Abstract
Heat shock protein 70s (Hsp70s) are a highly conserved class of molecular chaperones that fold a large proportion of the proteome. Nematostella vectensis (Nv) is an estuarine sea anemone that has emerged as a model species to characterize molecular responses to physiological stressors due to its exposure to diverse, extreme abiotic conditions. Previous transcriptional data has shown dramatic differences among expression profiles of three NvHsp70 isoforms (NvHsp70A, B and D) under stress but it is unknown if, and to what extent, the client proteins for these chaperones differ. In order to determine client specificity, NvHsp70A, B and D were expressed in Saccharomyces cerevisiae budding yeast lacking native Hsp70 and interacting proteins for each Hsp70 were determined with mass spectrometry in yeast ambient and heat shock conditions. Our analyses showed <50% of identified interacting proteins were common to all three anemone Hsp70s and 3–18% were unique to an individual Hsp70. Mapping of temperature induced interactions suggest that under stress a proportion of clients are transferred from NvHsp70A and NvHsp70D to NvHsp70B. Together, these data suggest a diverse set of interacting proteins for Hsp70 isoforms that likely determines the precise functions for Hsp70s in organismal acclimation and potentially adaptation.
Keywords: Hsp70, Molecular chaperones, Global interactomes, Yeast, Nematostella
1. Introduction
Heat shock proteins, particularly those from the Hsp70 and Hsp90 families, are molecular chaperones that engage in various cellular processes to maintain cellular homeostasis. When under stressors such as heat shock, anoxia, and changes chemical exposures, these chaperones can be upregulated or activated to assist in the folding of newly synthesized and denatured proteins [1–3]. Their high sequence similarity and abundance in all living organism has made these chaperones excellent proteins to serve as biological markers in the ecology and evolution of stress responses [4]. In particular, Hsp70 has become a common target due to its extensive conservation across most eukaryotes and distinctive cellular localization patterns [5,6]. Over the past few decades, Hsp70 has consequently developed as an early response biomarker for determination of organismal stress in a variety of animals [5,6]. For example, prior studies of Hsp70 in reef building corals determined that different abiotic stressors elicit upregulation of various isoforms of the chaperone [7,8]. Similar results were found when studying the Hsp70 isoforms of sea anemones and hydrozoans [9,10]. Furthermore, populations of particular species living in different environments or geographic locations exhibit distinct Hsp70 expression patterns, which may indicate adaptation to more physiologically challenging conditions and stressors [11,12].
Climate models of marine habitats predict these habitats will undergo considerable changes in temperature in the coming decades, leading to higher average annual temperatures and more dramatic seasonal fluctuations [13,14]. Studying the mechanisms by which organisms respond and potentially adapt to environmental stressors like temperature may provide insights into how species may respond to ongoing climate change. While mechanisms of adaptation are surely diverse, HSP expression and diversification remain at the forefront of candidate biomarkers for both acute and chronic thermal stress.
Although taxonomic sampling and environmental conditions inducing Hsp expression in cnidarians has increased in recent years, much of this research has been restricted to analyses of transcription or protein abundance with polyclonal antibodies. There are no studies that we are aware of that have studied protein-level functions for the HSPs, which limits our ability to determine why these proteins may be upregulated for particular stressors and how different Hsps in the same size class may be involved in unique pathways. A general limitation for cnidarians and most other groups of animals is the lack of in vivo protein technologies.
The starlet sea anemone, Nematostella vectensis, is a marine cnidarian that has developed into a model organism to understand the physiological and molecular responses of cnidarians to environmental variation [15,16]. N. vectensis specializes in shallow coastal waters, usually salt marshes and lagoons, with a distribution in North America along the Atlantic coast, spanning a large latitudinal cline (Nova Scotia to Florida) and the Pacific coast [17,18]. These acute and chronic temperature differences in exposures for N. vectensis, along with the anemone’s limited migratory ability, have likely resulted in populations with different thermal tolerances due to selection pressure for physiological plasticity and genetic adaptations [19,20]. Previous studies on the different N. vectensis isoforms of Hsp70 (NvHsp70s) have focused solely on transcriptional analysis, resulting in a lack of knowledge regarding isoform-specific functions of NvHsp70 and its protein-protein interactions. Hsp isoforms in N. vectensis remain largely uncharacterized. Early genomic surveys suggested perhaps a dozen members of the Hsp20 family, 5 Hsp70s, and 3 Hsp90s are present [21,22]. Transcriptome-wide surveys from adult N. vectensis cultured under various stressors have shown particular Hsps in each size class to have significant changes in expression [23,24]. Our previous studies demonstrate that when expressed as the sole Hsp70 in yeast, the NvHsp70 isoforms present unique cellular phenotypes suggesting they possess distinct biological functions [25].
To address these gaps in the current understanding of proteome homeostasis in N. vectensis, we performed a comparative interactome analysis of three cytosolic HSP70 proteins (NvHsp70 A, B and D) to quantify chaperone-client complexes when expressed in yeast. Interestingly, we observed differences in client and co-chaperone specificity, as well as contrasting interactome changes upon heat shock. These results, taken together with previous studies showing distinct transcriptional patterns for NvHsp70A, B and D provide an evolutionary rationale into why N. vectensis has maintained and utilizes three cytosolic Hsp70s.
2. Materials and methods
2.1. Yeast strains and cloning
Yeast cultures were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or in SD (0.67% yeast nitrogen base, 2% glucose) supplemented with the appropriate nutrients to select for plasmids and gene replacements. NvHsp70A, B, and D were amplified from cDNA synthesized from RNA isolated from Nematostella vectensis adults originally collected from Sippewissett, MA and inserted into the pAG415GPDccdB vector for expression in yeast using In-Fusion cloning as in [25]. All inserts were sequence confirmed with Sanger sequencing. For HIS-tagged versions of NvHsp70A, B, and D, the Hsp70 isoforms were PCR amplified in a similar manner to the untagged versions except using a Forward primer that contained an N-terminal 6xHIS tag and GG linker (primer sequence available on request). The resulting PCR product was cloned via In-Fusion cloning into pAG415GPD-ccdB. Each NvHsp70 isoform construct was transformed into yeast strain ssa1–4Δ [26] using PEG/lithium acetate. After restreaking onto media lacking leucine, transformants were streaked again onto media lacking leucine and containing 5-fluoro-orotic acid (5-FOA), resulting in yeast that expressed NvHsp70 isoforms as the sole cytoplasmic Hsp70 in the cell.
2.2. Purification of NvHsp70 interactomes from yeast
Yeast clones expressing each NvHsp70 isoform were grown to an OD600 of 0.5 in YPD media. Cells were then incubated at 39 °C for two hours (heat shock) or incubated at 30 °C (untreated). His-tagged proteins were purified as follows: 200 μg of protein extract was incubated with 30 μL His-Tag Dynabeads (Invitrogen) at 4 °C for 20 min with gentle agitation. Dynabeads were collected by magnet then washed five times with wash buffer (50 mM Na-phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20). After the final wash, the buffer was aspirated and the Dynabeads were incubated with 100 μL elution buffer (300 mM imidazole, 50 mM Na-phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20) for 20 min and collected via magnet. Supernatant was combined with 25 μl of 5× SDS-PAGE sample buffer and denatured for five minutes at 95 °C. 20 μL of sample was analyzed by SDS-PAGE. For mass spectrometry analysis, samples were processed as in below method sections. For validation of NvHsp70-Ydj1 interaction, 20 μl of sample was analyzed by SDS-PAGE and analyzed in a Western blot using a pan-Hsp70 antibody (Enzo, #AID-SPA-822-F) and Ydj1 antibody (StressMarq #SMC-166D). GAPDH was used as a loading control (Thermo #MA15738). Each experiment was performed in biological triplicate.
