Abstract
Background and Aims
Contrasting life-history traits can evolve through generations of dwarf plant ecotypes, yet such phenotypic changes often involve decreased plant size and reproductive allocation, which can configure seed dispersal patterns and, subsequently, population demography. Therefore, evolutionary transitions to dwarfism can represent good study systems to test the roles of life-history traits in population demography by comparing genetic structure between related but phenotypically divergent ecotypes.
Methods
In this study, we examined an ecotypic taxon pair of the world’s smallest goldenrod (stem height 2.6 cm) in alpine habitats and its closely related lowland taxon (30–40 cm) found on Yakushima Island, Japan. Genetic variation in chloroplast DNA sequences, nuclear microsatellites and genome-wide single-nucleotide polymorphisms were used to investigate 197 samples from 16 populations, to infer the population genetic demography and compare local genetic structure of the ecotypes.
Key Results
We found a pronounced level of genetic differentiation among alpine dwarf populations, which were much less geographically isolated than their lowland counterparts. In particular, several neighbouring dwarf populations (located ~500 m apart) harboured completely different sets of chloroplast haplotypes and nuclear genetic clusters. Demographic modelling revealed that the dwarf populations have not exchanged genes at significant levels after population divergence.
Conclusions
These lines of evidence suggest that substantial effects of genetic drift have operated on these dwarf populations. The low-growing stature and reduced fecundity (only 3.1 heads per plant) of the dwarf plants may have reduced gene flow and rare long-distance seed dispersal among habitat patches, although the effects of life-history traits require further evaluation using ecological approaches.
Keywords: Alpine, dwarfism, flowering time, landscape genetics, seed dispersal, Solidago
INTRODUCTION
Understanding the factors and processes that shape early population divergence is one of the major objectives of evolutionary biology. Although large-scale spatial genetic structure often reflects historical isolation (e.g. climate change, geological evolution and land bridge configurations) (Magri et al., 2006; Qiu et al., 2011; Sork et al., 2016), for local genetic structure the roles of gene flow and ecological differentiation become more influential. The pattern of gene flow often depends on landscape heterogeneity more than geographical distance, which configures the connectivity of suitable habitats and thus individual dispersal patterns (Manel et al., 2003; Holderegger et al., 2010). Life-history traits (e.g. dispersal mode, growth form and size), which are inherent to each population/species, can also influence gene exchanges between populations (Vekemans and Hardy, 2004; Hardy et al., 2006). Meta-analyses of population genetic statistics over a range of plant lineages have revealed the phenotypic trait sets of significance for predicting genetic structure (Hamrick 1996; Nybom and Bartish, 2000; Aguinagalde et al., 2005; Duminil et al., 2007, 2009). Among these, the way in which seeds are dispersed can be an important determinant in shaping population genetic structure when it is a good representation of the realized dispersal distance (Takeuchi et al., 2004; Aguinagalde et al., 2005; Duminil et al., 2007). Hence, for non-zoochorous plants seed dispersal distance can be formulated as a function of release height (Tamme et al., 2014), with seed dispersal distance increasing with increasing release height (De Frenne et al., 2011). However, the effects of release height (or plant height) on genetic structure are not always straightforward, partly because life-history traits affect genetic structure in ways that depend on context (e.g. historical, ecological and especially taxonomic context) (Duminil et al., 2007).
To isolate the effect of life-history traits, studies can benefit from comparison of the genetic structure of phylogenetically related species that differ in particular traits but share most evolutionary contexts. Contrasting life-history traits can evolve through the generation of compact or dwarf plant ecotypes, which are repeatedly selected in various evolutionary contexts, including coastal habitats (Lowry, 2012), alpine environments (Billings and Mooney, 1968), and under grazing pressure by mammals (McNaughton, 1984; Suzuki et al., 2009). Such phenotypic changes are thought to be a local adaptation to harsh environments by avoiding abiotic or biotic stress, and are sometimes accompanied by life-history trait changes, including a markedly decreased number of inflorescences and lower reproductive allocation (Suzuki, 2008; Hirano et al., 2017). Therefore, evolutionary transitions to dwarf ecotypes can offer opportunities to test the roles of changes in life-history traits by comparing genetic structure between related but phenotypically divergent ecotypes.
A hotspot of dwarf plants is located in the alpine area of Yakushima Island, a continental island of the Japanese archipelago in East Asia. More than 80 taxa with miniaturized plant body size have been identified on the island (Hatsushima, 1991). Comparative analyses of the dwarf taxa on the island and their counterpart species with larger body size have revealed evidence for the recent emergence of parallel dwarfism (Huziwara, 1965; Fujishima et al., 1990; Yokoyama et al., 2003; Tsukaya, 2005; Ishikawa et al., 2006; Shinohara and Murakami, 2006; Yano et al., 2007; Kokubugata et al., 2010; Higashi et al., 2013; Okuyama, 2016; Sun et al., 2016; Kakezawa et al., 2017). Similarly, the Eurasian goldenrod (Solidago virgaurea L. complex) evolved into an extremely dwarf taxon [Solidago minutissima (Makino) Kitamura] in the central alpine area of Yakushima Island, which includes populations with an average stem height of 2.6 cm (Fig. 1). In lowlands of the island, larger Solidago populations, which are genetically very close to the dwarf type, are parapatrically distributed (Sakaguchi et al., 2018). Stem height and reproductive efforts as represented by the number of flower heads differ considerably between the Solidago ecotypes (Fig. 1).
Fig. 1.
(A) Location of Yakushima Island in the southernmost area of the main islands of the Japanese Archipelago and the border of the Ryukyu Archipelago. (B) Geographical map of Yakushima Island. The altitudinal zonation and topography of the island are shown. The localities of lowland populations (L1–L5) and alpine dwarf populations (R1–R7, W1–W4) are shown as black circles. Two other known localities for lowland populations are indicated by open circles. (C) Stature of a flowering plant from the lowland population (KYO 00071707). (D) Scanned images of living plants from four alpine populations. (E) Alpine dwarf plants in divergent habitats: (E1) W3 population in wetland habitat; (E2) R1 population in rock habitat.
In general, plant height has been shown to be one of the most influential factors for predicting dispersal distance (Thomson et al., 2011). Also, reduced fecundity or number of seeds of dwarf plants can have negative effects on the number of seeds being uplifted and dispersed across long distances (Nathan et al., 2002), which could reduce gene flow between populations. In this study, according to these predictions we investigated gene flow in divergent ecotypic populations of Solidago on Yakushima Island to test the hypothesis of whether the reduced probability of propagule dispersal in dwarf populations decreases gene flow among local populations. Under this scenario, genetic drift is expected to have larger roles in isolating dwarf populations, resulting in a stronger genetic structure than that of larger populations. The hypothesized evolutionary process represents an important feedback effect of being a dwarf on population demography, and testing this hypothesis using molecular markers can provide insights into the genetic consequences of life-history trait evolution in plants.
MATERIALS AND METHODS
Distribution of Solidago populations on Yakushima Island
Since the dwarf taxon Solidago minutissima has long been of evolutionary interest to botanists, the localities are well represented by specimens, which show that dwarf populations occur in the high-elevation area of 1700–1930 m a.s.l. and rarely around 1300 m (KYO 00071694) (Fig. 1). In August 2014, we visited 11 alpine populations to collect leaf material along a route from Yodo-go River to Mt Nagata (1886 m) via the major peaks of Mt Kuromi (1831 m), Mt Kurio (1867 m) and Mt Miyanoura (1936 m). Because alpine dwarf populations were found in both wetlands and crevasses in rock surfaces, we recorded the habitat type for each population. The lowland populations are known to inhabit the southern slopes of the mountain ranges (Fig. 1). Although there are relatively few localities of the lowland populations, most occur in sunny places along the main rivers or around waterfalls, indicative of a fragmented distribution. We collected lowland population samples from five localities in November 2013 and August 2014.
Molecular experiments
The collected leaf samples were dried immediately using silica gel and kept in the dark at room temperature before DNA extraction. Plant DNA was extracted using a slightly modified cetyltrimethylammonium bromide method (Murray and Thompson, 1980).
Chloroplast fragment sequencing.
The population samples (197 samples from 16 populations; Table 1) were sequenced in one intergenic region (trnH-psbA) of the chloroplast genome using universal primers (Shaw et al., 2005). The PCR was performed in a 10-µL volume containing 20 ng of template DNA, 1 µL of 10× PCR buffer, 0.8 µL of 2.5 mm dNTP, 0.05 µL of 50 µm each primer, and 0.25 units of TaKaRa Ex Taq (TaKaRa, Otsu, Japan). The PCR cycle was as follows: template denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 7 min. The PCR products were sequenced using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit v3.1 (Applied Biosystems, Foster City, CA, USA), and electrophoresed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Table 1.
