg19 reference genome using Torrent Mapping Alignment Program (TMAP) and gene counts were quantified to the RefSeq annotation model. Differential gene expression was determined using DESeq2 release 3.7 in R version 3.5.0. Significant differential expression was determined based on P-value < 0.05, adjusted P-value < 0.1, and fold change of <−1.5 or >1.5. The online tool WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) (Zhang et al. 2005; Wang et al. 2013a) was used to identify overrepresented biological processes and gene ontology pathways.
Microbiome Sequencing
Using 3 ηg of DNA as input, 16S hypervariable regions were amplified using both primer sets and reagents in the Ion 16S Metagenomics Kit (ThermoFisher Scientific) following manufacturer's recommendations and 18 PCR cycles on an Applied Biosystems GeneAmp PCR System 9700. PCR products were ligated with Ion Xpress Barcode Adapters 1-16 Kit (ThermoFisher Scientific) to allow for multiplexing, and libraries were quantified using the Ion Universal Library Quantitation Kit (ThermoFisher Scientific) on a Viia7 Real-Time PCR machine (ThermoFisher Scientific). Using 10 pM of library as input, template preparation was performed using the Ion OneTouch 2 System and the Ion PGM Hi-Q OT2 Kit, before loading on an Ion 318 chip v2 and sequencing on the Ion PGM System using the Ion PGM Hi-Q Sequencing Kit (ThermoFisher Scientific). Raw sequence data files were uploaded to ThermoFisher's Ion Reporter and analyzed using the Metagenomics 16S 21.1 version 5.10 workflow. The consensus file for each sample was imported into R version 3.5.2 and figures generated using ggplot2 version 3.1.0. In addition, both boys completed a survey collecting information regarding birth delivery method, early childhood health and diet information, current dietary habits, and gastrointestinal history.
ADDITIONAL INFORMATION
Data Deposition and Access
Whole-exome, RNA-seq, and microbiota data are available under the NCBI Sequence Read Archive (SRA) study accession numbers PRJNA625607 and PRJNA626020.
Ethics Statement
Informed written consent was obtained from all participants or their legal caregivers prior to the study. All experimental protocols were approved by and conducted in accordance with the Queen's University Health Sciences and Affiliated Teaching Hospitals Research Ethics Board (PSIY-074-99 and PSIY-121-01).
Acknowledgments
We thank the family for their participation in this study.
Author Contributions
C.P.S. and X.L. designed the study; C.P.S., B.K., A.J.M.M., and M.L.H. recruited the participants and collected samples; A.G., M.L.H., B.K., and M.A. collected the phenotype data; C.P.S. performed next-generation sequencing; C.P.S., B.K., L.H.-L., and K.L. analyzed data and performed validation; C.P.S. drafted the manuscript; and all authors reviewed the manuscript.
Funding
This work was supported by Ongwanada Special Operating Research Fund for the genetic study of autism and related disorders, private donations, and the Ontario Association on Developmental Disabilities (B.K.).
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Aligianis IA, Johnson CA, Gissen P, Chen D, Hampshire D, Hoffmann K, Maina EN, Morgan NV, Tee L, Morton J, et al. 2005. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet 37: 221–224. 10.1038/ng1517 [DOI] [PubMed] [Google Scholar]
- Bernier R, Steinman KJ, Reilly B, Wallace AS, Sherr EH, Pojman N, Mefford HC, Gerdts J, Earl R, Hanson E, et al. 2016. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet Med 18: 341–349. 10.1038/gim.2015.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhoven HV. 2011. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 45: 81–104. 10.1146/annurev-genet-110410-132512 [DOI] [PubMed] [Google Scholar]
- Chien WH, Gau SS, Liao HM, Chiu YN, Wu YY, Huang YS, Tsai WC, Tsai HM, Chen CH. 2013. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Mol Autism 4: 26 10.1186/2040-2392-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, et al. 2014. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 46: 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, et al. 2019. An open resource of structural variation for medical and population genetics. bioRxiv 10.1101/578674 [DOI] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczes´niak MW, Gaffney DJ, Elo LL, Zhang X, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. 2011. A copy number variation morbidity map of developmental delay. Nat Genet 43: 838–846. 10.1038/ng.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano CR, Mazefsky CA, White SW, Dichter GS. 2014. Future directions for research in autism spectrum disorders. J Clin Child Adolesc Psychol 43: 828–843. 10.1080/15374416.2014.945214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. 2016. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis 22: 1137–1150. 10.1097/MIB.0000000000000750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Buxbaum JD. 2015. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet 24: R24–R31. 