Abstract
Background and Aims
Efficient biological nitrogen fixation (BNF) requires leghaemoglobin (Lb) to modulate oxygen pressure in nodules. Excess N supply severely inhibits BNF through effects on Lb during nodulation. As yet, a systematic identification and characterization of Lb-encoding genes in soybean has not been reported.
Methods
The effects of N on BNF were studied in soybean plants inoculated with rhizobia and exposed to excess or low N availability in hydroponic cultures. To identify soybean Lb proteins, BLAST searches were performed on the Phytozome website. Bioinformatic analysis of identified GmLbs was then carried out to investigate gene structure, protein homology and phylogenetic relationships. Finally, quantitative real-time PCR was employed to analyse the expression patterns of soybean Lb genes in various tissues and in response to high N availability.
Key Results
Excess N significantly accelerated nodule senescence and the production of green Lb in nodules. In total, seven haemoglobin (Hb) genes were identified from the soybean genome, with these Hb genes readily split into two distinct clades containing predominantly symbiosis-associated or non-symbiotic Hb members. Expression analysis revealed that all of the symbiosis-associated Lbs except GmLb5 were specifically expressed in nodules, while the non-symbiotic GmHbs, GmHb1 and GmHb2, were predominantly expressed in leaves and roots, respectively. Among identified GmLbs, GmLb1–4 are the major Lb genes acting in soybean nodulation, and each one is also significantly suppressed by exposure to excess N.
Conclusions
Taken together, the results show that excess N inhibits BNF by reducing nodule formation, Lb concentration and nitrogenase activity. The characteristics of the entire Hb family were analysed, and we found that GmLb1–4 are closely associated with nodule development and N2 fixation. This works forms the basis for further investigations of the role of Lbs in soybean nodulation.
Keywords: Soybean, leghaemoglobin, excess N, biological nitrogen fixation, phylogenetic analysis, expression patterns, nodule development
INTRODUCTION
Biological nitrogen fixation (BNF) is a prominent natural source of nitrogen (N) in agro-ecosystems. In leguminous plants, BNF occurs in nodules, highly specialized organs in which N2 is fixed into biologically available ammonia through symbiotic interactions with rhizobia (Oldroyd and Downie, 2008; Oldroyd et al., 2011). Soybean (Glycine max), the most commonly cultivated legume species, fixes ~16.4 million tons of N2 annually, which represents about 77 % of the N2 fixed by the leguminous crop species (Herridge et al., 2008). Due to its high BNF capacity, soybean serves as a central player in sustainable agriculture.
Within the symbiosome that houses N2-fixing rhizobia, the nitrogenase complex is sensitive to oxygen (O2) and must be kept under anaerobic conditions. Legumes have evolved to meet this condition by making leghaemoglobin (Lb) an essential component of N2 fixation in order to regulate O2 pressure in bacterial symbionts (Appleby and Bergersen, 1980; Hoy and Hargrove, 2008). Therefore, identification and characterization of Lb protein family members in soybean might prove to be an important component of developing new approaches to enhance biological N2 fixation.
Haemoglobins (Hbs) are a superfamily of haem-containing proteins that contain a conserved three-on-three or two-on-two arrangement of α-helical globin fold structures that are involved in binding and/or transporting oxygen via a haem prosthetic group (Garrocho-Villegas et al., 2007; Hoy and Hargrove, 2008). Haemoglobin was originally identified and studied in mammals, but now has been found to exist in nearly all organisms, including Archaeobacteria, Eubacteria and eukaryotes (Vinogradov et al., 2005, 2006). Plants have evolved to harbour up to three types of Hb, delimited as symbiotic, non-symbiotic and truncated Hbs (Gupta et al., 2011). Symbiotic Hbs are known as leghaemoglobins and were first isolated from the red pigment of soybean nodules (Baulcombe and Verma, 1978). The primary function of these Lbs is to deliver O2 to bacteroids at low but steady rates that allow efficient bacterial respiration while also preventing O2-mediated inactivation of nitrogenase in infected cells (Appleby and Bergersen, 1980; Hoy and Hargrove, 2008). Non-symbiotic Hbs are found in a large variety of plants other than legumes and their main function seems to be related to scavenging nitric oxide (NO), which is an important molecule in responses to abiotic and biotic stresses in plants (Baudouin, 2011; Gupta and Igamberdiev, 2011). The third type of plant Hbs is a set of truncated molecules that are 20–40 amino acid residues shorter than normal Hbs (Vuletich and Lecomte, 2006). As yet, the functions of truncated Hbs remain unclear, though there is some evidence that they may be involved in regulating oxygen delivery under high O2 concentration conditions (Watts et al., 2001).
In mature nodules, Lb is the most abundant protein. Yet, despite this abundance and the vital role Lb plays in BNF, Lb-encoding genes in legumes remain poorly characterized. In Lotus japonicus, three symbiotic and two non-symbiotic Lbs have been identified, with the knockdown of symbiotic Lbs via RNA interference resulting in increased free O2 concentrations within nodules, significant declines in bacterial nitrogenase activity, and complete loss of BNF (Ott et al., 2005, 2009). In Medicago, accumulation of MtCAS31 (cold-acclimation-specific 31) delays drought-induced nodule senescence and maintains nitrogenase activity through a direct protein–protein interaction that protects the Lb MtLb120-1 (Li et al., 2018a). An Lb gene in Vicia faba, VfLb29, is a late nodulin that is specifically activated both in infected root nodule cells and in arbuscule-containing cortical cells in mycorrhizal roots, which suggests that Lb might play diverse roles in plant–microbe symbioses (Vieweg et al., 2004). Soybean nodules contain four major Lb genes, known as Lba (Glyma.10G199100), Lbc1 (Glyma.10G199000), Lbc2 (Glyma.20G191200) and Lbc3 (Glyma.10G198800) (Brisson and Verma, 1982; Marcker et al., 1984). Accumulation of Lbc1 and Lbc3 mRNA has been found in very young nodules on soybean roots prior to Lba expression (Marcker et al., 1984). This indicates that differentially expressed Lbs might fulfil distinct functions during nodule formation and development. However, the precise nature of these physiological functions remains to be determined. In many legumes, pink- and/or red-pigment Lbs perform critical functions in N2 fixation. Later, during nodule senescence, whether natural or stress-induced, green Lbs are associated with decreased N2 fixation capacity (Navascués et al., 2012).
In agriculture, the application of N fertilizer or the presence of a high level of residual N severely limits both the formation of nodules and N2 fixation (Streeter, 1998; Miller et al., 2007). More importantly, excess N accelerates nodule senescence, which is accompanied by the generation of green Lb proteins in bacteroids. This observation of green Lb in nodules has been associated with the presence of divalent iron, and might be a product of the breaking of the tetrapyrrole ring in Lb complexes (Virtanen and Laine, 1946). Moreover, ferrous, ferric and ferryl Lbs can also react with reactive nitrogen species. Green Lbs and red Lbs have identical globins, but the haem groups of green Lbs are nitrated with a nitro (NO2) group on the 4-vinyl of the haem complex (Navascués et al., 2012). Beyond this, little is known about how green Lbs are involved in N-induced nodule senescence.