2.3. Trypsin digestion of samples from SDS-PAGE gel plugs
The gel plugs for each sample to be analyzed were excised by sterile razor blade, divided into 2 sections ~1 cm each, and chopped into ~ 1 mm3 pieces. Each section was washed in dH2O and destained using 100 mM NH4HCO3 pH 7.5 in 50% acetonitrile. A reduction step was performed by addition of 100 μl 50 mM NH4HCO3 pH 7.5 and 10 μl of 200 mM tris(2-carboxyethyl)phosphine HCl at 37 °C for 30 min. The proteins were alkylated by addition of 100 μl of 50 mM iodoacetamide prepared fresh in 50 mM NH4HCO3 pH 7.5 buffer, and allowed to react in the dark at 20 °C for 30 min. Gel sections were washed in water, then acetonitrile, and vacuum dried. Trypsin digestion was carried out overnight at 37 °C with 1:50–1:100 enzyme–protein ratio of sequencing grade-modified trypsin (Promega) in 50 mM NH4HCO3 pH 7.5, and 20 mM CaCl2. Peptides were extracted with 5% formic acid and vacuum dried and sent to the Mayo Clinic Proteomics Core facility for HPLC and LC-MS/MS data acquisition.
2.4. HPLC for mass spectrometry
All samples were re-suspended in Burdick & Jackson HPLC-grade water containing 0.2% formic acid (Fluka), 0.1% TFA (Pierce), and 0.002% Zwittergent 3–16 (Calbiochem), a sulfobetaine detergent that contributes the following distinct peaks at the end of chromatograms: MH+ at 392, and in-source dimer [2 M + H+] at 783, and some minor impurities of Zwittergent 3–12 seen as MH+ at 336. The peptide samples were loaded to a 0.25 μl C8 OptiPak trapping cartridge custompacked with Michrom Magic (Optimize Technologies) C8, washed, then switched in-line with a 20 cm by 75 μm C18 packed spray tip nano column packed with Michrom Magic C18AQ, for a 2-step gradient. Mobile phase A was water/acetonitrile/formic acid (98/2/0.2) and mobile phase B was acetonitrile/isopropanol/water/formic acid (80/10/10/0.2). Using a flow rate of 350 nl/min, a 90 min, 2-step LC gradient was run from 5% B to 50% B in 60 min, followed by 50%–95% B over the next 10 min, hold 10 min at 95% B, back to starting conditions and re-equilibrated.
2.5. LC-MS/MS analysis
Electrospray tandem mass spectrometry (LC–MS/MS) was performed at the Mayo Clinic Proteomics Core on a Thermo Q-Exactive Orbitrap mass spectrometer, using a 70,000 RP survey scan in profile mode, m/z 340–2000 Da, with lockmasses, followed by 20 MSMS HCD fragmentation scans at 17,500 resolution on doubly and triply charged precursors. Single charged ions were excluded, and ions selected for MS/MS were placed on an exclusion list for 60 s.
2.6. LC–MS/MS data analysis, statistical analysis
All LC-MS/MS *.raw data files were analyzed with MaxQuant version 1.5.2.8, searching against the SPROT Saccharomyces cerevisiae database downloaded 1/9/2018, and modified by the removal of Sc HSP70 proteins Ssa1, Ssa2, Ssa3, Ssa4 and with the addition of the Nematostella vectensis HSP70 bait proteins (NvHsp70A, NvHsp70B, and NvHsp70D), and searched using the following criteria: LFQ quantification with a min of 1 high confidence peptide. Trypsin was selected as the protease with max miss cleavage set to 2.
Carbamiodomethyl (C) was selected as a fixed modification. Variable modifications were set to Deamidation (NQ), Oxidization (M), Formylation (n-term), and Phosphorylation (STY). Orbitrap mass spectrometer was selected using a MS error of 20 ppm and a MS/MS error of 0.5 Da. A 1% FDR cutoff was selected for peptide, protein, and site identifications.
LFQ intensities were reported based on the MS level peak areas determined by MaxQuant and reported in the proteinGroups.txt file as. Proteins were removed from this results file if they were flagged by MaxQuant as “Contaminants”, “Reverse” or “Only identified by site”. Complete three biological replicates were performed. The abundance data from each biological replicate were normalized to the ratio of the N. vectensis HSP70 isoform bait protein in that run (e.g., normalized to the NvHSP70A, NvHSP70B, or NvHSP70D, respectively). LFQ Peak intensities were analyzed in each run to determine protein hits that fell into the category of either Unstressed only hits or Stressed only hits and retained if they confirmed to this state across all 3 runs. LFQ Significance cutoffs are Significantly Up > 1.2 ratio (Log2 0.26) and Significantly Down < 0.8 ratio (Log2–0.32).
Any hits that were not observed in at least two replicates each were labeled ‘no quant’ (a normalized ratio was still calculated but not included in final data set analysis).
A list of proteins identified and corresponding ratios can be found in Supplemental Table T1. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) [27] via the PRIDE partner repository [28,29] with the dataset identifier PXD012144.
2.7. Data analysis, statistical analysis and visualization
Venn diagrams were performed with Venny 2.1. Gene Ontology analysis was performed using GO Slim Mapper on the Saccharomyces Genome Database (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl). Interactome visualization was performed using Prism 7 graphing software for individual interactomes and comparative interactomes. Data points indicating an interaction change of greater or less than a two-fold change, calculated by an average Log2 ratio of > 1 or < −1, were considered significant. In order to determine common interactors among the NvHsp70 isoforms, comparative interactomes were plotted to demonstrate binding activity of interactors between the Hsp70 isoforms. The interaction changes for each isoform are represented by a two-Log ratio. The associations and dissociations between isoforms and their suggested clients in Saccharomyces cerevisiae with > 1 or < −1 were clustered using the String Database Version 10.5.
