Abstract
Backgrounds and Aims
Dimorphandra wilsonii Rizzini, a critically endangered and protected tree, has a restricted distribution in the ecotone between the Cerrado and the Atlantic Forest in south-eastern Brazil. In this area, it co-occurs with D. mollis Benth., a common tree from the Cerrado, and D. exaltata Schott., a rare tree from the Atlantic Forest. Previous studies of D. wilsonii indicated heterozygosity excess at the individual level. Field observation of some intermediate phenotypes between D. wilsonii and both congeners suggests hybridization of D. wilsonii with D. mollis and/or D. exaltata. Here, we tested the hypothesis that D. wilsonii may have originated from hybridization between D. exaltata and D. mollis. We also performed cytogenetic analysis to examine if the heterozygosity excess could be explained by polyploidy in D. wilsonii.
Methods
We evaluated the genetic diversity and population structure of D. wilsonii using 11 nuclear simple sequence repeats (SSRs) genotyped in 152 individuals sampled across the taxon’s range. We performed comparative genetic analyses using overlapping SSR markers between D. wilsonii and previously published SSR data in D. mollis and D. exaltata to subsequently perform a series of allelic comparisons, multivariate and Bayesian analysis.
Key Results
Our results suggest that D. wilsonii individuals are most likely to correspond to F1 hybrids between D. exaltata and D. mollis. Cytogenetic analysis indicated that D. wilsonii is diploid with the same chromosome number as D. mollis (2n = 2x = 28).
Conclusions
Our study raises questions about the taxonomic status and the evolutionary future of D. wilsonii. We suggest that the conservation and management strategy for D. wilsonii should be revised and that it should take into account both parental Dimorphandra species in the ecotone, with special emphasis on the threatened D. exaltata. Finally, this study highlights the value of genetic information for the design of conservation strategies.
Keywords: Atlantic Forest, Cerrado, conservation genetics, Dimorphandra, Dimorphandra wilsonii, ecotone, hybridization
INTRODUCTION
Interspecific hybridization is widely accepted as an important evolutionary process in plants, occurring in about 25 % of plant species (Mallet, 2005) with diverse evolutionary outcomes (Paun et al., 2009; Soltis and Soltis, 2009). Hybridization can enhance biodiversity by generating novel gene combinations in hybrids (Rieseberg, 1997; Rieseberg and Carney, 1998) and through the formation of new taxa by homoploid or allopolyploid hybrid speciation (Paun et al., 2009; Abbott et al., 2013; Li et al., 2016; Maguilla and Escudero, 2016; Abbott, 2017; Nieto Feliner et al., 2017). The novel gene combinations in hybrids can generate new phenotypes via transgressive segregation which can allow hybrids to outcompete their parents in some habitats (Rieseberg et al., 1999, 2003). Hybridization can also influence the genetic and phenotypic variability of parents through introgression, i.e. the transfer of genomic regions between species due to hybridization and recurrent backcrosses (Suarez-Gonzalez et al., 2018). Introgression can be especially relevant for conservation and management of small, inbred and threatened populations as it can affect the recipient species’ evolutionary potential by transfer of genetic adaptations, by an increase in genetic diversity and by masking deleterious mutations (Hamilton and Miller, 2016). Alternatively to these biodiversity-enhancing effects, hybridization can increase the risk of extinction because of genetic or demographic swamping (Rhymer and Simberloff, 1996; Todesco et al., 2016). Demographic swamping occurs when hybrids have strongly reduced fitness relative to parents, which leads to waste of reproductive effort and falling of population growth rates of one parental species below replacement rates (Levin et al., 1996; Gibson et al., 2019). Genetic swamping occurs when one or both parental lineages are replaced by hybrids, i.e. when pure parental genomes are replaced by genomes of hybrid ancestry, although in some cases the individuals will phenotypically resemble the parents due to the decoupling of genotype and phenotype (Muhlfeld et al., 2014; Todesco et al., 2016).
The areas where hybridizing lineages interbreed, called hybrid zones, can serve as ‘natural laboratories’ for evolutionary studies (Hewitt, 1988; Field et al., 2011; Taylor et al., 2015; Bariotakis et al., 2016). Hybrid zones are characterized by clines in the genetic constitution of individuals from one parental taxon to another; they can evolve in situ or arise at secondary contact of diverged taxa, and they can be temporary or long lasting in time (Barton and Hewitt, 1985; Abbott, 2017). In hybrid zones, hybridization may enhance the proportion of individual heterozygosity and the genetic diversity of populations through admixture (Zalapa et al., 2010; Li et al., 2016; Marques et al., 2016). The relative fitness of hybrids in relation to their parents is especially relevant because it determines the evolutionary outcomes of hybridization between the involved taxa (Gompert et al., 2017). For example, when hybrids are less fit than both parents, a temporally stable tension zone consisting mainly of recurrently formed F1 individuals can establish (Barton and Hewitt, 1985; Abbott, 2017). Conversely, if hybrids show higher fitness than either parent, typically in intermediate habitats, the outcome may be hybrid speciation, or a stable hybrid zone along an environmental cline (Rieseberg et al., 2003; Abbott, 2017). Genetic studies in hybrid zones can thus target a diversity of pure and hybrid genotypes to infer patterns of gene flow and reproductive isolation as well as the adaptive factors shaping the evolution of divergence between species.
The ecotonal area between the Cerrado and the Atlantic Forest biomes in south-eastern Brazil is a highly heterogeneous environment characterized by a mosaic of forests and savannas (Durigan and Ratter, 2006), and thus constitutes a natural laboratory for evolutionary studies. The Cerrado is the most biodiverse savanna in the world (Brandon et al., 2005). It is mostly constituted of grasslands with sparse trees, but also shows forest physiognomies such as the xerophytic ‘Cerradão’ and gallery forests (Silva et al., 2006). The Atlantic Forest is one of the most diverse and threatened tropical forests (Fiaschi and Pirani, 2009) and comprises a range of physiognomies consisting of evergreen, semi-deciduous and mixed forests (araucaria forests) (Oliveira-Filho and Fontes, 2000). The Cerrado has a predominantly seasonal climate with a marked dry season and its soils have low fertility and high aluminium concentrations (Motta et al., 2002). In comparison, the Atlantic Forest has generally more fertile soils and higher annual precipitation than the Cerrado, although the semi-deciduous forest also shows high seasonality (Eisenlohr and de Oliveira-Filho, 2015). In addition, natural wildfires have been proposed as a key factor in the establishment of species in the Cerrado, whereas, in the forests, light availability seems to be a more important factor (Hoffmann and Franco, 2003; Goulart et al., 2011). As a consequence, the species of each biome show distinct morphological and physiological traits as well as ecological strategies in response to the specific environmental conditions of these heterogeneous habitats.
The proximity of both habitat types has favoured the migration of evolutionary lineages between Atlantic Forest and Cerrado through time. For example, groups of species adapted to the Cerrado have evolved from ancestral forest lineages in the last 4 million years (Simon et al., 2009; Simon and Pennington, 2012). In addition, paleopalynological studies showed that Pleistocene climatic fluctuations led to cycles of forest replacement by sub-tropical grasslands and savannas during colder and drier conditions, and of forest expansion into savannas under wetter and warmer conditions (Behling, 1995, 2002; Behling and Negrelle, 2001). These effects of Pleistocene climatic fluctuations on distribution range dynamics, including retreat, expansion and recolonization, shaped the patterns of genetic structure of plant species of both biomes (Novaes et al., 2010; Ribeiro et al., 2011, 2016; Buzatti et al., 2017, 2018; Souza et al., 2017). In addition, during the Last Glacial Maximum (LGM), species of the Cerrado and the Atlantic Forest might have co-occurred, allowing ancient hybridization events among these species (Resende-Moreira et al., 2017). Furthermore, a few genetic studies have suggested more recent gene flow between closely related savanna and forest tree species or ecotypes occurring in the savanna/forest ecotone (Lacerda et al., 2002; Cavallari et al., 2010; Resende-Moreira et al., 2017). Thus, this ecotone is a valuable region for the study of hybridization, intra-specific divergence and speciation, and can yield insights into the origin and evolution of species of these biomes and their evolutionary relationships.
Dimorphandra wilsonii Rizzini, known as faveiro-de-Wilson, is a long-lived tree that can reach 17 m in height and 1.2 m in diameter. Up until now, it has been listed in the IUCN Red List as a critically endangered species (Fernandes, 2006) that occurs mainly in the Cerrado/Atlantic Forest ecotone, in an area of approx. 5000 km2 in south-eastern Brazil (figure 1 of Fernandes and Rego, 2014). Because of its rarity and the level of threat it faces, a National Plan of Action (PAN of faveiro-de-Wilson) was launched in 2014 to establish measures for its conservation. This includes the evaluation of the genetic status of populations and the selection of trees/populations to produce progenies for ex situ conservation and for reintroduction and restoration of populations. After the extensive search performed in the frame of the conservation programme for D. wilsonii, about 420 individuals are currently known and there are most probably very few unknown individuals in nature (Fernando Moreira Fernandes, pers. commun.). Until now, only two studies have been performed to investigate the genetic diversity of D. wilsonii (Souza and Lovato, 2010; Vinson et al., 2015), based on the sampling of only 20–22 adult trees known until then. The study of Vinson et al. (2015) analysed D. wilsonii progenies and found that they carried alleles unobserved in known adults, leading to a suspicion of hybridization with the co-occurring D. mollis Benth. Furthermore, D. wilsonii can produced fruits with viable seeds from selfed and cross-pollinated flowers Martins et al., (2014).