Although Hbs are widespread in the plant kingdom, little is known about their genes and gene products in most legumes, including soybean. The objectives of this research were to conduct a systematic identification and characterization of soybean Hb genes, and to use this information to identify soybean Lb family members involved in the development of nodules and regulation of N2 fixation. Firstly, we investigated BNF capacity during nodule development as affected by high N supply, and in the process we were further able to conduct a systematic investigation of the entire Hb gene family. Analysis included the identification of Hb gene and protein sequences in soybean, along with comparison of soybean Hbs with Hbs from other species in phylogenetic and gene structural analysis. In addition, tissue-specific expression patterns and responses to excess N supply were also assessed using quantitative real-time PCR (qPCR). In the end, many avenues for continued research were opened, many of which might impact nutrient management strategies and efforts to more efficiently maintain soybean yields under a range of N availability conditions.
MATERIALS AND METHODS
Identification and bioinformatics analysis of GmLb genes
A BLAST search was performed with the known soybean (Glycine max) symbiotic (accession number V00453) and non-symbiotic (accession number U47143) Hb (Gopalasubramaniam et al., 2008) amino acid sequences at the Phytozome website (http://www.phytozome.net). This yielded a total of seven predicted Hb genes, including five symbiotic Hbs known as Lbs, named GmLb1 to GmLb5, and two non-symbiotic Hbs named GmHb1 and GmHb2 based on amino acid sequence similarity. General information for each Hb member (e.g. number of exons and introns, length of open reading frame) was also retrieved from Phytozome. Calculation of protein size was performed using the Compute pI/Mw tool (http://www.expasy.org/tools/pi_tool.html). A phylogenetic tree of Hbs from soybean (GmLb1–5/GmHb1–2), common bean (PvLbs/PvHbs), Medicago truncatula (MtLbs/MtHbs), Arabidopsis thaliana (AtHbs), rice (OsHbs) and maize (ZmHbs) was constructed using the neighbour-joining method with 1000 bootstrap replicates in the MEGA 6.1 program. The corresponding locus names derived from the above plant species are shown in Supplementary Data Table S1. The genomic structures of soybean Hb genes were analysed at the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php).
Plant materials and growth conditions
Seeds from the soybean genotype HN66 were surface-sterilized in 10 % H2O2 for 1 min, rinsed with distilled water, and germinated in sand-irrigated with a low-N (LN) nutrient solution containing 530 µm N (Li et al., 2018b). One week after germination, uniform seedlings were inoculated with rhizobium strain Bradyrhizobium sp. (BXYD3) by immersing roots in a rhizobial suspension for 2 h prior to transplanting into LN hydroponic cultures as previously described (Li et al., 2018b). Plants were reared in growth chambers (day/night: 14 h/10 h, 26 °C/24 °C) for 7, 14, 20, 30, 40 and 50 d after inoculation (dai). Moreover, the soybean plants were transferred to high-N (HN, 20 mm N added) or a refreshed LN solution at their peak of N2 fixation (30 dai) for 9 d. The nodules at different growth stages or under the HN condition were separately harvested and further classified into two groups based on nodule diameter (D), specifically large (D ≥ 2 mm) and small (D < 2 mm) nodules, and then the number and weight of nodules were determined for each group. Large nodules were also used for RNA extraction, determination of Lb concentration, nitrogenase activity analysis and infected cell surveys.
For expression analysis of Hb members in various tissues, seeds of the soybean genotype Williams 82 were germinated in sand and pre-treated as described above. After inoculation with BXYD3, seedlings were grown in a low-N solution for 30 d prior to harvesting roots, nodules, stems, leaves and flowers. Pods ~1 cm long and immature seeds were harvested separately 40 and 50 d after inoculation. All samples were stored at −80 °C for RNA extraction and qPCR analysis.
Observation of infected cells in nodules and nitrogenase activity
To better evaluate how N supply affects nodule growth and development, nodules were embedded in paraffin and sectioned transversely to a thickness of 18 µm with a microtome (Leica RM2235). After dewaxing, nodule sections were stained with 0.1 % toluidine blue and observed by light microscopy (Axio Imager A2m; Carl Zeiss). Ten nodules from LN or HN treatments were selected randomly and measured. Three cross-sections were made on each nodule, which led to a total of 30 cross-sections being observed for each treatment in determination of the number of infected cells per mm2 and the surface area of 100 infected cells in ImageJ (https://imagej.nih.gov/ij/).
The nitrogenase activity of nodules was analysed using an acetylene reduction assay (David et al., 1980). Nodules were separately sampled in 8-mL airtight glass bottles from which 1 mL of air was extracted prior to the injection of an equal volume of acetylene. After 2 h, the reaction was terminated by adding 0.5 m NaOH. Other research has found that downregulation of O2 concentration in the infected cells of nodules is able to cause an O2-related limitation of nitrogenase activity (Carroll et al., 1987; Kaiser et al., 1997). Because the O2 barrier will close down after 30 min or so in isolated nodules, a short (30 min) reaction assay could be appropriate for nitrogenase activity analysis. Given the equal treatment in all cases, subsequently in this study 0.3 mL of gas was analysed in gas chromatography with a flame ionization detector (GC-2014AF). Nitrogenase activity was calculated as the amount of acetylene reduced by nitrogenase in nodules per hour and per gram of nodule fresh weight.
Measurement of Lb concentration
For the measurement of Lb concentration in nodules, about 50 mg of fresh nodules was first ground and homogenized in 16-fold volumes of 0.1 m precooled PBS (Na2HPO4-NaH2PO4 buffer at 5 °C, pH 6.8). The resulting slurry was then centrifuged at 12 000 g for 15 min prior to assaying the supernatant by spectrophotometry at a wavelength of 540, 520 and 560 nm. The Lb, including red and green pigment, was calculated separately as described previously (Larue and Child, 1979). Bovine Hb was used as a protein standard in this study.
RNA extraction and qPCR
High-quality total RNA was isolated from soybean roots, nodules, stems, leaves, flowers, young pods and seeds using the RNAiso Plus reagent (TaKaRa Japan) according to the manufacturer’s instructions. Subsequently, RNA samples were treated with RNase-free DNase I (TaKaRa, Japan) to remove genomic DNA contamination. First-strand cDNA was synthesized using oligo(dT), deoxyribonucleotide triphosphates and MMLV reverse transcriptase (Promega, USA) based on protocols from the supplier. Quantitative real-time PCR was performed in 20-μL volumes containing 2 μL of 1:50 diluted cDNA, 0.6 μL of specific primers, 6.8 μL of distilled, deionized water and 10 μL of TransStart Top Green qPCR SuperMix (Trans) using a LightCycler 96 (Roche Diagnostics) PCR system. Reaction conditions for thermal cycling were: 95 °C for 1 min, 40 cycles of 95 °C for 15 s, 56–60 °C for 15 s and 72 °C for 30 s. The annealing temperature (56–60 °C) was adjusted to suit the amplification of individual Hb genes. The elongation factor 1α gene (TefS1, accession number X56856) from soybean was used as an endogenous control to evaluate relative transcript abundance. Relative expression values were calculated as the ratio of the expression value of the target gene to that of TefS1 using the 2−ΔΔCT method. The specific primers used for detecting target genes are listed in Supplementary Data Table S2.