3. Results
3.1. Quantitative mass spectrometry analysis of NvHsp70 interactomes during heat shock
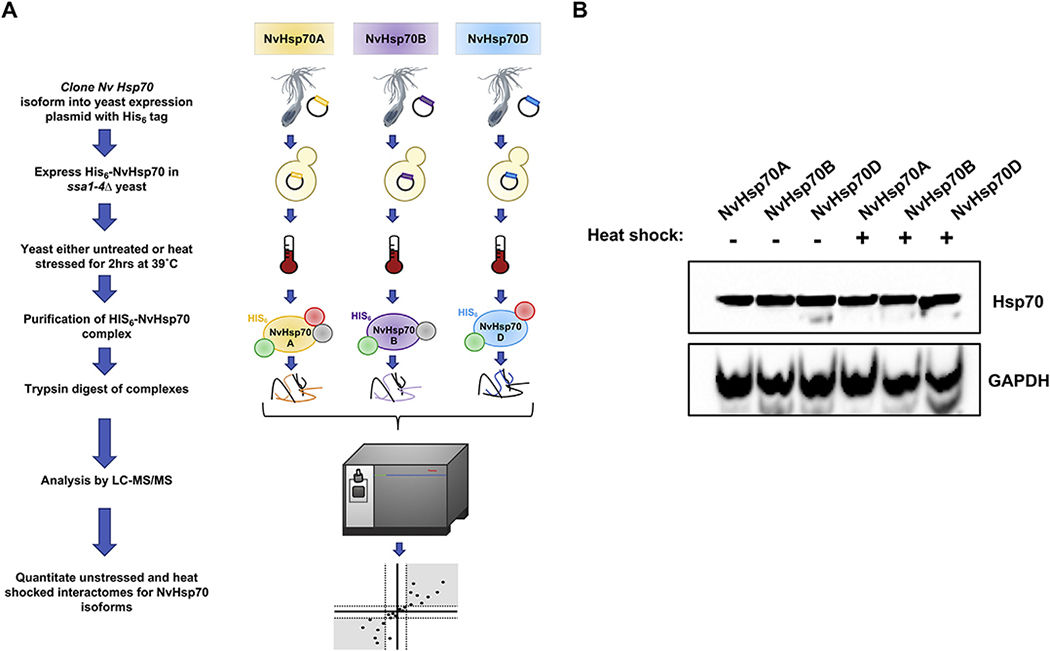
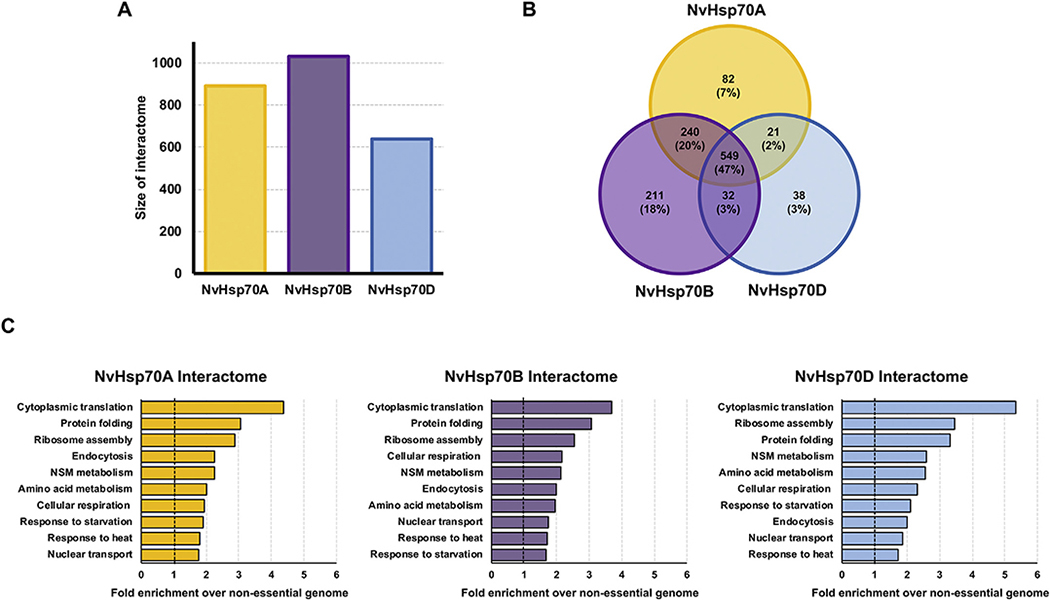
Previous studies revealed differences in function of NvHsp70 isoforms at both the transcriptional level (in N. vectensis) and at the phenotypic level when expressed in yeast [25]. We considered the possibility that the functional differences between NvHsp70 isoforms may arise from differences in client and co-chaperone binding. We employed quantitative affinity purification followed by mass spectrometry (APMS) to assess the dynamics of the NvHsp70A, B and D interactomes upon heat stress. We expressed HIS-tagged versions of NvHsp70 (NvHsp70A, B and D) in ssa1–4Δ, a yeast strain in which all four SSA (Hsp70) genes have been deleted [26]. We purified His-NvHsp70 A, B and D complexes by immobilized metal affinity chromatography (IMAC) before and after heat shock (39 °C for 2 h) and performed quantitative LC–MS/MS proteomics, Fig. 1A). This approach facilitated identification of the respective interactomes of NvHsp70 without competition from other native Hsp70 isoforms. To ensure that the NvHsp70 isoforms were expressed as expected, we analyzed cell lysate from NvHsp70A, B and D-expressing cells under unstressed and heat shock conditions. NvHsp70A, B and D were equally expressed under all conditions (Fig. 1B). Quantitative proteomics identified a total of 1172 interactors (in unstressed and heat shock) associated with the three NvHsp70 isoforms (Fig. 2A). NvHsp70B interacted with the most proteins, 1031 (88%) in total, of which 211 (18%) were unique to this isoform. NvHsp70A interacted with a total of 891 proteins (76%), of which 82 (7%) were unique. Lastly, NvHsp70D interacted with a total of 639 (55%), of which 38 (3%) were unique to the D isoform (Fig. 2B). NvHsp70A and NvHsp70B shared 240 interactors (21%), whereas NvHsp70A and NvHsp70D only shared 21 interactors (1.8%) and NvHsp70B and NvHsp70D 32 interactors (3%) (Fig. 2B). Consistent with the high sequence similarity of the isoforms, 549 (47%) of the interactors were found to interact with all three NvHsp70 proteins (Fig. 2B).
Fig. 1.
Quantitative affinity purification mass spectrometry for comparing NvHsp70 isoform interactions under heat stress. (A) NvHsp70 isoforms were expressed with an N-terminal 6xHIS tag in yeast lacking native Hsp70s. After exposing cells to heat stress (39 °C for 2 h), chaperone interactomes were isolated by nickel-NTA magnetic bead affinity purification. Following PAGE and in-gel trypsin digestion, peptides were analyzed by Orbitrap LC–MS/MS and MassQuant informatic analysis, allowing identification of interacting proteins and determination of their relative enrichment after heat stress. Each experiment was performed in biological triplicate. (B) Expression of NvHsp70 isoforms in yeast. Lysate of cells treated as in (A) were analyzed by SDS-PAGE/Western Blotting using pan-Hsp70 and GAPDH antibodies.
Fig. 2.
Gene Ontology (GO) term analysis of NvHsp70 interactors. (A) Total size of interactomes (unstressed plus heat shock) obtained from each NvHsp70 isoform. (B) Venn diagram of NvHsp70A, B and D interactors (unstressed plus heat shock) remaining after applying statistical filters. (C) Functional classification of the NvHsp70A, B and D interactome. Interactors were categorized by cellular function using GO Slim analysis and relative enrichment compared to occurrence in the non-essential genome was calculated. The top 10 enriched cellular processes are shown for each interactomes. (B) Venn diagram of NvHsp70A, B and D interactors (unstressed plus heat shock) remaining after applying statistical filters.
3.2. Gene ontology analysis (GO) of NvHsp70 isoform interactomes
Gene ontology (GO) analysis of unique candidate interactors of each NvHsp70 isoform revealed significant enrichment of multiple cellular functions (Fig. 2C). In all three isoforms, the GO term cytoplasmic translation was the most enriched, with ribosome assembly present in second or third most enriched term, correlating with the established role of Hsp70 in the folding of newly synthesized client proteins (Fig. 2C). Given the well characterized function of Hsp70, it was unsurprising that protein folding and response to heat were also highly enriched. Interestingly, despite the phenotypic differences observed when expressing NvHsp70 isoforms in yeast [24], the three interactomes did not differ greatly overall in GO term composition (Fig. 2C).