Dimorphandra wilsonii co-occurs with two Dimorphandra species, D. mollis and D. exaltata Schott, throughout their ecotonal distribution range; these are the only Dimorphandra species in the distribution range of D. wilsonii. Dimorphandra mollis is a common species from the Cerrado and D. exaltata is a rare and threatened tree species from the Atlantic Forest (Muniz et al., 2019). The three taxa are distinguishable mainly based on leaflet features and on trunk morphology. Dimorphandra mollis has leaves composed of 6–14 leaflets with a dense indumentum, D. wilsonii has 6–12 leaflets with a moderately dense indumentum and D. exaltata has 4–6 glabrous leaflets. Dimorphandra wilsonii and D. exaltata have straighter trunks and thinner rhytidomes, similar to other forest-adapted trees, while D. mollis has a more twisted trunk and a thicker rhytidome, characteristic of Cerrado-adapted trees (da Silva, 1986). Pollination is assured by bees of the Apidae family in D. wilsonii (Martins et al., 2014), by small insects in D. mollis (Panegassi et al., 2000; Gonçalves et al., 2010) and has not been described for D. exaltata. However, due to similar floral morphology of Dimorphandra species, their pollination syndromes are likely to be similar (da Silva, 1986). The three taxa in our study display flowers synchronously in the rainy season (November–February) (da Silva, 1986).
During fieldwork, we observed some putative D. wilsonii individuals with intermediate phenotypic characteristics between D. wilsonii and D. mollis or between D. wilsonii and D. exaltata, suggesting that hybridization may be occurring between D. wilsonii and the other two species. Additionally, an unexpected pattern of heterozygosity excess at the individual level was previously observed in small samples of D. wilsonii genotyped with two different sets of simple sequence repeat (SSR) loci (Souza, 2012; Vinson et al., 2015). This pattern could represent a consequence of ongoing hybridization, as suggested in recent studies that observed high individual heterozygosity and negative inbreeding coefficients (representing a heterozygosity excess) in populations dominated by hybrids of recent origin, i.e. F1s and few F2s or recombinant hybrids (Zalapa et al., 2010; Marques et al., 2016; Zeng et al., 2016).
In this study, our initial aim was to use SSR markers and larger sample sizes than in the previous studies (Souza and Lovato, 2010; Vinson et al., 2015) to evaluate the genetic diversity, the pattern of genotypic heterozygosity and the genetic structure of D. wilsonii, and to use this information in the conservation programme. However, after confirming high levels of individual heterozygosity, we hypothesized that D. wilsonii may have originated from hybridization between D. exaltata and D. mollis. Specifically, we suspected D. wilsonii individuals to represent mainly F1s between D. exaltata and D. mollis as well as some later-generation hybrids and/or backcrosses. Assuming this hypothesis, our study addressed the following main questions. (1) What are the hybridization patterns among the three Dimorphandra taxa co-occurring in this ecotonal region? (2) Can a model in which D. exaltata and D. mollis are the parental species of D. wilsonii explain the patterns of allelic and genotypic variability in D. wilsonii? (3) Which hybrid classes occur, and in which proportions, in the D. wilsonii population? (4) What are the consequences of hybridization for the conservation of D. wilsonii and its congeners? To answer these questions, we performed a series of comparative genetic analyses involving the three Dimorphandra taxa, using overlapping markers between the novel SSR data generated in D. wilsonii (this study) and previously published genotypic data of D. mollis (Souza et al., 2017) and D. exaltata (Muniz et al., 2019). As the chromosome number in D. wilsonii was not known, we also performed cytogenetic analysis in this taxon to verify whether the heterozygote excess could be explained by polyploidy. Our study offers insights about the evolutionary origin of D. wilsonii and increases our understanding of the evolutionary processes that contribute to the high biodiversity of the Cerrado and the Brazilian Atlantic Forest. It also provides useful information for the conservation and management of Dimorphandra taxa that are facing habitat loss, fragmentation and climate change.
MATERIALS AND METHODS
Study taxa and sampling
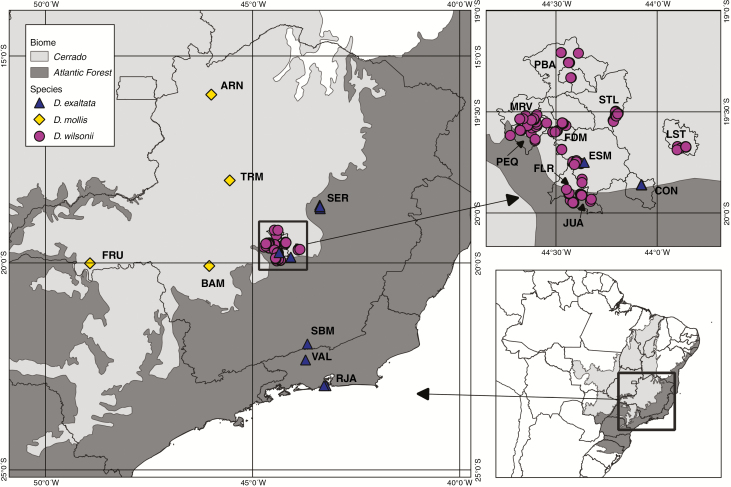
Genetic diversity, genotypic heterozygosity and genetic structure of D. wilsonii were estimated based on the genotyping of 11 SSR markers (see below) in 152 georeferenced individuals, sampled in ten municipalities of central Minas Gerais state, south-eastern Brazil (Fig. 1; Table 1; a small number of individuals sampled south of PBA municipality were merged with PBA for analysis). The individuals were selected to cover most of the D. wilsonii distribution area (Martins et al., 2014). Dimorphandra wilsonii individuals were found mainly in pasture areas, where cattle eat their fruits and where only few seedlings can establish or, alternatively, in small and isolated patches of forested areas such as the Cerradão (forested savanna) or semi-deciduous forest.
Fig. 1.
Study area with the sampled individuals of Dimorphandra wilsonii, D. mollis and D. exaltata.
Table 1.
Population genetic diversity parameters and inbreeding coefficients by locus genotyped and per municipality in Dimorphandra wilsonii
| n | A | A R | H O | H E | F IS | |
|---|---|---|---|---|---|---|
| Loci | ||||||
| Dmo5 | 151 | 7 | 2.6 | 0.940 | 0.542 | –0.740 |
| Dmo7 | 151 | 13 | 5.5 | 0.868 | 0.762 | –0.139 |
| Dmo13 | 148 | 7 | 4.7 | 0.939 | 0.690 | –0.364 |
| Dmo20 | 149 | 9 | 2.4 | 0.175 | 0.219 | 0.202 |
| Dmo21 | 152 | 4 | 3.0 | 0.908 | 0.593 | –0.533 |
| Dw21 | 150 | 4 | 2.2 | 0.113 | 0.417 | 0.729 |
| Dw33 | 151 | 5 | 3.5 | 0.556 | 0.537 | –0.036 |
| Dw105 | 150 | 10 | 4.9 | 0.933 | 0.730 | –0.280 |
| Dw28 | 151 | 8 | 5.1 | 0.430 | 0.796 | 0.460 |
| Dw52 | 149 | 10 | 4.6 | 0.752 | 0.744 | –0.010 |
| Dw103 | 149 | 5 | 3.8 | 0.926 | 0.699 | –0.327 |
| Municipalities | ||||||
| PBA | 12 | 3.5 | 3.3 | 0.750 | 0.572 | –0.202 |
| MRV | 20 | 4.8 | 3.5 | 0.754 | 0.582 | –0.307 |
| PEQ | 18 | 4.4 | 3.6 | 0.701 | 0.600 | –0.175 |
| FDM | 24 | 3.8 | 3.2 | 0.730 | 0.580 | –0.265 |
| ESM | 16 | 4.2 | 3.4 | 0.700 | 0.600 | –0.173 |
| FLR | 8 | 3.4 | 3.3 | 0.667 | 0.606 | –0.109 |
| JUA | 21 | 4.3 | 3.3 | 0.666 | 0.567 | –0.180 |
| STL | 18 | 4.0 | 3.1 | 0.609 | 0.482 | –0.154 |
| LST | 15 | 2.7 | 2.4 | 0.686 | 0.446 | –0.568 |
| Overall | 152 | 7.5 | 3.8 | 0.686 | 0.612 | –0.121 |
n = sample size, A = number of alleles, AR = allele richness for a sample size of n = 7 individuals, HO = observed heterozygosity, HE = expected heterozygosity, FIS = inbreeding coefficient. FIS values significantly different from zero at P < 0.05 after multiple test correction are indicated in bold. The overall values were calculated based on all individuals of D. wilsonii pooled.