Data analysis
All of the data were analysed statistically using Sigma Plot (version 12.0) for calculating means and standard errors. Comparisons between groups were performed using Student’s t-test or univariate analysis in SPSS (version 16.0).
RESULTS
Analysis of the relationship between nodule development and BNF capacity
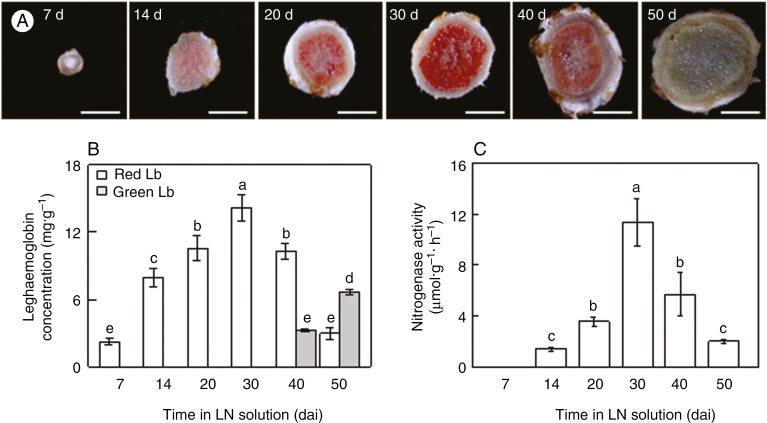
Because soybean nodules belong to the determinate type, the size of nodules could represent the age of nodules. Therefore, we harvested the biggest nodules at 7, 14, 20, 30, 40 and 50 dai to investigate the relationship between nodule development and BNF capacity under LN conditions (530 µm N added). The colour of the nodules changed incrementally from white (7 dai) to pink (14, 20 and 40 dai) and red (30 dai), then to green (50 dai) at different growth stages (Fig. 1A). Consistently, the red Lb concentration increased significantly with nodule growth, peaked at 30 dai, and subsequently declined with age, while the green Lb concentration was increased at 40 dai and significantly elevated at 50 dai (Fig. 1B); interestingly, nitrogenase activity showed similar changes (Fig. 1C). These results indicated that the green derivatives of Lbs generated during nodule senescence, and the development of nodules with pink or red Lbs, are closely related to soybean BNF capacity.
Fig. 1.
Leghaemoglobin concentration and nitrogenase activity in nodules at different growth stages. (A) Nodule colour at different growth stages. Scale bars = 2 mm. (B) Leghaemoglobin concentration. (C) Nitrogenase activity. Seven-day-old soybean seedlings were inoculated with rhizobia and grown in LN (530 μM N added) nutrient solution for 7, 14, 20, 30, 40 and 50 dai. Values are mean ± s.e. (n = 5). Different letters indicate a significant difference at the 0.05 level.
Effects of excess N supply on BNF capacity
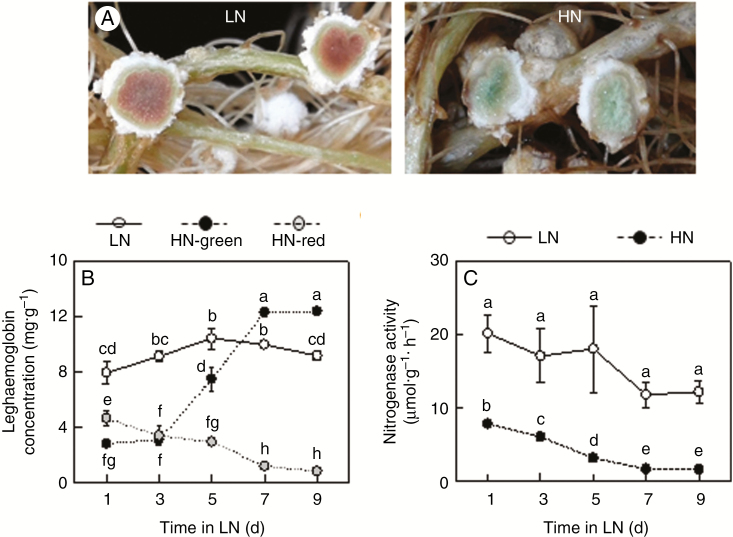
To gain insight into the effect of excess N on biological nitrogen fixation capacity, rhizobia-inoculated soybeans were grown in hydroponic cultures for 30 d, at which point the nodules had their highest N2-fixation capacity, and were then transferred to either HN (20 mm N added) or LN solution for 9 d. In these conditions, excess N application resulted in the production of large amounts of green Lb derivatives in nodules (Fig. 2A). These green Lbs are typically generated during nodule senescence, when they severely inhibited BNF. Consistent with this role, in this study we found that the green Lb concentration increased significantly with increasing HN treatment duration, and its highest concentration was 12.37 mg g−1 FW at 9 d (Fig. 2B). However, the red Lb concentration and nitrogenase activity were significantly reduced 1.70- and 2.56-fold, respectively, in HN treatments compared with LN treatments as soon as 1 d. Then, they gradually declined with additional HN days (Fig. 2B, C). This indicates that excess N may accelerate nodule senescence, which means that appropriate N supplies are quite important for proper management of soybean nodulation.
Fig. 2.
Leghemoglobin concentration and nitrogenase activity as affected by excess N supply in nodules on soybean roots reared in hydroponic culture. (A) Photographs showing nodule growth. (B) Leghemoglobin concentration. (C) Nitrogenase activity. Seven-day-old soybean seedlings were inoculated with rhizobia and grown in LN (530 μm N added) nutrient solution for 30 d and the inoculated plants were then transferred into LN or HN (20 mm N added) solution for 9 d. Values are mean ± s.e. (n = 10). Different letters indicate a significant difference at the 0.05 level.
Nodule growth and development of infected cells are affected by excess N
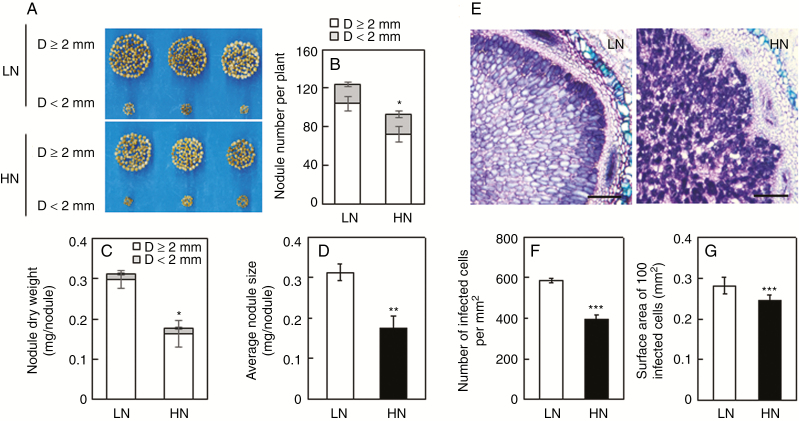
In order to evaluate the effects of excess N supply on nodule growth, nodules were harvested separately and classified into two groups based on nodule diameter (D) under LN and HN conditions. The results showed that excess N supply not only limited nodule development, particularly the formation of big nodules, but also suppressed nodule initiation (Fig. 3A). The number of large nodules (D ≥ 2 mm), nodule mass and individual nodule size were 30.7 %, 45.2 % and 30.3 % less in the HN than in the LN condition, respectively (Fig. 3B–D). In addition, toluidine blue staining was employed in observations of infected cells. As shown by the morphology of typical nodule slices, infected cells within nodules were bigger and more numerous under LN conditions than under HN conditions, which inhibited infected cell growth enough to produce declines in infected cell size and number (Fig. 3E). Quantitatively, the number of infected cells (Fig. 3F) and the surface area per 100 infected cells (Fig. 3G) were 21.2 % and 52.7 % less, respectively, in HN than in LN. These results suggest that HN suppression of BNF might be caused by inhibition of nodule formation and infected cell development in nodules through acceleration of nodule senescence.