Information and genetic variation of the Solidago populations analysed in this study. For the haplotype frequency of chloroplast DNA (cpDNA), the number of each haplotype within the population is shown in parentheses. Abbreviations: n, number of samples analysed; Na, number of alleles averaged over loci for nuclear SSR and double-digest restriction-site-associated DNA sequencing (ddRAD-seq) SNP markers; AR, allelic richness, HE, expected heterozygosity; HO, observed heterozygosity. Asterisks indicate populations with significant deviation from Hardy–Weinberg equilibrium (P < 0.01)
| Population code | Population no. | Habitat type | Flowering time | Altitude (m) | cpDNA | Nuclear SSR | ddRAD-seq | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Haplotype frequency | n | Na | AR | H E | H O | n | Na | Ar | H E | |||||
| R1 | POP_163 | Rock | Late July to early September | 1828 | 24 | H1 (1), H3 (23) | 24 | 3.3 | 1.8 | 0.391 | 0.328 * | 17 | 1.7 | 1.1 | 0.217 |
| R2 | POP_164 | Rock | Late July to early September | 1747 | 3 | H3 (3) | 3 | 1.8 | 1.6 | 0.238 | 0.304 | 3 | 1.2 | 1.0 | 0.263 |
| R3 | POP_165 | Rock | Late July to early September | 1927 | 2 | H3 (2) | 2 | 1.8 | 1.8 | 0.287 | 0.382 | 2 | 1.1 | 0.9 | 0.314 |
| R4 | POP_166 | Rock | Late July to early September | 1864 | 4 | H3 (4) | 4 | 1.9 | 1.6 | 0.263 | 0.279 | 2 | 1.0 | 0.9 | 0.329 |
| R5 | POP_167 | Rock | Late July to early September | 1856 | 11 | H3 (11) | 11 | 2.6 | 1.7 | 0.328 | 0.327 | 10 | 1.6 | 1.0 | 0.203 |
| R6 | POP_168 | Rock | Late July to early September | 1823 | 17 | H3 (17) | 17 | 3.2 | 1.6 | 0.286 | 0.266 | 12 | 1.7 | 1.1 | 0.212 |
| R7 | POP_169 | Rock | Late July to early September | 1695 | 3 | H3 (3) | 3 | 1.8 | 1.6 | 0.271 | 0.294 | 3 | 1.2 | 0.9 | 0.269 |
| W1 | POP_170 | Wetland | Late July to early September | 1753 | 19 | H2 (14), H3 (5) | 19 | 3.4 | 1.8 | 0.368 | 0.331 | 17 | 1.7 | 1.1 | 0.218 |
| W2 | POP_171 | Wetland | Late July to early September | 1784 | 19 | H3 (19) | 19 | 3.2 | 1.6 | 0.292 | 0.241 | 12 | 1.6 | 1.0 | 0.206 |
| W3 | POP_172 | Wetland | Late July to early September | 1781 | 21 | H3 (21) | 21 | 3.2 | 1.7 | 0.328 | 0.294 | 10 | 1.6 | 1.1 | 0.205 |
| W4 | POP_173 | Wetland | Late July to early September | 1729 | 21 | H3 (21) | 21 | 3.3 | 1.7 | 0.311 | 0.265 | 14 | 1.6 | 1.0 | 0.210 |
| L1 | POP_160 | Rock | Late October to November | 410 | 14 | H4 (1), H5 (13) | 14 | 3.4 | 2.0 | 0.454 | 0.408 | 8 | 1.5 | 1.0 | 0.211 |
| L2 | POP_161 | Riverside | Late October to November | 31 | 14 | H4 (14) | 14 | 3.1 | 1.9 | 0.398 | 0.401 | 9 | 1.5 | 1.0 | 0.215 |
| L3 | POP_159 | Riverside | Late October to November | 62 | 14 | H4 (14) | 14 | 3.0 | 1.9 | 0.398 | 0.408 | 10 | 1.6 | 1.0 | 0.215 |
| L4 | POP_162 | Riverside | Late October to November | 21 | 6 | H4 (6) | 6 | 2.1 | 1.6 | 0.276 | 0.225 | 3 | 1.1 | 0.8 | 0.278 |
| L5 | POP_099 | Riverside | Late October to November | 337 | 5 | H4 (5) | 5 | 2.2 | 1.9 | 0.342 | 0.353 | 0 | - | - | - |
| Mean (alpine) | 13 | 13.1 | 2.7 | 1.7 | 0.306 | 0.301 | 9.3 | 1.5 | 1.0 | 0.241 | |||||
| Mean (lowland) | 10 | 9.8 | 2.6 | 1.8 | 0.354 | 0.359 | 5.5 | 1.4 | 1.0 | 0.236 | |||||
| Mean (all) | 12.3 | 12.3 | 2.7 | 1.7 | 0.327 | 0.319 | 8.3 | 1.4 | 1.0 | 0.238 |
Nuclear microsatellite genotyping.
The population samples (197 samples from 16 populations; Table 1) were genotyped using 17 microsatellite markers, consisting of expressed sequence tag simple sequence repeat (SSR) markers developed for this complex (Sakaguchi and Ito, 2014) and three genomic SSR markers (Salt1, Salt3 and Salt17) of Solidago altissima (Sakata et al., 2013). For all loci, the forward primer was synthesized with one of three different M13 sequences (5′-CACGACGTTGTAAAACGAC-3′/5′-TGTGGAA TTGTGAGCGG-3′/5′- CTATAGGGCACGCGTGGT-3′), and the reverse primer was tagged with a pig-tail sequence (5′-GTTTCTT-3′) to promote full adenylation (Brownstein et al., 1996). The PCR reaction was performed following the methods of Sakaguchi and Ito (2014). The PCR products were loaded onto an autosequencer (GenomeLab GeXP; Beckman Coulter, Brea, CA, USA) to assess the fragment lengths using Fragment Analysis Software v. 8.0 (Beckman Coulter).
Double digest restriction-site-associated DNA sequencing
A double digest restriction-site associated DNA (ddRAD) library was prepared for the population genetic samples (132 samples from 15 populations; Table 1) using the protocol by Peterson et al. (2012) with minor modifications. Briefly, 10 ng of genomic DNA was digested with EcoRI and BglII (New England Biolabs, Ipswich, MA, USA) and adapters were ligated at 37 °C overnight in a 10-µL volume containing 1 µL of 10× NEBuffer 2, 0.1 µL of 100× BSA (New England Biolabs), 0.4 µL of 5 µm EcoRI adapter and BglII adapter, 0.1 µL of 100 mm ATP and 0.5 µL of T4 DNA ligase (Enzymatics, Beverly, MA, USA). The reaction solution was purified with AMPure XP (Beckman Coulter). Next, 3 µL of purified DNA was used in the PCR amplifications in a 10-µL volume containing 1 µL of each 10 µm index and TruSeq universal primer, 0.3 µL of KOD-Plus-Neo enzyme, 1 µL of 10× PCR buffer (Toyobo, Osaka, Japan), 0.6 µL of 25 mm MgSO4 and 1 µL of 10 mm dNTP. For several of the DNA samples used in the phylogenetic analysis, the PCR amplification included 1 µL of each 10 µm index and TruSeq universal primer and 5 µL of 2× KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA). Thermal cycling was initiated with a 94 °C step for 2 min, which was followed by 20 cycles of 98 °C for 10 s, 65 °C for 30 s and 68 °C for 30 s. The PCR products were pooled and purified again with AMPure XP. The purified DNA was loaded onto 2.0 % agarose gel and fragments of around 320 bp were retrieved using E-Gel SizeSelect (Life Technologies, Carlsbad, CA, USA). After quality assessment using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), the library was sequenced with 51-bp single-end reads in one lane of an Illumina HiSeq2000 (Illumina, San Diego, CA, USA). The primer sequences used in this study are described in Sakaguchi et al. (2015).
Data analysis of chloroplast sequences and nuclear microsatellite genotypes
The chloroplast sequence data were edited and aligned using BioEdit v. 7.0.8.0 software (Hall, 1999). The alignment file was imported into GENALEX 6.3 software (Peakall and Smouse, 2006) to recover the chloroplast haplotypes. The relationships among the haplotypes were inferred based on a median-joining network using Network v. 5 software (Bandelt et al., 1999).
The microsatellite genotype data were used to characterize genetic diversity and infer the genetic structure of the Solidago populations. Number of alleles per locus (Na), allelic richness (AR) (El Mousadik and Petit, 1996) and expected and observed heterozygosity (HE and HO) (Nei, 1987) were calculated for each population using FSTAT 2.9.3 software (Goudet, 1995). FSTAT 2.9.3 was also used to test significant deviation from Hardy–Weinberg equilibrium for populations with a sufficient number of samples (n > 15) and compare genetic differentiation FST between the alpine and lowland groups, including 1000 permutations to obtain a two-tailed P value. To explore the population genetic structure, a population genetic model implemented in the STRUCTURE software (Pritchard et al., 2000) was applied, which assumed Hardy–Weinberg disequilibrium within genetic clusters, because the species complex has self-incompatibility (Kühn et al., 2004) and inbreeding is uncommon (Sakaguchi and Ito, 2014). It was also assumed that each individual had admixed ancestral origins, that different gene pools that had retained their allele frequency were correlated (Falush et al., 2003), and that the sampling locations were designated as prior information to obtain improved parameter estimates (Hubisz et al., 2009). Then, 20 independent STRUCTURE simulations were run for each K (K = 1–15), with 100 000 burn-in steps followed by 100 000 Markov chain Monte Carlo steps. To complement the model-based STRUCTURE analysis, a neighbour-joining phylogenetic tree (Saitou and Nei, 1987) was constructed using the ape R package (Paradis et al., 2004) based on a distance matrix calculated using the stats R package (R Development Core Team, 2018) in R v. 3.2.2 (R Development Core Team, 2015).