We selected 76 individuals from four populations of D. mollis which were previously genotyped at five out of the 11 SSR loci analysed in D. wilsonii (Dmo5, Dmo7, Dmo13, Dmo20 and Dmo21; Souza et al., 2017). These four populations were located in Cerrado areas of Minas Gerais state in Brazil, with distances to D. wilsonii trees ranging from approx. 100 km to 500 km (Fig. 1); genotypes of sympatric D. mollis populations were not available. For D. exaltata, we selected 62 individuals from six sites, two in the Cerrado/Atlantic forest ecotone and four in the Atlantic Forest, which were previously genotyped at the same 11 loci as D. wilsonii (Muniz et al., 2019; Fig. 1). Three of the Atlantic Forest sites (VAL, SBM and RJA; Fig. 1) were considered as a single population named RJA because they had small sample sizes and constituted a distinct gene pool (Muniz et al., 2019). The distance of sampled individuals of D. exaltata to D. wilsonii ranged from 5 km to 320 km. For the inter-specific analyses, we included an additional 15 putative D. wilsonii individuals that were not included in the D. wilsonii population analyses due to the uncertainty of their botanical determination; these were genotyped at 11 SSR loci like the other D. wilsonii individuals (see below).
DNA isolation and microsatellite genotyping
Dimorphandra wilsonii leaves or cambium tissue were dried in silica gel and stored at –20 °C after sampling in the field. DNA isolation was performed according to the protocol published in Souza et al. (2012a). Dimorphandra wilsonii individuals were genotyped at 11 SSR markers, six of them isolated in D. wilsonii (Dw21, Dw28, Dw33, Dw105, Dw52 and Dw103; Aksoy et al., 2013) and five isolated in D. mollis (Dmo5, Dmo7, Dmo13, Dmo20 and Dmo21; Souza et al., 2012b). The amplifications were performed according to Souza et al. (2012b) for all markers. Amplification fragments were separated through capillary electrophoresis on an ABI prism 3500xl automated sequencer including the GeneScan™ ROX 500™ size standard. Alleles were scored using GeneMapper version 5.0 (Applied Biosystems); fragments not assigned to alleles by the software were assigned manually. MicroChecker version 2.2.0.2 (Van Oosterhout et al., 2004) was used to evaluate scoring errors due to null alleles, stuttering or large allele dropout in D. wilsonii. The previously published SSR data in D. exaltata and D. mollis were obtained with the same protocols in the same laboratory, and allele binning was cross-validated between the three taxa (Souza et al., 2017; Muniz et al., 2019).
Genetic diversity, heterozygosity and structure of D. wilsonii
We used ARLEQUIN 3.5 (Excoffier and Lischer, 2010) to estimate the mean number of alleles per locus (A) and the expected and observed heterozygosities (HE and HO, respectively) in D. wilsonii. We used FSTAT version 2.9.3.2 (Goudet, 2002) to estimate the allelic richness (AR) with the rarefaction method of El Mousadik and Petit (1996), the inbreeding coefficient (FIS), and to test for departures from Hardy–Weinberg equilibrium with exact tests, with significance assessed after sequential Bonferroni correction for multiple comparisons. Genetic diversity parameters were estimated (1) at the level of municipalities (Fig. 1) because legal protection and management of D. wilsonii are co-ordinated by municipalities (Martins et al., 2014), and (2) in the total data set. Randomization-based tests were used to evaluate linkage disequilibrium for all pairs of loci in the full data set using FSTAT version 2.9.3.2 (Goudet, 2002). Genetic structure was evaluated using the spatial Principal Component Analysis (sPCA) implemented in the adegenet package (Jombart, 2008). The method uses georeferenced genotypes to investigate the patterns of genetic variance between individuals while controlling for effects of spatial autocorrelation between them. This method has a wide applicability to explore genetic data sets because it does not rely on assumptions of Hardy–Weinberg equilibrium or linkage equilibrium among loci (Jombart et al., 2008). Spatial autocorrelation was assessed using Moran’s I spatial autocorrelation coefficient estimated using a connection network based on the K nearest neighbours, with K set to 15. To assess the significance of global and local genetic structures, we used Monte Carlo tests in the spca_randtest function implemented in the adegenet package (Jombart, 2008).
Evolutionary relationships among D. wilsonii, D. mollis and D. exaltata
As we observed a strong departure from Hardy–Weinberg genotypic proportions in D. wilsonii with observed heterozygosity higher than expected heterozygosity and some loci showing HO very close to 1, we conducted a series of analyses to investigate a possible hybrid origin for D. wilsonii. First, we computed the genotypic and allele frequencies of the three Dimorphandra taxa using GenALEx version 6.503 (Peakall and Smouse, 2006, 2012) to evaluate the sharing of alleles among them and to detect putative hybrid genotypes. We then performed a PCA with the three taxa together using the adegenet package (Jombart, 2008) and estimated genetic differentiation between them using pairwise FST in ARLEQUIN 3.5 (Excoffier and Lischer, 2010). We also analysed the individuals of the three taxa together using the Bayesian clustering method implemented in STRUCTURE software version 2.3.4 (Pritchard et al., 2000; Hubisz et al., 2009). We ran five repetitions for each number of clusters, K, with K comprised between 1 and 5, using the admixture model with correlated allele frequencies and 1 000 0000 Markov Chain Monte Carlo (MCMC) iterations after discarding 100 000 iterations as burn-in. We used the ‘print credible regions’ option with default parameters to estimate posterior confidence intervals of individual ancestry proportions Q in each cluster. The optimal number of clusters was determined using Evanno’s ΔK method (Evanno et al., 2005) in STRUCTURE HARVESTER software 0.6.7 (Earl and vonHoldt, 2012). We averaged the results of individual runs for a given number of clusters using CLUMPP version 1.1.2 (Jakobsson and Rosenberg, 2007).
We also investigated the hybrid origin and the hybrid class of D. wilsonii individuals using the hybrid index estimated with the introgress package in R (Buerkle, 2005; Gompert and Buerkle, 2010). The hybrid index is computed using a maximum likelihood method to estimate the genetic contribution of hybridizing ‘parental’ populations or species to individuals of unknown ancestry (Buerkle, 2005). Because D. wilsonii was the only taxon to display an excess of observed heterozygosity (see the Results; Souza et al., 2017; Muniz et al., 2019), we used D. mollis and D. exaltata as putative parental populations of D. wilsonii. For each D. wilsonii individual, a hybrid index was obtained, with a value of zero representing a D. mollis genome and a value of one a D. exaltata genome. We used the software NewHybrids version 1.1 to calculate the posterior probability of individuals belonging to pre-determined genotypic classes (Anderson and Thompson, 2002). We used six genotypic classes comprising two purebred parental classes, F1 and F2 hybrids and two backcrosses, one to each parental class, to evaluate the classification of D. wilsonii, D. mollis and D. exaltata individuals without using any prior information on assignment to genotypic class. We performed five repeated runs in NewHybrids using Jeffreys-like priors for 1 000 000 MCMC iterations with a burn-in period of 100 000 iterations. Lastly, to evaluate the efficiency in the identification of hybrids in both STRUCTURE and NewHybrids, we simulated parental populations and hybrid individuals using the function hybridize in the adegenet package (Jombart, 2008). We simulated five data sets using allele frequencies (five SSR loci) of D. mollis and D. exaltata individuals as parental populations. The simulated data sets comprised 60 individuals of each parental class, 75 F1 hybrids, 25 F2 hybrids and 25 individuals of each backcross type to resemble the sample sizes of our original data set for the three taxa. Using these data sets, we conducted STRUCTURE and NewHybrids analyses as described above and calculated the efficiency, accuracy and the overall performance of the analyses as defined by Vähä and Primmer (2006).
Cytogenetic analysis in D. wilsonii
To exclude that the unexpected heterozygosity excess in D. wilsonii might be due to polyploidy, we studied chromosome numbers in root meristems. Seeds collected in three municipalities (ESM, MRV and PBA; Fig. 1) were germinated on vermiculite in plastic boxes and watered as necessary. The radicles were pre-treated in a solution of 0.002 m 8-hydroxyquinoline for 4 h at 16–18 °C. Radicles were then fixed in Carnoy’s solution (3:1 ethanol:acetic acid v/v) for at least 24 h at room temperature and stored in 70 % ethanol at −20 °C. Root meristems were rinsed, softened in 5 n HCl for 20 min and rinsed three times in distilled water. The meristems were then squashed in 45 % acetic acid to spread the cells. The slides were frozen in liquid nitrogen to remove the coverslip, stained with 2 % Giemsa and sealed with Entellan mounting solution (Merck Millipore, Darmstadt, Germany). Metaphase mitotic cells were observed using an Olympus BX51 microscope to count chromosomes. The best metaphases plates were photographed using an Olympus DP70 digital camera.