Fig. 3.
Effects of excess N supply on nodule growth and infected cell development. (A) Phenotypes of nodules. (B) Nodule number. (C) Nodule dry weight. (D) Average nodule size. (E) Toluidine blue-stained nodule cross-sections. Scale bars = 200 µm. (F) Number of infected cells. (G) Surface area of 100 infected cells. Seven-day-old soybean seedlings were inoculated with rhizobia and grown in LN (530 μm N added) nutrient solution for 30 d prior to transferring inoculated plants into LN or HN (20 mm N added) solution for 9 d. Nodules were classified into two groups according to diameter (D): large (D ≥ 2 mm) and small (D < 2 mm). Values are mean ± s.e. (n = 3). Ten nodules were selected from each replicate for infected cell analysis. Asterisks represent significant differences in a single trait between plants grown in LN and HN treatments: *0.01 < P ≤ 0.05; **0.001< P ≤0.01; ***P ≤ 0.001 (Student’s t-test).
Identification of GmLb members in soybean
Upon observing the effects of excess N on nodule colour and Lb concentration as described above (Fig. 2), we attempted to investigate the soybean Lb gene family more comprehensively. In total, five putative Lb and two non-symbiotic Hb genes were identified according to BLASTP searches of the protein database of soybean in the Phytozome web portal (http://www.phytozome.net/). General information on the distinguishing characteristics of Lbs and Hbs is summarized in Table 1. They were located on chromosomes 10, 11 and 20, with four found on chromosome 10, two on chromosome 11 and one on chromosome 20. The open reading frames of identified Lbs/Hbs ranged from 435 to 507 bp in length and were predicted to encode proteins containing 144–168 amino acids (Table 1).
Table 1.
General information for the seven identified Hb members
| Genes | Locus | Chromosomal location | Exon/intron number | Length of ORF (bp) | Number of amino acids | Protein size (kDa) |
|---|---|---|---|---|---|---|
| GmLb1 | Glyma.10G198800.1 | 10 | 4/3 | 438 | 145 | 15.6 |
| GmLb2 | Glyma.10G199000.1 | 10 | 4/3 | 435 | 144 | 15.4 |
| GmLb3 | Glyma.10G199100.1 | 10 | 4/3 | 435 | 144 | 15.4 |
| GmLb4 | Glyma.20G191200.1 | 20 | 4/3 | 438 | 145 | 15.5 |
| GmLb5 | Glyma.10G198900.1 | 10 | 4/3 | 507 | 168 | 18.1 |
| GmHb1 | Glyma.11G121700.1 | 11 | 4/3 | 474 | 157 | 17.6 |
| GmHb2 | Glyma.11G121800.1 | 11 | 4/3 | 486 | 161 | 18.0 |
Gene locus, exon and intron number and open reading frame (ORF) length were determined from sequences deposited at the Phytozome website (http://www.phytozome.net). Protein size was calculated using the Compute pI/Mw tool (http://www.expasy.org/tools/pi_tool.html).
Phylogenetic analysis and classification of soybean Hbs
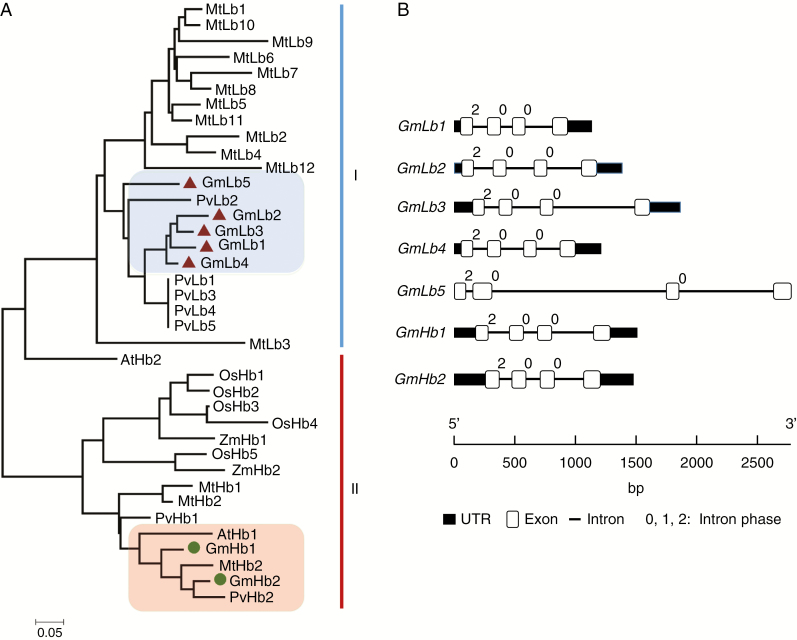
To determine evolutionary relationships among Hb and similar proteins from other species, a phylogenetic tree was constructed by neighbour-joining analysis in MEGA 6.1 using the full Hb protein sequences from soybean, common bean, M. truncatula, A. thaliana, rice and maize. As shown in Fig. 4A, the analysed plant Hb proteins can be divided into two clades, labelled clade I and clade II. Clade I contains most of the tested legume Lbs and includes five GmLbs and PvLbs along with 12 Medicago Lb members. Within clade I, four soybean Lbs, GmLb1, GmLb2, GmLb3 and GmLb4, aligned very closely, while GmLb5 sorted into a subgroup with common bean PvLb2. Other than AtHb2 aligning closely with other clade I Lbs, the remaining legume Hbs and the Hbs from non-legume plant species all belonged to clade II (Fig. 4A).
Fig. 4.
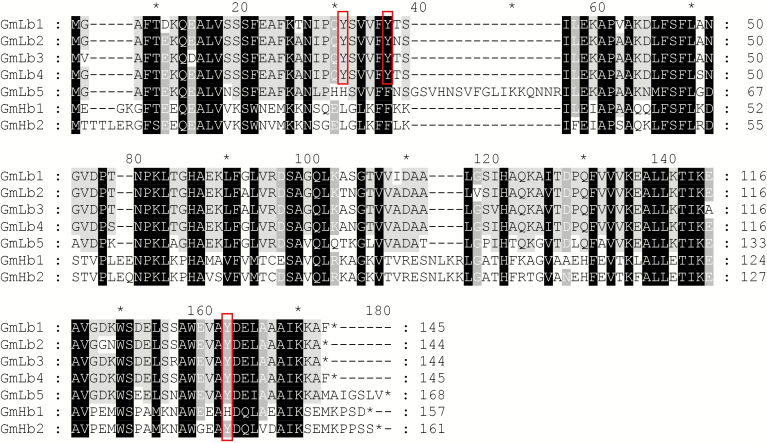
General characteristics of the five identified GmLbs and two non-symbiotic GmHbs in soybean. (A) Phylogenetic analysis of Hb from different plant species. This phylogenetic tree includes seven Hbs from soybean (Glycine max), seven from common bean (Phaseolus vulgaris), 15 from Medicago truncatula, two from Arabidopsis thaliana, five from rice (Oryza sativa) and two from maize (Zea mays). It was constructed by the neighbour-joining method with 1000 bootstrap replicates in the MEGA 6.1 program. Haemoglobin proteins sorted into two clades, labelled clade I and clade II. Five GmLbs are highlighted by red triangles in the blue box, and two GmHbs are indicated by green circles in the red box. (B) Gene structures of symbiotic Lbs and non-symbiotic Hbs from soybean. White boxes represent exons, thin lines represent introns and black boxes represent 5′-/3′-UTRs. Numbers above introns indicate intron phases. Phase 0, 1 and 2 introns were defined as introns starting before the first, after the first and after the second nucleotide of a codon, respectively.