10.1093/hmg/ddv273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas RB, Lovejoy C, Cassidy S, Allison C, Smith P, Baron-Cohen S. 2017. The overlap between autistic spectrum conditions and borderline personality disorder. PLoS ONE 12: e0184447 10.1371/journal.pone.018444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi RS, Deth RC, Mittal R, Aranke M, Kay SS, Moshiree B, Eshraghi AA. 2018. Early disruption of the microbiome leading to decreased antioxidant capacity and epigenetic changes: implications for the rise in autism. Front Cell Neurosci 12: 256 10.3389/fncel.2018.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli M, Petretto E, Aitman TJ. 2010. Gene copy number variation and common human disease. Clin Genet 77: 201–213. 10.1111/j.1399-0004.2009.01342.x [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van VS, Moreau Y, Pettett RM, Carter NP. 2009. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet 84: 524–533. 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. 2009. Human genetic variation and its contribution to complex traits. Nat Rev Genet 10: 241–251. 10.1038/nrg2554 [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Stranger BE. 2015. The impact of human copy number variation on gene expression. Brief Funct Genomics 14: 352–357. 10.1093/bfgp/elv017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin T, Yuan B, Bi W, Liu P, Rosenfeld JA, Coban-Akdemir Z, Pursley AN, Nagamani SCS, Marom R, Golla S, et al. 2017. Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med 9: 83 10.1186/s13073-017-0472-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Rees E, Walters JT, Smith KG, Forty L, Grozeva D, Moran JL, Sklar P, Ripke S, Chambert KD, et al. 2016. Copy number variation in bipolar disorder. Mol Psychiatry 21: 89–93. 10.1038/mp.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, et al. 2009. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet 41: 160–162. 10.1038/ng.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, Beckmann JS, Rosenfeld JA, Eichler EE. 2014. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet 94: 415–425. 10.1016/j.ajhg.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K, Francioli L, Tiao G, Cummings B, Alföldi J, Wang Q, Collins R, Laricchia K, Ganna A, Birnbaum D, et al. 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 [DOI] [Google Scholar]
- Lang BT, Cregg JM, DePaul MA, Tran AP, Xu K, Dyck SM, Madalena KM, Brown BP, Weng YL, Li S, et al. 2015. Modulation of the proteoglycan receptor PTPσ promotes recovery after spinal cord injury. Nature 518: 404–408. 10.1038/nature13974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerer E, Levi S, Israel S, Yaari M, Nemanov L, Mankuta D, Nurit Y, Ebstein RP. 2010. Low CD38 expression in lymphoblastoid cells and haplotypes are both associated with autism in a family-based study. Autism Res 3: 293–302. 10.1002/aur.156 [DOI] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. 2011. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70: 886–897. 10.1016/j.neuron.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Loranger AW, Sartorius N, Andreoli A, Berger P, Buchheim P, Channabasavanna SM, Coid B, Dahl A, Diekstra RFW, Ferguson B, et al. 1994. The International Personality Disorder Examination: the World Health Organization/Alcohol, Drug Abuse, and Mental Health Administration International Pilot Study of Personality Disorders. Arch Gen Psychiatry 51: 215–224. 10.1001/archpsyc.1994.03950030051005. [DOI] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. 2014. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42: D986–D992. 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. 2008. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82: 477–488. 10.1016/j.ajhg.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martucci LL, Amar M, Chaussenot R, Benet G, Bauer O, de Zélicourt A, Nosjean A, Launay JM, Callebert J, Sebrie C, et al. 2019. A multiscale analysis in CD38−/− mice unveils major prefrontal cortex dysfunctions. FASEB J 33: 5823–5835. 10.1096/fj.201800489R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson JL, Nebel-Schwalm MS. 2007. Comorbid psychopathology with autism spectrum disorder in children: an overview. Res Dev Disabil 28: 341–352. 10.1016/j.ridd.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Munesue T, Yokoyama S, Nakamura K, Anitha A, Yamada K, Hayashi K, Asaka T, Liu HX, Jin D, Koizumi K, et al. 2010. Two genetic variants of CD38 in subjects with autism spectrum disorder and controls. Neurosci Res 67: 181–191. 10.1016/j.neures.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Newschaffer CJ, Croen LA, Daniels J, Giarelli E, Grether JK, Levy SE, Mandell DS, Miller LA, Pinto-Martin J, Reaven J, et al. 2007. The epidemiology of autism spectrum disorders. Annu Rev Public Health 28: 235–258. 10.1146/annurev.publhealth.28.021406.144007 [DOI] [PubMed] [Google Scholar]