RESULTS
Genetic diversity and structure of D. wilsonii
We found no evidence of stutter bands or genotyping errors across all loci genotyped in D. wilsonii. Significant frequencies of null alleles were found in the loci Dm20, Dw21 and Dw58 across all individuals (Supplementary data Table S1). We found significant pairwise linkage disequilibrium in 13 of 55 comparisons across all loci when considering all samples.
The total number of alleles per locus (A) ranged from 4 to 13 in D. wilsonii, with a mean A = 7.5 (Table 1). Heterozygosity values per locus ranged from 0.113 to 0.940 for HO and from 0.219 to 0.796 for HE (Table 1). Six loci showed significant negative FIS values, ranging from –0.740 to –0.139, reflecting an excess of heterozygotes in relation to Hardy–Weinberg genotypic proportions. Three loci showed significant positive FIS values, ranging from 0.202 to 0.729, i.e. a deficit of heterozygotes, and two loci showed FIS values not significantly different from zero. The overall FIS across all individuals and loci was significantly negative, showing a value of –0.121 (Table 1).
At the municipality level, A ranged from 2.7 to 4.8, and AR, based on a minimum sample size of seven individuals, ranged from 2.4 to 3.6 (Table 1). The HE values of municipalities ranged from 0.446 to 0.606 (Table 1). Based on AR and HE, the most diverse municipalities were PEQ, ESM and FLR, and the least diverse was LST.
The sPCA revealed significant global structure among individuals of D. wilsonii (P = 0.001). The test for local structure was not significant, indicating that neighbouring individuals were not significantly dissimilar (P = 1.000). The first sPCA axis identified LST in the very eastern part of the sampling range as the most divergent municipality, with individuals displaying the highest negative sPCA scores (Supplementary data Fig. S1). The western part of the sampling range exhibited a gradient of sPCA scores, with the highest positive scores in southern municipalities (JUA and FLR), intermediate values in the central municipalities (PEQ, FDM and STL) and a few high scores in the northern municipality PBA (Supplementary data Fig. S1).
Evolutionary relationships among D. wilsonii and the two co-occurring species, D. exaltata and D. mollis
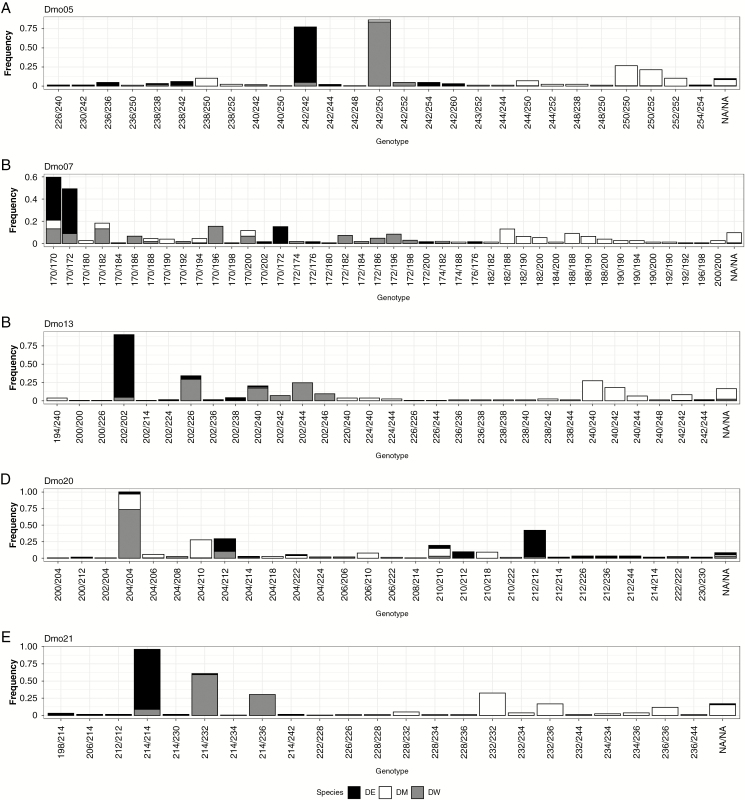
The comparison of alleles of the three Dimorphandra taxa revealed that 45 alleles were observed in D. wilsonii at the five loci genotyped in the three taxa with 21 alleles shared with D. exaltata and 30 alleles shared with D. mollis. Dimorphandra mollis and D. exaltata showed similar total numbers of alleles for the sampled populations, with 44 alleles in D. mollis and 38 alleles in D. exaltata, but only 17 alleles were shared between the two species (Supplementary data Table S2).
Dimorphandra mollis and D. exaltata showed lower HO than HE, and had significant positive overall FIS values, i.e. a deficit of heterozygotes, in contrast to D. wilsonii that displayed a heterozygote excess (Table 2). Patterns of allele and genotype frequencies in the three taxa (Supplementary data Table S2; Fig. 2) allowed us to formulate hypotheses about their evolutionary relationships, which can be illustrated mainly with the loci that showed higher heterozygote excess in D. wilsonii: at Dmo05 (FIS = –0.740), the most common alleles in D. exaltata (242) and D. mollis (250) are frequently found as a heterozygous genotype in D. wilsonii (242–250) (Fig. 2); similarly, at Dmo21 (FIS = –0.533), alleles 214 frequent in D. exaltata, and 232 or 236 frequent in D. mollis combine into common heterozygous genotypes (214–232; 214–236) in D. wilsonii (Fig. 2). These patterns suggest that D. wilsonii individuals could represent recent hybrids (mainly F1) between D. mollis and D. exaltata.
Table 2.
Population genetic diversity parameters and inbreeding coefficients estimated for municipalities or populations and for species (overall) for Dimorphandra wilsonii, D. exaltata and D. mollis based on the five loci genotyped in the three species
| n | A | A R | H O | H E | F IS | |
|---|---|---|---|---|---|---|
| D. wilsonii | ||||||
| PBA | 12 | 3.2 | 2.9 | 0.800 | 0.525 | –0.560 |
| FDM | 25 | 4.0 | 3.1 | 0.808 | 0.563 | –0.448 |
| JUA | 21 | 4.0 | 3.1 | 0.771 | 0.532 | –0.467 |
| LST | 15 | 2.4 | 2.3 | 0.815 | 0.481 | –0.740 |
| PEQ | 22 | 4.8 | 3.6 | 0.785 | 0.606 | –0.305 |
| STL | 20 | 4.2 | 3.0 | 0.580 | 0.456 | –0.280 |
| MRV | 22 | 4.6 | 3.4 | 0.745 | 0.549 | –0.368 |
| ESM | 22 | 5.0 | 3.7 | 0.817 | 0.623 | –0.321 |
| FLR | 8 | 3.4 | 3.4 | 0.746 | 0.586 | –0.299 |
| Overall | 167 | 9.0 | 3.8 | 0.763 | 0.577 | –0.324 |
| D. exaltata | ||||||
| SER | 20 | 3.8 | 2.4 | 0.273 | 0.302 | 0.098 |
| CON | 22 | 4.0 | 2.5 | 0.326 | 0.313 | –0.040 |
| ESM | 13 | 2.8 | 2.2 | 0.177 | 0.199 | 0.113 |
| RJA | 7 | 2.8 | 2.8 | 0.257 | 0.451 | 0.449 |
| Overall | 62 | 7.8 | 2.8 | 0.268 | 0.338 | 0.208 |
| D. mollis | ||||||
| FRU | 21 | 4.8 | 4.0 | 0.664 | 0.634 | 0.046 |
| TRM | 15 | 4.2 | 3.6 | 0.627 | 0.552 | 0.123 |
| ARN | 20 | 5.4 | 4.2 | 0.665 | 0.698 | –0.050 |
| BAM | 20 | 4.2 | 3.2 | 0.495 | 0.455 | 0.081 |
| Overall | 76 | 8.2 | 4.4 | 0.588 | 0.680 | 0.136 |
n = sample size, A = number of alleles, AR = allele richness for a sample size of n = 7 individuals, HO = observed heterozygosity, HE = expected heterozygosity, FIS = inbreeding coefficient. FIS values significantly different from zero at P < 0.05 after multiple test correction are indicated in bold. The overall values were calculated based on all populations pooled for each species.
Fig. 2.
Bar plot showing the distribution of genotypic frequencies for the five overlapping loci genotyped in D. exaltata (DE; black), D. mollis (DM; white) and D. wilsonii (DW; grey). Loci: (A) Dmo5, (B) Dmo7, (C) Dmo13, (D) Dmo20 and (E) Dmo21.