Demographic inference of population divergence of alpine and lowland groups
To infer the past demography of the alpine and lowland populations, approximate Bayesian computation (ABC) was applied to simulate genetic polymorphism data of the microsatellite loci for the two population groups. ABC is a flexible computational algorithm designed to perform complex model-based inferences, and can bypass exact likelihood calculations using summary statistics and large computer simulations (Cornuet et al., 2008; Csillery et al., 2010). In this study, we assumed five alternative demographic models, in which population size was assumed to change from the ancestral population (NANC) to the current sizes (NALP and NLOW) during time TDIV, with different combinations of migration parameters: (1) full isolation with migration (IM) model (NANC, NALP, NLOW, TDIV, MLOW>ALP and MLOW<ALP); (2) a bidirectional migration model with the same migration rate (NANC, NALP, NLOW, TDIV and M); (3) directional migration model 1, in which post-divergence migration occurred in only one direction from the alpine to the lowland population (NANC, NALP, NLOW, TDIV and MLOW<ALP); (4) directional migration model 2, in which migration occurred from the lowland to the alpine population (NANC, NALP, NLOW, TDIV and MLOW>ALP); and (5) a divergence model, where the diverged populations were isolated without migration (NANC, NALP, NLOW and TDIV). The prior distributions employed were as follows: population size parameters (NANC, NALP and NLOW) in log scales were uniform [2, 5] with no constraints among the size parameters; the time parameter TDIV was uniform [100, 10 000] among generations; the mutation rate per generation in log scale was uniform [–6, –4]. The prior information for the migration parameters was set as uniform [0.001, 2] for each complexed parameter of (N × M). The prior distribution for the time parameter was set so that the divergence would have taken place in a relatively recent timeframe, since the population divergence on Yakushima Island has been suggested to have followed the divergence of regional lineages of Japan in the late Pleistocene era (Sakaguchi et al., 2018). Each one million data simulations for the 144 alpine and 53 lowland individuals genotyped at 17 unlinked microsatellite loci was performed by drawing parameter values from the prior distribution combinations using fastsimcoal2 v. 2.5.2.21 software (Excoffier et al., 2013) and ABCtoolbox v. 2 (Wegmann et al., 2010). Seven summary statistics [mean number of alleles and heterozygosity over loci for each of the alpine and lowland populations, and the modified M ratio (Garza and Williamson, 2001) over the loci for each of the alpine and lowland populations, and FST between populations] of the genetic data were calculated with the arlsumstat v. 3.5.2 software (Excoffier and Lischer, 2010). The summary statistics were used for the model selection procedure based on the posterior probability of the models using the abc R package (Csilléry et al., 2012) in R v. 3.2.2. Model selection was performed with a tolerance value (i.e. percentage of accepted simulations) of 0.0001, and non-linear regression correction based on the neuralnet method was applied to correct imperfect matches between the observed and simulated summary statistics. The goodness of fit of a selected model was assessed by checking that the observed data were surrounded by simulated data, which were displayed as principle component analysis (PCA) envelopes containing a specific percentage of simulations (here, three percentages were tested, 10, 20 and 30 %), using the abc R package. In addition, the rejection algorithm implemented in the gfit function of the abc package was performed to obtain a value of the goodness-of-fit statistic for 100 pseudo-observed datasets with a tolerance value of 0.01 as the default. Then, the posterior distribution of the demographic parameters was estimated for each demographic scenario based on 0.01 % of the simulated data closest to the observed data with the neuralnet corrections using the abc R package.
Data analysis of ddRAD-seq-derived short reads
After read-trimming using Trimmomatic v. 0.32 software (Bolger et al., 2014) following the methods of Sakaguchi et al. (2017), Stacks 1.08 (Catchen et al., 2011) was used to de novo assemble the ddRAD-seq reads, as no genome reference of Solidago virgaurea complex is currently available. The assembly was performed with the following parameter settings: minimum number of identical reads required to create a stack, m = 3; nucleotide mismatches between loci within a single individual, M = 2; and mismatches between loci when building the catalogue, n = 1. The single nucleotide polymorphism (SNP) genotype for each individual was exported with a minimum read depth of 6 using the ‘populations’ command (Catchen et al., 2011). The exported genotype data were processed using PLINK v. 1.07 software (Purcell et al., 2007); markers with minor allele frequency <0.03, missing individual rate >0.6, missing locus rate >0.6 and significant deviation from Hardy–Weinberg equilibrium (P < 0.01) were filtered out (Sakaguchi et al., 2017). The SNPs were then analysed using BayeScan v2.0 software (Foll and Gaggiotti, 2008) (Supplementary Data Fig. S1), and the markers with a log-transformed posterior probability exceeding a threshold of 1.5 were considered to be candidate outliers. The outliers were removed from the dataset to generate the SNP datasets consisting of candidate neutral markers. For the filtered SNP data, we performed STRUCTURE and neighbour-joining phylogenetic tree analyses, as was done for the nuclear microsatellite genotype data.
Landscape genetics of the alpine populations
To examine the association between individual genetic variation and geography and habitat type, PCA was applied to the microsatellite and SNP data of the alpine populations. For the SNP dataset, BayeScan v2.0 was used to detect outlier markers displaying allele frequencies unusually diverged between the different habitat types (‘rock’ or ‘wetland’) in alpine areas (Table 1). The candidate outliers were removed from the subsequent landscape genetic analysis, because we were interested in estimating the neural population dynamics in the alpine area. The R package adegenet (Jombart, 2012) was used to perform the PCA analysis in R v. 3.1.0 to extract synthetic variables in which genetic variation between individuals was maximized. The resultant axis scores of the first 20 PCA axes were used to find the multivariate redundancy analysis (RDA) axes explicitly in association with the habitat difference, while the effect of geography was partialled out. RDA is a multivariate regression method based on constrained ordination. In brief, RDA summarizes the generic variation matrix, which can be constrained by a matrix of explanatory variables (e.g. environmental factors in the case of landscape genetics). Then, the remaining genetic variance, which was not constrained by explanatory variables, was analysed with a standard PCA. Accordingly, the total variance in the genetic matrix can be partitioned into constrained and unconstrained variances. Furthermore, it can deal with individual metrics without transforming distances, which could reduce the power of distance-based analyses to detect associations, and can incorporate multiple variables simultaneously and is relatively robust to collinearity between predictors (Wagner and Fortin, 2016). Using RDA for individual-based landscape genetics can provide high accuracy in detecting environmental associations and offer robust performance when applied to various landscapes (Kierepka and Latch, 2015). The RDA and subsequent model selection based on Akaike’s information criterion values were performed using the vegan package in R v. 3.1.0.
Correlation analysis
The pairwise genetic distance between individuals was calculated following the method of Kosman and Lenard (2005) using the popgenreport R package. Euclidean distance matrices were calculated for the variables for correlation with genetic distance, including geographical distance and habitat type difference (0 or 1), using the R packages stats and geosphere (Hijmans, 2016). The statistical correlation between the genetic matrix and four distance matrices was analysed using Mantel tests and partial Mantel tests to control for the effect of geography. The correlation analyses were performed for both the alpine and the lowland population. An estimation of the parameters with 999 permutations was conducted using the vegan R package (Oksanen et al., 2012).
Demographic analysis of alpine populations
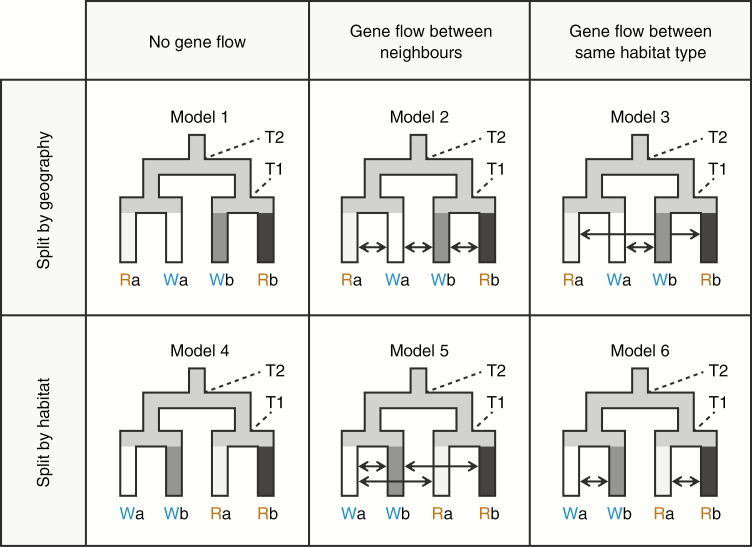
We compared population demographic models for alpine Solidago populations, in which geography, habitat type and gene flow were considered to have shaped the observed genetic polymorphisms (Fig. 4). For this analysis, alpine populations were grouped into four populations [Ra (consisting of R1 and R2 populations; n = 27), Wa (W1 and W2; n = 38), Wb (W3 and W4; n = 42) and Rb (R4–R7; n = 35)], based on their geographical positions, habitat types and genetic structure (see Results section). The R3 population was excluded from this analysis because of its small sample size (n = 2) and difficulty in group assignment. The common ancestral population (size NANC) was assumed to branch consecutively at T2 and T1, to generate the four current populations (sizes NRa, NRb, NWa, NWb). Models 1–3 represented a branching pattern in which populations split according to geography, while models 4–6 assumed that first divergence took place according to habitat type (i.e. rock versus wetland divergence). In models 2 and 5, bidirectional gene flow was considered only between adjacent populations (Ra ⟷ Wa, Wa ⟷ Wb and Wb ⟷ Rb), while gene flow was assumed to occur between the same habitat types (Ra ⟷ Rb and Wa ⟷ Wb) in models 3 and 6. The prior distributions were set as in the modelling for alpine and lowland populations, except that the time parameters T1 and T2 followed uniform [10, 3000] and uniform [10, 5000], respectively. Each million of the data simulations was performed using fastsimcoal2 v. 2.5.2.21 software (Excoffier et al., 2013) and ABCtoolbox v. 2 (Wegmann et al., 2010). In addition to the within-population summary statistics used in alpine–lowland analysis, pairwise FST between populations was calculated with arlsumstat v. 3.5.2 software (Excoffier and Lischer, 2010). The 18 summary statistics were used for the model selection procedure and parameter inference using the abc R package (Csilléry et al., 2012) in R v. 3.2.2.