3.3. Analysis of the dynamic NvHSP70 interactomes
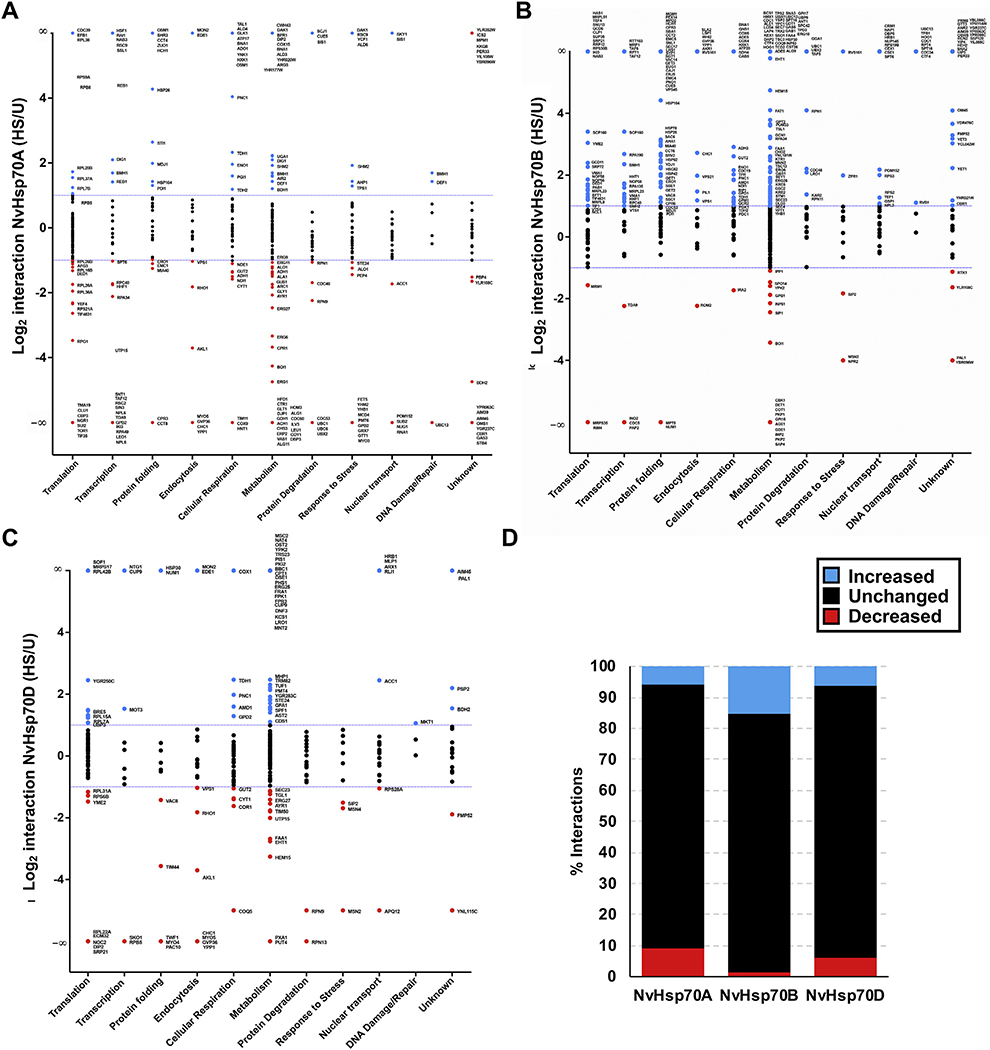
To represent chaperone dynamics, the average Log2 ratio change in interaction upon heat stress treatment for each quantitated interacting protein was examined (Fig. 3A–D). Changes in interaction’s expression levels, > 1 or < −1 (Log2), were considered significant. Values between those significant parameters were defined as unchanged. Interactors were sorted by non-redundant GO terms and graphed by average change in interaction with the NvHsp70A, B, or D isoform upon heat stress compared with untreated conditions. Each NvHsp70 isoform displayed different patterns of interactions upon heat stress. Among NvHsp70A clients, 331 out of 644 quantified (51%) interactions remained unchanged, 109 (17%) increased in interaction and 204 (32%) decreased (Fig. 3A, D). Among NvHsp70B clients, 311 out of 735 quantified (42%) interactions remained unchanged, 378 (51%) increased, and 46 (6%) decreased (Fig. 3B, D). Among NvHsp70D clients, 309 out of 492 quantified (63%) interactions remained unchanged, 100 (20%) increased, and 83 (17%) deceased (Fig. 3C, D).
Fig. 3.
The dynamic NvHsp70A, B, and D interactomes during heat shock. Interactors were organized into functional categories following GO terms and plotted against interaction change (Log2 ratio) with either (A) NvHsp70A, (B) NvHsp70B, or (C) NvHsp70D following heat stress treatment (39 °C for 2 h). The dotted lines represent an interaction change of Log2 > 1 or Log2 < −1. Interactors are colored according to change in interaction as follows: blue (significant increase), red (significant decrease), or black (no significant change) upon heat stress. (D) Summary of each NvHsp70 interactome showing total number of increased, decreased and unchanged interactions upon heat stress. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Analysis of unique interactors of NvHsp70 isoforms
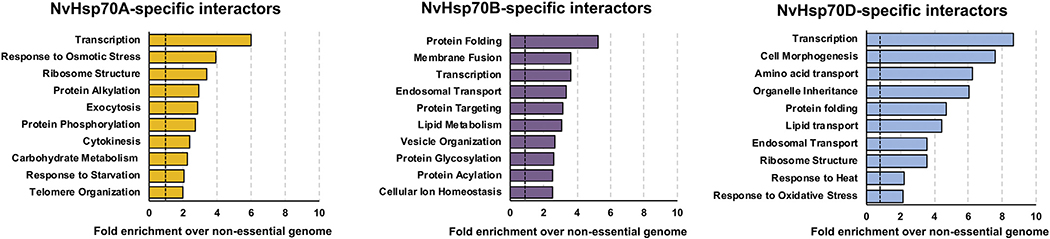
While GO comparison of all three NvHsp70 interactomes did not reveal any global preference for specific classes of clients, we entertained the possibility that studying unique interactors of each Hsp70 isoform may provide insight into NvHsp70 isoform-client specificity. Proteins related to transcription were highly enriched in all three NvHsp70 isoforms (Fig. 4). This was interesting as it implies that the NvHsp70 isoforms are together contributing to the activation of cellular transcription by binding specific, non-overlapping proteins. Aside from transcription, each isoform had almost completely mutually exclusive GO profiles. Together with data in Fig. 1B and C it appears that while the isoforms overlap on core function, they do bind unique clients in specific biological pathways.
Fig. 4.
Gene Ontology (GO) term analysis of NvHsp70 isoform-specific interactors. NvHsp70 isoform-specific interactors were categorized by cellular function using GO Slim analysis and relative enrichment compared to occurrence in the non-essential genome was calculated. The top 10 enriched cellular processes are shown for each interactomes.