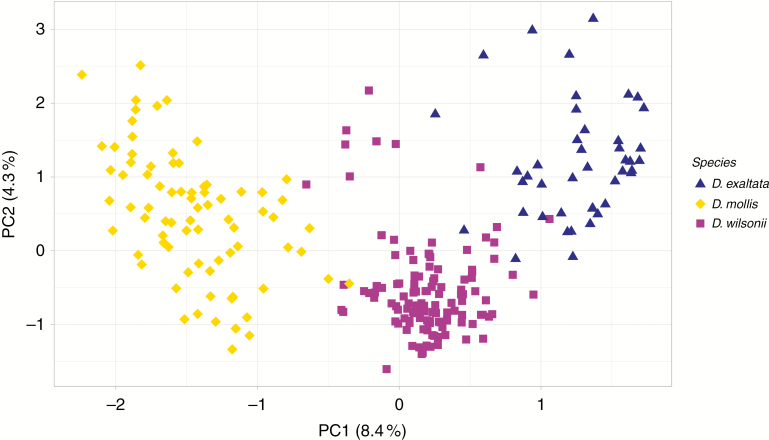
In a PCA of the three Dimorphandra taxa, the first axis explained 8.4 % of the total variation and separated each taxon as a distinct group of individuals. Dimorphandra wilsonii individuals occupied intermediate positions in relation to D. mollis and D. exaltata (Fig. 3). The pairwise FST between D. mollis and D. exaltata was 0.46, indicating a high divergence between the two species, while the levels of divergence between D. wilsonii and D. mollis or D. exaltata were lower, with FST equal to 0.20 and 0.24, respectively.
Fig. 3.
Principal component analysis showing the genetic variation among Dimorphandra wilsonii, D. exaltata and D. mollis.
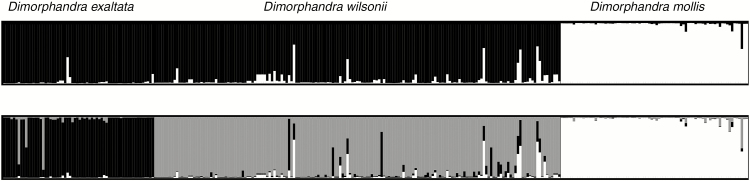
The optimal number of genetic clusters present in the three taxa data set as inferred using the Bayesian clustering method STRUCTURE and the ΔK criterion of Evanno et al. (2005) was K = 2, but the ΔK value for K = 3 was similar to that of K = 2 (Supplementary data Fig. S2). K = 2 separated D. mollis from a group formed by D. wilsonii and D. exaltata (Fig. 4A). K = 3 placed each Dimorphandra taxon in a distinct genetic group (Fig. 4B). For K = 2, only 6 % of D. wilsonii individuals were admixed (Q < 0.85) and none was classified as belonging to another Dimorphandra taxon (Fig. 4A). For K = 3, 12 % of D. wilsonii individuals were admixed (Fig. 4B).
Fig. 4.
Bar plots of admixture coefficients estimated for Dimorphandra wilsonii, D. mollis and D. exaltata using the Bayesian clustering method implemented in STRUCTURE for (A) K = 2, (B) K = 3.
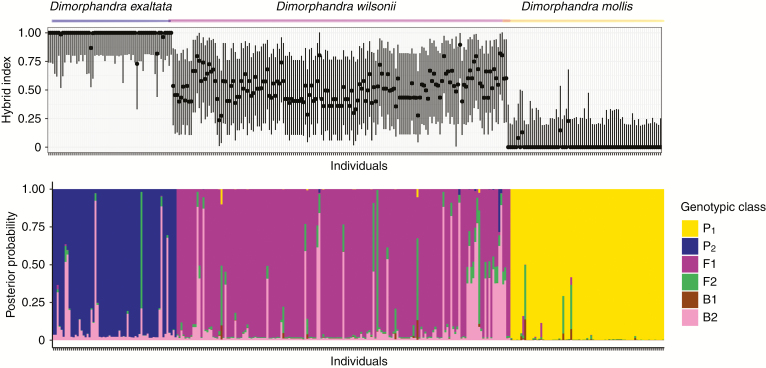
Hybrid index values of the 167 samples of D. wilsonii ranged from 0.235 to 0.896, with 67 % of individuals showing a hybrid index between 0.400 and 0.600, and 95 % confidence intervals (CIs) always comprised 0.5 (Fig. 5A). Conversely, only two individuals of each D. mollis and D. exaltata had hybrid index values with CIs comprising 0.5 (Fig. 5A). NewHybrids classified 73 % of D. wilsonii individuals as F1 using 0.9 as a threshold for the posterior probability (Fig. 5B). The sum of posterior probabilities of falling into any of the four hybrid classes was >0.9 for 98 % of D. wilsonii individuals (Fig. 5B). On the other hand, 97 % of D. mollis and 77 % of D. exaltata individuals were classified as purebred parental with a posterior probability >0.9. Only three D. exaltata individuals showed a cumulated posterior probability >0.9 of falling into any hybrid class, whereas no D. mollis individual was classified as hybrid according to these criteria.
Fig. 5.
Assessment of hybridization between Dimorphandra species. (A) Estimated hybrid index with the 95 % confidence intervals for individuals of D. wilsonii, D. mollis and D. exaltata. (B) Posterior probability of genotypic assignment in two purebred parental (P1 and P2) and four hybrid classes, F1, F2 and two backcrosses with parentals (B1 and B2), based on NewHybrids analysis for individuals of D. wilsonii, D. mollis and D. exaltata.
The analysis of simulated data sets showed that STRUCTURE correctly assigned purebred parental individuals with Q values lower than 0.150 (higher than 0.850) with an efficiency higher than 90 % (Supplementary data Table S3; Fig. S3). Furthermore, STRUCTURE showed a mean efficiency of 97 % in the classification of hybrid individuals in general but poorly assigned hybrids to specific hybrid classes (Supplementary data Table S3; Fig. S3). These results indicate a good overall performance of STRUCTURE for the distinction between parental and hybrid individuals. NewHybrids correctly classified an average of 93 % and 85 % of parental individuals as belonging to P1 or P2, corresponding, respectively, to D. mollis and D. exaltata. The assignment of hybrids to genotypic classes was in general poorer, e.g. the classification of F1 showing an accuracy of 48 % (Supplementary data Table S3). However, only 5 % of the simulated hybrids, mostly backcrosses with P1, were assigned as parental individuals, which indicates a high capacity to differentiate between pure parental individuals and hybrids (Supplementary data Table S3; Fig. S4).
Chromosome counts of D. wilsonii
All 16 metaphase cells analysed showed 2n = 28 chromosomes (Supplementary data Fig. S5). It was possible to analyse samples from three populations; 14 counts were made in individuals of ESM and only one in PBA and one in MRV. This chromosome number was identical to the previous description by Bandel (1974) in D. mollis which showed that the most common haploid chromosome number in Caesalpinioideae was n = 14. Based on the basic chromosome numbers described for tribe Caesalpinieae (x = 13, x = 14) (Goldblatt, 1981), we suggest that D. wilsonii individuals are diploid (2n = 2x = 28).
DISCUSSION
The analyses gathered here indicated that D. exaltata and D. mollis are genetically well-differentiated species and suggest that D. wilsonii individuals most probably correspond to F1 hybrids between the parental species D. exaltata and D. mollis. The three taxa co-occur in a small ecotonal area between the Cerrado and the Atlantic Forest biomes, in south-eastern Brazil. Dimorphandra mollis is a common and economically important species from the Cerrado (Ratter et al., 2003), and D. exaltata is a rare and threatened species from the Atlantic Forest (Muniz et al., 2019). The putative hybrid zone indicates that the Cerrado/Atlantic Forest ecotone can be a valuable region for studying evolutionary relationships in tropical trees. We discuss our findings in the light of the literature, including the implications of the hybridization process for the conservation of the three Dimorphandra taxa.
Evolutionary relationships among D. wilsonii, D. mollis and D. exaltata
A striking genetic feature in D. wilsonii is a large excess of heterozygotes in relation to Hardy–Weinberg genotypic expectations, reflected in high negative fixation indexes (FIS). Several processes can generate an increase in the frequency of heterozygotes and produce negative FIS, including asexual reproduction, self-incompatibility, polyploidy, natural selection and hybridization (Stoeckel et al., 2006). Our extensive sampling across the distribution range of D. wilsonii revealed only two individuals with the same multilocus genotype at 11 SSRs, suggesting that asexual reproduction is absent or very rare. Additionally, reproductive studies showed that D. wilsonii is self-compatible (Martins et al., 2014). Furthermore, the SSRs exhibited a maximum of two alleles per single-locus genotype, in agreement with cytogenetic analysis which indicated that D. wilsonii is diploid and exhibits the same chromosome number as D. mollis (Bandel, 1974).