Phylogenetic relationships among soybean Lbs and Hbs were further distinguished by variation in amino acid identity between members of this family (Supplementary Data Table S3). Clade I GmLbs all exhibited 69–95 % amino acid identity, with alignments between GmLb1, GmLb2, GmLb3 and GmLb4 displaying the highest identities (92–95 %). The two non-symbiotic GmHbs in clade II were 86 % identical in their amino acid sequences. Between clade I and clade II, sequence identity dropped to 41–45 %. Taking sequence identity and phylogenetic tree analysis together, these results suggest that symbiotic Lbs (clade I) have diverged significantly from non-symbiotic Hbs (clade II) through the course of soybean evolution.
Full-length cDNAs and genome sequences of GmLbs/GmHbs were used to investigate gene structures on the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php). In this analysis, all Hbs were found to have four exons and three introns, with an intron phase pattern of 2, 0 and 0 from the 5′ to the 3′ end (Table 1, Fig. 4B). The symbiotic Lbs in clade I, namely GmLb1, GmLb2, GmLb3, GmLb4 and GmLb5, each contain one or more long introns, especially GmLb5, which has a total intron length of 2415 bp (Fig. 4B and Supplementary Data Table S4). This gene structure pattern stands in contrast to the gene structure in the non-symbiotic clade II members GmHb1 and GmHb2, which have similar gene structures with total intron lengths of 769 and 596 bp, respectively. It is also notable that the first three exons of clade I GmLbs are of equal length, while both clade II GmHbs have the same second and third exon and second intron lengths (Supplementary Data Table S4). This difference in gene structure indicates that the symbiosis-related Lb genes evolved from these non-symbiotic Hbs.
Conserved amino acids of GmLbs
Multiple sequence alignment was conducted to examine amino acid sequence similarity and to identify conserved motifs across families of plants, with identical and similar amino acid sequences being shaded black and grey, respectively, in Fig. 5. All GmLb/GmHb members possess a typical globin domain (http://www.phytozome.net/). Symbiotic Lbs play important roles in BNF through modulation of O2 diffusion into nodules (Appleby, 1992). During senescence, these same Lbs tend to generate the green derivatives observed within nodules, which is caused by haem nitration (Navascués et al., 2012). Further study revealed that Tyr30 located in the haem distal pocket is the major target of nitration. Otherwise, Tyr25 and a small proportion of C-terminal Tyr133 were also capable of being converted into the nitrated species NO2-Tyr25 and NO2-Tyr133 in GmLbs (Sainz et al., 2015). Here, multiple sequence alignment revealed that the symbiotic GmLb1, 2, 3 and 4 with the highest protein homology to each other contain Tyr25 and Tyr30 amino acid residues (red box in Fig. 5 and Supplementary Data Table S3). Excepting GmHb1, other Hbs have a Tyr133 residue, suggesting that these residues are conserved in soybean symbiotic Lbs, which imparts the potential to form NO2-Tyr25, NO2-Tyr30 and/or NO2-Tyr133 in nodules during senescence.
Fig. 5.
Alignment of GmLb and GmHb amino acid sequences. Sequence alignment was performed with the ClustalW and GeneDoc programs. Identical and similar amino acids are shaded in black and grey, respectively. The red box represents the tyrosine amino acid residue primarily targeted for nitration to form 3-nitrotyrosine (NO2-Tyr), which then generates the green derivatives of Lbs during nodule senescence.
Tissue-specific expression of GmLbs/GmHbs
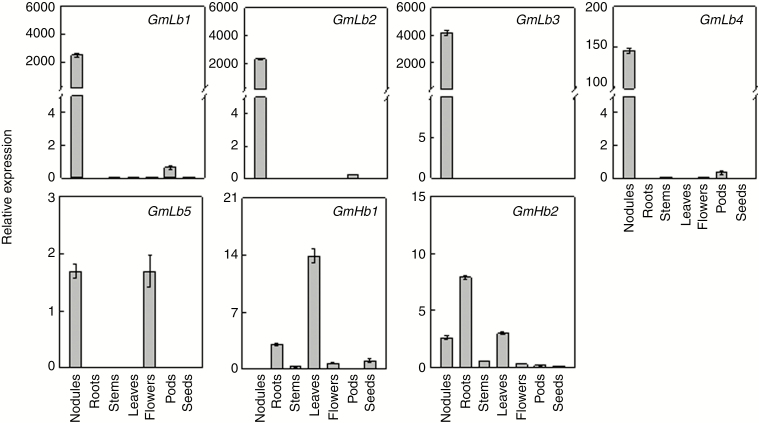
To clarify GmLb/GmHb expression patterns among soybean tissues, qPCR analysis was performed to determine relative transcript abundances in seven tissues, namely roots, nodules, stems, young leaves, flowers, pods and young seeds (Fig. 6). The results showed that four symbiotic Lb genes, GmLb1 to GmLb4, were most abundant in nodules, while GmLb5 was also highly expressed in flowers, indicating that GmLb1, 2, 3 and 4 might be the key Lb genes involved in BNF in soybean. In contrast, the non-symbiotic Hbs in clade II, GmHb1 and GmHb2, were predominantly expressed in leaves and roots, respectively, suggesting other roles, possibly still modulating O2 levels, for clade II genes in response to environmental stress during plant growth and development.
Fig. 6.
Tissue-specific expression patterns of GmLb and GmHb members. Seven-day-old seedlings were inoculated with rhizobia and transplanted into LN (530 μm N added) nutrient solution. qPCR was conducted with transcripts extracted from nodules, roots, stems, leaves and flowers sampled after 30 d of soybean growth, along with pods and young seeds sampled after 40 and 50 d, respectively. Relative expression values were calculated as the ratio of GmLb or GmHb expression to that of the soybean housekeeping gene TefS1 (accession number X56856). Values are mean ± s.e. (n = 4).