3.5. Decoding inter-Hsp70 isoform client transport under stress
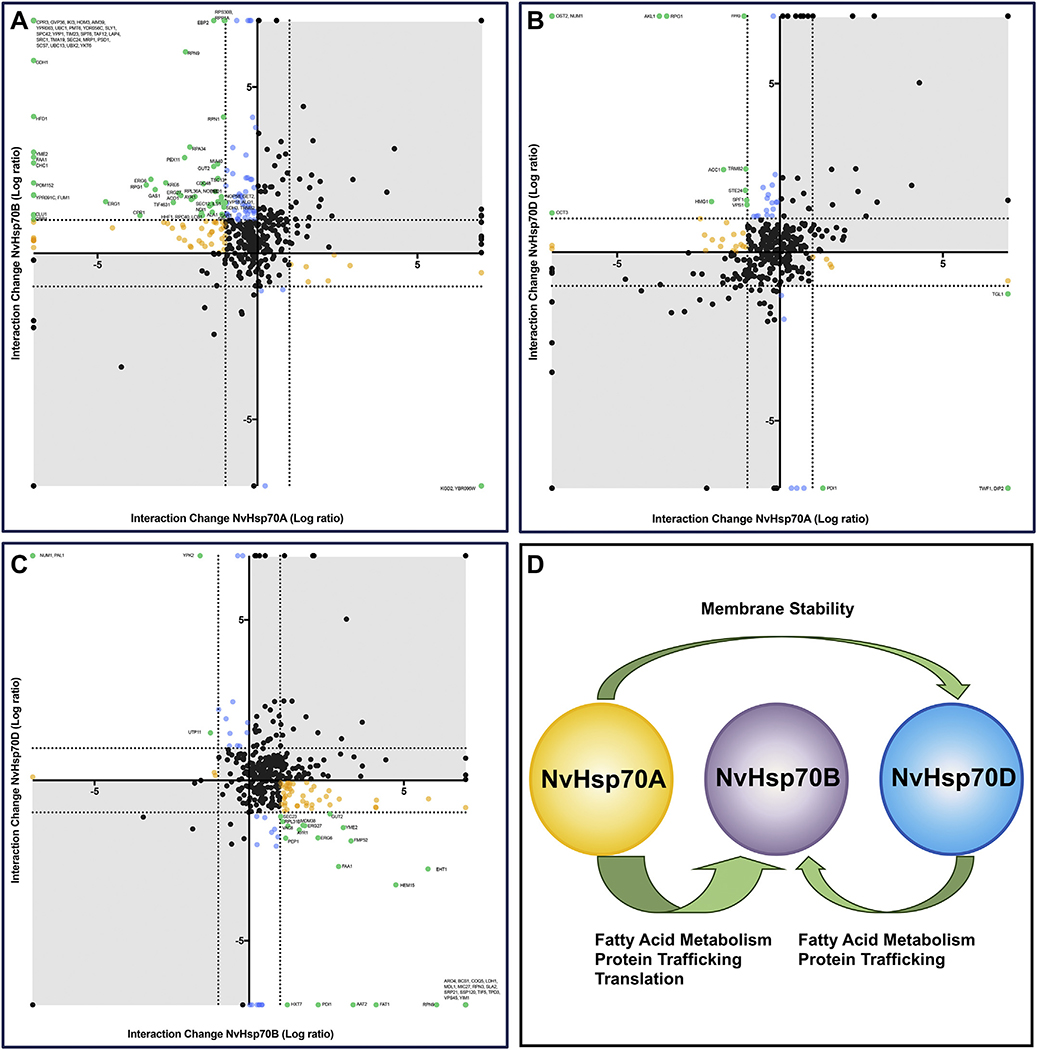
Chaperone interactions are dynamic, associating and disassociating in order to maintain stability, proper activity, and correct cellular localization of clients. While transfer of clients between Hsp70 and Hsp90 has been previously observed, inter-Hsp70 isoform transport of clients has not [30,31]. To determine whether inter-NvHsp70 isoform client transport occurs under stress, pairwise comparisons of NvHsp70 isoform associations with clients were performed. NvHsp70A and NvHsp70B shared the largest number of interactors with a total of 789, of which 9.8% (n = 77) were significant (Fig. 5A, represented in green) for inverse changes (e.g., lower in Hsp70A and higher in Hsp70B) in expression between isoforms. Among these significant interactions, 75 (97%) lost interaction with NvHsp70A yet gained interaction with NvHsp70B (Fig. 5A). GO analysis of proteins transferred from NvHsp70A to NvHsp70B under heat stress revealed an enrichment for fatty acid metabolism, protein trafficking and protein synthesis.
Fig. 5.
Understanding movement of clients between Hsp70 isoforms. (A) For each interactor observed to bind both NvHsp70A and NvHsp70B, change in association for NvHsp70A (Log2 ratio, X-axis) vs. change in association for NvHsp70B (Log2 ratio, Y-axis) was plotted. The dotted lines represent an interaction change of up or down two-fold. Interactors are colored according to significant change in chaperone association upon heat stress as follows: increased interaction with NvHsp70A, decreased for interaction with NvHsp70B (green); significantly altered interaction for NvHsp70B, but not NvHsp70A (blue); significantly altered interaction for NvHsp70A, but not NvHsp70B (yellow) and black (no significant change with either chaperone). (B) Interactions changes of NvHsp70A vs NvHsp70D upon heat stress. (C) Interactions changes of NvHsp70B vs NvHsp70D. (D). Summary of A-C. A large number of clients are transferred from NvHsp70A and NvHsp70D to NvHsp70B under heat stress. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Conversely, clients seen to transfer from NvHsp70B to HvHsp70A only comprised 3% of the interactions (Fig. 4A). Only two, Kgd2 (a TCA cycle protein) and YBR096W (a protein of unknown function that localizes to the ER), were observed to make a complete association with NvHsp70A and complete dissociation with NvHsp70B under stress (Fig. 5A).
NvHsp70A and NvHsp70D shared a total of 570 proteins, of which only 2.8% (n = 16) were significant (Fig. 5B, represented in green) for inverse changes in associations between isoforms. Among these significant interactions, four were associated with NvHsp70A while dissociated with NvHsp70D. The proteins that completely associated with A and dissociated with D were Twf1 and Dip1. Twelve proteins were dissociated with NvHsp70A but associated with HvHsp70D, of which Num1 and Ost2 were completely associated with D and dissociated with NvHsp70B and NvHsp70D shared a total of 580 proteins, of which 6.4% (n = 37) were significant (Fig. 5C, represented in green) for inverse changes in associations between isoforms. Among these significant interactions, 89% were associated for NvHsp70B and dissociated in NvHsp70D. The proteins that completely associated with NvHsp70B and dissociated with HvHsp70 D were Tif5, Ssp120, Srp21, Aro4, Yim1, Bcs1, Vps45, Coq5, Ldh1, Rpn33, Tdp3, Mdl1, Sla2, and Mic27. GO analysis revealed a clustering for proteins involved in fatty acid metabolism and protein synthesis.
The remaining 11% were negatively expressed for NvHsp70B and positively expressed for HvHsp70D, of which two interactors, Num1 and Pal1, were completely associated with NvHsp70D and dissociated with NvHsp70B. Taken together these results indicate that under heat stress, there is inter-Hsp70 isoform client transference, with clients involved in fatty acid metabolism and protein synthesis transferring from NvHsp70A and NvHsp70D to NvHsp70B (Fig. 5D).