Despite the small number of SSRs, our genetic data were very informative due to contrasting allele frequencies between D. exaltata and D. mollis, and showed several lines of evidence suggesting that the excess of heterozygotes in D. wilsonii is probably due to hybridization. First, D. wilsonii shows allele frequencies intermediate between D. mollis and D. exaltata. Secondly, abundant heterozygous genotypes in D. wilsonii are concordant with representing F1 hybrids between D. mollis and D. exaltata, or with some individuals being possibly later-generation hybrids. Thirdly, PCA situated D. wilsonii individuals between D. mollis and D. exaltata, concordant with its intermediate allele frequencies. Finally, the hybrid index and NewHybrids analysis indicated that D. wilsonii individuals most commonly carry genotypes congruent with being F1 hybrids and less commonly genotypes congruent with later-generation hybrids. The latter included possible F2s and backcrosses, but these hybrid classes were difficult to distinguish because of the small number of markers used. A hybrid population may show negative inbreeding coefficients and intermediate allele frequencies in relation to the parental species when it shows a high proportion of F1 in relation to later-generation hybrids, a pattern that was also observed in Eucalyptus (Field et al., 2011), Salix (Gramlich et al., 2016) and Populus (Zeng et al., 2016). So, our data suggest that D. wilsonii individuals are recent hybrids formed between the parental species D. mollis and D. exaltata.
Although most analyses supported the origin of D. wilsonii through hybridization, the Bayesian clustering method STRUCTURE grouped D. wilsonii individuals in the same genetic cluster as D. exaltata. While our simulations suggested a good performance of STRUCTURE to detect pure gene pools and hybrids (Supplementary data Table S3), it is known that STRUCTURE can produce erroneous clustering results because of stochasticity in genealogical lineage sorting when using a small number of markers (Orozco-terWengel et al., 2011). STRUCTURE’s performance to reveal the optimal K or estimate admixture proportions is also affected by uneven samples sizes and levels of genetic divergence among populations (Vähä and Primmer, 2006; Kalinowski, 2011; Neophytou, 2014; Wang, 2017). This could be the case for D. mollis and D. exaltata, which are very divergent at the species level and can show moderate to high genetic divergence at the population level (Souza et al., 2017; Muniz et al., 2019). Also, D. wilsonii shows a strong departure from STRUCTURE expectations of linkage equilibrium and Hardy–Weinberg equilibrium within inferred clusters. We suggest that the low number of loci, the lack of a strict sympatric population of D. mollis with D. wilsonii and the strong departure from Hardy–Weinberg genotypic proportions in D. wilsonii may have negatively influenced the performance of STRUCTURE in our analysis.
A few studies have revealed hybridization between closely related lineages in the ecotone between the Atlantic Forest and the Cerrado (Lacerda et al., 2002; Cavallari et al., 2010), but D. wilsonii is the first known case where probable hybrids have been described as a separate species. Areas such as the Cerrado/Atlantic Forest ecotone, where closely related species can be found in adjacent divergent environments, can harbour several types of hybrid zones which are mainly determined by the fitness of hybrids (Abbott et al., 2013; Abbott, 2017). On one hand, strong selection against hybrids may lead to tension zones where formation of hybrids is maintained by the recurrent crossing of parental populations (Gompert et al., 2017). On the other hand, the hybrids may show higher fitness than their parents, in intermediate or new environments in mosaic hybrid zones (Abbott, 2017). Dimorphandra wilsonii individuals produce abundant seeds in nature which germinate and grow well under greenhouse conditions (Fernandes and Rego 2014). Moreover, the observation of D. wilsonii individuals assigned to F2 or backcross classes suggests the presence of later-generation hybrids in our data and thus indicates that hybrids can successfully interbreed, although we have to be cautious about the correct genotypic class assignment of these individuals. However, juveniles are rarely found in the wild, generally highly disturbed environments, and the fitness of D. wilsonii offspring has never been investigated in nature. Recently, the conservation programme of D. wilsonii started to produce saplings in nurseries for reintroduction in the wild (Martins et al., 2014). The evaluation of fitness components of saplings, such as growth and survival, in the nursery and after reintroduction in nature may add further understanding on the nature of the Dimorphandra hybrid zone.
Our study raises several important questions about the origin, the taxonomic status and the evolutionary future of D. wilsonii. Is the hybrid formation ancient and recurrent? Is the hybrid formation recent and driven by anthropogenic interference? As we have pointed out, the area where D. wilsonii occurs is highly disturbed by anthropogenic activities. An in-depth analysis using genomic data in D. wilsonii and its two proposed parental species would allow better characterization of the genomic structure of the proposed hybrid origin, thus providing partial answers to these questions. The evaluation of the type and the evolutionary outcomes of the hybridization, such as levels of backcrossing with parental species and introgression, and the fitness of hybrids in nature represent valuable information for the conservation aims for these species, as discussed below. Furthermore, such in-depth information on evolutionary processes operating in the ecotonal area between the Cerrado and the Atlantic Forest can contribute to understanding the evolutionary dynamics between these biomes and to illuminating the consequences of ongoing climatic changes in this area.
Implications for conservation of D. wilsonii, D. exaltata and D. mollis in the Cerrado/Atlantic Forest ecotone
The definition of the actual taxonomic status of D. wilsonii and the structure of the putative hybrid zone are needed since hybrids are not protected based on most legislations about threatened species (vonHoldt et al., 2018). In the case of D. wilsonii this is especially important because it already has a national plan for its conservation with scientific research, legal instruments, human action and financial resources oriented for its protection (Martins et al., 2014). Moreover, the determination of the taxa involved in hybridization, the type, the age and the likely evolutionary outcomes in hybrid zones can have consequences for the design of conservation strategies and the management of the species involved (Allendorf et al., 2001; Jackiw et al., 2015; Gompert and Buerkle, 2016; Hamilton and Miller, 2016). Although our study raised doubts about the taxonomic status of D. wilsonii, it revealed that D. wilsonii harbours moderate genetic diversity, with significant variation in allele distribution across the sampling area. Dimorphandra wilsonii showed levels of allelic richness and HE similar to D. mollis (Table 2; Souza et al., 2017) and higher levels than D. exaltata (Table 2; Muniz et al., 2019). Dimorphandra exaltata is a very threatened Atlantic Forest species with most of its records collected in sympatric areas with D. wilsonii with a predicted loss of suitable areas in the future due to climate change (Muniz et al., 2019). Although D. mollis is widespread and not considered threatened currently, the species is economically important and its populations are heavily exploited due to its content of the flavonoid rutin, used as an antioxidant in the pharmaceutical industry (Panegassi et al., 2000; Gonçalves et al., 2010), and which is also found in D. wilsonii (Martins et al., 2014). Furthermore, despite the high historical effective populations sizes of D. mollis, a reduction in its suitable areas and in effective populations sizes was detected (Souza et al., 2017). In general, hybrids are considered threat factors for endangered species as hybrids may be less fit than their parents and introgressive backcrosses can lead to the invasion of the genome of species and ultimately to extinction by hybridization (Levin et al., 1996; Rhymer and Simberloff, 1996; Todesco et al., 2016). However, hybridization can have a creative evolutionary potential increasing standing genetic variation (Marques et al., 2019), the probability of adaptive radiations (Kagawa and Takimoto, 2018) and by hybrid speciation (Abbott et al., 2013). Additionally, some authors have argued that hybrids may have high conservation value and also can contribute in the management of the related species involved (Allendorf et al., 2001; Thompson et al., 2010; Hamilton and Miller, 2016). Therefore, conservation efforts should be directed to both parental species in the Atlantic Forest/Cerrado ecotone besides D. wilsonii, with special attention paid to the evaluation of evolutionary outcomes of the putative hybridization. Specifically, D. wilsonii may be a source of allelic diversity and adaptive alleles for D. exaltata if viable backcross offspring could be obtained with this species, thus increasing D. exaltata’s genetic diversity and its evolutionary potential. The current management strategy for D. wilsonii conservation has focused on a demographic increase of populations through reintroduction of saplings. The results of our study suggest that this approach should be re-evaluated because of unknown outcomes of the disproportional increase of hybrid individuals in relation to parental species, especially the threatened D. exaltata. Above all, our data on Dimorphandra species highlight the importance of genetic information for adequate design of conservation strategies.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: test for null alleles and estimates of null allele frequencies for each locus across all individuals of D. wilsonii. Table S2: allele frequencies of D. wilsonii (DW), D. exaltata (DE) and D. mollis (DM). Table S3: evaluation of accuracy, efficiency and overall performance for STRUCTURE and NewHybrids using five simulated data sets. Figure S1: map showing the scores for the first axis of a spatial principal component analysis for individuals of Dimorphandra wilsonii. Figure S2: Evanno’s ΔK values for the number of clusters K assumed in the analysis using the Bayesian clustering method STRUCTURE for the three species together. Figure S3: box plots showing the observed distribution of admixture coefficients. Figure S4: box plots showing the distribution of the posterior probability values estimated for each genotypic class in NewHybrids. Figure S5: bitotic metaphase cells from root meristem of D. wilsonii.