Fig. 4.
Graphic representation of alternative demographic models for alpine Solidago populations in Yakushima Island. Four population groups were defined based on geographic proximity and habitat type (R, rock; W, wetland). Different colours in population trees indicate different population sizes, and arrows represent bidirectional gene flow.
Predicting seed dispersal distance
To compare the influences of stem height difference between alpine dwarf and lowland populations, the mean seed dispersal distance was predicted based on the model proposed by Tamme et al. (2014). Here, dispersal distance is expressed as a function of four fixed terms:
where DS is dispersal syndrome, GF is growth form, SM is seed mass and RH is release height.
DS and GF were set to ‘wind.special’ and ‘herb’, considering the traits of S. virgaurea. For the SM variable, seed mass was obtained based on 30 mature seeds from individuals of S. minutissima under cultivation in Kyoto University, to obtain an estimate of single-seed mass of 0.4 mg. In this study, release height was assumed to be stem height when the Solidago plants were in bloom. We measured the stem height of natural Solidago populations in alpine wetland habitats in August 2014. The stem height data for the lowland populations were collected from voucher specimens deposited in the Kyoto University herbarium (KYO). The average stem height values were used to represent the values for the alpine and lowland populations. We used the R package dispeRsal (Tamme et al., 2014) in R v. 3.4.0 (R Development Core Team, 2017) to predict the mean dispersal distances, with a linear mixed model with the family [‘Compositae’ (= Asteraceae)] and order (‘Asterales’) as nested random variables. More information on this package can be found in the supplementary material of Tamme et al. (2014). In addition, we counted the number of flower heads per stem on the alpine and lowland plants used to measure the stem heights.
RESULTS
Divergence between alpine dwarf and lowland populations
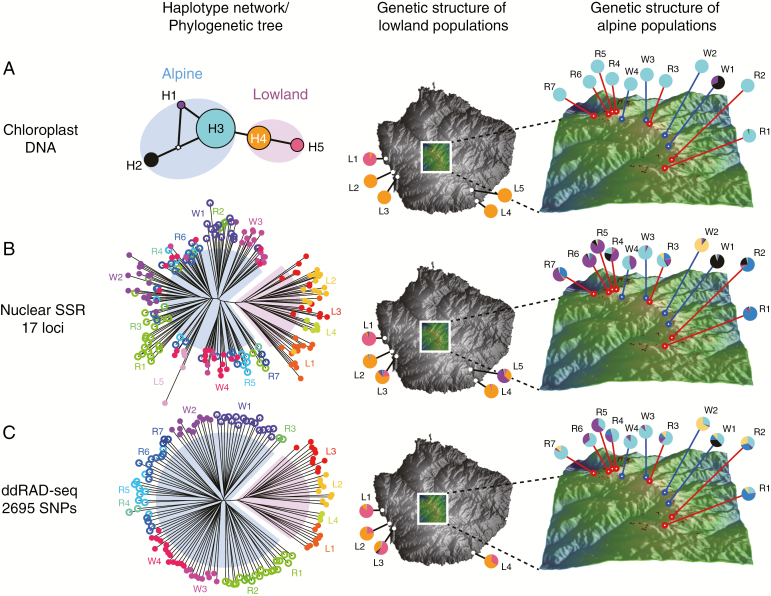
Chloroplast DNA sequencing detected five haplotypes, which were distinguished by the length of polymorphisms of SSRs (GenBank accession numbers LC318477–LC318481). The alpine and lowland populations showed complete differentiation in haplotype frequency, where H1–H3 were specific to alpine plants and H4–H5 to lowland plants (Fig. 2). The two closely related haplotypes H3 and H4, separated by only one mutational step, were the most frequent within each group, indicating they may represent more ancestral states (see haplotype network in Fig. 2A), while the remaining haplotypes were detected in particular populations (W1 and L1).
Fig. 2.
Population genetic structure of Solidago populations inferred by (A) chloroplast DNA sequencing, (B) nuclear SSRs and (C) ddRAD-seq of SNP markers. The haplotype network based on chloroplast DNA variations and phylogenetic trees based on nuclear markers are shown, where the pale blue and pink blobs indicate the population assignment based on altitudinal difference (blue, alpine; pink, lowland). To illustrate the spatial genetic structure, (A) the haplotype frequency of each population and (B, C) the portion of genetic clusters inferred by the STRUCTURE analysis (Pritchard et al., 2000) for each population are shown. Note that the colours used for the population origins in phylogenetic trees and the STRUCTURE clusters are not comparable (B, C).
Nuclear markers of microsatellites (127 alleles detected over 197 samples genotyped at a rate of 0.998) and genome-wide SNPs from ddRAD-seq (2695 SNPs over 132 samples genotyped at a rate of 0.646) revealed clear lineage divergence patterns between the alpine and lowland populations (FST = 0.207 for microsatellites and GST = 0.041 for SNPs), which was consistent between the different marker types and clustering methods (Fig. 2B, C). When the degree of population differentiation was compared between the alpine and lowland population groups, no significant group difference in FST values (alpine group, FST = 0.134; lowland, FST = 0.168; P = 0.634) was detected. The phylogenetic tree based on SNPs clustered the individual samples well according to population origin, while the tree based on microsatellites failed to do so, due to a lack of genetic polymorphisms for each sample. However, at the population level, the microsatellite data revealed clearer clustering patterns in the STRUCTURE analysis compared with SNP data, from which more ‘admixture-like’ assignment probabilities were inferred for each population. The clustering patterns at K = 7 are shown in Fig. 2B and C, because the log-likelihood of the data in STRUCTURE runs reached a plateau around K = 7 and no further meaningful clusters appeared after K = 7 (see more clustering patterns for K = 1–10 in Supplementary Data Fig. S2). The population genetic structure in the alpine area was complex; the two wetland populations (W1 and W2) were assigned to unique genetic clusters (black and pale yellow colours in Fig. 2B, C), while some clusters (e.g. light blue and purple) were shared among geographically close populations. Compared with the alpine area, the lowland populations were far more geographically isolated, but we detected a genetic cluster (orange colour in Fig. 2B, C) shared by the L2–L4 populations. Population L1 was genetically diverged from the remaining populations in terms of the dominance of the H5 chloroplast haplotype and pink genetic clusters in the nuclear marker analysis (Fig. 2).
Population demographic inference
The nuclear microsatellite data were used to model the lineage divergence pattern of the alpine and lowland populations based on the ABC approach. While acknowledging that scenario selection based on the ABC method should be considered with caution owing to its approximation error in the posterior probabilities (Robert et al., 2011), the simple divergence model received overwhelming support over the alternative models. The posterior probability of the divergence model was 0.981 versus full isolation with the migration model (0.006), bidirectional migration model (0.002) and directional migration models (0.001 and 0.016, respectively). The Bayes factors were 1.000 for the divergence model, 0.001 for the full model, 0.002 for the bidirectional migration model, and 0.007 and 0.134 for the alternative directional migration models. The model fit was evaluated based on PCA visualization, which confirmed that the observed data were well surrounded by envelopes of simulated data (Supplementary Data Fig. S3). The goodness-of-fit test for the divergence model yielded a P value of 0.446, which indicated that the mean distance between the observations and accepted simulations did not differ significantly. The posterior distributions of demographic parameters of the selected model had unimodal peaks, distinct from the uniform prior distributions (Supplementary Data Fig. S4). The population sizes of the two populations were estimated to have decreased from their common ancestral population after divergence (Table 2). The absolute time estimate of population divergence was calculated as 22 628 (15 099–29 596) years before present, an event during the last glacial maximum, by converting the posterior estimate of TDIV of 6465 (4314–8456) generations (Table 2) with an assumed generation time of 3.5 years (Kawano, 1988).
Table 2.