Transcription of GmLbs in nodules at different growth phases and in response to excess N application
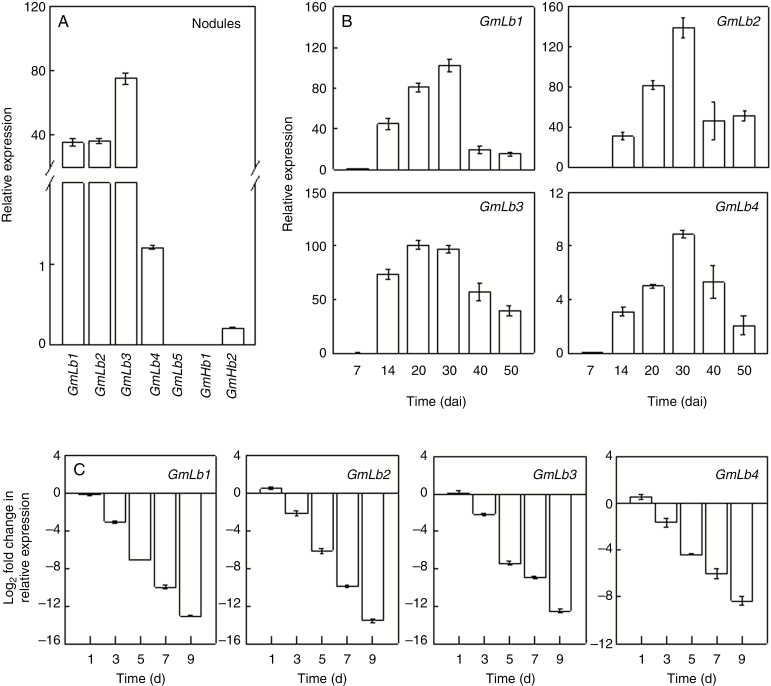
To evaluate the expression of soybean Lb genes in nodules at different growth stages and how it might respond to excess N application, we first investigated the transcription of all GmLb members in nodules, with the result that GmLb1 to GmLb4 were the most abundantly expressed GmLbs in nodules (Fig. 7A). Therefore, GmLb1 to GmLb4 were then selected to be analysed separately in further experiments. During nodule growth and development, the transcripts of GmLb1, 2, 3 and 4 all gradually increased and peaked at 30 dai, except GmLb3, which was most abundant in nodules at 20 and 30 dai; all transcripts then declined with nodule senescence (Fig. 7B). In the HN treatment, expression levels of GmLb1, 2, 3 and 4 were all downregulated in nodules by more than 2-fold at 3 d in comparison with expression in nodules produced under LN conditions (Fig. 7C). Then, the transcription of four GmLbs obviously decreased with increasing HN duration. At 9 d, GmLb1–4 exhibited a 13.1, 13.5, 12.5 and 8.3-fold decrease, respectively, in transcript level in response to HN. Taken together, these results suggest that GmLb1 to GmLb4 are the major Lb genes required for the binding and release of oxygen in order to maintain oxygen gradients during N2 fixation in nodules.
Fig. 7.
Transcription of symbiotic GmLbs and non-symbiotic GmHbs in nodules. (A) Relative expression of GmLbs/GmHbs in soybean nodules under LN conditions. (B) Expression of nodule-specific GmLb genes in nodules at 7, 14, 20, 30, 40 and 50 dai. Relative expression values were calculated as the ratio of GmLb expression to that of the soybean housekeeping gene TefS1 (accession number X56856). Relative expression was normalized against the mean of each GmLb transcription level at 7 dai. (C) Expression of nodule-specific GmLb genes in response to excess N supply, shown as the binary logarithm of fold changes in relative expression of GmLb members under HN (20 mm N added) compared with LN (530 μm N added) conditions. Values are mean ± s.e. (n = 4).
DISCUSSION
Soybean, the most widely grown legume crop globally, possesses a superior capacity to fix N2. A large proportion of the N2 fixed by soybean nodules is also available for the growth of neighbouring or subsequent crops in intercropping/rotation systems, and thereby contributes to increases in grain production while also reducing the demand for N fertilizers in agro-ecosystems (Meng et al., 2015; Wang et al., 2016). However, several environmental factors can restrict BNF in legumes, most notably the common practice of applying high rates of N to soils. Excess N facilitates nodule senescence through the production of green Lb derivatives (Navascués et al., 2012; Saito et al., 2014). In legumes, the absence of red Lb in bacteroids of root nodules precedes the abortion of BNF (Ott et al., 2005, 2009). To better understand the possible functions and mechanism of Lb activity in BNF, members of the Lb gene family were identified and investigated in a systematic fashion.
Nitrogen has been shown to inhibit BNF through decreases in nodule number, along with declines in nodule mass and N2 fixation activity, as well as acceleration of nodule senescence in green nodules (Saito et al., 2014; Ferguson et al., 2018). This homeostatic activity presumably allows soybeans to balance the high cost of N2 fixation with tight modulation of O2 levels, while also impacting the translocation of carbon among root sinks capable of contributing to N acquisition from soils or symbiotic relationships with mycorrhizal fungi or nodulating rhizobial species (Fujikake et al., 2003). Previous work demonstrated that high NO3− availability not only leads to Lb degradation, as reflected by declines in both tissue Lb concentrations and the ratio of Lb to soluble proteins, but also represses the expression of the Lb genes (Bisseling et al., 1978; Becana and Sprent, 1989; de Billy et al., 1991; Gallusci et al., 1991; Sainz et al., 2015). In the current work, we found that red Lb biosynthesis is closely correlated with soybean BNF capacity and that vigorous nodules with red Lb pigments turn green during nodule senescence (Fig. 1). Furthermore, our results also revealed that excess N inhibited BNF, as indicated by limited nodule formation and development, and reduced Lb levels as well, which was consistent with the previous reports (Saito et al., 2014). Furthermore, high N supply also accelerated nodule senescence and the production of green Lb in nodules (Figs 2 and 3). Green Lbs usually contain nitrated haems, the Tyr30 located in the haem distal pocket being the major target of nitration in soybean, followed by Tyr25 (Navascués et al., 2012; Sainz et al., 2015).
Previous analysis has also identified four major Lb genes associated with symbiotic BNF and which encode products possessing remarkably high affinities for O2, specifically Lba (Glyma.10G199100), Lbc1 (Glyma.10G199000), Lbc2 (Glyma.20G191200) and Lbc3 (Glyma.10G198800) (Brisson and Verma, 1982; Marcker et al., 1984). Here, we confirmed the production of these soybean Lbs and renamed them as GmLb1, GmLb2, GmLb3 and GmLb4 (Table 1). Multiple sequence alignment revealed that Tyr25 and Tyr30 are only found in the four major Lbs associated with symbiosis, GmLb1 to GmLb4 (Fig. 5), which, together with the potential for tyrosine to be nitrated, suggests that Tyr25 and Tyr30 might be key residues in BNF. Furthermore, GmLb1, 2, 3 and 4 were all severely downregulated during nodule senescence as well as in nodules exposed to excess N (Fig. 7B, C). This led to significant declines in Lb concentration and thus limited infected cell development and nitrogenase activity in nodules (Figs 1–3). For further details of the mechanisms of Lb biosynthesis and their individual functions in nodules and responses to various environmental stresses during nodulation, we require further study focused on these specific topics.