3.6. NvHSP70 isoforms display unique regulatory chaperone-co-chaperone protein interaction profiles
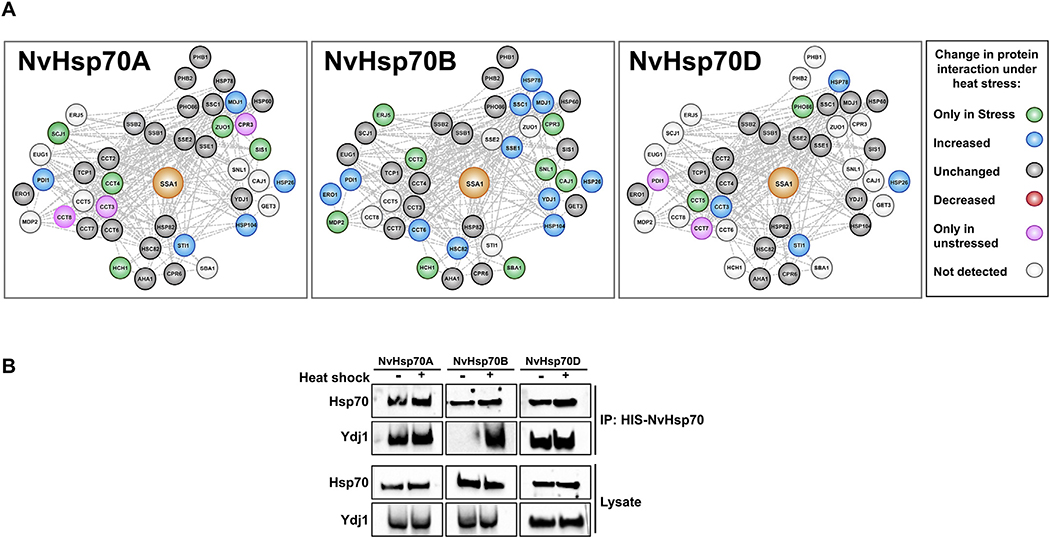
Hsp70 is regulated by a suite of “co-chaperone” helper proteins that regulate both intrinsic ATPase activity as well as client substrate specificity [32,33]. These co-chaperones comprise of J-proteins (containing J-domains), nucleotide exchange factors (NEFs) and other chaperones such as Hsp90 that regulate Hsp70 function. Given their critical role in client specificity, we decided to examine whether NvHsp70 clientomes may be explained by unique chaperone and/or co-chaperone binding preferences. NvHsp70B uniquely displayed a preference for mitochondrial chaperones/co-chaperones (Hsp78, Ssc1, Mdj1) as well as the major chaperones Hsc82, Hsp104 and Hsp26 under stress (Fig. 6A). There appeared to be a selectivity for the CCT chaperone complex. Under stress NvHsp70A increased binding to Cct3 and Cct4 but dissociated from Cct3 and Cct8. Stress promoted a greater NvHsp70B-Cct6 interaction, whereas NvHsp70D displayed a unique stress induced Cct interaction pattern. Under heat shock, NvHsp70DCct3/5 interaction increased, but interaction with Cct7 was completely inhibited (Fig. 6A). Two key regulators of Hsp70 activity in yeast are the Hsp40 isoforms Ydj1 and Sis1. Although they demonstrate partial functional redundancy, loss of Sis1 in yeast is lethal, whereas loss of Ydj1 is not [34,35]. Suggesting an Hsp70 isoform-co-chaperone selectivity, Sis1 interaction only increased with NvHsp70A under stress. In contrast, Ydj1 interaction increased only with NvHsp70B (Fig. 6A and B).
Fig. 6.
Analysis of co-chaperone/chaperone interactors of NvHsp70 isoforms. (A) Networks of 42 co-chaperones and chaperones that were detected by mass spectrometry as interactors of NvHsp70 isoforms were generated using Cytoscape. The nodes were colored based on NvHsp70 interaction change upon heat stress. (B) Validation of NvHsp70 isoform-Ydj1 interaction. Cells expressing NvHsp70 isoforms were grown and treated as in Fig. 1B. NvHsp70 complexes were purified using HIS-dynabeads and analyzed via SDS-PAGE followed by Western Blotting using antisera to Hsp70 or Ydj1.
4. Discussion
4.1. Investigating chaperone interactions in “non-model” organisms
Cells rely on stress regulated signaling pathways to maintain protein homeostasis under stressful cellular conditions. Crucial regulatory elements of the stress response are expression of chaperones like Hsp70 and their dynamic interactions with client proteins. These chaperone-client complexes associate and disassociate in order to maintain stability, proper activity, and correct cellular localization. The heat shock response (HSR) is an ancient and broadly conserved transcriptional regulatory program that results in immediate induction of Hsp’s to protect and repair the cellular proteome [36]. When Hsp70 binds and folds denatured proteins to help them regain lost activities, there are instances where proteins are too damaged for the chaperone to repair and are then targeted for degradation by Hsp70 via the ubiquitin proteasomal system. Therefore, the loss of Hsp70 and client interaction could influence the overall cellular stability or destruction depending on the stress conditions [37–39]. Historically. Hsp70 isoforms have been regarded to be functionally identical, differing only in their cellular localization or expression pattern. Experiments on Ssa isoforms in yeast have hinted that there may be isoform specific function and client specificity [3,40–44]. The advent of genomic epitope tagging of chaperones via CRISPR has made it possible to study chaperones complexes in their native stoichiometry but such approaches remain in their infancy in non-model organisms like Nematostella vectensis [45,46].
The ease of phenotypic screening (both high and low throughput), large range of expression plasmids and ability to edit and delete genes make yeast an ideal model system for expressing and studying foreign proteins. In our ssa1–4Δ yeast system, yeast have their genomic Hsp70s removed and are kept viable via expression of Ssa1 on a URA3-based plasmid. Expression of chaperones from other organisms is then easily achieved via a 5-FOA plasmid swapping system. Given the essential nature of chaperones, we know that any cells that survive the 5-FOA swap have maintained essential chaperone function. This strategy has been successfully employed in multiple studies [40,42,43]. Recent studies from our lab have identified differences in both the pattern of transcription of NvHsp70 isoforms in Nematostella and in function when expressed in yeast [25]. We considered that the unique phenotypes observed when the isoforms were expressed in yeast may be due to unique chaperone-client/co-chaperone interactions. We bypassed the issues generated by the lack of molecular tools to analyze protein interactions in Nematostella by successfully completing a high-resolution analysis of NvHsp70 isoform interactomes when expressed in yeast. This is the first instance of NvHsp70 proteins being studied at the protein/protein interactome level and provides an intellectual framework for future more complex studies that may take place in the native organism.
4.2. NvHsp70 isoforms possess different client binding capacities
The major yeast Hsp70 isoform Ssa1 binds to a substantial number of proteins and is capable of remodeling this interactome upon exposure to external stresses such as DNA damage and internal conditions such as cell cycle progression [30,31,44]. Analysis of the NvHsp70 interactomes shows that a large proportion of interactions (549, representing 47% of total interactions) were shared between all three isoforms. This is unsurprising given the high amino acid similarity between these isoforms and evidence from work in yeast suggesting that there is significant functional redundancy between the Ssa proteins [41]. Interestingly, there was substantial variation in the number of unique interactors for each isoform. NvHsp70B had the most total interactions at 1031 with 211 of these being unique to NvHsp70B. The higher binding capacity seen in NvHsp70B may suggest that NvHsp70B has unique roles in Nematostella, binding unique clients under stress conditions. There are several lines of evidence that support this hypothesis. The transcription of NvHsp70B is induced under heat stress in Nematostella and yeast expressing NvHsp70B as their sole Hsp70 isoform are substantially more stress resistant [25].
4.3. NvHsp70 isoforms display differential plasticity upon heat shock
Chaperone interactions are highly dynamic and change upon stresses such as DNA damage, cell cycle and heat stress [30,31,44,47,48]. All isoforms displayed a core of interactions that remained unchanged upon exposure to heat stress. This has been seen in other chaperone studies and suggests that these are interactions that either change under other stresses or are housekeeping proteins required to keep the cell viable [30,31,44,47,48].
Despite the amino acid similarity of the NvHsp70 isoforms (Fig. S1), we observed significant differences in the way the clientomes of each NvHsp70 isoform responded under heat stress. NvHsp70B interactions underwent greater remodeling upon exposure to heat stress. The increased number of interactions with the NvHsp70B isoform during heat stress may suggest that it plays a role in stress tolerance, an idea corroborated by the fact that cells expressing NvHsp70B are resistant to a range of stressors [25].