ACKNOWLEDGEMENTS
We thank the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) and the Sistema de Autorização e Informação em Biodiversidade (SISBIO) for providing the licence (ICMBio: 47925-2) to collect genetic samples. We also thank Fernando M. Fernandes for locating and identifying individuals and collecting samples in the field. We thank Rosangela L. Brandão for technical assistance in the laboratory. A.C.M., J.P.L.-F., M.H. and M.B.L. conceived and designed the research. A.C.M. and J.P.L.-F. performed field trips to collect samples. A.C.M., R.S.O.B. and H.A.V.S. conducted the experiments. R.C.M. conducted the cytogenetics analysis and its interpretation. A.C.M. and M.H. analysed the data. M.B.L. and J.P.L.-F. provided financial resources for the research. A.C.M., J.P.L.-F., M.H. and M.B.L. wrote the manuscript. All authors reviewed the manuscript. The authors declare no conflict of interest. Genotype data will be submitted to Dryad.
FUNDING
This work was supported by Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG); Fundação Grupo Boticário de Proteção à Natureza and has benefited from an ‘Investissement d’Avenir’ grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-25-01). Also, we thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for their support of the first author’s PhD scholarship and research fellowships to J.P.L.-F. and M.B.L., respectively.
LITERATURE CITED
- Abbott RJ. 2017. Plant speciation across environmental gradients and the occurrence and nature of hybrid zones. Journal of Systematics and Evolution 55: 238–258. [Google Scholar]
- Abbott R, Albach D, Ansell S, et al. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26: 229–246. [DOI] [PubMed] [Google Scholar]
- Aksoy S, Almeida-Val VMF, Azevedo VCR, et al. 2013. Permanent genetic resources added to molecular ecology resources database 1 October 2012–30 November 2012. Molecular Ecology Resources 13: 341–343. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends in Ecology and Evolution 16: 613–622. [Google Scholar]
- Anderson EC, Thompson EA. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandel G. 1974. Chromosome numbers and evolution in the Leguminosae. Caryologia 27: 17–32. [Google Scholar]
- Bariotakis M, Koutroumpa K, Karousou R, Pirintsos SA. 2016. Environmental (in)dependence of a hybrid zone: insights from molecular markers and ecological niche modeling in a hybrid zone of Origanum (Lamiaceae) on the island of Crete. Ecology and Evolution 6: 8727–8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. 1985. Analysis of hybrid zones. Annual Review of Ecology and Systematics 16: 113–148. [Google Scholar]
- Behling H. 1995. A high resolution Holocene pollen record from Lago do Pires, SE Brazil: vegetation, climate and fire history. Journal of Paleolimnology 14: 253–268. [Google Scholar]
- Behling H. 2002. South and southeast Brazilian grasslands during Late Quaternary times: a synthesis. Palaeogeography, Palaeoclimatology, Palaeoecology 177: 19–27. [Google Scholar]
- Behling H, Negrelle RRB. 2001. Tropical rain forest and climate dynamics of the Atlantic Lowland, Southern Brazil, during the late quaternary. Quaternary Research 56: 383–389. [Google Scholar]
- Brandon K, Da Fonseca GAB, Rylands AB, Da Silva JMC. 2005. Introduction to special section: Brazilian conservation – challenges and opportunities. Conservation Biology 19: 595–600. [Google Scholar]
- Buerkle CA. 2005. Maximum-likelihood estimation of a hybrid index based on molecular markers. Molecular Ecology Notes 5: 684–687. [Google Scholar]
- Buzatti RSDO, Lemos-Filho JP, Bueno ML, Lovato MB. 2017. Multiple Pleistocene refugia in the Brazilian cerrado: evidence from phylogeography and climatic niche modelling of two Qualea species (Vochysiaceae). Botanical Journal of the Linnean Society 185: 307–320. [Google Scholar]
- Buzatti RSDO, Pfeilsticker TR, de Magalhães RF, Bueno ML, Lemos-Filho JP, Lovato MB. 2018. Genetic and historical colonization analyses of an endemic Savanna tree, Qualea grandiflora, reveal ancient connections between Amazonian Savannas and Cerrado core. Frontiers in Plant Science 9: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari MM, Gimenes MA, Billot C, et al. 2010. Population genetic relationships between Casearia sylvestris (Salicaceae) varieties occurring sympatrically and allopatrically in different ecosystems in south-east Brazil. Annals of Botany 106: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigan G, Ratter JA. 2006. Successional changes in cerrado and cerrado/forest ecotonal vegetation in western São Paulo State, Brazil, 1962–2000. Edinburgh Journal of Botany 63: 119–130. [Google Scholar]
- Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Eisenlohr PV, de Oliveira-Filho AT. 2015. Revisiting patterns of tree species composition and their driving forces in the Atlantic Forests of Southeastern Brazil. Biotropica 47: 689–701. [Google Scholar]
- El Mousadik A, Petit RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco Title. Theoretical and Applied Genetics 92: 832–839. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Fernandes FM. 2006. Dimorphandra wilsonii. The IUCN Red List of Threatened Species 2006: e.T61926A12574230 10.2305/IUCN.UK.2006.RLTS.T61926A12574230.en. [DOI] [Google Scholar]
- Fernandes FM, Rego JO. 2014. Dimorphandra wilsonii Rizzini (Fabaceae): distribution, habitat and conservation status. Acta Botanica Brasilica 28: 434–444. [Google Scholar]
- Fiaschi P, Pirani JR. 2009. Review of plant biogeographic studies in Brazil. Journal of Systematics and Evolution 47: 477–496. [Google Scholar]
- Field DL, Ayre DJ, Whelan RJ, Young AG. 2011. Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida. Heredity 106: 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson I, Welsh AB, Welsh SA, Cincotta DA. 2019. Genetic swamping and possible species collapse: tracking introgression between the native Candy Darter and introduced Variegate Darter. Conservation Genetics 20: 287–298. [Google Scholar]
- Goldblatt P. 1981. Cytology and phylogeny of Leguminosae. Advances in Legume Systematics 2: 427–463. [Google Scholar]
- Gompert Z, Buerkle CA. 2010. introgress: a software package for mapping components of isolation in hybrids. Molecular Ecology Resources 10: 378–384. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA. 2016. What, if anything, are hybrids: enduring truths and challenges associated with population structure and gene flow. Evolutionary Applications 9: 909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Mandeville EG, Buerkle CA. 2017. Analysis of population genomic data from hybrid zones. Annual Review of Ecology, Evolution, and Systematics 48: 207–229. [Google Scholar]
- Gonçalves AC, Reis CAF, de Almeida Vieira F, de Carvalho D. 2010. Spatial genetic structure in natural populations of Dimorphandra mollis (Fabaceae) in the north of Minas Gerais State, Brazil. Brazilian Journal of Botany 33: 325–332. [Google Scholar]
- Goudet J. 2002. FSTAT (version 2.9. 3.2): a program to estimate and test gene diversities and fixation indices http://www.unil.ch/izea/softwares/fstat.html.