In the current study, a total of seven soybean Hb genes were isolated in a genome-wide search based on sequence similarity. These Hbs could then be categorized into two distinct clades in a phylogenetic analysis of Hb proteins from monocot and dicot species (Fig. 4A), which was further supported by distinct patterns in gene structures among the two Hb clades, along with protein identities that were higher within clades than between clades (Fig. 4B, Supplementary Data Table S3). Interestingly, clade I includes GmLb1 to GmLb5, which appear active in root tissues involved in symbiotic interactions, along with most of the other tested legume Lbs. In contrast, clade II contains the soybean Hbs not involved in symbiosis, GmHb1 and GmHb2, as well as all of the tested monocot Hbs from rice and maize and Hbs from the non-symbiotic plant Arabidopsis (Fig. 4A). On the whole, these results indicate that Hbs can be found in monocot and dicot plant species, though those participating in symbiotic interactions have diverged significantly from other Hbs. This conclusion is further supported by expression profiles in which all symbiosis-associated clade I GmLbs except GmLb5 were specifically expressed in nodules (Figs 4 and 6). In short, it appears that specific biological functions associated with GmLbs/GmHbs might be grouped into categories that are in accordance with gene classification, though further experiments are required to test this hypothesis.
The qPCR results returned for the GmLbs/GmHbs observed in various soybean tissues in this study confirm the tissue-specific expression patterns reported previously in global transcription studies using next-generation sequencing (Libault et al., 2010). Similarly, the symbiosis-associated GmLb1, 2, 3 and 4 are specifically expressed in nodules (Figs 6 and 7A). Beyond the expression of GmLb1–4, transcripts of the symbiosis-associated GmLb5 were also detected in both nodules and flowers at approximately equal levels of expression, and the non-symbiotic GmHb2 was most abundant in roots (Fig. 6). This result stands in contrast to the report authored by Libault et al. (2010), in which GmLb5 was not detected in flowers and GmHb2 was highly expressed both in nodules and roots. These differences in expression patterns observed for the two Hbs between the two studies might result from differences between the tested genotypes, variations in growth conditions, including N availability, differences in sampling procedures, or differences in the timing of sampling relative to developmental stage. Notwithstanding these specific differences between studies, GmLb expression patterns appear to be stable and reproducible, and have been largely confirmed in three independent studies.
Expression profiles for GmLbs suggest potentially specific roles for individual members. Transcripts of the symbiosis-associated members of the GmLb family, GmLb1 to GmLb4, were found exclusively in nodules, where they were also the most highly expressed GmLbs (Fig. 7A), suggesting that these four GmLbs are the genes primarily responsible for Lb expression in nodules, which is consistent with previous studies (Brisson and Verma, 1982; Marcker et al., 1984). An implication of this conclusion is that symbiosis-associated GmLbs play vital roles in BNF.
Other Hb genes not associated with symbiotic interactions have been isolated from a number of both legume and non-leguminous plant species. These Hb genes exhibit a variety of O2-binding properties and gene expression patterns, many of which include responses to numerous stimuli. For example, the clade-I non-symbiotic Hbs from barley are expressed in aleurone and root tissues subjected to low oxygen stress (Taylor et al., 1994). Another Hb gene in cotton, GhHb1, is induced in roots challenged with the Verticillium wilt fungus. Moreover, ectopic overexpression of GhHb1 in Arabidopsis leads to constitutive expression of the defence genes PR-1 and PDF1.2, which confers enhanced disease tolerance on treated plants (Qu et al., 2006). In Lotus japonicus, the non-symbiotic Hb LjHb1 has been strongly induced in response to hypoxia, cold stress, and exposure to an exogenous NO donor. Observations using fluorescence microscopy demonstrated that induction of LjHb1 expression corresponds to the generation of NO (Shimoda et al., 2005). Altogether, these results suggest that non-symbiotic Hb genes are involved in various biotic and abiotic stress responses. In this study, the expression of two non-symbiotic Hbs, GmHb1 and GmHb2, varied among the seven tissues observed, with the highest expression occurring in leaves and roots (Fig. 6). This result indicates that each of these two genes might function in the development of organ-specific responses to different stresses. Further experiments with forward and reverse genetic approaches are needed to conclusively test this hypothesis and to determine specific roles for each GmLb and GmHb.
Conclusions
In this study, we found that red Lb biosynthesis is closely associated with soybean BNF, and excess N application significantly inhibited BNF and accelerated nodule senescence and production of green pigmented Lb. Subsequent systematic isolation and characterization of soybean GmLb protein family members was then performed, along with assessment of GmLb molecular features and expression patterns in vegetative and reproductive tissues. The results reveal the existence of seven Hb members that can be divided into two distinct subgroups based on sequence similarity and gene structural features, which is further supported by the observation of distinct expression profiles between the two subgroups. The first group, clade I, includes symbiotic-associated Lbs, GmLb1 to GmLb4 being the four major nodulation Lbs, each of which was further observed to be suppressed in nodules developing under excess N availability conditions. The non-symbiotic GmHbs, GmHb1 and GmHb2, sorted into clade II and are predominantly expressed in leaves and roots, respectively, which suggests that Hb genes fill diverse roles in plant growth and development beyond acting in nodulation on soybean roots. Further understanding of the molecular mechanisms of Lb biosynthesis may lead to insights that can be applied to improving BNF and, thus, soybean production.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Table S1: Hb gene loci from different plant species included in phylogenetic tree analysis.
Table S2: gene-specific primers used for qPCR analysis.
Table S3: amino acid sequence identity within the coding regions of haemoglobin protein family members.
TableS4: length of exons and introns in each soybean GmLb and GmHb gene.
ACKNOWLEDGEMENTS
We thank Miss Jiakun Zheng of the Fujian Agriculture and Forestry University for sampling assistance, and Tom Walk of Golden Fidelity LLC for critical reading. The authors declare no conflict of interest.
FUNDING
This work was financially supported by the China National Key Program for Research and Development (2016YFD0100700) and the National Natural Science Foundation of China (31601814).