4.4. Client transfer between NvHsp70 isoforms upon heat stress
Chaperone isoforms are very dynamic; some clients need to associate with chaperones upon stress to maintain stability, whereas other clients need to lose chaperone interactions allowing correct higher order complex association. Combinations of in vivo and in vitro studies have shown that Hsp70 often folds newly synthesized polypeptide chains into a semi-folded structure. This partially folded client is then transferred to Hsp90 for full folding. Our previous analysis of yeast Hsp70 and Hsp90 interactions under DNA damage confirmed this, but also demonstrated that the reverse can happen-clients can be transferred back from Hsp90 to Hsp70 [30,31]. Fascinatingly, in this study, our data suggests for the first time that clients may shuttle from one Hsp70 isoform to another upon stress. A large number of clients that dissociate from NvHsp70A and NvHsp70D under stress increase association with NvHsp70B. Given the extensive inducibility of NvHsp70B expression (> 100-fold increase in transcription) in Nematostella it may be that at high temperatures more NvHsp70B protein is present in relation to the other isoforms, providing a greater capacity to bind and fold clients. However, the experiments in this study were performed using constitutively expressed isoforms and suggest that client selectivity by isoforms is a real phenomenon. Although it remains unclear why clients may prefer interaction with NvHsp70B under stress, the unique amino acid sequence of NvHsp70B may afford extra stability at high temperatures. Several studies have demonstrated that post-translational modifications (PTMs) on Hsp70 alter client interactions [44,48–50]. It may be that the unique pattern of NvHsp70 interactions under stress is explained by the presence of unique PTMs present or absent on these isoforms.
4.5. NvHsp70 isoforms may exert differential control over the heat shock response
Several Hsp70s (human Hsc70, yeast Ssa2 and NvHsp70D) are expressed constitutively while others such as human Hsp72, yeast Ssa3 and NvHsp70B are induced under stresses that include heat shock, DNA damage and oxidative stress. The level of Hsp70s in cells are tightly controlled by activity of Heat Shock Factor 1 (HSF1). Under conditions that promote protein unfolding, Hsp70 (which usually binds and inhibits HSF activity) dissociates from HSF1 allowing HSF to trimerize and bind to specific genomic regions known as Heat Shock Elements (HSEs). HSF binding to HSEs triggers expression of a variety of proteins, many of which are co-chaperones or chaperones themselves, completing a feedback loop that dynamically balances chaperone expression against unfolded/misfolded proteins [36,51]. Interestingly in our study, only NvHsp70A interacted with yeast HSF1 and did so under both unstressed and heat shock conditions. It is likely then that NvHsp70A acts as a specific negative regulator of the heat shock response. This may explain the poor growth phenotype of yeast cells solely expressing NvHsp70A [25]. While NvHsp70A may act uniquely as an HSF regulator, it is worth noting that there are three HSF variants in Nematostella (unpublished data, Reitzel lab). It is thus possible that each NvHsp70 isoform has a different HSF isoform preference creating a complicated and interconnecting pattern of HSP-HSF-client interactions at any one time.
4.6. Unique NvHsp70-co-chaperone interactions may explain client specificity
Hsp70s are regulated by co-chaperone proteins that determine client binding specificity, Hsp70 ATP binding and hydrolysis and stability of disease-related clients [32,34,47,52,53]. Two key Hsp70 co-chaperones in yeast are Ydj1 and Sis1. Studies utilizing Ydj1-Sis chimeras clearly show that these two proteins have overlapping but distinct functions in the cell [35,54–56]. While no studies have been done to investigate Ssa isoform specificity for co-chaperone binding, Ssa1 phosphorylation at T36 alters Ydj1 binding while leaving Sis1 interaction unchanged [44]. Under heat stress, we have observed a difference in NvHsp70 isoform-Ydj1/Sis1 interaction. While beyond the scope of this study, it will be interesting to examine whether this specificity is conserved in both yeast native Ssa isoforms and related human Hsp70s.
5. Conclusion
Hsp70s are evolutionarily conserved chaperones that interact with numerous proteins for proteostasis. Most organisms have numerous Hsp70s encoded in their genome but it is not understood how similar the client proteins are for these Hsp70 homologs in ambient or stressful conditions. Here we provide the first description of the isoform-specific diversity of Hsp70 clients for a marine invertebrate species, the cnidarian Nematostella vectensis, through heterologous expression in yeast. Despite high sequence similarity for the three Hsp70s from this sea anemone, comparisons of the client proteins revealed that less than half were found for all three and the unique associations for each Hsp70 increased when exposed to temperature stress. Our study provides the first data set defining the potential “interactome” of Hsp70s for a marine cnidarian and suggest numerous specific interactions in Hsp70 homologs, including for the first time shuttling of proteins between Hsp70 isoforms.
Supplementary Material
Biological significance.
Although the Hsp70 family of molecular chaperones has been studied for > 50 years, it is still not fully understood why organisms encode and express many highly-similar Hsp70 isoforms. The prevailing theory is that these isoforms have identical function, but are expressed under unique cellular conditions that include heat shock to cope with increased number of unfolded/misfolded proteins. The sea anemone Nematostella vectensis encodes three Hsp70 isoforms A, B and D that when expressed in yeast demonstrate unique functionalities. This study provides the interactome of NvHsp70s A, B and D and demonstrates that Hsp70 isoforms, while highly similar in sequence, have unique co-chaperone and client interactors.
Acknowledgements
We acknowledge the PRIDE team for the deposition of our data to the ProteomeXchange consortium. This project was supported by NCI R15 CA208773 (AWT) and NSF Award 1545539 (AMR).
Abbreviations
- HSP
Heat Shock Protein
- 5-FOA
5-Fluoro-orotic acid
- Nv
Nematostella vectensis
- UV
ultraviolet
- NBD
Nucleotide-binding domain
- SBD
Substratebinding domain
- HS
Heat Shock
- HSF
Heat Shock Factor
- HU
Hydroxyurea
Footnotes
Declaration of Competing Interests
The authors declare no conflict of interest.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/archive/) partner repository with the dataset identifier PXD012144.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jprot.2019.103416.