Prior and posterior distributions of demographic parameters for the simple split model, which was selected based on the posterior probability of models for the Solidago population in alpine and lowland areas of Yakushima Island
| Statistics | Log(NALP) | Log(NLOW) | Log(NANC) | T DIV | Log(mutation rate) | |
|---|---|---|---|---|---|---|
| Prior | Uniform [2, 5] | Uniform [2, 5] | Uniform [2, 5] | Uniform [10, 10 000] | Uniform [−6, −4] | |
| Posterior | 2.5 % percentile | 1.90 | 2.30 | 3.64 | 4314 | −5.22 |
| Median | 2.65 | 2.64 | 4.42 | 6465 | −4.55 | |
| Mean | 2.75 | 2.70 | 4.40 | 6502 | −4.62 | |
| Mode | 2.47 | 2.56 | 4.59 | 6364 | −4.48 | |
| 97.5 % percentile | 4.39 | 3.15 | 5.05 | 8456 | −4.15 |
Landscape factors associated with population divergence
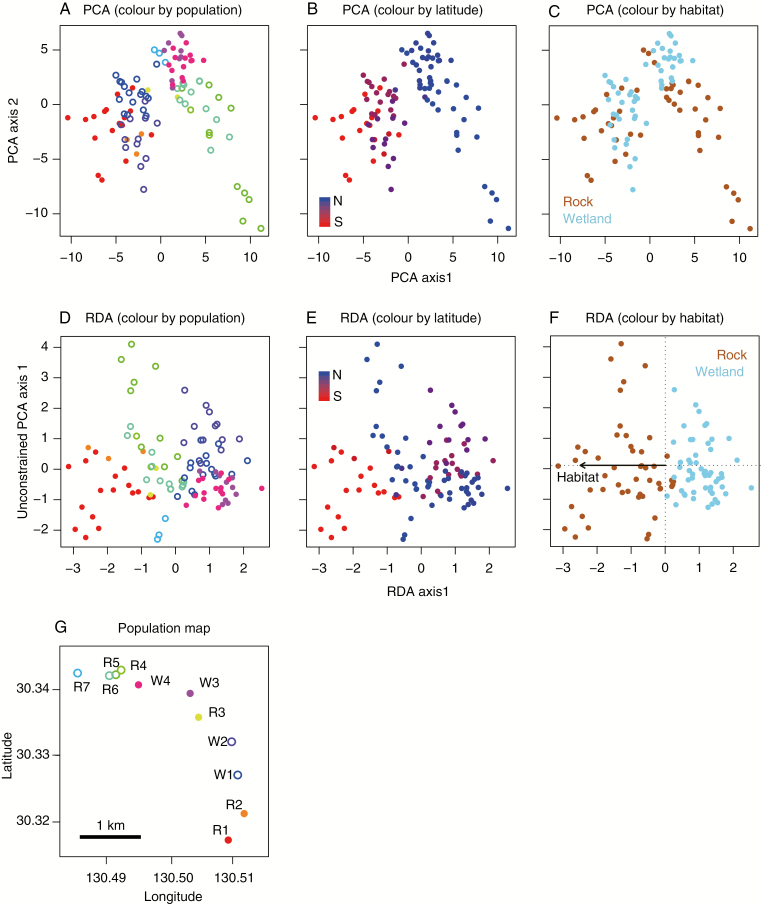
The alpine dwarf populations were distributed across open habitats in rock crevasses or wetlands along mountain ridges (Figs 2 and 3G). PCA was applied to summarize the genome-wide SNP data to estimate individual genetic structure in the dwarf populations (Fig. 3). Along the first two PCA axes, the individuals were scored in the order of the geographical position of the populations (Fig. 3A, B) and not with habitat type (Fig. 3C). The first 20 PCA axis scores were then correlated with geography and habitat variables using the RDA method. The model selection result suggested that most of the genetic variation in alpine dwarf populations was explained by a combination of the two explanatory variables (geography and habitat), which was consistent with the analyses based on genome-wide SNPs and nuclear SSRs (nSSRs) (selected full models’ Akaike weights: 0.842 and 0.920 for SNPs and SSRs, respectively; Table 3). The RDA first-axis score and the constraining variable (habitat type) showed strong correlations for both genetic markers [SNPs, r = −0.991; nSSRs, r = −0.944] (Fig. 3F). Individual correlations using Mantel tests revealed significant correlations between genetics and geography and habitat type in the alpine populations, and the correlation between genetics and habitat was still significant when the effect of geography was partialled out (Table 4, Supplementary Data Fig. 5). The geographic dependency of genetic divergence was also detected in the lowland populations, which was significant in both the SNP and nSSR datasets (Table 4).
Fig. 3.
Genetic structure of alpine Solidago individuals as inferred by (A–C) PCA and (D–F) RDA based on ddRAD-seq of SNP markers. The different colours represent (A, D, G) population origins, (B, E) spatial arrangement of populations and (C, F) habitat differences (rock or wetland type).
Table 3.
Model selection results for the RDAs of the PCA scores of alpine Solidago populations for the ddRAD-seq of SNP and nSSR markers. The analysis is constrained by habitat variables and conditioned with a geographical term. AIC, Akaike information criterion; con, conditioned; wi, Akaike weight
| Marker | Model | AIC | ΔAIC | wi |
|---|---|---|---|---|
| SNPs | Habitat + con(Geography) | 531.7 | 0 | 0.842 |
| con(Geography) | 535.6 | 3.89 | 0.120 | |
| Habitat | 538.3 | 6.58 | 0.031 | |
| Null | 541.6 | 9.93 | 0.006 | |
| nSSR | Habitat + con(Geography) | 362.9 | 0 | 0.920 |
| Habitat | 368.6 | 5.69 | 0.053 | |
| con(Geography) | 370.2 | 7.30 | 0.024 | |
| Null | 374.7 | 11.75 | 0.003 |
Table 4.
Mantel test results of the correlation between genetic distance and spatial and environmental isolation factors in alpine Solidago populations. Partial Mantel tests were performed to detect the correlation with the habitat factor by controlling for a partial effect of geography. The results are shown for the datasets of the ddRAD-seq of SNP and nSSR markers
| Populations | Marker | Model | r | P value |
|---|---|---|---|---|
| Alpine | SNPs | Genetic ~ Geography | 0.440 | < 0.001 |
| Genetic ~ Habitat | 0.176 | < 0.001 | ||
| Genetic ~ Habitat | Geography | 0.106 | < 0.001 | ||
| Lowland | SNPs | Genetic ~ Geography | 0.438 | < 0.001 |
| Alpine | nSSR | Genetic ~ Geography | 0.245 | < 0.001 |
| Genetic ~ Habitat | 0.136 | < 0.001 | ||
| Genetic ~ Habitat | Geography | 0.084 | < 0.001 | ||
| Lowland | nSSR | Genetic ~ Geography | 0.354 | < 0.001 |
Population demography of alpine dwarf populations
The posterior probability for model 1 and model 4 was 0.469 and 0.385, respectively, and these two demographic models without gene flow terms attained higher support than the other models (Table 5). This indicates that gene flow between adjacent (assumed in models 2 and 4) or same-habitat (models 3 and 6) populations is at non-significant levels (Fig. 4). At the same time, the comparable levels of support for models 1 and 4 suggest that our genetic data may not have enough information to distinguish the branching patterns, or that hierarchical population divergence may have occurred in short periods of time (T1 and T2 posterior distributions overlapped largely in both models; Table 5). The posteriors of population size parameters showed similar distributions in these two models (medians of current sizes ranging from 3.92 to 4.24 for model 1 and from 3.79 to 4.61 for model 4; ancestral size of 2.42 for model 1 and 2.44 for model 4) (Table 5). Population divergence was inferred to have occurred 2701 (1289–4333) and 2745 (850–4609) generations ago for models 1 and 4, respectively. The goodness-of-fit test yielded P values of 0.826 for model 1 and 0.551 for model 4, and the observed data were well surrounded by envelopes of simulated data in PCA plots (Supplementary Data Fig. S6).
Table 5.