LITERATURE CITED
- Appleby CA. 1992. The origin and functions of haemoglobin in plants. Science Progress 76: 365–398. [Google Scholar]
- Appleby CA, Bergersen FJ. 1980. Preparation and experimental use of leghaemoglobin. In: Bergersen FJ, ed. Methods for evaluating biological nitrogen fixation. Chichester: Wiley, 315–335. [Google Scholar]
- Baudouin E. 2011. The language of nitric oxide signalling. Plant Biology 13: 233–242. [DOI] [PubMed] [Google Scholar]
- Baulcombe D, Verma DPS. 1978. Preparation of a complementary DNA for leghaemoglobin and direct demonstration that leghaemoglobin is encoded by the soybean genome. Nucleic Acids Research 5: 4141–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becana M, Sprent JI. 1989. Effect of nitrate on components of nodule leghaemoglobins. Journal of Experimental Botany 40: 725–731. [Google Scholar]
- de Billy F, Barker DG, Gallusci P, et al. . 1991. Leghaemoglobin gene transcription is triggered in a single cell layer in the indeterminate nitrogen-fixing root nodule of alfalfa. Plant Journal 1: 27–35. [Google Scholar]
- Bisseling T, Van Den Bos RC, Van Kanmmen A. 1978. The effect of ammonium nitrate on the synthesis of nitrogenase and the concentration of leghemoglobin in pea root nodules induced by Rhizobium leguminosarum. Biochimica et Biophysica Acta 539: 1–11. [DOI] [PubMed] [Google Scholar]
- Brisson N, Verma DP. 1982. Soybean leghemoglobin gene family: normal, pseudo, and truncated genes. Proceedings of the National Academy of Sciences of the USA 79: 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, Hansen AP, McNeil DL, et al. . 1987. Effect of oxygen supply on nitrogenase activity of nitrate- and dark-stressed soybean (Glycine max (L.) Merr.) plants. Australian Journal of Plant Physiology 14: 679–687. [Google Scholar]
- David KA, Apte SK, Banerji A, et al. . 1980. Acetylene reduction assay for nitrogenase activity: gas chromatographic determination of ethylene per sample in less than one minute. Applied and Environmental Microbiology 39: 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Mens C, Hastwell AH, et al. . 2018. Legume nodulation: the host controls the party. Plant Cell Environment 42: 1–11. [DOI] [PubMed] [Google Scholar]
- Fujikake H, Yamazaki A, Ohtake N, et al. . 2003. Quick and reversible inhibition of soybean root nodule growth by nitrate involves a decrease in sucrose supply to nodules. Journal of Experimental Botany 54: 1379–1388. [DOI] [PubMed] [Google Scholar]
- Gallusci P, Dedieu A, Journet EP, et al. . 1991. Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen-fixing root nodule development and response to exogenously supplied nitrate. Plant Molecular Biology 17: 335–349. [DOI] [PubMed] [Google Scholar]
- Garrocho-Villegas V, Gopalasubramaniam SK, Arredondo-Peter R. 2007. Plant hemoglobins: what we know six decades after their discovery. Gene 398: 78–85. [DOI] [PubMed] [Google Scholar]
- Gopalasubramaniam SK, Kovacs F, Violante-Mota F, et al. . 2008. Cloning and characterization of a caesalpinoid (Chamaecrista fasciculata) hemoglobin: the structural transition from a nonsymbiotic hemoglobin to a leghemoglobin. Proteins Structure Function and Bioinformatics 72: 252–260. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Igamberdiev AU. 2011. The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 11: 537–543. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Hebelstrup KH, Mur LAJ, et al. . 2011. Plant hemoglobins: important players at the crossroads between oxygen and nitric oxide. FEBS Letters 585: 3843–3849. [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM. 2008. Global inputs of biological nitrogen fixation in agricultural systems. Plant and Soil 311: 1–18. [Google Scholar]
- Hoy JA, Hargrove MS. 2008. The structure and function of plant hemoglobins. Plant Physiology and Biochemistry 46: 371–379. [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Layzell DB, Shelp BJ. 1997. Role of oxygen limitation and nitrate metabolism in the nitrate inhibition of nitrogen fixation by pea. Physiologia Plantarum 101: 45–50. [Google Scholar]
- Larue TA, Child JJ. 1979. Sensitive fluorometric assay for leghemoglobin. Analytical Biochemistry 92: 11–15. [DOI] [PubMed] [Google Scholar]
- Li X, Feng H, Wen JQ, et al. . 2018. a MtCAS31 aids symbiotic nitrogen fixation by protecting the leghemoglobin MtLb120-1 under drought stress in Medicago truncatula. Frontiers in Plant Science 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XX, Zheng JK, Yang YQ, et al. . 2018. b INCREASING NODULE SIZE1 expression is required for normal rhizobial symbiosis and nodule development. Plant Physiology 178: 1233–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, et al. . 2010. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant Journal 63: 86–99. [DOI] [PubMed] [Google Scholar]
- Marcker A, Lund M, Jensen EO, et al. . 1984. Transcription of the soybean leghemoglobin genes during nodule development. EMBO Journal 3: 1691–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Zhang A, Wang F, et al. . 2015. Arbuscular mycorrhizal fungi and rhizobium facilitate nitrogen uptake and transfer in soybean/maize intercropping system. Frontiers in Plant Science 6: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Fan XR, Orsel M, et al. . 2007. Nitrate transport and signalling. Journal of Experimental Botany 58: 2297–2306. [DOI] [PubMed] [Google Scholar]
- Navascués J, Pérez-Rontomé C, Gay M, et al. . 2012. Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proceedings of the National Academy of Sciences of the USA 109: 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JM. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual Review of Plant Biology 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, et al. . 2011. The rules of engagement in the legume-rhizobial symbiosis. Annual of Review of Genetics 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, et al. . 2005. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Current Biology 15: 531–535. [DOI] [PubMed] [Google Scholar]
- Ott T, Sullivan J, James EK, et al. . 2009. Absence of symbiotic leghemoglobins alters bacteroid and plant cell differentiation during development of Lotus japonicus root nodules. Molecular Plant-Microbe Interactions 22: 800–808. [DOI] [PubMed] [Google Scholar]
- Qu ZL, Zhong NQ, Wang HY, et al. . 2006. Ectopic expression of the cotton non–symbiotic hemoglobin gene GhHb1 triggers defense responses and increases disease tolerance in Arabidopsis. Plant and Cell Physiology 47: 1058–1068. [DOI] [PubMed] [Google Scholar]
- Sainz M, Calvo-Begueria L, Pérez-Rontomé C, et al. . 2015. Leghemoglobin is nitrated in functional legume nodules in a tyrosine residue within the heme cavity by a nitrite/peroxide-dependent mechanism. Plant Journal 81: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Tanabata S, Tanabata T, et al. . 2014. Effect of nitrate on nodule and root growth of soybean (Glycine max (L.) Merr.). International Journal of Molecular Sciences 15: 4464–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Nagata M, Suzuki A, et al. . 2005. Symbiotic rhizobium and nitric oxide induce gene expression of non-symbiotic hemoglobin in Lotus japonicus. Plant and Cell Physiology 46: 99–107. [DOI] [PubMed] [Google Scholar]
- Streeter J, Wong PP. 1998. Inhibition of legume nodule formation and N2 fixation by nitrate. Critical Reviews in Plant Sciences 7: 1–23. [Google Scholar]
- Taylor ER, Nie XZ, MacGregor AW, et al. . 1994. A cereal haemoglobin gene is expressed in seed and root tissues under anaerobic conditions. Plant Molecular Biology 24: 853–862. [DOI] [PubMed] [Google Scholar]
- Vieweg MF, Frühling M, Quandt HJ, et al. . 2004. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Molecular Plant-Microbe Interactions 17: 62–69. [DOI] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Bailly X, et al. . 2005. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proceedings of the National Academy of Sciences of the USA 102: 11385–11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Bailly X, et al. . 2006. A phylogenomic profile of globins. BMC Evolutionary Biology 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen AL, Laine T. 1946. Red, brown and green pigments in leguminous root nodules. Nature 157: 25–26. [DOI] [PubMed] [Google Scholar]
- Vuletich DA, Lecomte JTJ. 2006. A phylogenetic and structural analysis of truncated hemoglobins. Journal of Molecular Evolution 62: 196–210. [DOI] [PubMed] [Google Scholar]
- Wang GH, Sheng LC, Zhao D, et al. . 2016. Allocation of nitrogen and carbon is regulated by nodulation and mycorrhizal networks in soybean/maize intercropping system. Frontiers in Plant Science 7: 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RA, Hunt PW, Hvitved AN, et al. . 2001. A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proceedings of the National Academy of Sciences of the USA 98: 10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.