- Goulart MF, Lovato MB, de Vasconcellos Barros F, Valladares F, Lemos-Filho JP. 2011. Which extent is plasticity to light involved in the ecotypic differentiation of a tree species from savanna and forest? Biotropica 43: 695–703. [Google Scholar]
- Gramlich S, Sagmeister P, Dullinger S, Hadacek F, Hörandl E. 2016. Evolution in situ: hybrid origin and establishment of willows (Salix L.) on alpine glacier forefields. Heredity 116: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JA, Miller JM. 2016. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conservation Biology 30: 33–41. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. 1988. Hybrid zones – natural laboratories for evolutionary studies. Trends in Ecology & Evolution 3: 158–167. [DOI] [PubMed] [Google Scholar]
- Hoffmann WA, Franco AC. 2003. Comparative growth analysis of tropical forest and savanna woody plants using phylogenetically independent contrasts. Journal of Ecology 91: 475–484. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. 2009. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackiw RN, Mandil G, Hager HA. 2015. A framework to guide the conservation of species hybrids based on ethical and ecological considerations. Conservation Biologyy 29: 1040–1051. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour AB, Pontier D. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101: 92–103. [DOI] [PubMed] [Google Scholar]
- Kagawa K, Takimoto G. 2018. Hybridization can promote adaptive radiation by means of transgressive segregation. Ecology Letters 21: 264–274. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. 2011. The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda DR, Lemos-Filho JP, Lovato MB, Lemos Filho JP, Acedo MDP, Lovato MB. 2002. Molecular differentiation of two vicariant neotropical tree species, Plathymenia foliolosa and P. reticulata (Mimosoideae), inferred using RAPD markers. Plant Systematics and Evolution 235: 67–77. [Google Scholar]
- Levin DA, Francisco-Ortega J, Jansen RK. 1996. Hybridization and the extinction of rare plant species. Conservation Biology 10: 10–16. [Google Scholar]
- Li Y, Tada F, Yamashiro T, Maki M. 2016. Long-term persisting hybrid swarm and geographic difference in hybridization pattern: genetic consequences of secondary contact between two Vincetoxicum species (Apocynaceae-Asclepiadoideae). BMC Evolutionary Biology 16: 20. doi: 10.1186/s12862-016-0587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguilla E, Escudero M. 2016. Cryptic species due to hybridization: a combined approach to describe a new species (carex: Cyperaceae). PLoS One 11: e0166949. doi: 10.1371/journal.pone.0166949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20: 229–237. [DOI] [PubMed] [Google Scholar]
- Marques I, Draper D, López-Herranz ML, Garnatje T, Segarra-Moragues JG, Catalán P. 2016. Past climate changes facilitated homoploid speciation in three mountain spiny fescues (Festuca, Poaceae). Scientific Reports 6: 36283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques DA, Meier JI, Seehausen O. 2019. A combinatorial view on speciation and adaptive radiation. Trends in Ecology & Evolution 34: 531–544. [DOI] [PubMed] [Google Scholar]
- Martins EM, Fernandes FM, Maurenza D, Pougy N, Loyola R, Martinelli G. 2014. Plano de ação nacional para a conservação do Faveiro-de-wilson (Dimorphandra wilsonii Rizzini). Brazil: Instituto de Pesquisas Jardim Botânico do Rio de Janeiro [Google Scholar]
- Motta PEF, Curi N, Franzmeier DP. 2002. Relation of soils and geomorphic surfaces in the Brazilian Cerrado. In: Oliveira PS, Marquis RJ, eds. The cerrados of Brazil: ecology and natural history of a neotropical savanna. New York: Columbia University Press, 13–32. [Google Scholar]
- Muhlfeld CC, Kovach RP, Jones LA, et al. 2014. Invasive hybridization in a threatened species is accelerated by climate change. Nature Climate Change 4: 620–624. [Google Scholar]
- Muniz AC, Lemos-Filho JP, Buzatti RSO, Ribeiro PCC, Fernandes FM, Lovato MB. 2019. Genetic data improve the assessment of the conservation status based only on herbarium records of a Neotropical tree. Scientific Reports 9: 5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou C. 2014. Bayesian clustering analyses for genetic assignment and study of hybridization in oaks: effects of asymmetric phylogenies and asymmetric sampling schemes. Tree Genetics and Genomes 10: 273–285. [Google Scholar]
- Nieto Feliner G, Álvarez I, Fuertes-Aguilar J, et al. 2017. Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity 118: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaes RM, De Lemos Filho JP, Ribeiro RA, Lovato MB. 2010. Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in Eastern Tropical South America. Molecular Ecology 19: 985–998. [DOI] [PubMed] [Google Scholar]
- Oliveira-Filho AT, Fontes MAL. 2000. Patterns of floristic differentiation among Atlantic Forests in Southeastern Brazil and the influence of climate. Biotropica 32: 793–810. [Google Scholar]
- Orozco-terWengel P, Corander J, Schlötterer C. 2011. Genealogical lineage sorting leads to significant, but incorrect Bayesian multilocus inference of population structure. Molecular Ecology 20: 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panegassi VR, Serra GE, Buckeridge MS, et al. 2000. Potencial tecnológico do galactomanano de sementes de faveiro (Dimorphandra mollis) para uso na indústria de alimentos. Food Science and Technology 20: 406–415. [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. 2009. Hybrid speciation in angiosperms: parental divergence drives ploidy. The New Phytologist 182: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2006. Genalex 6: genetic analysis in Excel. Molecular Ecology Notes 6: 288–295. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratter JA, Bridgewater S, Ribeiro JF. 2003. Analysis of the floristic composition of the Brazilian cerrado vegetation III: comparison of the woody vegetation of 376 areas. Edinburgh Journal of Botany 60: 57–109. [Google Scholar]
- Resende-Moreira LC, Ramos ACS, Scliar MO, et al. 2017. Gene flow between vicariant tree species: insights into savanna-forest evolutionary relationships. Tree Genetics and Genomes 13: 36. [Google Scholar]
- Rhymer JM, Simberloff D. 1996. Extinction by hybridization and introgression. Annual Review of Ecology and Systematics 27: 83–109. [Google Scholar]
- Ribeiro PCC, de Lemos-Filho JP, Buzatti RS de O, Lovato MB, Heuertz M. 2016. Species-specific phylogeographical patterns and Pleistocene east–west divergence in Annona (Annonaceae) in the Brazilian Cerrado. Botanical Journal of the Linnean Society 181: 21–36. [Google Scholar]
- Ribeiro RA, Lemos-Filho JP, Ramos AC, Lovato MB. 2011. Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity 106: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. 1997. Hybrid origins of plant species. Annual Review of Ecology and Systematics 28: 359–389. [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Carney SE. 1998. Plant hybridization. New Phytologist 140: 599–624. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- da Silva MF. 1986. Flora Neotropica monograph. Dimorphandra 44: 1–128. [Google Scholar]
- Silva JF, Fariñas MR, Felfili JM, Klink CA. 2006. Spatial heterogeneity, land use and conservation in the cerrado region of Brazil. Journal of Biogeography 33: 536–548. [Google Scholar]
- Simon MF, Pennington T. 2012. Evidence for adaptation to fire regimes in the tropical Savannas of the Brazilian Cerrado. International Journal of Plant Sciences 173: 711–723. [Google Scholar]
- Simon MF, Grether R, de Queiroz LP, Skema C, Pennington RT, Hughes CE. 2009. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proceedings of the National Academy of Sciences, USA 106: 20359–20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2009. The role of hybridization in plant speciation. Annual Review of Plant Biology 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Souza HA. 2012. Diversidade genética e filogeografia de duas espécies de faveiro (Dimorphandra mollis e D. wilsonii): implicações para conservação e manejo. PhD Thesis, Universidade Federal de Minas Gerais, Brazil. [Google Scholar]
- Souza HAV, Lovato MB. 2010. Genetic diversity and structure of the critically endangered tree Dimorphandra wilsonii and of the widespread in the Brazilian Cerrado Dimorphandra mollis: implications for conservation. Biochemical Systematics and Ecology 38: 49–56. [Google Scholar]
- Souza HA, Muller LA, Brandão RL, Lovato MB. 2012a Isolation of high quality and polysaccharide-free DNA from leaves of Dimorphandra mollis (Leguminosae), a tree from the Brazilian Cerrado. Genetics and Molecular Research 11: 756–764. [DOI] [PubMed] [Google Scholar]
- Souza HA, Collevatti RG, Lemos-Filho JP, Santos FR, Lovato MB. 2012b Development of microsatellite markers for Dimorphandra mollis (Leguminosae), a widespread tree from the Brazilian cerrado. American Journal of Botany 99: e102–e104. [DOI] [PubMed] [Google Scholar]
- Souza HA, Collevatti RG, Lima-Ribeiro MS, Lemos-Filho JP, Lovato MB. 2017. A large historical refugium explains spatial patterns of genetic diversity in a Neotropical savanna tree species. Annals of Botany 119: 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel S, Grange J, Fernández-Manjarres JF, Bilger I, Frascaria-Lacoste N, Mariette S. 2006. Heterozygote excess in a self-incompatible and partially clonal forest tree species – Prunus avium L. Molecular Ecology 15: 2109–2118. [DOI] [PubMed] [Google Scholar]
- Suarez-Gonzalez A, Lexer C, Cronk QCB. 2018. Adaptive introgression: a plant perspective. Biology Letters 14: pii: 20170688. doi: 10.1098/rsbl.2017.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL, Harrison RG. 2015. Hybrid zones: windows on climate change. Trends in Ecology & Evolution 30: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gaudeul M, Debussche M. 2010. Conservation value of sites of hybridization in peripheral populations of rare plant species. Conservation Biology 24: 236–245. [DOI] [PubMed] [Google Scholar]
- Todesco M, Pascual MA, Owens GL, et al. 2016. Hybridization and extinction. Evolutionary Applications 9: 892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä JP, Primmer CR. 2006. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology 15: 63–72. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. [Google Scholar]
- Vinson CC, Dal’Sasso TCS, Sudré CP, Mangaravite E, de Oliveira LO. 2015. Population genetics of the naturally rare tree Dimorphandra wilsonii (Caesalpinioideae) of the Brazilian Cerrado. Tree Genetics & Genomes 11: 46. [Google Scholar]
- vonHoldt BM, Brzeski KE, Wilcove DS, Rutledge LY. 2018. Redefining the role of admixture and genomics in species conservation. Conservation Letters 11: e12371. [Google Scholar]
- Wang J. 2017. The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Molecular Ecology Resources 17: 981–990. [DOI] [PubMed] [Google Scholar]
- Zalapa JE, Brunet J, Guries RP. 2010. The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evolutionary Applications 3: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YF, Zhang JG, Duan AG, Abuduhamiti B. 2016. Genetic structure of Populus hybrid zone along the Irtysh River provides insight into plastid–nuclear incompatibility. Scientific Reports 6: 28043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.