Summary of population demographic analysis of alpine Solidago populations in Yakushima Island. Posterior probabilities (PP) and Bayes factors (BF) of models, prior and posterior distributions of the demographic parameters for five alternative models are shown [median (2.5 %, 95 % percentiles) are reported for each parameter]. NM, effective number of migrants
| Model | PP | Demographic parameter | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BF | log(NRa) | log(NRb) | log(NWa) | log(NWb) | log(NANC) | T 1 | T 2 | log (NM) | log (mutation rate) | |
| unif [2, 5] | unif [2, 5] | unif [2, 5] | unif [2, 5] | unif [2, 5] | unif [10, 3000] | unif [10, 5000] | unif [0.001, 2] | unif [-6, -4] | ||
| Model 1 | 0.469 | 4.07 | 3.92 | 4.11 | 4.24 | 2.42 | 1483 | 2701 | – | -4.89 |
| 1.000 | (2.63, 4.96) | (2.76, 4.31) | (3.52, 4.81) | (4.04, 5.13) | (2.02, 3.56) | (1202, 1813) | (1289, 4333) | – | (-5.20, -4.85) | |
| Model 2 | 0.002 | 3.97 | 3.04 | 3.26 | 4.15 | 2.02 | 1871 | 2796 | 1.065 | -5.38 |
| 0.003 | (3.96, 4.98) | (3.29, 4.51) | (3.59, 4.42) | (3.94, 4.87) | (2.06, 3.20) | (590, 2747) | (834, 4339) | (0.183, 1.972) | (-5.45, -4.64) | |
| Model 3 | 0.102 | 4.07 | 3.92 | 4.11 | 4.24 | 2.42 | 1084 | 2178 | 0.527 | -4.89 |
| 0.218 | (3.39, 4.32) | (3.04, 4.34) | (3.55, 4.39) | (3.66, 4.60) | (2.12, 3.05) | (717, 1651) | (−41, 5812) | (0.851, 1.163) | (-5.04, -4.77) | |
| Model 4 | 0.385 | 4.55 | 3.79 | 3.94 | 4.61 | 2.44 | 1138 | 2745 | – | -5.00 |
| 0.820 | (3.97, 4.95) | (3.04, 4.27) | (3.26, 4.38) | (4.15, 4.86) | (2.02, 3.14) | (399, 2724) | (850, 4609) | – | (-5.38, -4.50) | |
| Model 5 | 0.022 | 3.76 | 3.90 | 3.81 | 4.10 | 2.30 | 1045 | 3229 | −0.182 | -4.93 |
| 0.046 | (2.62, 4.43) | (2.75, 4.45) | (3.27, 4.11) | (3.68, 4.24) | (2.00, 3.15) | (−159, 2289) | (1964, 4337) | (0.775, 1.901) | (-5.23, -4.49) | |
| Model 6 | 0.021 | 3.95 | 3.52 | 3.92 | 4.40 | 2.46 | 1523 | 2543 | 0.354 | -5.05 |
| 0.044 | (3.44, 4.17) | (3.11, 3.73) | (3.36, 4.42) | (3.10, 5.17) | (1.85, 3.38) | (744, 2395) | (725, 4448) | (0.665, 1.177) | (-5.79, -4.13) |
Simulated seed dispersal distance and flower head production
The average stem length of alpine wetland Solidago plants was 2.6 cm (s.d. = 1.6 cm) (see flowering plant images in Fig. 1), which was much shorter than that of the lowland plants [38.6 cm (s.d. = 16.0 cm)]. Based on the linear mixed effect model as a function of life-history traits, the mean seed dispersal distances were predicted to be 4.8 m for alpine plants and 14.8 m for lowland plants. The average number of flower heads produced per stem was 3.1 (s.d. = 2.7) for the alpine wetland populations and 17.0 (s.d. = 9.1) for the lowland populations.
DISCUSSION
Historical divergence of the alpine dwarf and lowland populations
The genetic analyses based on three types of molecular markers consistently revealed a clear divergence between the alpine dwarf and lowland Solidago populations. No chloroplast haplotypes were shared between the population groups (Fig. 2A), and the individuals were clustered separately for each of the alpine and lowland populations based on nuclear microsatellite and SNP analyses, except for one population, L5, showing an ‘admixed’ origin based on nuclear microsatellites (Fig. 2B, C). The population demographic analysis offered further insights into their lineage divergence history, suggesting that the alpine populations evolved independently of the lowland without extensive genetic exchange (Table 2).
This finding is in contrast to the previously reported population dynamics of alpine S. virgaurea populations in other mountain ranges, which repeatedly revealed little genetic differentiation from lowland populations (Nakamura et al., 1997; Hirano et al., 2017; Sakaguchi et al., 2017). The S. virgaurea complex generally occupies a wide range of elevations in other temperate regions (Nishizawa et al., 2001; Bergstern, 2009; Kiełtyk and Mirek, 2014; Takahashi and Matsuki, 2017), and active gene flow along elevational gradients has been suggested to link local populations, resulting in low population differentiation (e.g. FST = 0.000–0.027) (Hirano et al., 2017). However, on Yakushima Island, under a humid climate with an annual mean precipitation reaching 7400 mm in the inland mountains (Takahara and Matsumoto, 2002), dense cloud forests develop along the middle elevation range, under which light-demanding Solidago plants cannot survive. Hence, the populations are distributed only in alpine open habitats or lowland areas with sparse canopies maintained by natural disturbances such as waterfalls or droughts on rock tops. In addition, temporal isolation likely contributed to the limited gene flow, as there is a difference in flowering time of nearly 2 months between the alpine and lowland populations (Table 1), serving as a prezygotic reproductive isolation mechanism. These altitudinal and temporal isolation factors, and probably also ecological local adaptation, would have reduced the probability of gene flow between the two population groups.
Pronounced genetic structuring in the alpine dwarf populations
The group comparison of genetic differentiation showed that the dwarf populations differentiated from each other at levels comparable to those of the lowland populations (P = 0.634 in permutation test). The STRUCTURE analysis showed that the allelic constitutions of several dwarf populations, particularly the wetland populations, drifted away even from neighbouring populations (Fig. 2, Supplementary Data Fig. S1). Also, demographic modelling revealed that gene flow after the divergence of dwarf populations was not at significant levels (Table 5). These findings are noteworthy because the dwarf populations included relatively large numbers of plants (at least several hundreds of ramets) and are less geographically isolated than the lowland populations. These characteristics of dwarf populations are suggestive that they may be less prone to drifts/bottlenecks than the lowland populations. In fact, the population genetic diversity of the dwarf populations was comparable to and sometimes higher than that of the lowland populations (Table 1), which occupy a much broader range and are far more isolated due to dense evergreen forests (Fig. 1). Hence, the pronounced population divergence in the alpine area would have been caused by factors specific to the dwarf populations.
Life-history traits of the dwarf populations may have affected the levels of gene flow between local populations. Stem length of dwarf Solidago plants was only 2.6 cm on average, from which the seed dispersal distance was predicted to be 4.8 m, a much shorter distance than the geographical distances between isolated populations. In addition, the decreased number of propagules due to dwarfism (3.1 heads for alpine plants versus 17.0 for lowland plants) could further reduce the probability of dwarf populations exchanging genes via rare long-distance dispersal (Tackenberg et al., 2003; Nathan, 2006). However, it should be noted that expected seed dispersal distance may not fully predict the realized gene flow pattern; unpredictable long-distance dispersal, variations in weather conditions and variation in recruitment of dispersed seeds under natural habitat conditions [e.g. topographic effects in complex alpine environments (Hirao and Kudo, 2004; Cortés et al., 2014)] may be equally important factors. Further research is needed to assess these possibilities, for example by assessing fine-scale gene dispersal (Hardy et al., 2004) and by simulating the seed dispersal probability under various weather conditions in alpine and lowland populations (Tackenberg et al., 2003).
Factors shaping genetic structure within dwarf populations
In landscape genetics theory, one ultimate driver of spatial genetic structure is the effect of landscape heterogeneity on gene flow and the resulting local populations (Manel et al., 2003; Wagner and Fortin, 2016). The alpine landscape is characterized by dynamic changes of topography, which generate diverse microhabitats for alpine plants (e.g. dry and windy ridges, rock outcrops, snowbeds in topographic depressions, and scattered wetland). Across the environmental matrix, light and water conditions (Körner, 2003), nutrient availability (Little et al., 2016), plant–soil interactions (Sedlacek et al., 2014) and community interactions (Wheeler et al., 2015) are key factors affecting seedling establishment and subsequent growth, and therefore gene flow between microhabitats in alpine complex environments. The apparent association between genetic variation and habitat type in the dwarf Solidago populations (Fig. 3, Tables 3 and 4) may be explained by a scenario in which gene flow predominantly links the populations in similar environments via selection against maladapted individuals that have migrated from alternative habitats (Rundle and Nosil, 2005; Nosil et al., 2009). In fact, however, we found strong population isolation even between neighbouring wetland habitats, as evidenced by complete fixation to different chloroplast haplotypes and a predominance of different genetic clusters in similar habitat types (e.g. W1 and W2) (Fig. 2). Also, habitat-specific gene flow models (models 3 and 6) were not preferred in demographic model analysis for the dwarf populations (Table 5). These lines of evidence suggest that the apparent association would be brought about by a non-adaptive process of genetic drift in these dwarf populations leading to substantial genetic differentiation (Fig. 3D). This explanation, in turn, highlights the importance of reduced gene flow in isolated dwarf populations, shaping strong population genetic structure (Wright, 1931; Pannell and Fields, 2014; Sexton et al., 2014).