References
- [1].Balchin D, Hayer-Hartl M, Hartl FU, In vivo aspects of protein folding and quality control, Science 353 (6294) (2016) aac4354. [DOI] [PubMed] [Google Scholar]
- [2].Kim YE, et al. , Molecular chaperone functions in protein folding and proteostasis, Annu. Rev. Biochem. 82 (2013) 323–355. [DOI] [PubMed] [Google Scholar]
- [3].Lotz SK, et al. , Not quite the SSAme: unique roles for the yeast cytosolic Hsp70s, Curr. Genet. (2019), 10.1007/s00294-019-00978-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Feder ME, Hofmann GE, Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology, Annu. Rev. Physiol. 61 (1999) 243–282. [DOI] [PubMed] [Google Scholar]
- [5].Kominek J, et al. , The complex evolutionary dynamics of Hsp70ss: a genomic and functional perspective, Genome Biol Evol 5 (12) (2013) 2460–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kultz D, Molecular and evolutionary basis of the cellular stress chaperone, Annu. Rev. Physiol. 67 (2005) 225–257. [DOI] [PubMed] [Google Scholar]
- [7].Louis YD, et al. , Gene expression biomarkers of heat stress in scleractinian corals: promises and limitations, Comp Biochem Physiol C Toxicol Pharmacol 191 (2017) 63–77. [DOI] [PubMed] [Google Scholar]
- [8].Nakamura M, et al. , Expression of hsp70, hsp90 and hsf1 in the reef coral Acropora digitifera under prospective acidified conditions over the next several decades, Biol Open 1 (2) (2012) 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gellner K, Praetzel G, Bosch TC, Cloning and expression of a heat-inducible hsp70 gene in two species of Hydra which differ in their stress response, Eur. J. Biochem. 210 (3) (1992) 683–691. [DOI] [PubMed] [Google Scholar]
- [10].Meyer E, Weis VM, Study of cnidarian-algal symbiosis in the “omics” age, Biol. Bull. 223 (1) (2012) 44–65. [DOI] [PubMed] [Google Scholar]
- [11].Barshis DJ, et al. , Genomic basis for coral resilience to climate change, Proc. Natl. Acad. Sci. U. S. A. 110 (4) (2013) 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bellantuono AJ, et al. , Coral thermal tolerance: turning gene expression to resist thermal stress, PLoS One 7 (11) (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lima FP, Wethey DS, Three decades of high-resolution coastal sea surface temperatures reveal more than warming, Nat. Commun. 3 (2012) 704. [DOI] [PubMed] [Google Scholar]
- [14].Solomon S, et al. , Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, Cambridge University Press, 2007, p. 2007. [Google Scholar]
- [15].Darling JA, et al. , Rising starlet: the starlet sea anemone, Nematostella vectensis, Bioessays 27 (2) (2005) 211–221. [DOI] [PubMed] [Google Scholar]
- [16].Pandolfi JM, et al. , Global trajectories of the long-term decline of coral reef ecosystems, Science 15 (301) (2003) 955–958. [DOI] [PubMed] [Google Scholar]
- [17].Hand C, Uhlinger KR, The unique, widely distributed, estuarine sea aneome, Nematostella vectensis Stephenson: a review, new facts, and questions, Estuar. Coasts 17 (2) (1994) 501–508. [Google Scholar]
- [18].Reitzel AM, et al. , Global population genetic structure of the starlet anemone Nematostella vectensis: multiple introductions and implications for conservation policy, Biol. Invasions 10 (2008) 1197–1213. [Google Scholar]
- [19].Parmesan C, Ecological and evolutionary responses to recent climate change, Annu. Rev. Ecol. Evol. Syst. 37 (2006) 637–669. [Google Scholar]
- [20].Visser ME, Keeping up with a warming world; assessing the rate of adaptation to climate change, Proc. Biol. Sci. 275 (1635) (2008) 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Goldstone JV, Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis, Cell Biol. Toxicol. 24 (6) (2008) 483–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reitzel AM, et al. , Aryl hydrocarbon receptor (AHR) in the cnidarian Nematostella vectensis: comparative expression, protein interactions, and ligand binding, Dev. Genes Evol. 224 (1) (2014) 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Elran R, et al. , Early and late response of Nematostella vectensis transcriptome to heavy metals, Mol. Ecol. 23 (19) (2014) 4722–4736. [DOI] [PubMed] [Google Scholar]
- [24].Oren M, et al. , Profiling moelcular and behavioral circadian rhythms in the nonsymbiotic sea anemone Nematostella vectensis, Sci. Rep 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Waller SJ, et al. , Characterizing functional differences in sea anemone Hsp70 isoforms using budding yeast, Cell Stress Chaperones 23 (5) (September 2018) 933–941, 10.1007/s12192-018-0900-7 (Epub 2018 Apr 25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jaiswal H, et al. , The chaperone network connected to human ribosome-associated complex, Mol. Cell. Biol. 31 (6) (2011) 1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Deutsch EW, et al. , The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition, Nucleic Acids Res. 45 (D1) (2017) D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vizcaino JA, et al. , The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013, Nucleic Acids Res 41 (2013) D1063–D1069 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vizcaino JA, et al. , 2016 update of the PRIDE database and its related tools, Nucleic Acids Res. 44 (D1) (2016) D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Truman AW, et al. , Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase, J. Proteome 112 (2015) 285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Truman AW, et al. , The quantitative changes in the yeast Hsp70 and Hsp90 interactomes upon DNA damage, Data Brief 2 (2015) 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Craig EA, Marszalek J, How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42 (5) (2017) 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kampinga HH, Craig EA, The HSP70 chaperone machinery: J proteins as drivers of functional specificity, Nat Rev Mol Cell Biol 11 (8) (2011) 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Reidy M, et al. , Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines, PLoS Genet 10 (10) (2014) e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kirkland PA, Reidy M, Masison DC, Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity, Genetics 188 (3) (2011) 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Verghese J, et al. , Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system, Microbiol. Mol. Biol. Rev. 76 (2) (2012) 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wegele H, et al. , Substrate transfer from the chaperone Hsp70 to Hsp90, J. Mol. Biol. 356 (3) (2006) 802–811. [DOI] [PubMed] [Google Scholar]
- [38].Wegele H, Muller L, Buchner J, Hsp70 and Hsp90–a relay team for protein folding, Rev Physiol Biochem Pharmacol 151 (2004) 1–44. [DOI] [PubMed] [Google Scholar]
- [39].Cyr DM, Hohfeld J, Patterson C, Protein quality control: U-box-containing E3 ubiquitin ligases join the fold, Trends Biochem. Sci. 27 (7) (2002) 368–375. [DOI] [PubMed] [Google Scholar]
- [40].Gupta A, et al. , The yeast stress inducible Ssa Hsp70 reduces alpha-synuclein toxicity by promoting its degradation through autophagy, PLoS Genet. 14 (10) (2018) e1007751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hasin N, et al. , Global transcript and phenotypic analysis of yeast cells expressing Ssa1, Ssa2, Ssa3 or Ssa4 as sole source of cytosolic Hsp70-Ssa chaperone activity, BMC Genomics 15 (2014) 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sharma D, Masison DC, Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects, Genetics 179 (3) (2008) 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sharma D, Masison DC, Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p, Proc. Natl. Acad. Sci. U. S. A. 108 (33) (2011) 13665–13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Truman AW, et al. , CDK-dependent Hsp70 phosphorylation controls G1 cyclin abundance and cell-cycle progression, Cell 151 (6) (2012) 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nitika AW Truman, Endogenous epitope tagging of heat shock protein 70 isoform Hsc70 using CRISPR/Cas9, Cell Stress Chaperones 23 (3) (2018) 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ikmi A, et al. , TALEN and CRISPR/Cas9-mediated genome editing in the earlybranching metazoan Nematostella vectensis, Nat. Commun 5 (2014) 5486. [DOI] [PubMed] [Google Scholar]
- [47].Sluder IT, et al. , The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity, PLoS Genet 14 (11) (2018) e1007462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nitika AW Truman, Cracking the chaperone code: cellular roles for Hsp70 phosphorylation, Trends Biochem. Sci. 42 (12) (2017) 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Muller P, et al. , C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/ degradation balances, Oncogene 32 (25) (2013) 3101–3110. [DOI] [PubMed] [Google Scholar]
- [50].Zemanovic S, et al. , Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance, Cell Rep 25 (9) (2018) (2605–2616 e7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Y, et al. , The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds, Mol. Biol. Cell 23 (17) (2012) 3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Knighton LE, Delgado LE, Truman AW, Novel insights into molecular chaperone regulation of ribonucleotide reductase, Curr. Genet. 65 (2) (April 2019) 477–482, 10.1007/s00294-018-0916-7 (Epub 2018 Dec 5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Moses MA, et al. , Targeting the Hsp40/Hsp70 chaperone Axis as a novel strategy to treat castration-resistant prostate Cancer, Cancer Res. 78 (14) (2018) 4022–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Borges JC, et al. , Identification of regions involved in substrate binding and dimer stabilization within the central domains of yeast Hsp40 Sis1, PLoS One 7 (12) (2012) e50927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tutar L, Tutar Y, Ydj1 but not Sis1 stabilizes Hsp70 protein under prolonged stress in vitro, Biopolymers 89 (3) (2008) 171–174. [DOI] [PubMed] [Google Scholar]
- [56].Higurashi T, et al. , Specificity of the J-protein Sis1 in the propagation of 3 yeast prions, Proc. Natl. Acad. Sci. U. S. A. 105 (43) (2008) 16596–16601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.