- Onore C, Careaga M, Ashwood P. 2012. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun 26: 383–392. 10.1016/j.bbi.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoui M, Gazzellone MJ, Thiruvahindrapuram B, Zarrei M, Andersen J, Wei J, Wang Z, Wintle RF, Marshall CR, Cohn RD, et al. 2015. Clinically relevant copy number variations detected in cerebral palsy. Nat Commun 6: 7949 10.1038/ncomms8949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet 39: 1256–1260. 10.1038/ng2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372. 10.1038/nature09146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Kendall K, Pardinas AF, Legge SE, Pocklington A, Escott-Price V, MacCabe JH, Collier DA, Holmans P, O'Donovan MC, et al. 2016. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry 73: 963–969. 10.1001/jamapsychiatry.2016.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, et al. 2015. ClinGen—the clinical genome resource. N Engl J Med 372: 2235–2242. 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rump P, Jazayeri O, van Dijk-Bos KK, Johansson LF, van Essen AJ, Verheij JBGM, Veenstra-Knol HE, Redeker EJW, Mannens MMAM, Swertz MA, et al. 2016. Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med Genomics 9: 7 10.1186/s12920-016-0167-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. 10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- Sauer C, Montag C, Worner C, Kirsch P, Reuter M. 2012. Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology 37: 1474–1482. 10.1038/npp.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. 2007. Challenges and standards in integrating surveys of structural variation. Nat Genet 39: S7–S15. 10.1038/ng2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, He Z, Silver J, Flanagan JG. 2009. PTPσ is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 326: 592 10.1126/science.1178310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaarda CP, Wood S, McNaughton AJM, Taylor S, Hudson ML, Liu X, Guerin A, Ayub M. 2019. Exome sequencing identifies de novo splicing variant in XRCC6 in sporadic case of autism. J Hum Genet 65: 287–296. 10.1038/s10038-019-0707-0. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, Bjornsdottir G, Walters GB, Jonsdottir GA, Doyle OM, et al. 2014. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505: 361–366. 10.1038/nature12818 [DOI] [PubMed] [Google Scholar]
- Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabro A, et al. 2017. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5: 24 10.1186/s40168-017-0242-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2005. A haplotype map of the human genome. Nature 437: 1299 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre-Ubieta L, Won H, Stein JL, Geschwind DH. 2016. Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22: 345–361. 10.1038/nm.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Thiruvahindrapuram B, Walker S, Wang Z, Hu P, Lamoureux S, Wei J, MacDonald JR, Pellecchia G, Lu C, et al. 2015. A high-resolution copy-number variation resource for clinical and population genetics. Genet Med 17: 747–752. 10.1038/gim.2014.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetani N, Bertozzi K, Chagnon MJ, Hendriks W, Tremblay ML, Bouchard M. 2009. Maturation of ureter-bladder connection in mice is controlled by LAR family receptor protein tyrosine phosphatases. J Clin Invest 119: 924–935. 10.1172/JCI37196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MJ, Batt J, Fladd CA, Henderson JT, Skarnes W, Rotin D. 1999. Neuronal defects and posterior pituitary hypoplasia in mice lacking the receptor tyrosine phosphatase PTPσ. Nat Genet 21: 334–338. 10.1038/6866 [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. 2008. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320: 539–543. 10.1126/science.1155174 [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. 2013a. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41: W77–W83. 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. 2013b. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism 4: 42 10.1186/2040-2392-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Parekh T, Lipkin WI. 2012. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio 3: e00261-11 10.1128/mBio.00261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM, Price ND. 2019. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol 37: 1217–1228. 10.1038/s41587-019-0233-9 [DOI] [PubMed] [Google Scholar]
- Wing L, Leekam SR, Libby SJ, Gould J, Larcombe M. 2002. The diagnostic interview for social and communication disorders: background, inter-rater reliability and clinical use. J Child Psychol Psychiatry 43: 307–325. 10.1111/1469-7610.00023 [DOI] [PubMed] [Google Scholar]
- Woodbury-Smith M, Scherer SW. 2018. Progress in the genetics of autism spectrum disorder. Dev Med Child Neurol 60: 445–451. 10.1111/dmcn.13717 [DOI] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW. 2015. A copy number variation map of the human genome. Nat Rev Genet 16: 172–183. 10.1038/nrg3871 [DOI] [PubMed] [Google Scholar]
- Zarrei M, Fehlings DL, Mawjee K, Switzer L, Thiruvahindrapuram B, Walker S, Merico D, Casallo G, Uddin M, MacDonald JR, et al. 2018. De novo and rare inherited copy-number variations in the hemiplegic form of cerebral palsy. Genet Med 20: 172–180. 10.1038/gim.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33: W741–W748. 10.1093/nar/gki475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-exome, RNA-seq, and microbiota data are available under the NCBI Sequence Read Archive (SRA) study accession numbers PRJNA625607 and PRJNA626020.