Although the genome-wide genetic differentiation in alpine populations would be largely caused by neutral population dynamics, this does not preclude the possibility that the populations showed adaptive responses to their native habitats, as often seen in the S. virgaurea complex (Kawano, 1988; Bergstern, 2009; Hirano et al., 2017; Sakaguchi et al., 2019). Given local adaptations to either habitat, the introduction of locally maladapted genes and phenotypes due to strong and asymmetrical gene flow can affect local populations via hybrid inviability (Edmands, 1999). Hence, the reduced gene flow among the alpine dwarf populations is hypothesized to have contributed to the population divergence by promoting local adaptation to alternative habitats, serving as divergent substrates for growth. Transplants (Sedlacek et al., 2015) and other approaches, such as common garden experiments (e.g. Sambatti and Rice, 2007; Hirst et al., 2017) and the space-by-time substitution (Sedlacek et al., 2016; Wheeler et al., 2016), and genomic screening of adaptive genes (e.g. Turner et al., 2010; Kubota et al., 2015) are desirable in future studies to understand the respective roles of neutral processes (i.e. the balance of gene flow and drifts) and natural selection in population isolation in these dwarf populations.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: results of the BayeScan analysis of genome-wide SNP markers obtained in Solidago populations. Figure S2: results of the STRUCTURE analysis of genome-wide SNP markers in Solidago populations. The log-likelihood of the data [LnP(D)] and delta K values are plotted against the number of genetic clusters (K = 1–15) for nuclear microsatellite and genome-wide SNP markers. The clustering patterns for each dataset are shown from K = 1 to K = 10. Figure S3: principal component analysis representing the goodness of fit of the approximate Bayesian computation for the population demographic analysis. The colour blobs indicate the density distribution of the simulated data, while the observed data are shown as black crosses. Figure S4: posterior (solid lines) and prior (dotted lines) distribution of each demographic parameter inferred from the selected divergent model. Figure S5: isolation by distance (IBD) and isolation by habitat plots for lowland and alpine populations based on (a–c) nSSRs and (d–f) ddRAD-seq of SNP markers. The solid lines are based on predictions from linear regression models. Figure S6: PCA representing the goodness of fit of the approximate Bayesian computation for the demographic analysis of alpine dwarf populations. The colour blobs indicate the density distribution of the simulated data, while the observed data are shown as black crosses. Results are shown for the two models with high posterior support.
FUNDING
This research was supported by the Japan Society for the Promotion of Science (KAKENHI 16H04827, 18K14778 and 25291085) and by the Japan Science and Technology Agency ‘Strategic International Collaborative Research Program’ (grant 4-1403).
ACKNOWLEDGEMENTS
The authors are grateful to K. Fuse for collecting the plant material and to T. Yahara, S. Kanetani, R. Tsujino and H. Nagamasu for providing the distribution records of Solidago populations on Yakushima Island. We also thank two anonymous reviewers and editors for comments that greatly improved manuscript. Permission to conduct research was provided by the Yakushima National Park, Ministry of Agriculture, Forestry and Fisheries, and by Yakushima Town. The restriction-site-associated DNA sequencing experiment was conducted by the Joint Usage/Research Program of the Center for Ecological Research, Kyoto University. The chloroplast DNA sequences are registered in GenBank (accession numbers LC318477–LC318481). The ddRAD-seq read data can be accessed using the following accession numbers (BioProject, PRJDB6307; BioProject Submission, PSUB007746; DRA Submission, DRA006122; BioSample Submission, SSUB008374) in the DDBJ database. The nuclear microsatellite genotype data will be deposited in Dryad upon acceptance of this manuscript.
LITERATURE CITED
- Aguinagalde I, Hampe A, Mohanty A, Martín JP, Duminil J, Petit RJ. 2005. Effects of life‐history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. Journal of Biogeography 32: 329–339. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bergstern A. 2009. Population differentiation in Solidago virgaurea along altitudinal gradients. Doctoral Thesis, Uppsala University, Sweden. [Google Scholar]
- Billings WD, Mooney HA. 1968. The ecology of arctic and alpine plants. Biological Reviews 43: 481–529. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20: 1004–6, 1008–10. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 1: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, et al. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24: 2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés AJ, Waeber S, Lexer C, et al. 2014. Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix herbacea. Heredity 113: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csilléry K, Blum MG, Gaggiotti OE, François O. 2010. Approximate Bayesian computation (ABC) in practice. Trends in Ecology & Evolution 25: 410–418. [DOI] [PubMed] [Google Scholar]
- Csilléry K, François O, Blum MGB. 2012. abc: an R package for approximate Bayesian computation (ABC). Methods in Ecology and Evolution 3: 475–479. [DOI] [PubMed] [Google Scholar]
- Duminil J, Fineschi S, Hampe A, et al. 2007. Can population genetic structure be predicted from life-history traits? American Naturalist 169: 662–672. [DOI] [PubMed] [Google Scholar]
- Duminil J, Hardy OJ, Petit RJ. 2009. Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evolutionary Biology 9: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S. 1999. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53: 1757–1768. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (L) Skeels endemic to Morocco. Theoretical and Applied Genetics 92: 832–839. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genetics 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Frenne P, Baeten L, Graae BJ, et al. 2011. Interregional variation in the floristic recovery of post‐agricultural forests. Journal of Ecology 99: 600–609. [Google Scholar]
- Fujishima H, Okada H, Horio Y, Yahara T. 1990. The cytotaxonomy and origin of Ranunculus yaegatakensis, an endemic taxon of Yakushima Island. Journal of Plant Research 103: 49–56. [Google Scholar]
- Garza JC, Williamson EG. 2001. Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318. [DOI] [PubMed] [Google Scholar]
- Goudet J. 1995. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity 86: 485–486. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hamrick J. 1996. Plant life histories: ecological correlates and phylogenetic constraints – effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London B: Biological Sciences 351: 1291–1298. [Google Scholar]
- Hardy OJ, González-Martínez SC, Colas B, Fréville H, Mignot A, Olivieri I. 2004. Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae). II. Correlated paternity within and among sibships. Genetics 168: 1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Maggia L, Bandou E, et al. 2006. Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Molecular Ecology 15: 559–571. [DOI] [PubMed] [Google Scholar]
- Hatsushima S. 1991. Flora of the northern Ryukyus. Kagoshima: Asahi-Insatsu-Shoseki. [Google Scholar]
- Higashi H, Sakaguchi S, Ikeda H, Isagi Y, Setoguchi H. 2013. Multiple introgression events and range shifts in Schizocodon (Diapensiaceae) during the Pleistocene. Botanical Journal of the Linnean Society 173: 46–63. [Google Scholar]
- Hijmans RJ. 2016. geosphere: Spherical Trigonometry. R package version 1.5-5. [Google Scholar]
- Hirano M, Sakaguchi S, Takahashi K. 2017. Phenotypic differentiation of the Solidago virgaurea complex along an elevational gradient: insights from a common garden experiment and population genetics. Ecology and Evolution 7: 6949–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao AS, Kudo G. 2004. Landscape genetics of alpine-snowbed plants: comparisons along geographic and snowmelt gradients. Heredity 93: 290–298. [DOI] [PubMed] [Google Scholar]
- Hirst MJ, Griffin PC, Sexton JP, Hoffmann AA. 2017. Testing the niche-breadth-range-size hypothesis: habitat specialization vs. performance in Australian alpine daisies. Ecology 98: 2708–2724. [DOI] [PubMed] [Google Scholar]
- Holderegger R, Buehler D, Gugerli F, Manel S. 2010. Landscape genetics of plants. Trends in Plant Science 15: 675–683. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. 2009. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huziwara Y. 1965. Chromosome analysis in the tribe Astereae. Japanese Journal of Genetics 40: 63–71. [Google Scholar]
- Ishikawa N, Yokoyama J, Ikeda H, Takabe E, Tsukaya H. 2006. Evaluation of morphological and molecular variation in Plantago asiatica var. densiuscula, with special reference to the systematic treatment of Plantago asiatica var. yakusimensis. Journal of Plant Research 119: 385–395. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2012. adegenet: an R package for the exploratory analysis of genetic and genomic data. [Google Scholar]
- Kakezawa A, Tamura MN, Agata K, Shinohara W. 2017. Crossability of a high-mountain dwarf variety of Lysimachia japonica (Primulaceae) endemic to Yakushima Island with its normal-sized lowland counterpart. Plant Systematics and Evolution 303: 807–813. [Google Scholar]
- Kawano S. 1988. Life history of Solidago virgaurea. Shokubutsu no Sekai 3. Tokyo: Kyoikusha. [Google Scholar]
- Kiełtyk P, Mirek Z. 2014. Taxonomy of the Solidago virgaurea group (Asteraceae) in Poland, with special reference to variability along an altitudinal gradient. Folia Geobotanica 49: 259–282. [Google Scholar]
- Kierepka EM, Latch EK. 2015. Performance of partial statistics in individual-based landscape genetics. Molecular Ecology Resources 15: 512–525. [DOI] [PubMed] [Google Scholar]
- Kokubugata G, Nakamura K, Shinohara W, Saito Y, Peng CI, Yokota M. 2010. Evidence of three parallel evolutions of leaf dwarfism and phytogeography in Lysimachia sect. Nummularia in Japan and Taiwan. Molecular Phylogenetics and Evolution 54: 657–663. [DOI] [PubMed] [Google Scholar]
- Körner C. 2003. Alpine plant life: functional plant ecology of high mountain ecosystems. Heidelberg: Springer Science & Business Media. [Google Scholar]
- Kosman E, Leonard KJ. 2005. Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Molecular Ecology 14: 415–424. [DOI] [PubMed] [Google Scholar]
- Kubota S, Iwasaki T, Hanada K, et al. 2015. A genome scan for genes underlying microgeographic-scale local adaptation in a wild Arabidopsis species. PLoS Genetics 11: e1005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn I, Durka W, Klotz S. 2004. BiolFlor: a new plant-trait database as a tool for plant invasion ecology. Diversity and Distributions 10: 363–365. [Google Scholar]
- Little CJ, Wheeler JA, Sedlacek J, Cortés AJ, Rixen C. 2016. Small-scale drivers: the importance of nutrient availability and snowmelt timing on performance of the alpine shrub Salix herbacea. Oecologia 180: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Lowry DB. 2012. Ecotypes and the controversy over stages in the formation of new species. Biological Journal of the Linnean Society 106: 241–257. [Google Scholar]
- Magri D, Vendramin GG, Comps B, et al. 2006. A new scenario for the quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytologist 171: 199–221. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology & Evolution 18: 189–197. [Google Scholar]
- McNaughton SJ. 1984. Grazing lawns: animals in herds, plant form, and coevolution. American Naturalist 124: 863–886. [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Miyamoto J, Murai K, Yamagishi M, Fukui K. 1997. The rDNA locus and the RAPDs analysis of Solidago virgaurea ssp. asiatica and ssp. leiocarpa (Compositae) in Mt. Hakusan. Journal of Phytogeography and Taxonomy 45: 65–73. [Google Scholar]
- Nathan R. 2006. Long-distance dispersal of plants. Science 313: 786–788. [DOI] [PubMed] [Google Scholar]
- Nathan R, Katul GG, Horn HS, et al. 2002. Mechanisms of long-distance dispersal of seeds by wind. Nature 418: 409–413. [DOI] [PubMed] [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Nishizawa T, Kinoshita E, Yakura K, Shimizu T. 2001. Morphological variation of the head characters in Solidago virgaurea L. inhabiting three mountains in central Honshu. Journal of Phytogeography and Taxonomy 49: 117–127. [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Molecular Ecology 18: 375–402. [DOI] [PubMed] [Google Scholar]
- Nybom H, Bartish IV. 2000. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3: 93–114. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2012. vegan: community ecology package. [Google Scholar]
- Okuyama Y. 2016. Mitella amamiana sp. nov., the first discovery of the genus Mitella (Saxifragaceae) in the Central Ryukyus. Acta Phytotaxonomica et Geobotanica 67: 17–27. [Google Scholar]
- Pannell JR, Fields PD. 2014. Evolution in subdivided plant populations: concepts, recent advances and future directions. New Phytologist 201: 417–432. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YX, Fu CX, Comes HP. 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution 59: 225–244. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2015. R version 3.2.2: A language and environment for statistical computing. [Google Scholar]
- R Development Core Team 2017. R version 3.4.0: A language and environment for statistical computing. [Google Scholar]
- Robert CP, Cornuet JM, Marin JM, Pillai NS. 2011. Lack of confidence in approximate Bayesian computation model choice. Proceedings of the National Academy of Sciences of the USA 108: 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. 2005. Ecological speciation. Ecology Letters 8: 336–352. [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Horie K, Ishikawa N, et al. 2017. Simultaneous evaluation of the effects of geographic, environmental and temporal isolation in ecotypic populations of Solidago virgaurea. New Phytologist 216: 1268–1280. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Horie K, Ishikawa N, et al. 2019. Maintenance of soil ecotypes of Solidago virgaurea in close parapatry via divergent flowering time and selection against immigrants. Journal of Ecology 107: 418–435. [Google Scholar]
- Sakaguchi S, Ito M. 2014. Development and characterization of EST-SSR markers for the Solidago virgaurea complex (Asteraceae) in the Japanese archipelago. Applications in Plant Sciences 1400035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Sugino T, Tsumura Y, et al. 2015. High-throughput linkage mapping of Australian white cypress pine (Callitris glaucophylla) and map transferability to related species. Tree Genetics & Genomes 11: 1–12. [Google Scholar]
- Sakaguchi S, Kimura T, Kyan R, et al. 2018. Phylogeographic analysis of the East Asian goldenrod (Solidago virgaurea complex, Asteraceae) reveals hidden ecological diversification with recurrent formation of ecotypes. Annals of Botany 121: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata Y, Kaneko S, Hayano A, Inoue-Murayama M, Ohgushi T, Isagi Y. 2013. Isolation and characterization of microsatellite loci in the invasive herb Solidago altissima (Asteraceae). Applications in Plant Sciences 1: 1200313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambatti JB, Rice KJ. 2007. Functional ecology of ecotypic differentiation in the Californian serpentine sunflower (Helianthus exilis). New Phytologist 175: 107–119. [DOI] [PubMed] [Google Scholar]
- Sedlacek J, Wheeler JA, Cortés AJ, et al. 2015. The response of the alpine dwarf shrub Salix herbacea to altered snowmelt timing: lessons from a multi-site transplant experiment. PLoS ONE 10: e0122395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek J, Cortés AJ, Wheeler J, et al. 2016. Evolutionary potential in the alpine: trait heritabilities and performance variation of the dwarf willow Salix herbacea from different elevations and microhabitats. Ecology and Evolution 6: 3940–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek JF, Bossdorf O, Cortés AJ, Wheeler JA, van Kleunen M. 2014. What role do plant–soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic and Applied Ecology 15: 305–315. [Google Scholar]
- Sexton JP, Hangartner SB, Hoffmann AA. 2014. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68: 1–15. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J. 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany 92: 142–166. [DOI] [PubMed] [Google Scholar]
- Shinohara W, Murakami N. 2006. How have the alpine dwarf plants in Yakushima been miniaturized? A comparative study of two alpine dwarf species in Yakushima, Blechnum niponicum (Blechnaceae) and Lysimachia japonica (Primulaceae). Journal of Plant Research 119: 571–580. [DOI] [PubMed] [Google Scholar]
- Sork VL, Gugger PF, Chen JM, Werth S. 2016. Evolutionary lessons from California plant phylogeography. Proceedings of the National Academy of Sciences of the USA 113: 8064–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z-S, Zhou W, Jin X-J, Ohi-Toma T, Li P, Fu C-X. 2016. A tale of two islands: parallel evolution of dwarfism in Smilax biflora (Smilacaceae). Phytotaxa 245: 89–106. [Google Scholar]
- Suzuki RO. 2008. Dwarf morphology of the annual plant Persicaria longiseta as a local adaptation to a grazed habitat, Nara Park, Japan. Plant Species Biology 23: 174–182. [Google Scholar]
- Suzuki RO, Kato T, Maesako Y, Furukawa A. 2009. Morphological and population responses to deer grazing for herbaceous species in Nara Park, western Japan. Plant Species Biology 24: 145–155. [Google Scholar]
- Tackenberg O, Poschlod P, Kahmen S. 2003. Dandelion seed dispersal: the horizontal wind speed does not matter for long-distance dispersal – it is updraft! Plant Biology 5: 451–454. [Google Scholar]
- Takahara H, Matsumoto J. 2002. Climatological study of precipitation distribution in Yaku-shima Island, southern Japan. Journal of Geography (Chigaku Zasshi) 111: 726–746. [Google Scholar]
- Takahashi K, Matsuki S. 2017. Morphological variations of the Solidago virgaurea L. complex along an elevational gradient on Mt Norikura, central Japan. Plant Species Biology 32: 238–246. [Google Scholar]
- Takeuchi Y, Ichikawa S, Konuma A, et al. 2004. Comparison of the fine-scale genetic structure of three dipterocarp species. Heredity 92: 323–328. [DOI] [PubMed] [Google Scholar]
- Tamme R, Götzenberger L, Zobel M, et al. 2014. Predicting species’ maximum dispersal distances from simple plant traits. Ecology 95: 505–513. [DOI] [PubMed] [Google Scholar]
- Thomson FJ, Moles AT, Auld TD, Kingsford RT. 2011. Seed dispersal distance is more strongly correlated with plant height than with seed mass. Journal of Ecology 99: 1299–1307. [Google Scholar]
- Tsukaya H. 2005. Molecular variation of Spiranthes sinensis (Orchidaceae) in Japan, with special reference to systematic treatment of seasonally differentiated groups and a dwarf form, f. gracilis, from Yakushima Island. Journal of Plant Research 118: 13–18. [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics 42: 260–263. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Hardy OJ. 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology 13: 921–935. [DOI] [PubMed] [Google Scholar]
- Wagner HH, Fortin M-J. 2016. Basics of spatial data analysis: linking landscape and genetic data for landscape genetic studies. In: Balkenhol N, Cushman S, Storfer A, Waits L, eds. Landscape genetics: concepts, methods, applications. Oxford: John Wiley, 77–98. [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JA, Schnider F, Sedlacek J, et al. 2015. With a little help from my friends: community facilitation increases performance in the dwarf shrub Salix herbacea. Basic and Applied Ecology 16: 202–209. [Google Scholar]
- Wheeler JA, Cortés AJ, Sedlacek J, et al. 2016. The snow and the willows: earlier spring snowmelt reduces performance in the low-lying alpine shrub Salix herbacea. Journal of Ecology 104: 1041–1050. [Google Scholar]
- Wright SJ. 1931. Evolution in Mendelian populations. Genetics 16: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano O, Nanami T, Ito K, Ikeda H, Hoshino T. 2007. Cytological studies of the genus Carex (Cyperaceae) in the Osumi Islands (Kagoshima Prefecture) I. Chromosome counts of five species collected from Yakushima Island. Journal of Japanese Botany 82: 29–33. [Google Scholar]
- Yokoyama J, Fukuda T, Tsukaya H. 2003. Morphological and molecular variation in Mitchella undulata, with special reference to the systematic treatment of the dwarf form from Yakushima. Journal of Plant Research 116: 309–315. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2018. The R Stats Package.
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. 2012. Double digest RADseq: an inexpensive method for De Novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.